Abstract
Protein dimerization controls many physiological processes in the body. Proteins form homo-, hetero-, or oligomerization in the cellular environment to regulate the cellular processes. Any deregulation of these processes may result in a disease state. Protein–protein interactions (PPIs) can be inhibited by antibodies, small molecules, or peptides, and inhibition of PPI has therapeutic value. PPI drug discovery research has steadily increased in the last decade, and a few PPI inhibitors have already reached the pharmaceutical market. Several PPI inhibitors are in clinical trials. With advancements in structural and molecular biology methods, several methods are now available to study protein homo- and heterodimerization and their inhibition by drug-like molecules. Recently developed methods to study PPI such as proximity ligation assay and enzyme-fragment complementation assay that detect the PPI in the cellular environment are described with examples. At present, the methods used to design PPI inhibitors can be classified into three major groups: (1) structure-based drug design, (2) high-throughput screening, and (3) fragment-based drug design. In this chapter, we have described some of the experimental methods to study PPIs and their inhibition. Examples of homo- and heterodimers of proteins, their structural and functional aspects, and some of the inhibitors that have clinical importance are discussed. The design of PPI inhibitors of epidermal growth factor receptor heterodimers and CD2–CD58 is discussed in detail.
1. INTRODUCTION
Most of the physiological processes in the body are controlled by cellular interactions that, in turn, are controlled by interacting bio-molecules. Among the biomolecules, proteins are responsible for most of the biochemical pathways that control the physiological processes. Proteins seldom act alone; they bind to other proteins or biomolecules, eliciting a physiological response. It is estimated that there are nearly 650,000 such interactions that control actions that enable the human body to function normally (Stumpf et al., 2008). The complex network of protein–protein interactions (PPIs) that carries out the biological process in an organism is termed “interactome” (Bogan & Thorn, 1998). A number of proteins self-associate to form dimers or oligomers and also form heterodimers. Homodimerization and heterodimerization of proteins regulate several of the biochemical pathways, and any deregulation of this process leads to disease states.
The association of two proteins could result in homodimers (complexation of identical monomers) or heterodimers (complexation of nonidentical monomers) in the cellular environment. Apart from these complexes, proteins can also form oligomers either to perform functions in cells or, in some cases, to control the functions of these proteins. Protein homo–hetero- and oligomerizations can be classified as stable or transient, depending on the timescale used, and the method of detection used. Based on their affinity, a lifetime of the complex, and composition, PPIs are classified as (i) homo-and hetero-oligomeric complexes, (ii) nonobligate and obligate complexes, and (iii) transient and permanent complexes (Acuner Ozbabacan, Engin, Gursoy, & Keskin, 2011). As described earlier, homo- and hetero complexation depends on identical or nonidentical monomers that form the complex. Whether they are classified as obligate or nonobligate is based on whether the monomers of the complex exist in the stable form in vivo on their own or not. An example of obligate homodimers is DNA-binding homodimer Ku proteins (Krishna & Aravind, 2010). Nonobligate proteins dissociate after they carry out a biochemical process. Protein complexes that participate in the signaling process form transient dimers, and after signaling, they dissociate and hence are examples of nonobligate interacting proteins. Whether they are transient or permanent PPI depends on the lifetime of the complex. Permanent interactions have Kd values <μM, whereas strong transient interactions have Kd values in the nM range, and weak transient interactions have Kd values in the μM range. Enzymes are extensively studied in terms of structure and multimerization; among the reported 452 human enzymes, only one-third are monomers, and the remaining enzymes are known to form homomultimers (Marianayagam, Sunde, & Matthews, 2004). Heterodimeric interactions are commonly found in enzyme inhibitors, enzyme complexes, antibody–antigen, signal proteins, and cell cycle proteins (Sowmya, Breen, & Ranganathan, 2015). The well-known G-protein-coupled receptors (GPCRs) are known to form dimers and oligomers. For proteins that undergo oligomerization, the equilibration between monomer–dimer and oligomer kinetics seems to control the physiological activity. In this chapter, we have provided some of the methods used for detecting PPI and its inhibition with examples. Most of the PPI inhibitors we described here are used for the purpose of modulation of biochemical pathways and for therapeutic purposes. It is not our intention to exhaustively cover the PPI inhibitors. We have highlighted some well-known examples of PPIs that are described in the literature and have covered some recent examples of PPIs and their inhibition. Readers can refer to reviews described in the literature for more extensive coverage (Arkin, Tang, & Wells, 2014; Bakail & Ochsenbein, 2016; Fry et al., 2013; Guo, Wisniewski, & Ji, 2014; Iyer, 2016; Jin, Wang, & Fang, 2014; London, Raveh, & Schueler-Furman, 2013; Morelli, Bourgeas, & Roche, 2011; Petta, Lievens, Libert, Tavernier, & De Bosscher, 2016; Sable & Jois, 2015; Skwarczynska & Ottmann, 2015; Zhang, Ben-David, & Sidhu, 2016). Examples of PPI inhibition related to epidermal growth factor receptor (EGFR) and CD2–CD58 that we have been working on in our laboratory are provided in detail.
2. METHODS TO STUDY PPIs AND THEIR INHIBITION
Most cellular assays used to screen compounds for biological activity or inhibitors of proteins employ either enzymatic assays or drug-like molecules binding to one of the proteins of interest, or measure the IC50 values of inhibition. These screening methods do not provide information about whether the designed compounds inhibit PPI. When PPI inhibitors are designed, pharmacological assays do not provide direct evidence of PPI inhibition and hence experimental methods that provide evidence of PPI inhibition have to be provided. We have highlighted some of the most commonly used methods of PPI and its inhibition. Coimmunoprecipitation and pull-down assays are used in the early stages of PPI inhibitor design, whereas assays such as surface plasmon resonance (SPR), proximity ligation assay (PLA), and enzyme fragment complementation assays are used in the later stages.
2.1. Coimmunoprecipitation
Coimmunoprecipitation (Hall, 2005) is an assay that is used to analyze PPIs from cells and provide physiologically relevant information concerning PPI since proteins are extracted from their native environment in most cases. In principle, the assay is similar to immunoprecipitation (IP), with modifications for studying PPI. In an IP assay, an antibody forms an immune complex with the protein of interest. This complex is captured using an immobilized protein (on a bead) ligand that binds to the antibody; hence the immune complex is captured on the bead. Washing the beads will wash away any proteins not precipitated on the beads. The antigen–antibody complex is eluted from the support, analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detected by Western blotting. In co-IP assays, the antibody–antigen complex will have a binding partner of the antigen; this means that the target antigen precipitated by the antibody “coprecipitates” with a binding partner/protein complex from a lysate. The detection of the coprecipitates provides information about the interacting partner protein. The two proteins can be identified by their respective antibodies in a Western blot assay, which can be performed in any laboratory equipped to perform biochemistry or molecular biology techniques. Although the assay is quite straightforward and sounds simple, there are some limitations. Antibody contamination and nonspecific binding are the major factors related to negative results of the assay. Apart from this, the stability of the complex of two proteins poses a challenge during the performance of the experiments. Since the assay involves several washing steps, the condition of the washing buffers and the way the precipitate is handled can cause the complex to dissociate, resulting in only antibody–one protein complex detection. In addition, if the two proteins interact with low affinity or if the interactions are transient, the proteins that take part in PPI may not be detected. Many protein complexes will remain intact after lysis using standard nondenaturing lysis buffers such as buffers with low ionic strength (i.e., <120mM NaCl) that contain nonionic detergents (NP-40 and Triton X-100). To prevent the disruption of the interaction between the proteins and loss of PPI in the sample, harsh conditions such as cell lysing by sonication or vortexing should be avoided. The samples should be handled gently to prevent the loss of bound complex proteins during centrifugation and washing. A different version of the same assay using a cross-linking technique can be used to stabilize the PPI. Depending on the size of the cross-linking agent, proteins that are interacting at a particular distance will be cross-linked, and these can be immunoprecipitated to detect a particular protein pair. More specific methods using highly specific association between streptavidin and biotin can be used. A wide range of affinity resins, magnetic beads, and coated plates based on immobilized avidin, streptavidin, are available commercially to be used in co-IP (Adams, Seeholzer, & Ohh, 2002; Hall, 2005).
2.2. Pull-Down Assay
This assay can be used as an initial screening assay to identify previously unknown PPI. It can be used along with co-IP to prove that two proteins interact with one another in a hypothesized biochemical pathway. In the pull-down assay, to capture the proteins that interact with one another, a “bait” protein is used (Nguyen & Goodrich, 2006). The bait protein is tagged with affinity tags such as histidine, glutathione, or biotin. These tags will bind to an immobilized affinity ligand in a column. The bait protein and another protein that interacts with the bait protein that forms PPI will both be captured on the affinity column and purified, and the proteins that interact with one another can be characterized. Tags can be attached during protein expression or after protein purification, depending on the type of affinity tag used.
The complex formed is eluted from the column and detected to confirm the presence of proteins that interact with one another. Stable protein complex formed can be washed with high ionic strength to eliminate any non-specific interactions. If the complex has weak interactions, assay conditions can be modified by pH and salt concentration. SDS-PAGE is a harsh treatment that denatures all proteins in the sample and thus restricts analysis. A nondenaturing method called competitive analyte elution can elute a biologically functional protein complex. In all pull-down assays, control experiments are necessary to generate biologically significant results.
2.3. Proximity Ligation Assay
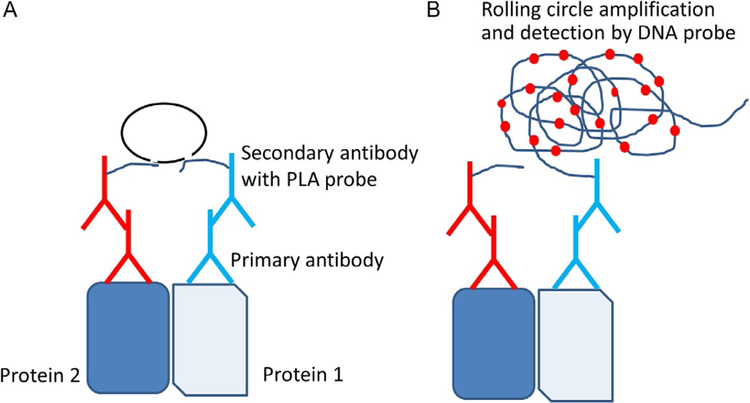
PLA is an assay that can be used to study endogenous PPIs (Fredriksson et al., 2002; Trifilieff et al., 2011). It can be used to study PPI in in vitro or in vivo tissue samples. There are different versions of the PLA; here, we describe antibody-based and fluorescent visualization assays. This assay is based on the principle that, if two proteins interact and the distance between the proteins is within 16nm, the interaction between the proteins can be detected by specific antibodies against these proteins. The primary antibodies used should be from different species so that secondary antibodies with probes can be detected. Furthermore, using this assay, one can quantify PPI in cells, and PPI inhibition can be studied based on the decrease in the number of PPI probes detected. The important consideration in this assay is the availability of antibodies to proteins of interest. The overall assay principle is depicted in Fig. 1.
Fig. 1.
The principle of proximity ligation assay (PLA). (A) Two proteins of interest are targeted by primary antibody from different species. Corresponding secondary antibodies with DNA probes are added. If the two proteins are in proximity the hybridized DNA will be used for rolling circle amplification. (B) Amplified DNA will be detected by a DNA probe. For visualization, a DNA probe with red fluorescence is used. Each dimer of protein in cells is viewed as a red dot with a high-resolution microscope. Adapted from Trifilieff, P., Rives, M. L., Urizar, E., Piskorowski, R. A., Vishwasrao, H. D., Castrillon, J., et al. (2011). Detection of antigen interactions ex vivo by proximity ligation assay: Endogenous dopamine D2-adenosine A2A receptor complexes in the striatum. BioTechniques, 51(2), 111–118. https://doi.org/10.2144/000113719.
The two proteins of interest are targeted with primary antibodies, one from mouse and another from rabbit. These antibodies bind to two proteins of interest, and secondary antibodies (probes) to the primary antibodies are added. The secondary antibodies that bind to primary antibodies have DNA probes. Two oligonucleotides are added that bind to DNA probes on the primary antibodies, and then ligase is added. The DNA forms a circle if the two primary antibodies are within a certain distance (Fig. 1). Once polymerase and nucleotides are added, a rolling circle amplification of probe DNA takes place. The amplified probe DNA is detected with a fluorescent probe. When a high-resolution microscope is used, each PPI is visible as red dot due to rolling circle amplification of the probe DNA. Thus, each red dot/fluorescent dot corresponds to a dimer pair of proteins. The assay can be carried out on fixed cells or tissues. As an example, we provide the HER2:HER3 interaction. PLA was used to demonstrate that HER2: HER3 PPI is present in SKBR-3 breast cancer cells. Addition of a peptidomimetic to cells such as SKBR-3 results in inhibition of PPI, as shown by PLA (Fig. 2). The PPI is shown as red fluorescent dots in the cells without any treatment; upon addition of a PPI inhibitory compound at different concentrations, the number of red fluorescent dots decreased, indicating inhibition of a particular set of PPI (in the figure, the PPI is HER2: HER3). The results can also be quantified for dose—response curves.
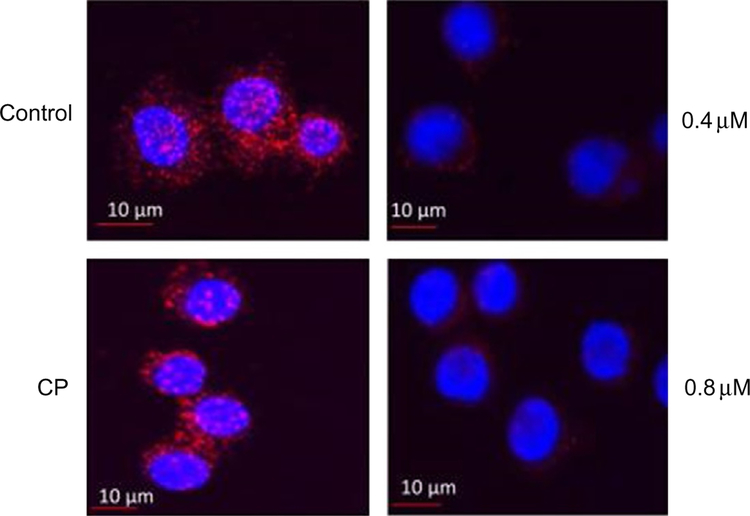
Fig. 2.
HER2:HER3 heterodimerization and its inhibition by compound 9 observed by PLA. Concentration-dependent inhibition of HER2:HER3 heterodimers was observed. Control-SKBR-3 cells showing only HER2:HER3 heterodimerization as red spots. CP-A control compound did not inhibit HER2:HER3 heterodimerization. At a sub-optimum dose of compound 9 (0.4μM), HER2:HER3 heterodimerization was inhibited to a lesser extent. At an optimum dose of compound 9 (0.8 μM), HER2:HER3 heterodimerization was significantly inhibited. Reproduced with permission from Kanthala, S., Banappagari, S., Gokhale, A., Liu, Y. Y., Xin, G., Zhao, Y., & Jois, S. (2015). Novel peptidomimetics for inhibition of HER2:HER3 heterodimerization in HER2-positive breast cancer. Chemical Biology & Drug Design, 85, 702–714. https://doi.org/10.1111/cbdd.12453. Copyright (2014) John Wiley and Sons.
2.4. Enzyme Fragment Complementation Assay (PathHunter Assay)
PathHunter™ assay (DiscoveRx, Fremont, CA) or enzyme fragment complementation assay can be used to confirm the interaction between two proteins and to evaluate whether the ligand designed inhibits the PPIs. These assays may only be applicable to certain proteins since the method requires the expression of proteins of interest in model cell lines, and the two proteins are attached with tags of fragments of beta-galactosidase. The assay is widely used for dimerization of receptor tyrosine kinase (RTK) proteins. The enzyme beta-galactosidase is split into two inactive fragments. The two fragments are expressed as tags in proteins of interest using engineered cells. When the two proteins interact with one another, the tags from the two proteins which were inactive form an active complex. The activity of the complex can be detected by an enzymatic assay using luminescence. The relative luminescence intensity can be quantified as the formation of PPI (Yin et al., 2009). In cells, RTK activation results in dimerization of receptors, and this leads to phosphorylation and binding of the SH2 domain to the kinase domain. When RTK and SH2 domains come into proximity, the inactive galactosidase fragments come together to form the active enzyme. The active enzyme produces a chemiluminescent signal that can be detected and directly correlated to kinase activation, phosphorylation, and interaction with SH2 domain-containing proteins. This assay is specific for RTK dimerization. The limitation of this method in the present form is that the assay is carried out on engineered cells and not on native forms. However, the assay is used to evaluate the inhibition of dimerization also. The application of this method is used in studying G-protein-coupled receptor (GPCR) binding by different proteins. Since GPCR is the target for most of the drugs, the assay is useful in designing drug-like molecules (Yin et al., 2009). In our laboratory, we have used the enzyme fragment complementation assay to evaluate the ability of peptidomimetics to inhibit HER2–HER3 dimerization in U2OS cell lines (Fig. 3).
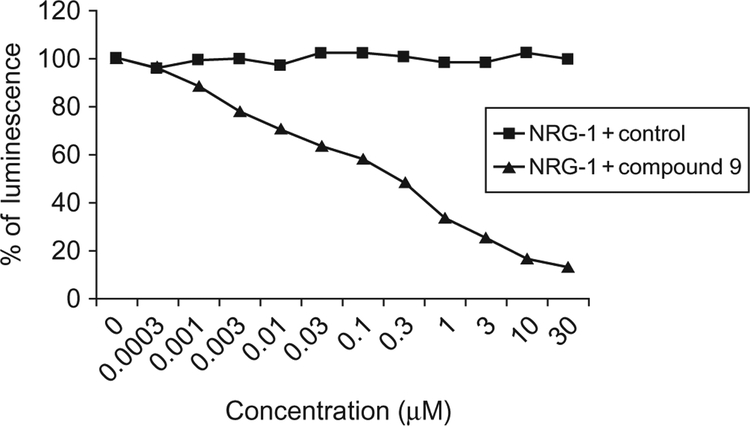
Fig. 3.
Inhibition of heterodimerization of HER2:HER3 in HER2, HER3 transfected U2OS cells by compound 9 at different concentrations using enzyme fragment complementation assay (DiscoveRx). Dose–response curve for inhibition of HER2:HER3 heterodimerization by compound 9 in the presence of 0.3μM NRG1 (triangles). Control compound (CP) in the presence of 0.3μM NRG-1 (filled squares). Reproduced with permission from Kanthala, S., Banappagari, S., Gokhale, A., Liu, Y. Y., Xin, G., Zhao, Y., & Jois, S.(2015). Novel peptidomimetics for inhibition of HER2:HER3 heterodimerization in HER2-positive breast cancer. Chemical Biology & Drug Design, 85, 702–714. https://doi.org/10.1111/cbdd.12453. Copyright (2014) John Wiley and Sons.
2.5. Surface Plasmon Resonance
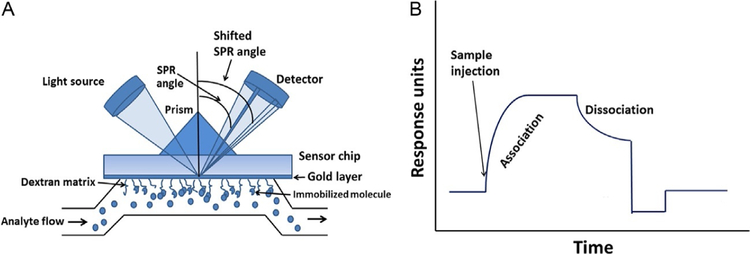
While many methods such as IP and PLA as well as enzyme fragment complementation assays provide information about PPI, the detection methods are indirect, and methods such as IP involve several steps. A technique that directly detects the interaction of two purified proteins is SPR (de Mol & Fischer, 2010; Wilson, 2002). In this method, one of the proteins is immobilized on a sensor chip surface, the other is made to flow over this surface, and the binding kinetics are detected by a change in the refractive index. The sensor chip is created by applying a thin layer of gold to a glass surface. A dextran matrix is applied to the gold to create an environment for biomolecular interactions, and this surface can be combined with or modified by different chemicals to provide linkers for different types of chemical reactions for immobilization. Most widely used sensor chips are carboxymethyl-dextran linked to a gold surface. Such layers are useful for immobilization of proteins and peptides that have free amine groups. SPR chips that are ready to use are available from commercial sources (GE Healthcare, Pittsburgh, PA). A source of light passes through a prism and strikes the surface of the flow cell at an angle such that the beam is totally reflected. This light beam creates surface plasmon wave of excited electrons on the gold surface (Fig. 4A). Although the light undergoes total reflection, an electromagnetic field component called an evanescent wave penetrates into the medium <500nm of the lower refractive index, in this case, into the dextran layer where PPIs can be studied. Depending on the material on the gold surface and dextran layer, the reflective beam will have a reduced intensity. The angle at which the light beam strikes the detector from a reference surface point is called the SPR angle. The SPR angle is sensitive to the composition of the layer at the surface of the gold. As two molecules interact with one another, there will be an accumulation of molecules at the dextran surface because of the binding of one protein to another; hence, the refractive index of the surface changes, causing a change in the SPR angle.
Fig. 4.
The principle of SPR analysis. (A) SPR chip with analyte flow and SPR angle. (B) SPR sensorgram indicating association and dissociation phases.
The SPR angle is directly proportional to the amount of bound molecules at the surface with respect to a reference surface. If the two molecules do not interact, there is no binding, and the SPR angle will not change; therefore, there will be no SPR signal. Thus, PPIs can be directly detected. A schematic diagram of SPR sensorgrams of the interaction of two proteins is shown in Fig. 4B. The advantages of this method are that no labeling is required for molecules and the interaction between the molecules is detected in real time. This means that the kinetics of association and dissociation can be measured, which is difficult in many other experiments that determine the binding affinity of two molecules. SPR technology requires a very small amount of sample. With BIACORE technology, 1000 resonance units (RU) corresponds to an SPR angle of 0.1 degrees. For most proteins, binding of 1ng/mm2 of protein at a dextran surface causes a change of 1000 RU. This technique also determines interaction affinity of molecules over a wide range of binding affinity (Table 1) and molecular weights. The area of detection of this interaction extends up to 300nm from the gold surface and, therefore, the interaction of an immobilized protein with a protein on a cell surface can be detected using the proper flow rate and buffer.
Table 1.
Range of Binding and Kinetics Parameters That Can Be Measured Using SPR Analysis
| Concentration Range | Kinetics | Affinity |
|---|---|---|
| 1 mM–l0pM | Association rate constant ka=103−107M−1s−1 |
Kd 100μM to 200pM |
| Dissociation rate constant kd=10−1−5×l0−6s−1 |
SPR chips have two- or three-channel flow cells. The volume of each flow cell is around 0.02μL and, hence, the amount of sample needed is very small. After the protein has been immobilized on the chip surface, the chip can be reused for only a certain period of time since the bound protein or ligand can be washed away; however, regeneration buffer can be used to make the chip surface available for a second analyte or another experiment. An SPR sensorgram consists of different phases—a baseline to start with and, after injection of the analyte if there is binding, an association phase and steady state where the binding equilibrium reaches saturation, and a dissociation phase. After dissociation, the chip can be regenerated, which is referred to as the regeneration phase. Binding kinetics can be obtained by titrating the analyte over a wide variety of concentration ranges against the protein immobilized on the chip surface. A typical binding experiment is shown in Fig. 4B. Apart from protein–protein or protein–ligand binding, PPI inhibition can be studied using the coinjection mode (Banappagari, Corti, Pincus, & Satyanarayanajois, 2012).
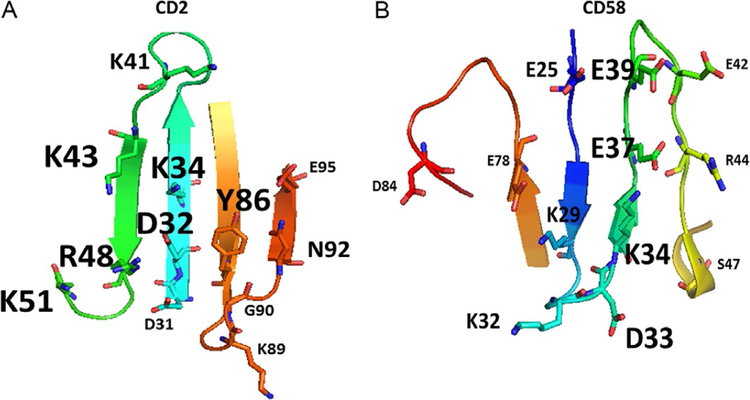
2.6. Mutational Studies to Identify Hot Spots in PPI
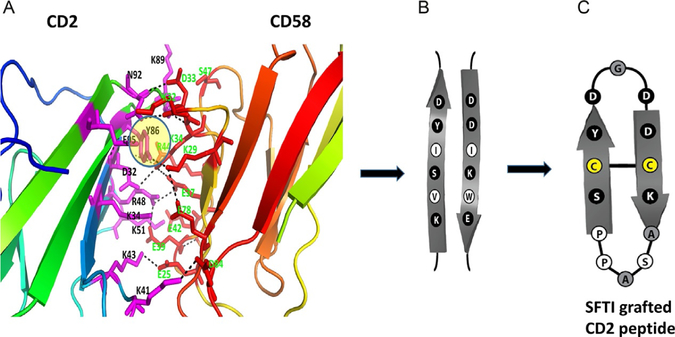
Most of the PPI inhibitors target the interface of proteins in small regions called hot spots. A region of protein surface is called a hot spot when replacement of an amino acid residue by alanine in that spot lowers the free energy of binding by at least 2kcal/mol (Clackson & Wells, 1995). Computational as well as experimental methods are available to identify hot spots on the surface of interacting proteins. Experimental study involves very tedious work where the two proteins of interest are expressed and purified, and binding of the two proteins is studied by isothermal calorimetry or any other method that can detect protein binding. Then, particular amino acids on each protein are mutated with alanine using site-directed mutagenesis, and binding studies on the mutated proteins are carried out. The change in the affinity of binding upon mutation is evaluated. One such example is the interaction between CD2 and CD58 proteins that are involved in cell adhesion and immune response. As an example, mutation studies carried out on CD2–CD58 protein pairs are illustrated in Fig. 5A and B. Point mutation was carried out on the proposed binding of the CD2 protein to CD58 protein (Kim et al., 2001). The crystal structure of CD2 complexed with CD58 has been reported (Wang et al., 1999). Detailed analysis of amino acid residues involved in PPI of CD2 and CD58 indicated that the interface interaction is mainly electrostatic in nature with 10 salt bridges and 5 hydrogen bonds (Wang et al., 1999) and that the interface area is around 1200Å2. When the amino acid residues that form salt bridges and hydrogen bonds in the two proteins were replaced with alanine by point mutation, the interaction between CD2 and CD58 was not significantly altered, suggesting that electrostatic interactions did not contribute significantly to form the heterodimer. When a hydrophobic amino acid Tyr86 in the interface region was subjected to point mutation by replacement of alanine, the binding affinity between the two proteins reduced nearly by 1000-fold, suggesting that the hot spot is a hydrophobic region. Based on this mutation study, the researchers proposed that hot spots on these proteins are at Tyr86 (Fig. 5A). Such hot spots can be used for the design of PPI inhibitors.
Fig. 5.
The structure of PPI domains of (A) CD2 and (B) CD58 with amino acids that form hydrogen bonding, salt bridges, and hydrophobic interaction (PDB ID 1QA9). Mutation studies suggested that the affinity of interaction between the two proteins is reduced when alanine is replaced by some of the residues in the proteins (Kim et al., 2001; Wang et al., 1999). Residues that have the most effect on affinity of binding are represented by large-sized letters for amino acid labels. Residues that partially affect the affinity are represented with medium-sized letters. Residues that do not affect the affinity by replacement with alanine are represented with small-sized letters.
Other experimental techniques such as fluorescence resonance energy transfer (FRET) (Karpova & McNally, 2006), bioluminescence resonance energy transfer (Kocan & Pfleger, 2011), mass spectrometry (Kaake et al., 2014), and isothermal calorimetry (Velazquez-Campoy, Leavitt, & Freire, 2004) are also available for studying PPI (Berg, 2005). In terms of investigating PPI in the pure form of the protein, NMR and X-ray crystallography are used. These methods provide detailed knowledge about the binding surface of the two proteins involved. However, protein purification, crystallization, and data collection are time consuming. Before proceeding with NMR or X-ray crystallography to determine details of PPI, relatively faster and physiologically more important methods such as Co-IP and pull-down assay are used.
3. INHIBITION OF DIMERIZATION OF PROTEIN AND PPI
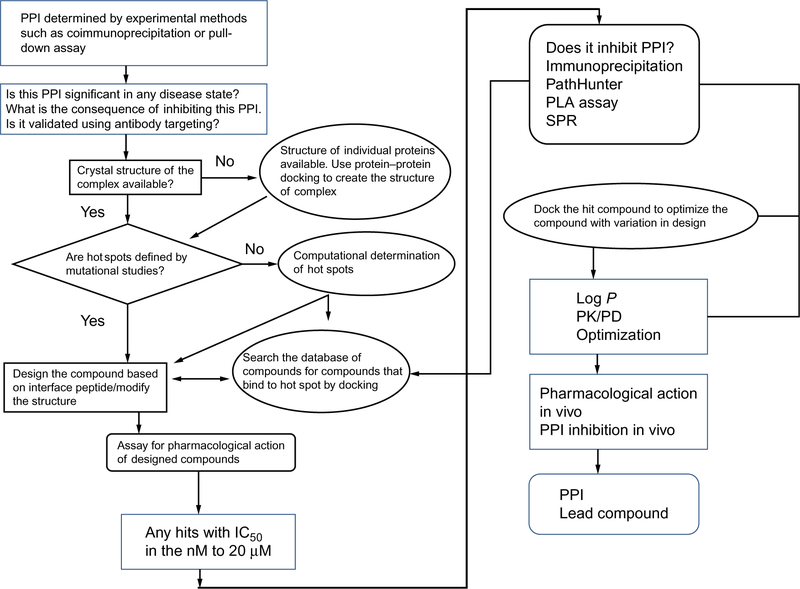
The first step in the design of PPI inhibitors is to evaluate whether the proteins that interact with one another and the biochemical pathway that results have any significance in any disease state. If they have importance in a disease state, will the inhibition of PPI lead to alleviation of this disease state? Most of the data related to the PPI importance in a disease state come from basic and clinical studies or genomics studies related to the disease. Once the proteins of interest that have significance in a disease state have been identified, the next question to ask is whether the inhibition of this particular PPI leads to any significant adverse effects on the general physiological processes in the body. If the PPI inhibition might result in significant effects on normal physiology, then that PPI should not be targeted for the drug discovery process. Once the target PPI partners are identified, is there a model system such as cell lines that is relevant for the disease state? If the cell lines chosen express the proteins of interest, they would serve as a good model, and molecular biology methods such as the coimmunoprecipitation assay and pull-down assay (described earlier in methods to study PPI) can be used to show that the two proteins of interest form heterodimers. On the other hand, if the crystal structures of the complex of proteins of interest have already been elucidated and are available in the literature, one can jump directly to the design of inhibitors. There are several steps in the design of PPI inhibitors. The overall process of the design of PPI inhibitors is schematically represented in Fig. 6.
Fig. 6.
A schematic diagram of a decision tree for the design of PPI inhibitors. Adapted from Sable, R., & Jois, S. (2015). Surfing the protein-protein interaction surface using docking methods: Application to the design of PPI inhibitors. Molecules, 20(6), 11569–11603. https://doi.org/10.3390/molecules200611569. MDPI.
As more and more crystal structures of the physiologically important proteins became available, and interactions of proteins were deciphered, it was realized that PPI surfaces are important in modulating the physiological processes for alleviating many disease states. PPIs control most of the physiological processes in the body, and any deregulation in PPI leads to disease states (Chene, 2006). This understanding led to a new area in drug design and discovery in which the molecules designed have to bind to PPI surfaces. In the early 1980s and 1990s, it was presumed that PPI surfaces were rather flat, and that inhibiting such surfaces could be done only by large molecules such as antibodies or fusion proteins (Wells & McClendon, 2007). However, this concept was overridden when peptides, peptidomimetics, and small molecules were designed to inhibit PPI. Typically, PPI surfaces have a span of 700–3000Å2 and have shallow grooves and channels or small ridges that can accommodate functional hydrophobic groups to interact with the receptor protein. For such large surface interactions, designing a drug seems difficult compared to designing traditional small molecules having drug-like properties (Lo Conte, Chothia, & Janin, 1999; Moreira, Fernandes, & Ramos, 2007; Reichmann, Rahat, Cohen, Neuvirth, & Schreiber, 2007; Wells & McClendon, 2007).
Detailed 3D-structure analyses of PPI revealed that PPIs are made up of secondary structure epitopes that are derived from a continuous or a discontinuous epitope. It is well documented in the literature that PPI surfaces are generally hydrophobic in nature and all of the interfaces between the two proteins do not necessarily contributes to binding energy. Small hydrophobic spots contribute to the free energy of binding and help to hold the two proteins together. Such regions on PPI interfaces are called hot spots (Clackson & Wells, 1995). These hot spots have a core region and a rim region; the amino acid composition of the core region contains aromatic residues, whereas, in the rim region, the amino acid composition is similar to that of the rest of the protein surface (Chakrabarti & Janin, 2002; Chene, 2006; DeLano, 2002). Amino acids that were frequently found in PPI hot spots were Trp, Tyr, Leu, Ile, Phe, and Arg (Bogan & Thorn, 1998). Amino acid Trp can form hydrophobic as well as hydrogen bonds with ligand molecules without the introduction of water at the PPI site. Among hydrophobic amino acids with similar structural properties, such as Leu and Ile, Ile seems to be preferred at PPI (Moreira et al., 2007). Based on the knowledge of PPI surface, compounds that can bind to PPI surface can be designed.
4. DESIGN OF COMPOUNDS BASED ON PROTEIN INTERFACE
At present the methods used to design the PPI inhibitors can be classified into three major groups: (1) structure-based drug design (SBDD), (2) high-throughput screening (HTS), and (3) fragment-based drug design (FBDD). Each of these methods is described briefly here.
4.1. Structure-Based Drug Design
SBDD uses the fact that peptide fragments that are involved in PPI surfaces can be mimicked by peptides, peptidomimetics, or small organic molecules. These molecules are called “proteomimics” (Jubb, Higueruelo, Winter, & Blundell, 2012). Since PPIs are concentrated in small regions, the interface contact is held by a few key residues that are arranged in a particular three-dimensional arrangement. The functional groups and geometric arrangement of these functional groups that exist in the interface of proteins can form a template for pharmacophores. These protein recognition sites usually consist of particular secondary structures that can be used in the design of PPI inhibitors. The most common are α-helical structures, extended or β-strand structures, loops, β-turns, and proline-rich motifs (Gokhale & Satyanarayanajois, 2014; Wilson, 2009). However, these small fragments of peptides acquire particular stable secondary structures in a globular protein because of different interactions within the protein structure. When such fragments of peptide structures are isolated, they do not acquire stable secondary structures that mimic those of the proteins. Hence, these secondary structures have to be constrained using different functional groups and different strategies. One of the best examples of constrained secondary structure is that of an α-helix mimic for p53–MDM2 PPI is important in the regulation of cancer development (Vassilev et al., 2004).
4.2. Fragment-Based Discovery in PPI Inhibition
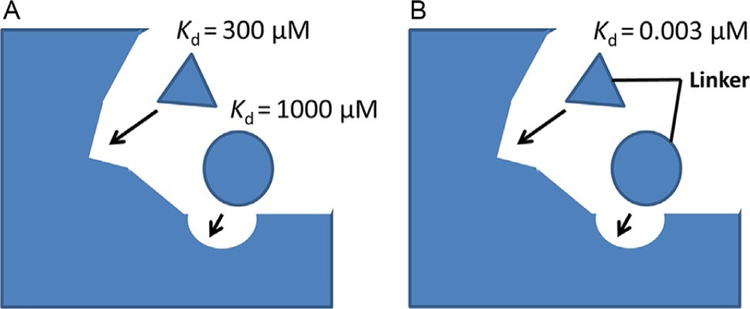
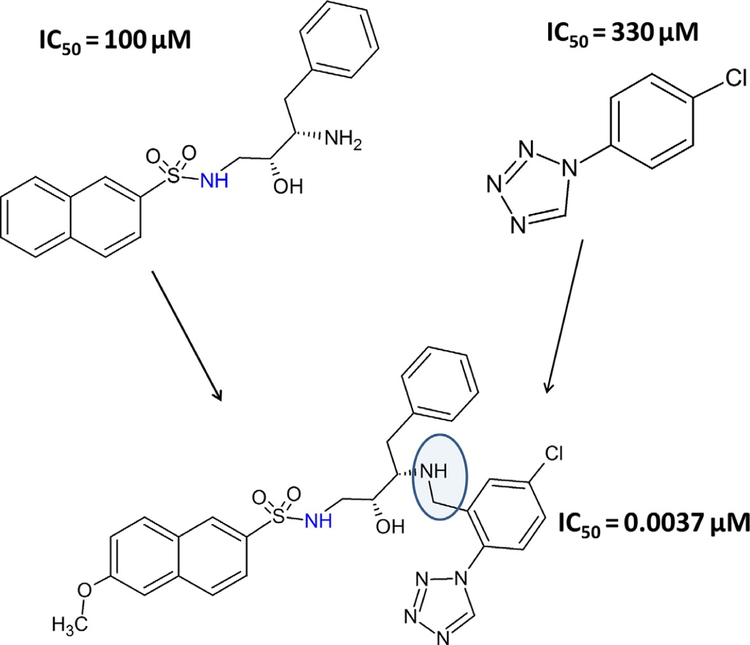
FBDD concept was initiated three decades ago (Jencks, 1981; Verlinde, Rudenko, & Hol, 1992); however, the method was restricted to enzyme-based drug discovery and, in particular, applied to kinase inhibitors. The first success of this method was achieved in 2011 after the approval by the Food and Drug Administration (FDA) of the drug Zelboraf (PLX4032) (Bollag et al., 2010), a B-Raf inhibitor. The same principle can be applied to the design of PPI inhibitors. In the first step in this method, a ligand is constructed from fragments of chemical building blocks that are optimized for binding to small subsites on protein surfaces. A library of low molecular weight fragments are screened for binding to proteins of interest (Scott, Coyne, Hudson, & Abell, 2012). Screening can be performed by different experimental techniques such as fluorescence assay or NMR. Using a method called structure–activity relationship (SAR) by NMR (Shuker, Hajduk, Meadows, & Fesik, 1996), a large number of small molecular fragments can be screened to bind to one subsite of a protein surface. In the second step, another set of compounds is screened to bind to another site (near the first site). Lead compounds that show relatively high-affinity binding are evaluated further to find the exact binding site of these two fragments on the protein surface. Then, using the medicinal chemistry knowledge, these two fragments are linked chemically to arrive at the new lead compound (Erlanson, 2006). Usually, the linked compound will have higher affinity to bind to protein compared to individual fragments that bind to different sites (Fig. 7A and B). Note that the fragments of compounds designed in the first and second steps bind with low affinity to the target protein (Fig. 8). When the fragments are linked, the affinity of the new molecule is significantly increased. The overall idea is to build a molecule that can fit into the shallow groove of the PPI using fragments of chemicals and knowledge of the binding site. An example of the SAR by the NMR method is the discovery of BH3-Bcl-XL inhibitor (Oltersdorf et al., 2005). Apart from these methods, natural product screening is also used for PPI inhibition (Sperl, Seifert, & Berg, 2009).
Fig. 7.
A schematic representation of the fragment-based drug design principle.(A) Fragments are screened from a database or are built based on PPI hot spots on the protein. Each fragment has a relatively weak dissociation constant. (B) The screened fragments are linked chemically to obtain a high-affinity binding ligand.
Fig. 8.
Example of fragment-based approach. Two molecules with IC50 values of 100 and 330μM, respectively, were linked to obtain a high-affinity ligand that has an IC50 value of 37nM. Adapted from Erlanson, D. A. (2006). Fragment-based lead discovery:A chemical update. Current Opinion in Biotechnology, 17(6), 643–652. https://doi.org/10.1016/j.copbio.2006.10.007. Copyright (2006) Elsevier.
4.3. High-Throughput Screening
Conventional tools used in drug discovery such as HTS (Fox et al., 2006; Mayr & Fuerst, 2008) can be used to discover PPI inhibitors. The first step in such cases is the creation of the structure of a complex of two proteins and identification of hot spots on one of the protein interfaces. If the hot spots have a cleft or a relatively deep pocket, the databases of the molecules are screened using experimental as well as computational analysis to find out whether any of the organic molecules bind to the cleft. A compound that binds to the groove in the lower nanomolar range will be used for lead compound generation. The HTS method involves screening large numbers of compounds in a chemical library to find a lead compound that binds to the target protein with high affinity (Kd<1μM). One important aspect of HTS is the development of a fast, reliable cell-based or enzyme-based assay to screen large numbers of compounds. Usually, these assays are carried out in microwell plates that have 1536 wells per plate and use 2.5–10μL of the medium. With such microplates, one can screen 200,000 compounds per day (Mayr & Fuerst, 2008). In the pharmaceutical and biotechnology industries, these systems are completely automated once the assay is established. For PPI inhibitor studies, such assays are modified to evaluate the binding of these compounds to one of the protein partners. Identification of a small molecule that inhibits the interaction of p53 with MDM2 provides an example of the use of HTS (Vassilev et al., 2004).
5. HOMODIMERS
It is reported that in eukaryotic organisms that contain a large number of self-interacting proteins and the ability to self-interact have several structural and functional advantages over proteins (Ispolatov, Yuryev, Mazo, & Maslov, 2005). Self-association of proteins is known to provide improved stability (Dunbar et al., 2004) and control over the accessibility and specificity of active sites in proteins (Marianayagam et al., 2004). Furthermore, self-association can help to minimize genome size. Apart from self-association, structurally similar proteins have statistically significant interaction propensity compared to structurally nonsimilar proteins (Ispolatov et al., 2005). Structural analysis of interface residues in homodimers using the available crystal structures has been reported, and it was found that hydrophobic residues such as Ala, Val, Leu, Met, Ile, and Phe are predominant at the homodimer interface (Zhanhua, Gan, Lei, Sakharkar, & Kangueane, 2005). Analysis of structures of homodimers and oligomers indicated that most of the homodimers and oligomers are symmetric (Dayhoff, Shoemaker, Bryant, & Panchenko, 2010; Levy, Pereira-Leal, Chothia, & Teichmann, 2006). Assymetric homodimers are also formed, but are known to perform certain specialized functions (Goodsell & Olson, 2000). Swapna, Srikeerthana, and Srinivasan (2012) have analyzed a non-redundant set of high-resolution crystal structures for symmetric and asymmetric homodimers and found that some functional proteins do form asymmetric structures. For the inhibition of drug design, symmetry in the dimerization is also important. Here we discuss some of the homodimers and their inhibition for drug discovery.
5.1. G-Protein-Coupled Receptors
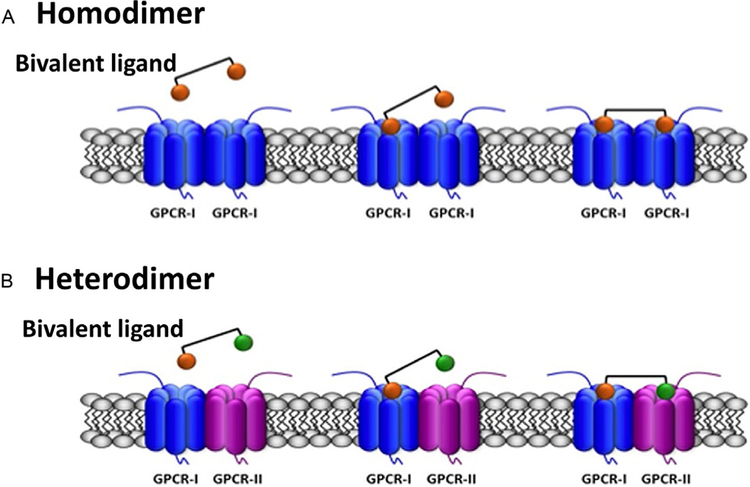
GPCRs are a major component of cell surface receptors, and there are more than 800 known GPCRs. These receptors are involved in the signal transduction process. The structure of GPCRs consists of an N-terminal region that is typically outside the cell membrane and seven transmembrane helices (TM-I–TM-VII). The C-terminus extends into the cytoplasm. GPCRs are involved in a variety of physiological processes, including visual sense, taste, smell, behavior regulation, autonomic nervous system, immune system and inflammation, homeostasis, and tumor formation (Luttrell, 2006). GPCRs are classified into glutamate, rhodopsin, adhesion, frizzled/taste2, and secretin families (Fredriksson, Lagerstrom, Lundin, & Schioth, 2003). Nearly half of the drugs used in clinical practice directly or indirectly target GPCRs (Jacoby, Bouhelal, Gerspacher, & Seuwen, 2006; Tyndall & Sandilya, 2005), clearly indicating the importance of these receptors in controlling physiological processes and drug targeting. GPCRs were initially considered to be monomers on the cell surface of neuronal and nonneuronal cells. However, because GPCR can form homodimers, heterodimers, and higher order oligomers on the plasma membrane, the concept of GPCRs being only monomers can no longer be substantiated (Bouvier, 2001; Franco et al., 2007; Szidonya, Cserzo, & Hunyady, 2008). In the past decade, several experimental reports that were available suggested that GPCR oligomerization may be important for receptor function, including agonist binding, potency, efficacy, and G-protein selectivity. One of the limitations of the biophysical experimental methods that were used to detect GPCR oligomers is that these methods could not distinguish between homodimers and oligomers (Chabre, Cone, & Saibil, 2003; James, Oliveira, Carmo, Iaboni, & Davis, 2006). For the past few years, several groups have designed “bivalent ligands” that bind to GPCR dimeric structure to prove the importance of dimerization as well as the possibility of drug design using the biva-lent ligand concept (Fig. 9). These molecules consist of two agonist/antagonist moieties separated by spacers of variable lengths. By bridging the pharmacophores at the two sites, a bivalent ligand can be designed that can bind to two sites, one on each homodimer or heterodimer. Such biva-lent ligands can also be used to detect protein dimers (Brogi, Tafi, Desaubry, & Nebigil, 2014; Franco, Martinez-Pinilla, Lanciego, & Navarro, 2016; Hiller, Kuhhorn, & Gmeiner, 2013; Xu et al., 2012).
Fig. 9.
Scheme of the simultaneous binding of bivalent ligands with linkers of appropriate length to two receptors in a GPCR dimer. The agonist/antagonist moieties of the bivalent ligand are selective for their respective receptors (orange for GPCR-I; green for GPCR-II) and are linked by a spacer. (A) Binding to two equal GPCRs forming a homo-dimer. (B) Binding to two different GPCRs forming a heterodimer. Reproduced with permission from Franco, R., Martinez-Pinilla, E., Lanciego, J. L., & Navarro, G. (2016). Basic pharmacological and structural evidence for class A g-protein-coupled receptor heteromerization. Frontiers in Pharmacology, 7, 76. https://doi.org/10.3389/fphar.2016.00076. Copyright (2016).
5.2. Receptor Tyrosine Kinase-Like Orphan Receptor 2
Receptor tyrosine kinase-like orphan receptors 1 and 2 (Ror1 and Ror2) are two members of Ror, which is a neurotrophic tyrosine kinase receptor within the RTK family. Ror receptors are very closely related to Trk neurotrophin (NT) receptors and muscle-specific kinase. Ror2 plays an important role in developmental morphogenesis, specifically of the cartilage-derived skeleton (Roarty, Shore, Creighton, & Rosen, 2015). It has been found that disruption of mouse Ror2 corresponds to extensive skeletal abnormalities in which all endochondrally derived bones are fore-shortened or misshapen, while in humans, the mutation in Ror2 gene accounts for short height, limb bone shortening, and segmental defects of the spine (Aglan et al., 2015). Receptor dimerization is induced by ligand binding to Ror2. Elucidation of molecular mechanism indicated that Ror2 binds to Wnt family glycoproteins and modulates the Wnt signaling. Ror2 is also known to interact with a bone morphogenetic protein receptor type Ib (BRI-b) that modulates cartilage development. The coexpression of Ror2 and casein kinase I is known to result in tyrosine phosphorylation of GPCR kinase (Liu, Ross, Bodine, & Billiard, 2007). Inhibition of Ror2 homodimerization can be useful in different types of cancer also via Wnt pathway. Recently, it has been shown that Ror2 is upregulated in renal cell carcinoma (RCC) tissues and cell lines. Knockdown of Ror2 also inhibits proliferation, migration, and invasion and induces G1 phase cell cycle arrest and apoptosis of RCC cell lines. Knockdown of Ror2 is also known to inhibit tumor growth in vivo. RCC represents one of the most resistant tumors to radiation and chemotherapy (Yang et al., 2017). Hence, the design of molecules to modulate Ror2 dimerization may lead to useful therapeutic agents. At present, there are no reports of known inhibitors of Ror2 homodimerization.
5.3. Glucocorticoid Receptor
Human glucocorticoid receptor (GR), a nuclear receptor superfamily receptor, is associated with many physiological processes such as immune regulation and metabolism. Homodimerization of GR is important for control of GR transcriptional activity (Oasa, Sasaki, Yamamoto, Mikuni, & Kinjo, 2015). GR generally binds to glucocorticoid response elements that modulate the transcription process as homodimers. GR consists of an N-terminal transactivation domain, a central DNA-binding domain (DBD), a C-terminal ligand-binding domain (LBD), and a flexible “hinge region” that separates the DBD and the LBD. Of several members of the nuclear receptor superfamily, the DBD is the most critical region. The two zinc-finger motifs present in the DBD recognize and bind specific DNA sequences on target response elements (Kadmiel & Cidlowski, 2013). GR is also known to participate in nongenomic signaling that does not require nuclear-GR-mediated transcription or translation. Nongenomic signaling effects of GR are rapid and have implications in various systems, including the cardiovascular, immune, and neuroendocrine systems. GR isoforms are expressed in nearly all tissue types, and glucocorticoid signaling is almost ubiquitously prevalent in the various organ systems (Oakley & Cidlowski, 2011). Due to their antiinflammatory, antiproliferative, proapoptotic, and antiangiogenic roles, glucocorticoids have been remarkably effective in treating various diseases. Although the details of dimerization of structural aspects of GR with ligand are relatively recent (Bledsoe et al., 2002; Madauss et al., 2008), GR has been a successful drug target for nearly 60 years. The first report of the clinical use of GR targeting was for rheumatoid arthritis (RA) in 1940, and since that time GR has been targeted for many conditions such as chronic inflammatory conditions, including asthma, skin infections, and ocular infections, as well as for immunosuppression in patients undergoing an organ transplant. In addition to their antiinflammatory properties, the antiproliferative and antiangiogenic actions of corticosteroids have been exploited for the treatment of cancers (Vilasco et al., 2011).
5.4. Amyloid Precursor Protein Dimers
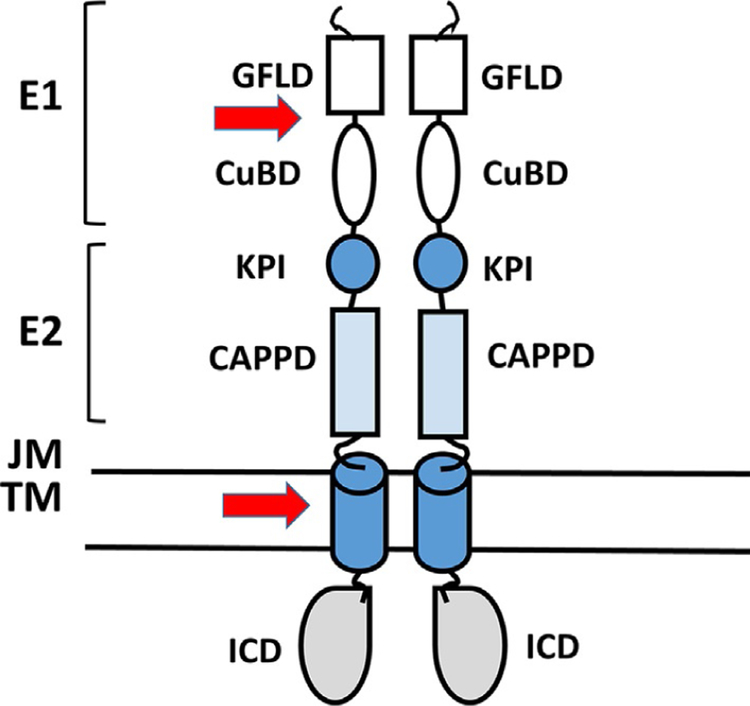
Amyloid precursor protein (APP) is a type I transmembrane protein expressed in many cell types, including neurons. APP is a 695 amino acid protein with a large ectodomain and relatively short intracellular region. APP has been shown to form homodimers (Khalifa et al., 2010). In APP, dimerization is known to be induced by the N-terminal region of APP, referred to as the E1 region (Fig. 10), which contains a growth factor-like domain and a copper-binding domain (CuBD) (Soba et al., 2005). A loop formed by disulfide bridges is required for the stabilization of the homodimeric state. Further, juxtamembrane (JM) and transmembrane (TM) regions also participate in homodimerization. APP is processed into smaller fragments, and there are two known catabolic pathways, namely, nonamyloidogenic and amyloidogenic pathways (Khalifa et al., 2010). APP processing seems to be a critical event in the onset and progression of Alzheimer’s disease (AD) (De Strooper, Vassar, & Golde, 2010) and hence homodimerization of APP and the details of domains and amino acid residues involved in particular domains are studied in detail. In AD, the main component of plaques is the amyloid beta (Aβ) peptides with 40–42 amino acids (Masters et al., 1985). These peptides are released from a precursor protein APP by sequential cleavage by beta-site APP-cleaving enzyme 1 (BACE1) and by the γ-secretase complex. Cleavage of APP that consists of 695 amino acids by BACE1 releases the large ectodomain of APP and membrane-anchored C-terminal APP fragment (CTF) of 99 amino acids. The 99 amino acid polypeptide will undergo further cleavage by γ-secretase resulting in Aβ peptides of various lengths. APP intracellular domain (ICD) is released into the cytosol (Eggert, Midthune, Cottrell, & Koo, 2009; Jung et al., 2014; Vassar et al., 1999). It was proposed that dimerization of TM domain and amino acids in the TM domain is important in this cleavage process. APP contains three glycine-xxx-glycine (GxxxG) motifs at the extra-cellular JM/TM boundary. It is reported that the GxxxG motifs in the APP TM domain participate in dimerization and this domain is located in the region where cleavage occurs. Structural studies on the APP JM/TM region in isolation showed that the GxxxG motifs mediate TM helix homodimerization of the protein in the lipid bilayer (Sato et al., 2009; Fig. 10). Mutational studies by the introduction of a cysteine residue at the junction of the JM/TM region were shown to form stable dimers linked by disulfide bridges. The stabilization of dimerization leads to increased Aβ production (Scheuermann et al., 2001). Aβ is produced as a stable dimer, indicating that the amyloidogenic secretases (β and γ) are able to process APP under its dimeric form. Thus, dimerization seems to help Aβ production. The motifs involved in dimerization of C-terminal APP fragments (CTFs) are also responsible for the packing of Aβ peptides into protofibrillar structures (Sato et al., 2006). The glycines present in GxxxG motifs are important in the PPI of TM helices as well as in the formation of the cross β-sheet structures found in the Aβ fibrils. The GxFxGxF framework seems to be the hot spot for designing drug-like molecules for AD. Peptides can be designed to disrupt sheet-to-sheet packing and inhibit the formation of mature toxic Aβ fibrils. Antibodies mapping to an epitope in this Aβ region are also able to significantly reduce the accumulation of intracellular Aβ, which is known to be highly neurotoxic (Tampellini et al., 2007). Thus, the dimerization process, the GxxxG motifs, the details of structure in the dimerization region, and the cleavage of this region by secretase are important in designing drugs for AD. Richter et al. (2010) have studied the molecular mechanism of γ-secretase modulators such as sulindac sulfide and indomethacin and, using molecular docking studies, have suggested that these compounds bind at the smooth surface provided by glycines arranged in GxxxG motifs (Richter et al., 2010).
Fig. 10.
Schematic representation of different domains of APP that form homodimers. Arrows indicate the proposed dimerization regions. CAPP, central APP domain; CuBD, copper-binding region; GFLD, growth factor-like domain; ICD, intracellular domain; JM, juxtamembrane region; KPI, Kunitz protease inhibitor domain; TM, transmembrane region. Schematic diagram is drawn based on Khalifa, N. B., Van Hees, J., Tasiaux, B., Huysseune, S., Smith, S. O., Constantinescu, S. N., et al. (2010). What is the role of amyloid precursor protein dimerization? Cell Adhesion & Migration, 4(2), 268–272; Eggert, S., Midthune, B., Cottrell, B., & Koo, E. H. (2009). Induced dimerization of the amyloid precursor protein leads to decreased amyloid-beta protein production. The Journal of Biological Chemistry, 284(42), 28943–28952. https://doi.org/10.1074/jbc.M109.038646.
Munter et al. (2007) have shown that γ-secretase processivity is reduced when CTFβ forms dimers, because of interactions of TM domain GxxxG motifs. This leads to the formation of fragments of Aβ isoforms which are larger in size compared to 40 amino acid Aβ. There are reports indicating that APP CTFβ dimers are not γ-secretase substrates. Jung et al. (2014) studied the importance of residues at the interface of APP ectodomain and TMD by mutating the lysine residues at the interface of the APP ectodomain and transmembrane domain (TMD) and evaluated the Aβ production. Based on their studies, they concluded that the monomeric form of the mutant increased long Aβ production without altering the initial ε-cleavage utilization, whereas dimeric forms of APP are not efficient γ-secretase substrates and primary sequence determinants within APP substrates alter γ-secretase processivity. Thus, there is controversy regarding the dimerization of APP and its link to cleavage of APP by γ-secretase. The design of inhibitors of APP has to be carefully considered when targeting a particular region of APP that helps for homodimerization.
5.5. EGFR Homodimers
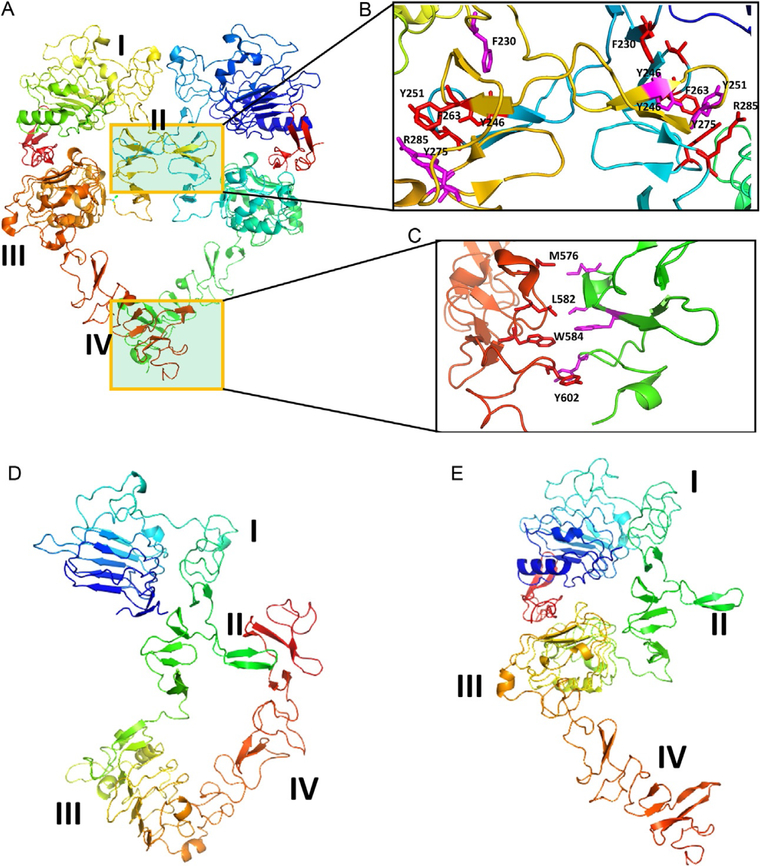
EGFR (also known as ErbB1 or HER1) is a well-known tyrosine kinase receptor involved in the signal transduction process. EGFR has importance in key stages of the development of organisms, such as cell proliferation, motility, differentiation, and tissue homeostasis. Overexpression of EGFR or enhancement of the receptor activity results in tumorigenesis. EGFR has an extracellular domain (ECD) consisting of 621 amino acids, a single TMD with 25 amino acids, and a cytoplasmic kinase domain with nearly 800 amino acids, forming the complete structure (Baselga & Swain, 2009; Ferguson, 2008). Peptide growth factor ligands such as epidermal growth factor (EGF), transforming growth factor α (TGFα), amphiregulin, betacellulin, epigen, epiregulin, and heparin-binding EGF-like growth factor are known to bind to EGFR. Binding of these ligands is known to induce change in the conformation of the ECD of EGFR. Among these, only EGF, TGFα, amphiregulin, and epigen associate specifically with the EGFR homodimer (Roskoski, 2014). The homodimer of EGFR ECD structure has been elucidated by X-ray crystallography (Lu et al., 2010) and electron microscopy (Mi et al., 2008, 2011). The ECD of EGFR consists of four domains, namely domains I–IV (domain I residues 1–165, domain II residues 166–309, domain III residues 310–481, and domain IV residues 482–621). In the homodimer, domains II and IV interact with one another, forming a PPI interface (Fig. 11A). Domain II and domain IV are composed of eight and seven disulfide modules, respectively. The homodimer crystal structure has a twofold symmetry around the dimerization arm of domain II. The ligand is known to bind within a cleft formed by domains I and III (Ogiso et al., 2002). Crystal structures of the monomeric EGFR with and without the ligand suggested that there is a substantial change in the conformation of the ECD between monomer and dimer. In the ligand unbound state, EGFR exists in so-called closed conformation (Fig. 11D) with domains II and IV interacting with one another. Domains III and IV undergo significant movement in their structure upon binding of the ligand (Ferguson, 2004, 2008; Fig. 11E). How this change in conformation results in the transmission of signaling from outside the cell into the cytoplasmic domain is not explained because the complete structure of EGFR molecule including ECD, TM, and kinase domain is difficult to elucidate. However, each domain structure is available as a fragment (Ferguson et al., 2003; Lu et al., 2010; Mineev et al., 2010; Stamos, Sliwkowski, & Eigenbrot, 2002). There have been attempts at modeling the complete 3D structures of EGFR and its homodimer. Molecular dynamics simulations have been carried out to explain the transmission of signaling from outside of the cell to inside the cell in terms of EGFR structure (Endres et al., 2013; Poger & Mark, 2014). In terms of PPI, domain II of EGFR has β-hairpins that interact with one another in hand-shaking fashion (Fig. 11B). It has been shown that deletions or mutations in domain II completely prevent ligand-induced EGFR activation (Garrett et al., 2002; Ogiso et al., 2002). Domain IV of EGFR extends out from domains I to II and seem to form PPI at the C-terminal part (Fig. 11C). Crystal structures revealed that domain IV is flexible and that the electron density around the C-terminal portion is not well defined. However, based on experimental data, the mode of interaction of domain IV was proposed (Lu et al., 2010).
Fig. 11.
EGFR extracellular domain (ECD). (A) Homodimer (PDB ID 1NJP). Domains II and IV participate in dimerization. (B) and (C) Expanded regions of PPI of domains II and IV. Domain II region has a β-turn structure in the PPI region, forming a hydrophobic core at the interaction site. Molecules that can mimic this region are designed to inhibit PPI.Domain IV also has hydrophobic interactions that can be used to target inhibitors.(D) and (E) EGFR open (PDB ID 1NJP) and closed conformations (1NQL). Domains I–IV are labeled. In the closed conformation, domains II and IV interact, blocking the dimerization arm of domain II and forming a dimer. Notice that in open conformation, the domain IV is moved away from domain II, allowing domain II to interact with other EGFR molecules. Figure was generated using PyMol software (Schrodinger LLC, OR).
EGFR homodimer formation and its inhibition can be detected by PLA assay as described by Fichter et al. (2014). Inhibition of dimerization of EGFR homodimers by small molecules and antibodies is reported. Based on the structure of dimerization arm β-loop peptide-based molecules were designed to inhibit the domain II of EGFR. These peptides were modified to peptidomimetic using a triazolyl bridge between the peptide strands to constrain the EGFR dimerization arm β-loop (Fig. 11B). The designed peptides have significantly improved proteolytic stability over the nonmodified peptide sequence, and their inhibitory effects are dependent on the number of the methylene units and orientation of the introduced triazolyl bridge (Hanold et al., 2015). Yang, Yang, Pike, and Marshall (2010) have reported a small molecule that targets the β-hairpin structure of domain II to inhibit EGFR homodimerization. Using chemical cross-linking methods, they have shown that the small molecule designed using a computational approach inhibits EGFR homodimer. The antibody cetuximab used for the treatment of colorectal cancer does not directly inhibit the dimerization of EGFR. It binds to an EGF-binding site and blocks the ligand-binding site, thus indirectly inhibiting the dimerization and downstream signaling process of EGFR (Graham, Muhsin, & Kirkpatrick, 2004). EGFR domain IV can also be targeted to design small molecules or peptides (Fig. 11C).
6. HETERODIMERIZATION OF PROTEINS AND INHIBITION
When PPI occurs between nonidentical chains, heterodimerization results. The stability of heterodimers can vary. For example, α/β tubulins form a stable dimer, and these dimers form long protofilaments, which are constituents of microtubules (Lowe, Li, Downing, & Nogales, 2001). The number of crystal structure complexes of heterodimers available from the Protein Data Bank is relatively small compared to those of homodimers. Sowmya et al. (2015) have analyzed a nonredundant set of 278 heterodimer complexes for interfacial structural features and found that there was a correlation between the interfacial surface area of PPI and the possible function of the protein. Computational methods are used to predict the heterodimeric complexes using the crystal structures of individual proteins based on binding studies and mutational data. However, a recent critical assessment of predicted interactions (CAPRI) report indicates that prediction of homodimers is easier and superior to the prediction of heterodimers of protein complexes (Lensink et al., 2016). Thus, there is still a long way to go in terms of the availability of structural biology information for PPI of heterodimers.
The most widely covered topics concerning PPI are p53–MDM2 (Vassilev et al., 2004), Bcl-Xl (Hikita et al., 2010; Oltersdorf et al., 2005), and IL-2–IL-2Rα interactions. In the case of the p53 and HDM2 interaction, the p53-binding site on HDM2 is a cleft rather than a flat surface. Identification of a small molecule that inhibits the interaction of p53 with MDM2 provides an example of the use of HTS (Vassilev et al., 2004). An example of modulation of PPI of proteins that interact via flat surfaces is seen in the case of IL-2 and its receptor IL-2Rα (Braisted et al., 2003; Tilley et al., 1997). Here we describe some of the heterodimers and their importance in physiological function and possible inhibition of dimerization for clinical applications.
6.1. p45–p75 Heterodimers
Injury to the brain and spinal cord results in major loss of physical and other functions. In many cases, these injuries are permanent because injured nerves cannot regrow to perform their function. The NTs are a family of neurotrophic factors that control multiple aspects of nervous system development and function. p75 is a member of the tumor necrosis factor receptor superfamily. The structure of p75 consists of four extracellular cysteine-rich domains, a single TM domain, and an ICD that consists of a JM and a death domain (DD) (Dechant & Barde, 2002; Lin et al., 2015). The p75 receptor has different effects, depending on its interactions with different partners and copartner proteins. For example, p75 interacts with Trk receptors (tropomyosin receptor kinase) to promote NT-dependent nerve growth. On the other hand, p75 inhibits nerve growth mediated by myelin-associated inhibitors via functioning in part as a coreceptor for the glycophosphatidylinositol-linked neuronal Nogo-66 receptor (NgR) or another non-NgR molecule (Gentry, Rutkoski, Burke, & Carter, 2004). The binding of p75 to proneurotrophins and with the coreceptor sortilin was shown to play a role in apoptosis (Nykjaer, Willnow, & Petersen, 2005). p75 is known to form homodimers in solution, and homodimerization (Nadezhdin et al., 2016) seems to be important for complexation with NgR that leads to inhibition of nerve growth. p45, an NT receptor homolog 2 (NRH2), NT receptor-like DD protein (NRADD). p45 exhibits vast sequence similarity to p75 in the TM, JM, and DD regions. p45 contains a short and truncated ECD with no NT-binding domain. p75 plays a role during injury to the brain and spinal cord. At the site of the injury in the brain and spinal cord, there are proteins that are released from the damaged myelin that binds to Nogo receptor (NgR) on the nerve and inhibits nerve growth. NgR has to form a complex with the p75 neurotropin receptor to inhibit the signaling. p45 can bind to p75 and impedes the formation of p75 homodimer that is required for p75/NgR complex formation and its downstream activation of RhoAGTPase. The complex formation of p75/NgR requires the binding of p75 through its TM and ICDs. Vilar et al. (2014) have shown that p45 binds specifically to conserved regions in the p75 TM and the ICD and that this blocks p75 dimerization along with its downstream signaling. Thus, modulation of oligomerization of p75 is a good strategy to overcome the effect of p75’s inhibitory effects on nerve regeneration, and hence the design of p75 inhibitors will have therapeutic applications for brain and spinal cord injury. In addition, p45 itself can be used as a therapeutic agent to injured neurons and can prevent the blocking of nerve growth by inhibiting p75 interactions in paralysis or spinal cord damage injuries (Vilar et al., 2014). At present, there are no known inhibitors of p75/NgR complex.
6.2. IL-6–IL-6Rα
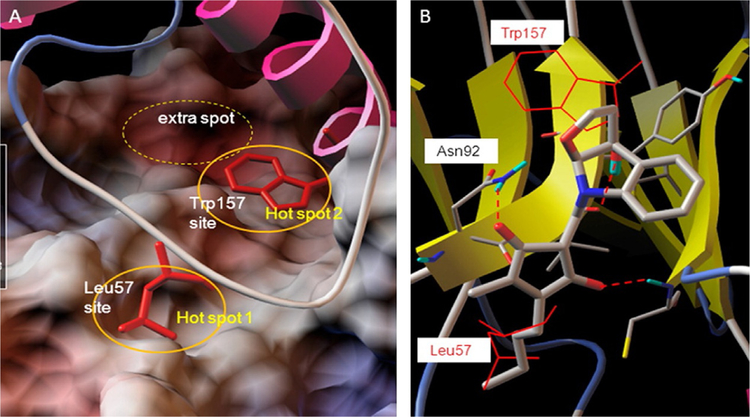
Interleukin 6 receptor, a cytokine receptor also known as CD126, interacts with IL-6 a cytokine and regulates cell growth, apoptosis, proliferation, and immune responses. IL-6 interacts with IL-6Rα and forms a binary complex and then guides glycoprotein GP130 to form the IL-6/IL-6Rα/GP130 heterotrimer. The IL-6/IL-6Rα/GP130 heterotrimers occur by the interaction between IL-6 of one trimer and the D1 domain of GP130 of the other trimer to form a hexamer. These IL-6/IL-6Rα/GP130 trimers trigger a signaling cascade of phosphorylation of Janus kinases (JAKs) and a downstream effector signal transducer and activator of transcription 3 (STAT3) that is followed by reciprocal dimerization of the Tyr705-phosphorylated STAT3, resulting in STAT3 nucleus translocation, DNA binding, and multiple oncogene transcriptions (Li et al., 2014). These JAKs and STAT3 pathways are very crucial for progression of various types of cancer. Madindoline A (MDL-A), a natural product, is known to bind to the ECD of GP130 and inhibit IL-6-dependent STAT3 tyrosine phosphorylation in hepatocellular carcinoma (HepS2) cells (Saleh, Greenman, Billings, Van Vranken, & Krolewski, 2005; Fig. 12). However, there are limitations in using MDL-A for therapeutic purposes because of several steps involved in the synthesis, weak-binding affinity to the receptor, and extraction of the natural product has very low yield. Li et al. (2014) used multiple ligand simultaneous docking (MLSD) and drug-repositioning approaches to identify compounds that inhibit PPI of IL-6 and GP130. Using this computational approach, Li et al. (2014) found two compounds, raloxifene and bazedoxifene, that were able to inhibit PPI. Raloxifene is a well-known oral selective estrogen receptor modulator (SERM) that has estrogenic actions on bone and antiestrogenic actions on the uterus and breast (Jones et al., 1984). Bazedoxifene, an analog of raloxifene, is also an SERM that is under development for the treatment of osteoporosis (Biskobing, 2007). Such drug molecules that are already approved for therapeutic purposes that are also PPI inhibitors have a high chance of becoming successful PPI inhibitor drugs as the drug is repurposed.
Fig. 12.
Hot spots at the interface of PPI in the IL-6/GP130 D1 domain. Modeling of binding hot spots at the IL-6/GP130 D1 domain interface. (A) D1 domain is represented as electrostatic potential surface (red, negatively charged; blue, positively charged; white, hydrophobic). IL-6 is in ribbon representation. The two larger yellow eclipses indicate the two main binding “hot spots,” Leu57-binding site and Trp157-binding site, between IL-6 and GP130. (B) Binding modeling of MDL-A to the GP130 D1 domain. D1 domain is in ribbon representation, and MDL-A is in thick ball-and-stick rendering. Hydrogen bonds are shown as red dotted lines. MDL-A disrupts both binding spots of the GP130 D1 domain. MDL-A forms three hydrogen bonds with Asn92, Cys6, and the carbonyl backbone of Val93 residues of GP130. The modeling indicates that the long butyl tail of MDL-A displaces Leu57 (thin red line), and the indoline moiety partially disrupts Trp157 (thin red line) of the helix D of IL-6. Reproduced with permission from Li, H., Xiao, H., Lin, L., Jou, D., Kumari, V., Lin, J., & Li, C. (2014). Drug design targeting protein-protein interactions (PPIs) using multiple ligand simultaneous docking (MLSD) and drug repositioning: Discovery of raloxifene and bazedoxifene as novel inhibitors of IL-6/GP130 interface. Journal of Medicinal Chemistry, 57(3), 632–641. https://doi.org/10.1021/jm401144z. Copyright (2014) American Chemical Society.
6.3. Epidermal Growth Factor Receptors
Here we provide an example of the design of a PPI inhibitor starting from the structure of a protein complex. The procedure described is a rational drug design or an SBDD method, which does not include any database screening or HTS. The description starts with the importance of proteins and PPI in normal and disease states and the significance of the biochemical pathway. The design information and the experimental results that indicate that the designed molecule inhibits PPI are described in detail to give the reader extensive knowledge of the way that PPI inhibitors are designed.
As mentioned earlier, the human epidermal growth factor receptor (HER) system of RTKs plays an important role in cell growth and differentiation in normal physiology (Ferguson, 2008). The receptor system consists of four members: HER1 or EGFR and HER2–4 (also called ErbB1–4). Among the EGFR, HER2 is known to always exist in the open conformation and is a preferred dimerization partner for other EGFR (Baselga & Swain, 2009). Deregulation of homo- and heterodimerization processes of these receptors or overexpression of receptors leads to different types of cancer and plays a key role in tumor progression (Lee-Hoeflich et al., 2008). Possible dimers such as EGFR–HER2, HER2–HER3, and HER2–HER4 have been proposed in the literature (Shankaran, Wiley, & Resat, 2006; Tao & Maruyama, 2008). Among these, EGFR–HER2 and HER2–HER3 are well known in different types of cancer. Mutation in EGFR seems to play a major role in breast and lung cancers. Because of the limitations of chemotherapy for cancer, EGFR-targeted therapy has attracted attention. Studies related to breast and non-small-cell lung cancer (NSCLC) have shown a link between HER2 expression and poor prognosis in patients with cancer (Hirsch et al., 2014; Hirsch, Varella-Garcia, & Cappuzzo, 2009). Approximately 18%–33% of breast and NSCLC tumors show a positive result for HER2 overexpression, suggesting the importance of HER2 in these types of cancers. The coexpression of EGFR and HER2 was associated with a significantly shortened overall survival rate in patients whose tumors expressed high levels of EGFR or HER2 (Brabender et al., 2001). Since HER2 protein is overexpressed in different cancer types, targeting the HER2 pathway will most likely target only cancer cells, and possible side effects on normal cells will be minimal. The kinase domain of EGFR has been targeted for cancer therapy using a tyrosine kinase inhibitor. However, most of these develop resistance within 5 years and, hence, ECDs are viable targets for cancer therapy (Oxnard et al., 2011).
6.3.1. Structure of ECDs of Proteins
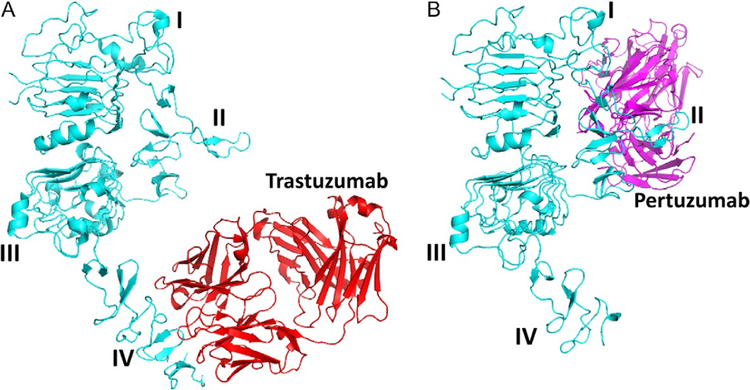
Based on the biochemical pathway, one can target the different dimerization and PPI sites on HER2 protein for developing therapeutic effects on cancer. Possible dimerization inhibition sites are domain II of ECD, domain IV of ECD, and a TMD. Detailed 3D structures of ECD of EGFR, HER2, and HER3 are all known. Structures of homodimers of EGFR ECD have been elucidated by X-ray crystallography (Lu et al., 2010; Fig. 11A). However, the structures of heterodimers of EGFR:HER2 or HER2:HER3 are not known. Since EGFRs have nearly 50% homology and similar domains, one can model the HER2:HER3 ECD using EGFR as a template structure. In the ECD of EGFR, domains II and IV are involved in PPI. The importance of domain II of the EGFR dimerization arm is well known (Burgess et al., 2003; Cho et al., 2003; Lu et al., 2010; Ogiso et al., 2002). The structure of HER2 monomer as well as HER2 complexed with antibodies trastuzumab and pertuzumab has been elucidated (Fig. 13A and B). HER2 domain IV is a clinically validated target since trastuzumab, an antibody, binds to domain IV of HER2 and has therapeutic value against HER2-positive breast cancer (Piccart-Gebhart et al., 2005). However, domain IV has not been well studied because of its flexibility. A homodimer of EGFR domain IV indicates the PPI and possible hot spots. Based on this, a heterodimer of HER2:HER3 was built (template-based modeling/docking), and possible hot spots were identified by FTMAP (Kozakov et al., 2015).
Fig. 13.
Binding of (A) trastuzumab to domain IV of ECD of HER2 protein (PDB ID 1N8Z).(B) Binding of pertuzumab (PDB ID 1S78) to domain II of ECD of HER2 protein. PyMol was used to generate the figures.
6.3.2. Design Concept
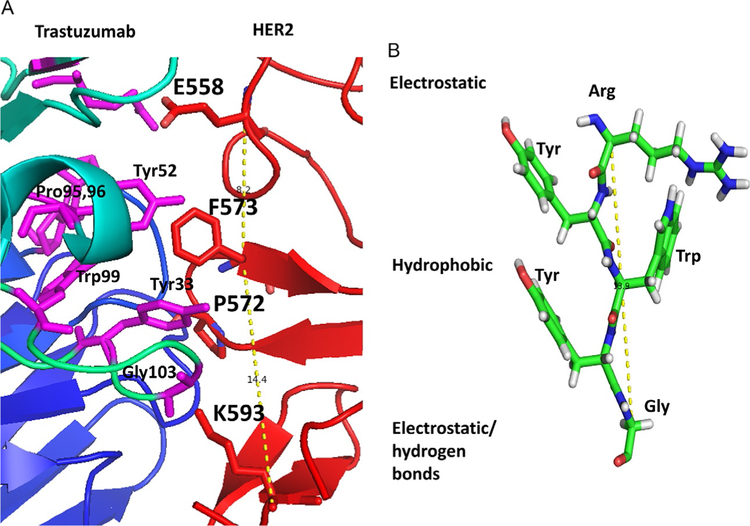
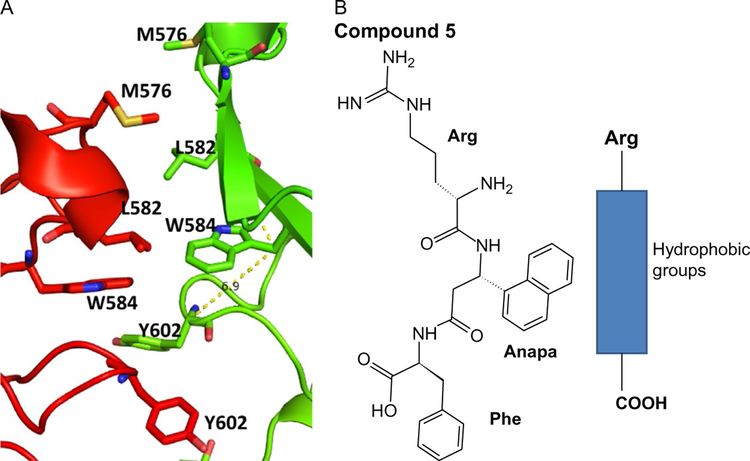
The idea here is to inhibit domain IV of the ECDs of EGFR:HER2 and HER2:EHR3. Inhibition of the ECD of these proteins inhibits the phosphorylation of the kinase domain and downstream signaling for cancer cell growth. Thus, the growth of cancer tumors can be reduced. Trastuzumab is known to bind to domain IV of HER2 protein (Fig. 13A). However, its exact mechanism of action is not clear. Examination of EGFR homodimer, the crystal structure of the complex of trastuzumab and HER2, indicates that domain IV has hydrophobic hot spots. We used the structure of a complex of HER2 protein with trastuzumab for the design of a template structure. Although the antibody structure is large, the binding region to HER2 protein is relatively small. The binding region has hydrophobic amino acid residues such as Tyr, Trp, and Phe (Fig. 14A). This hydrophobic region is surrounded by electrostatic and hydrogen-bonding interactions. By measuring the distance between Cα atoms of amino acid residues Glu558, Phe573, and Lys593 of HER2, a peptide template structure was built with amino acids Arg-Tyr-Trp-Tyr-Gly (Fig. 14B). These residues correspond to the antibody trastuzumab that interacts with HER2. Evaluation of these peptides for antiproliferative activity showed that they were not potent in inhibiting cell growth of HER2-positive cells BT-474 and SKBR-3 (Satyanarayanajois, Villalba, Jianchao, & Lin, 2009). The designed peptide can also interact with PPI site of domain IV of HER2 (Fig. 15A). The peptide design was modified using a peptidomimetic approach with the incorporation of a beta-naphthyl group to fit into the hydrophobic core of the HER2 protein PPI site with trastuzumab (Fig. 15B). Since Arg participates in salt bridge, Arg on the left side, a hydrophobic core at the center, and Phe at the C-terminal were used to build a peptidomimetic (Fig. 15B). This peptidomimetic was evaluated for its antiproliferative activity using a cellular assay such as MTT or CellTiter-glo assay in breast cancer cells that over-express HER2 protein. Furthermore, to find the specificity of the designed molecules for HER2-overexpressing cell lines, different cell lines that do not overexpress HER2 protein such as MCF-7 and HCT-116 were used. The molecule that has a beta-naphthyl group exhibited antiproliferative activity with an IC50 value of 0.4μM in HER2-overexpressing cancer cell lines. However, in MCF-7 and HCT-116, the activity was 40μM, suggesting the specificity of this compound for HER2-positive cancer cell lines. The binding of this compound was verified by fluorescence assay and SPR. It is known that other homologous proteins such as EGFR and HER3 are also important in different types of cancer. EGFR, HER3, and HER4 have a sequence homology of nearly 50%, and all of them have similar 3D structures. To show that the designed compound binds specifically to HER2, SPR studies were carried out; it was shown that the designed compound 5 binds only to HER2 protein ECD (Banappagari et al., 2012).
Fig. 14.
(A) PPI region of trastuzumab-HER2 domain IV complex (PDB ID 1N8Z). Hydro-phobic region of trastuzumab formed by amino acid residues Tyr52, Pro95,96, Trp99, and Tyr33 interacts with Phe573 and P572 of HER2 protein. On either side of the hydro-phobic region, there is electrostatic interaction formed by Glu558 of HER2 with Arg59 of trastuzumab and from K593 of HER2 to Gly103. (B) This interaction can be used to design a template structure for a PPI inhibitor. Figure was generated using PyMol (Schrodinger LLC, OR).
Fig. 15.
The structure of the designed template compound 5 and a peptidomimetic (B) along with the PPI site (A) of the EGFR homodimer of domain IV (PDB ID 1N8Z and 3NJP). Note that the PPI site is dominated by hydrophobic residues. Peptidomimetic can bind to this PPI site. PyMol (Schrodinger LLC, OR) was used to generate the structure of the protein complexes.
To improve the activity of the compound, several modifications were performed. An Asp residue was introduced at the C-terminal, making the compound more specific for HER2-overexpressed cells and giving it better activity (Banappagari, Ronald, & Satyanarayanajois, 2011; Kanthala et al., 2015; Kanthala, Gauthier, & Satyanarayanajois, 2014). In addition, using the PPI of domain IV of EGFR and HER2 proteins (compound 18, patent application: WO/2015/175299), a conformationally constrained cyclic peptidomimetic compound was designed. The resulting compound exhibited antiproliferative activity around 200nM in breast cancer cell lines and 18nM in HER2-positive lung cancer Calu-3 cell lines. Once the structural aspects of the compound were optimized for pharmacological action, it was investigated for PPI inhibition activity. Using SPR analysis, enzyme fragment complementation assay, and PLA assay, it was shown that the compound designed inhibited not only EGFR:HER2 dimerization but also HER2:HER3 dimerization (Banappagari, Ronald, & Satyanarayanajois, 2010; Kanthala et al., 2015, 2014). All of these studies are related to cell-based studies. To determine whether the compound inhibits the heterodimerization of HER2:HER3 and has any pharmacological action in vivo, a xenograft model of breast cancer mice was used. In vivo studies indicated that the compound was able to suppress the growth of a tumor in a xenograft model of breast cancer, inhibit the phosphorylation of HER2 kinase, and inhibit HER2:HER3 dimerization in breast cancer tumors in mice (Kanthala et al., 2015). The compound was shown to be stable for more than 24h in mouse serum. Overall, the example provided here illustrates the way that a PPI inhibitor can be designed and modified to improve selectivity and activity based on the structure of protein complexes.
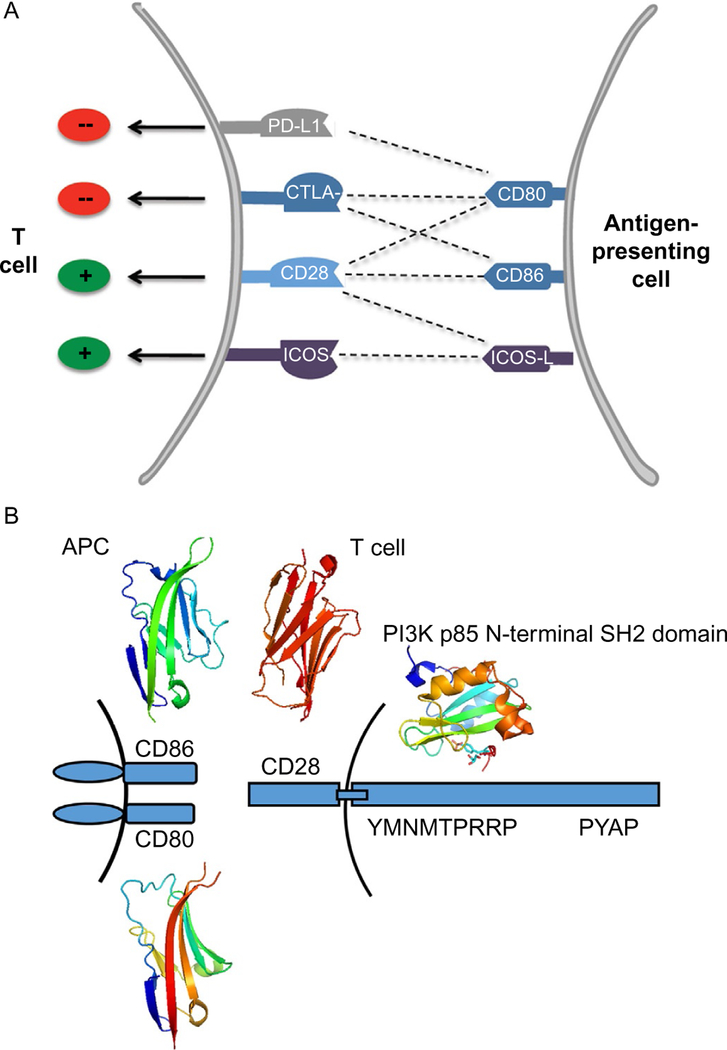
6.4. PD-1–PD-L1 Pathway
In the two-signal model, T cells interact with antigen-presenting cells (APCs) with two important signals—the first from the T-cell receptor (TCR) and major histocompatibility (MHC) complex and the second from costimulatory molecules (Bretscher, 1999). If these two signals are generated, immune response takes place. The strength and duration of the TCR–MHC signal depend on the costimulatory molecules and cytokines generated based on these signaling proteins. However, this signal, although necessary, is alone not sufficient to induce T-cell proliferation. There are several costimulatory molecules that generate signaling in an orchestrated process to assist the immunological synapse (IS) and control the immune function (Chen & Flies, 2013). In the case of Signal 2, important costimulatory molecules are of two receptors on T lymphocytes CD28 and cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) that interact with B7 family (CD80 and CD86) on APCs. If Signals 1 and 2 are provided, the result is T-cell clonal expansion and the induction of effector functions such as lymphokine production, T-cell proliferation, and cytokine secretion. Costimulation is necessary for a productive immune response to take place. The lack of costimulation after engagement of T-cell receptor by antigen results in a state of antigen-specific unresponsiveness termed anergy (Schwartz, 2003). Thus, a costimulatory pathway plays a potential role in the treatment of autoimmune diseases, organ rejection, and tumor immunity. CD28 is generally expressed on the surface of T cells and is one of the major costimulatory molecules for initial T-cell activation. Interaction of TCR CD28 ligation results in the augmentation of many aspects of T-cell-mediated immunity. The immune stimulatory features of the CD28 pathway are triggered by engagement of two well-described ligands found on APC. CD28 along with CTLA-4 and B7–1, B7–2 balance the costimulation and coinhibition process. When CD28 binds to its ligands B7–1 and B7–2, it delivers the T-cell stimulatory signal resulting in T-cell proliferation, expansion, and differentiation (Fig. 16). On the other hand, when CTLA-4 on activated T cells binds to B7 ligands, it delivers negative signals (Beyersdorf, Kerkau, & Hunig, 2015; Dilek et al., 2013; Gardner, Jeffery, & Sansom, 2014). Thus, these molecules maintain the immunological tolerance and response (Fig. 16). Programmed death-1 (PD-1) plays an important role in negatively regulating immune responses (Buchbinder & Desai, 2016; Greenwald, Freeman, & Sharpe, 2005). PD-1 is a CD28 family member expressed on activated T cells, B cells, and myeloid cells. PD-1 delivers a coinhibitory signal upon binding to ligands PD-L1 or PD-L2. PD-1 binding to its ligand produces tyrosine phosphorylation of the PD-1 cytoplasmic domain and recruitment of phosphatases, particularly Src homology phosphatase 2 (SHP2) (Fig. 16). This results in dephosphorylation of TCR-proximal signaling molecules, including ZAP70, PKC-θ, and CD3-ζ, leading to attenuation of the TCR/CD28 signal.
Fig. 16.
Structures of the B7/CD28 family. Structures are modeled on the crystal determinations. Loops have been added to one end of the IgV domains to emphasize the orientation of the CDR-like loops and their interaction with ligand or lack thereof. Reproduced with permission from Freeman, G. J. (2008). Structures of PD-1 with its ligands: Sideways and dancing cheek to cheek. Proceedings of the National Academy of Sciences of the United States of America, 105(30), 10275–10276. https://doi.org/10.1073/pnas.0805459105. Copyright (2008) National Academy of Sciences, USA.
The PD-1/PD-L1 pathway delivers inhibitory signals that regulate both peripheral and central tolerance. Disruption of the Pdcd1 gene can accelerate autoimmune diseases in mice, including a lupus-like disorder in lpr mice or diabetes in nonobese diabetic mice (Freeman, 2008). Cancer cells are known to escape the immune system in the body. PD-L1 is expressed on a wide variety of tumors and participates in the immunosuppressive activity of cancer cells (Sharpe, Wherry, Ahmed, & Freeman, 2007; Zang & Allison, 2007). PD-L1 on tumors inhibits T-cell activation and lysis of tumor cells; it is also known that PD-L1 interaction with its receptor leads to the death of tumor-specific T cells (Brown et al., 2003). Monoclonal antibodies have been developed against PD-L1 or its ligands for therapeutic purposes (Dolan & Gupta, 2014). The structures of PD-1–PDL-1 and PD-1–PDL2 have been elucidated using X-ray crystallography (Lazar-Molnar et al., 2008; Lin et al., 2008). PD-1, a type I transmembrane protein of the Ig superfamily, consists of an extracellular N-terminal IgV-like domain, a TMD, and a cytoplasmic tail (Ishida, Agata, Shibahara, & Honjo, 1992; Zhang et al., 2004) that is involved in inhibitory signal transmission. The PD-1 ectodomain contains a single IgV domain typical of the CD28 family, whereas PD-L1 and PD-L2 belong to the B7 family and are composed of IgV and IgC domains. In the 1:1 complex of PD-1 and PD-L2, a binding interface is formed by the front β-sheets of both the PD-1 and PD-L2 IgV domains. Interaction of PD-1 on activated T cells with PD-L1 or L2 diminishes the effector T-cell activity in peripheral organs and tissues during inflammation. This is an important step to protect against tissue damage when the immune system is activated in response to infection. However, in cancer, this pathway is used for cancer cell survival from immune surveil-lance to mask the cancer cells from the immune system. Thus, blocking PD-1–PD-L1 pathway can lead to T-cell activation against cancer cells (Tang & Heng, 2013). It is reported that in cancers such as melanoma, hepatocellular carcinoma, glioblastoma, lung, kidney, breast, ovarian, pancreatic, and esophageal, as well as hematological malignancies, positive expression of PD-L1 was seen clearly, indicating that cancer cells use the PD-1–PD-L1 pathway for their survival from T-cell immune response against them (Zitvogel & Kroemer, 2012). Thus, modulation of the PD-1–PD-L1 pathway by modulation of PPI of these proteins has therapeutic value.
Monoclonal antibodies targeted to PD-1 or PD-L1 can prevent PD-1-mediated T-cell inhibition, leading to antitumor immune responses. However, selecting a particular antibody for one of these molecules is important. If antibodies are directed against PD-1, they block PD-1 binding to both of its ligands, PD-L1 and PD-L2, whereas anti-PD-L1 antibodies should be selective in preventing PD-1 binding to PD-L1, maintaining the interactive and binding ability of PD-1 to PD-L2. Medina and Adams (2016) reported a study where both CTLA-4 and PD-1 blockade were used in a combined manner. Blocking of both the pathways, CTLA-2 and PD-1 resulted in stronger antitumor effect than blockade of either pathway alone.
Several antibody molecules, including MDX-1106/BMS-936558/ONO-4538, CT-011, MK-3475, MPDL3280A/RG7446, BMS-936559, and AMP-224, have been developed to modulate PPI of the PD-1–PDL1 pathway. Antibodies nivolumab and pembrolizumab that are targeted to PD1 are approved by FDA for the treatment of patients with unresectable or metastatic melanoma as well as metastatic squamous and nonsquamous NSCLC, with progression on or after platinum-based chemotherapy. However, administration of these antibodies to cancer patients needs careful monitoring and analysis of changes in the immune response in patients. Thus, clinicians will need to have an understanding of the science of how inhibition of PD-1 can lead to tumor reduction with associated immune-mediated adverse events.
6.5. CD-28/CD-80
CD28 is a stimulatory cell surface receptor of the Ig superfamily. Other members in this family include cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4), CD152-inducible costimulator (ICOS) (Boomer & Green, 2010; Tezuka et al., 2000), programmed death receptor 1 (PD-1) (Ishida et al., 1992), and B- and T-lymphocyte attenuator (Watanabe et al., 2003). The ligands for CD28 are CD80 (B7/BB1 or B7–1) and CD86 (B7–2). In addition to binding to CD28, both CD80 and CD86 also bind to the inhibitory protein CTLA-4, which is a CD28 homolog expressed on activated T cells and, especially, regulatory T cells (Tregs) (Walker, 2013). CD28 is also known to bind to ICOS. Studies have indicated that CD28 is involved in several functions, and that CD28 and CTL-4 have integrated functions (Fig. 17A). CD28 costimulation provides the T cells to control unwanted (antiself) signaling and triggering wanted (antimicrobial) immunity (Gardner et al., 2014). There are at least three mechanisms to ensure that CD28 is capable of generating only a costimulatory signal when the TCR is engaged, but does not activate the T cell by itself. Without TCR stimulation, CD28 binds to its ligand CD80 monovalently (low affinity). In the presence of TCR–MHC ligation, the CD28 homo-dimer form binds to its ligand CD80 bivalently. Overall, CD28 induces a costimulatory signal in the T-cell upon coligation together with the TCR and amplifies the TCR signal (Beyersdorf et al., 2015). Upon TCR and CD28 stimulation, the T cell forms an IS with the APC (Brzostek, Gascoigne, & Rybakin, 2016). CD28 signaling is important for the production of IL2, and it is known that CD28 costimulation increases the IL-2 production compared to without CD28 stimulation (Boomer & Green, 2010).
Fig. 17.
(A) Complexities of CD28 costimulatory pathways. The CD28 costimulatory receptor can be ligated by CD80, CD86, and ICOS-L (B7–H2). The CTLA-4 coinhibitor competes with CD28 for binding to CD80 and CD86. However, CD80 can also bind to PD-L1 (B7–H1) and deliver a coinhibitory signal. ICOS competes with CD28 for binding to B7–H2. (B) Schematic representation of PPI interactions of CD28 with CD80/CD86. CD28 cytoplasmic domain motifs YMNM and PYAP that are important in signaling are indicated. Crystal structures of domains of CD28 (1YJD), CD80 (4RWH), CD86 (1NCN), and p53 (5GJI) domain important in binding to CD28 signaling are shown. Panel (A): Reproduced with permission from Ford, M. L., Adams, A. B., & Pearson, T. C. (2014). Targeting co-stimulatory pathways: Transplantation and autoimmunity. Nature Reviews Nephrology, 10(1), 14–24. https://doi.org/10.1038/nrneph.2013.183. Copyright (2013) Nature Publishing Group.
CD2 binds to its ligands and initiates signaling in the cytoplasmic domain. Details of molecular mechanism and binding epitopes involved in CD28 signaling indicated that CD28 initiates two signaling cascades using two motifs in the cytoplasmic domain of CD28. Each of these motifs binds to distinct set of proteins. One of the signaling motifs consists of YMNM sequence that involves phosphorylation of a tyrosine residue (Fig. 17B). The other pathway is initiated by the more distal proline-rich regions (Boomer & Green, 2010; Rudd, Taylor, & Schneider, 2009; Sharpe, 2009). However, the involvement of these motifs in in vivo studies is not clear. In vivo studies using knockout mice studies suggested that mutation of the proximal YMNM motif did not have any effect on signaling (Dodson et al., 2009). However, mutation of the distal proline motif (PYAP) (Fig. 17B) resulted in impaired CD28-dependent functions (Friend et al., 2006). Thus, the distal proline motif is involved in a critical, nonredundant signaling pathway required for CD28 function, whereas the proximal tyrosine-based motif signaling is not clearly understood.
CD28 PPI inhibition represents an example of indirect inhibition of PPI for drug design. The mechanism of PPI of CD28 is quite complicated, and inhibition of CD28 can lead to unwanted effects. CTLA-4 is also known to bind to B7 ligands (CD80, CD86) and send inhibitory signaling to APCs by counteracting signaling generated by CD28. Thus, CTLA-4 binding to CD80 or CD86 (B7 molecules) inhibits T-cell activation and proliferation. This strategy can be used to target CD28 for designing immunosuppressive drugs. A fusion protein consisting of the ECD of human CTLA-4 linked to the Fc domain of human IgG1 (CTLA-4-Ig, abatacept) was designed to block CD28–CD80/CD86 interactions. Abatacept has a high affinity for CD80 and CD86, blocks CD28-dependent costimulation, and inhibits T-cell proliferation in vitro (Linsley & Nadler, 2009). Based on these encouraging results, abatacept was evaluated in preclinical animal models of autoimmune diseases. Evaluation of abatacept in collagen-induced arthritis (CIA) of the rat showed that it prevented the onset of CIA, indicating that the fusion protein mediated blockade of T-cell costimulation in vivo. The drug was approved for RA in 2006. Modification of abatacept with two amino acids resulted in 10-fold higher in vitro potency (Larsen et al., 2005). This modification resulted in a second-generation CTLA-Ig protein belatacept (Larsen et al., 2005). Belatacept selectively inhibits T-cell activation by preventing CD28 activation and by binding its ligands B7–1 and B7–2. This prevents the stimulation of CD28 antagonizing CD80 and CD86 on APCs and thus blocks the signals of the transduction pathway.
There were attempts to directly inhibit CD28 signaling with its ligands (Hunig, 2007; Poirier, Blancho, & Vanhove, 2011); however, such attempts were not successful. Ford, Adams, and Pearson (2014) describe the way a selective blockade of CD28 was attempted in a phase I trial of an agonistic anti-CD28 monoclonal antibody, TGN1412. However, a fatal immunological reaction called “cytokine storm” caused by significant T-cell activation was observed in patients, suggesting that direct manipulation of CD28 signaling is harmful because CD28 is involved in myriad signaling pathways. To avoid this overwhelming T-cell activation, novel domain antibodies, in which the Fc portion is completely removed, were produced. These antibodies to CD28 that lack a Fc region have permitted the development of novel blocking, nonactivating reagents that can safely and specifically block CD28 costimulatory signals but leave the coinhibitory signaling due to CTLA-4 intact. A monovalent CD28-specific fusion antibody sc28AT, a novel nonactivating single-chain Fv-based reagent, was developed (Zhang et al., 2011). Evaluation of this fusion protein in a nonhuman primate model indicated that sc28AT modestly prolongs cardiac and renal allograft survival (Poirier et al., 2010). To improve the pharmacokinetic profile (Poirier et al., 2012), a pegylated anti-CD28 SCFV named FR104, which was derived from the anti-CD28 mAb clone, was developed. The pegylated reagent has been shown to prevent graft rejection in monkeys. Furthermore, there are attempts to search for small molecules that can target CD28 and inhibit PPI of CD28 with its ligands (Uvebrant et al., 2007).
6.6. CD2–CD58 Interactions and the Design of Multicyclic Peptides
T-cell adhesion to APCs and the subsequent immune response is important in its pathogenesis. The two-signal hypothesis proposes that T-cell activation requires recognition of an antigen by the TCR (Signal 1) and a concomitant signal provided by adhesion/costimulatory molecules (Signal 2) to achieve full activation (Leitner et al., 2010). Among the adhesion/costimulatory molecules, the most abundant is CD2, a transmembrane protein in T cells, which binds to its ligand CD58 on APC. CD58, also known as leukocyte function-associated antigen 3 (LFA-3), is a cell adhesion molecule with only one known ligand, CD2 (Chen & Flies, 2013; Davis et al., 2003; van der Merwe & Davis, 2003). Ligation of CD2 on T cells to CD58 on APC facilitates T-cell–APC adhesion and is important in the early stages of immune response. This interaction results in the induction of IFN-γ and subsequent regulation of human leukocyte antigen–antigen D related (HLA-DR), intracellular adhesion molecule-1 (ICAM-1), and B-7 molecules on APC, causing an amplification of the signal for immune response. In vitro studies have indicated that inhibition of CD2 and CD58 interactions using an LFA-3 fusion protein (alefacept) inhibits T-cell activation (da Silva et al., 2002). Alefacept is a recombinant human CD58-Ig fusion protein that effectively binds to CD2 and prevents CD2 interaction with CD58. It has been successfully used clinically to treat plaque psoriasis (Chamian et al., 2005). Modulation of adhesion interaction between cell adhesion molecules has been shown to be effective in treating autoimmune diseases such as RA (Chen & Flies, 2013; Ford et al., 2014; Papoutsaki & Costanzo, 2013). Current treatments for autoimmune diseases with biologics include antibodies and fusion proteins (Papp et al., 2012; Webber, Hirose, & Vincenti, 2011). However, they have limitations in terms of stability (shelf-life), administration, and immunogenicity (Hansel, Kropshofer, Singer, Mitchell, & George, 2010). We have designed peptides that block cell adhesion between T cells expressing CD2 and Caco-2 cells expressing CD58 (Gokhale, Weldeghiorghis, Taneja, & Satyanarayanajois, 2011). The CD58-binding domain of CD2 consists of β-strands with charged residues (Fig. 18A and B). From our studies, it is very clear that peptides designed from β-strands exhibit cell adhesion inhibition activity between T cells and epithelial cells. The peptides could block the anti-CD58 binding to CD58 expressed on Caco-2 cells, indicating that peptides bind to the CD58 protein. In rodents, the homolog of CD58 is CD48. It is postulated that CD48 and CD58 have the same evolutionary origin. In mice, CD2 binds to its ligand CD48 to generate an immune response (Ianelli, Edson, & Thorley-Lawson, 1997). CD48 has a high degree of homology to CD58 and is similar in the 3D structure (Velikovsky et al., 2007; Wang et al., 1999). The adhesion domains of the two proteins overlap with backbone rmsd of 1.35Å. We have shown that peptides designed from human CD2 protein bind to mouse CD48. Furthermore, one of the designed peptide (peptide 6) that is known to bind to CD58 was able to suppress the progression of RA in the CIA model. We have also shown that peptide 6 is not immunogenic in mice (Gokhale, Kanthala, Latendresse, Taneja, & Satyanarayanajois, 2013; Gokhale et al., 2011).
Fig. 18.
(A) PPI interface of CD2–CD58 showing electrostatic and hydrophobic interactions. There are 10 salt bridges, 5 hydrogen bonds, and 1 hydrophobic interaction (PDB ID 1QA9). The design of peptides and multicyclic grafted peptide from CD2 adhesion domain based on the interface residues of CD2 of CD2–CD58 interactions. (B) F and C strands from the CD2 protein that interact with CD58. (C) Grafting of CD2 F and C strand residues onto sunflower trypsin inhibitor peptide to generate stable peptides that inhibit PPI of CD2–CD58 for immunomodulation (Sable et al., 2016 ACS Chemical Biology).
6.6.1. Multicyclic Peptide Approach
Although the designed peptides have shown in vitro and in vivo activity, in general, many of these peptides have limitations in terms of in vivo stability (Vlieghe, Lisowski, Martinez, & Khrestchatisky, 2010). Multicyclic structures have unusually stable properties; they are generally nonimmunogenic and can be used as templates for grafting (Poth, Chan, & Craik, 2013). In order to improve the stability of the peptides, we grafted the CD2 peptide epitope onto multicyclic structures (Fig. 18C). Structural comparisons using molecular modeling suggested that sunflower trypsin inhibitor (SFTI) is suitable for grafting the CD2 peptides that we designed. In the multicyclic SFTI structure, the two β-strands are placed nearly 5Å apart, which is suitable for grafting CD2 peptide 6 as we reported earlier (Gokhale et al., 2011). The β-strands are stabilized by disulfide bridges and cyclization that resembles peptide 6. We have grafted the CD2-derived peptides onto SFTI and have shown that the designed molecule inhibits CD2–CD58 interaction in cell adhesion assays at nanomolar concentrations. The peptide SFTI-1a was shown to bind to CD58 protein using SPR and inhibit PPI between CD2 and CD58. Furthermore, the peptide was shown to suppress T-cell immune response in an in vitro assay using the cells derived from a transgenic animal model of RA. Since these peptides are intended to exhibit thermal and enzymatic stability, SFTI-1a was evaluated for its thermal and serum stability. The peptide exhibited thermal stability up to 80°C and was stable in human serum up to 48h (Sable et al., 2016). This study illustrates the design of peptides to inhibit PPI and translation of the pharmacophore template from the peptide to multicyclic stable structure that can be used for PPI inhibition and as a therapeutic agent for autoimmune diseases such as RA.
PPI drug discovery has been slow in the last decade, but has steadily increased in the past few years. More than 40 PPI have been targeted for drug discovery, and some of them have reached clinical trials (Petta et al., 2016). Only a few drugs that are on the market are PPI inhibitors. For example, tirofiban, an antiplatelet drug that is an inhibitor of glycoprotein IIb/IIIa (Arkin et al., 2014), is already on the market. Lifitegrast inhibits the interaction between leukocyte function associated antigen-1 (LFA-1) (CD11a/α2, CD18/β2) and ICAM-1. LFA-1 is a β2 integrin receptor found on leukocytes and is involved in T-cell activation through binding to its ligand ICAM-1 (Long, 2011). Lifitegrast successfully passed clinical trials for the treatment of keratoconjunctivitis sicca inflammatory dry eye syndrome (Sheppard et al., 2014) and was approved by the FDA in 2016. Although there are challenges in PPI drug design, it is a rapidly growing area with many pharmaceutical companiesparticipating in this new area of drug discovery (Mullard, 2012). Table 2 provides a list of PPI inhibitors that are in clinical trials.
Table 2.
Some of the Protein–Protein Interaction Inhibitors That Are in the Clinical Trial
| Drug Name | Proprietary | Clinical Trial Stage | Indications | Website URLa |
|---|---|---|---|---|
| BRD4 inhibitors | ||||
| I-BET762 (GSK525762) |
GSK | Phase II | Solid tumor | https://clinicaltrials.gov/ct2/show/NCT01587703 |
| CPI-0610 | Constellation Pharmaceutical, Cambridge, MA | Phase I | Lymphoma | https://clinicaltrials.gov/ct2/show/NCT01949883?term=CPI-0610&rank=2 |
| Ten-010 | Hoffmann-La Roche | Phase I | Acute myeloid leukemia | https://clinicaltrials.gov/ct2/show/NCT02308761?term=Ten-010&rank=1 |
| OTX015 (MK-8628) |
OncoEthix | Phase I | Cancer | https://clinicaltrials.gov/ct2/show/NCT01713582?term=OTX015&rank=2 |
| BRD3 inhibitors | ||||
| RVX-208 | Resverlogix Corp | Phase II | Diabetes, atherosclerosis | https://clinicaltrials.gov/ct2/show/NCT01728467?term=RVX-208&rank=1 |
| IAP inhibitors | ||||
| LCL161 | Novartis | Phase I | Advanced solid tumors | https://clinicaltrials.gov/ct2/show/NCT01098838?term=LCLl61&rank=5 |
| GDC-0917/ CUDC-427 |
Genentech | Phase I | Solid cancer | https://clinicaltrials.gov/ct2/show/NCT01226277?term=GDC-0917&rank=l |
| GDC-0152 (RG7419) |
Genentech | Phase I | Solid cancer | https://clinicaltrials.gov/ct2/show/NCT00977067?term=GDC-0152&rank=l |
| Bivalent inhibitors-SMAC mimetics | ||||
| Birinapant (TL-32711) |
TetraLogic Pharmaceuticals | Phase II | Cancer | https://clinicaltrials.gov/ct2/show/NCT01188499?term=Birinapant&rank=5 |
Date accessed May 17, 2017.
REFERENCES
- Acuner Ozbabacan SE, Engin HB, Gursoy A, & Keskin O (2011). Transient protein-protein interactions. Protein Engineering, Design & Selection, 24(9), 635–648. 10.1093/protein/gzr025. [DOI] [PubMed] [Google Scholar]
- Adams PD, Seeholzer S, & Ohh M (2002). Identification of associated proteins by coimmunoprecipitation In Protein–protein interactions: A molecular cloning manual (pp. 59–74). New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [Google Scholar]
- Aglan M, Amr K, Ismail S, Ashour A, Otaify GA, Mehrez MA, et al. (2015). Clinical and molecular characterization of seven Egyptian families with autosomal recessive robinow syndrome: Identification of four novel ROR2 gene mutations. American Journal of Medical Genetics. Part A, 167A(12), 3054–3061. 10.1002/ajmg.a.37287. [DOI] [PubMed] [Google Scholar]
- Arkin MR, Tang Y, & Wells JA (2014). Small-molecule inhibitors of protein-protein interactions: Progressing toward the reality. Chemistry & Biology, 21(9), 1102–1114. 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakail M, & Ochsenbein F (2016). Targeting protein–protein interactions, a wide open field for drug design. Comptes Rendus Chimie, 19(1), 19–27. [Google Scholar]
- Banappagari S, Corti M, Pincus S, & Satyanarayanajois S (2012). Inhibition of protein-protein interaction of HER2-EGFR and HER2-HER3 by a rationally designed peptidomimetic. Journal of Biomolecular Structure & Dynamics, 30(5), 594–606. 10.1080/07391102.2012.687525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banappagari S, Ronald S, & Satyanarayanajois SD (2010). A conformationally constrained peptidomimetic binds to the extracellular region of HER2 protein. Journal of Bio-molecular Structure & Dynamics, 28(3), 289–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banappagari S, Ronald S, & Satyanarayanajois SD (2011). Structure-activity relationship of conformationally constrained peptidomimetics for antiproliferative activity in HER2-overexpressing breast cancer cell lines. MedChemComm, 2(8), 752–759. 10.1039/C1MD00126D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, & Swain SM (2009). Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nature Reviews. Cancer, 9(7), 463–475. 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- Berg T (2005). Methods in molecular biology In Fu H (Ed.), Protein–protein interactions:Methods and applications: Vol. 261 (pp. 446–447). Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA; 10.1002/cbic.200400235. Wiley Online Library. [DOI] [Google Scholar]
- Beyersdorf N, Kerkau T, & Hunig T (2015). CD28 co-stimulation in T-cell homeostasis: A recent perspective. ImmunoTargets and Therapy, 4, 111–122. 10.2147/ITT.S61647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskobing DM (2007). Update on bazedoxifene: A novel selective estrogen receptor modulator. Clinical Interventions in Aging, 2(3), 299–303. [PMC free article] [PubMed] [Google Scholar]
- Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, et al. (2002). Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell, 110(1), 93–105. [DOI] [PubMed] [Google Scholar]
- Bogan AA, & Thorn KS (1998). Anatomy of hot spots in protein interfaces. Journal of Molecular Biology, 280(1), 1–9. 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. (2010). Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature, 467(7315), 596–599. 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomer JS, & Green JM (2010). An enigmatic tail of CD28 signaling. Cold Spring Harbor Perspectives in Biology, 2(8), a002436 10.1101/cshperspect.a002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M (2001). Oligomerization of G-protein-coupled transmitter receptors. Nature Reviews. Neuroscience, 2(4), 274–286. 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- Brabender J, Danenberg KD, Metzger R, Schneider PM, Park J, Salonga D, et al. (2001). Epidermal growth factor receptor and HER2-neu mRNA expression in non-small cell lung cancer is correlated with survival. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 7(7), 1850–1855. [PubMed] [Google Scholar]
- Braisted AC, Oslob JD, Delano WL, Hyde J, McDowell RS, Waal N, et al. (2003). Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. Journal of the American Chemical Society, 125(13), 3714–3715. [DOI] [PubMed] [Google Scholar]
- Bretscher PA (1999). A two-step, two-signal model for the primary activation of precursor helper T cells. Proceedings of the National Academy of Sciences of the United States of America, 96(1), 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogi S, Tafi A, Desaubry L, & Nebigil CG (2014). Discovery of GPCR ligands for probing signal transduction pathways. Frontiers in Pharmacology, 5, 255 10.3389/fphar.2014.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Dorfman DM, Ma FR, Sullivan EL, Munoz O, Wood CR, et al. (2003). Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. Journal of Immunology, 170(3), 1257–1266. [DOI] [PubMed] [Google Scholar]
- Brzostek J, Gascoigne NR, & Rybakin V (2016). Cell type-specific regulation of immunological synapse dynamics by B7 ligand recognition. Frontiers in Immunology, 7, 24 10.3389/fimmu.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder EI, & Desai A (2016). CTLA-4 and PD-1 pathways: Similarities, differences, and implications of their inhibition. American Journal of Clinical Oncology, 39(1), 98–106. 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, et al. (2003). An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Molecular Cell, 12(3), 541–552. [DOI] [PubMed] [Google Scholar]
- Chabre M, Cone R, & Saibil H (2003). Biophysics: Is rhodopsin dimeric in native retinal rods? Nature, 426(6962), 30–31. discussion 31 10.1038/426030b. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P, & Janin J (2002). Dissecting protein-protein recognition sites. Proteins,47(3), 334–343. [DOI] [PubMed] [Google Scholar]
- Chamian F, Lowes MA, Lin SL, Lee E, Kikuchi T, Gilleaudeau P, et al. (2005). Alefacept reduces infiltrating T cells, activated dendritic cells, and inflammatory genes in psoriasis vulgaris. Proceedings of the National Academy of Sciences of the United States of America, 102(6), 2075–2080. 10.1073/pnas.0409569102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, & Flies DB (2013). Molecular mechanisms of T cell co-stimulation and co-inhibition. Nature Reviews. Immunology, 13(4), 227–242. 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chene P (2006). Drugs targeting protein-protein interactions. ChemMedChem, 1(4), 400–411. 10.1002/cmdc.200600004. [DOI] [PubMed] [Google Scholar]
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW Jr., et al. (2003). Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature, 421(6924), 756–760. 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- Clackson T, & Wells JA (1995). A hot spot of binding energy in a hormone-receptor interface. Science, 267(5196), 383–386. [DOI] [PubMed] [Google Scholar]
- da Silva AJ, Brickelmaier M, Majeau GR, Li Z, Su L, Hsu YM, et al. (2002). Alefacept, an immunomodulatory recombinant LFA-3/IgG1 fusion protein, induces CD16 signaling and CD2/CD16-dependent apoptosis of CD2(+) cells. Journal of Immunology, 168(9), 4462–4471. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Ikemizu S, Evans EJ, Fugger L, Bakker TR, & van der Merwe PA (2003). The nature of molecular recognition by T cells. Nature Immunology, 4(3), 217–224. 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- Dayhoff JE, Shoemaker BA, Bryant SH, & Panchenko AR (2010). Evolution of protein binding modes in homooligomers. Journal of Molecular Biology, 395(4), 860–870. 10.1016/j.jmb.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant G, & Barde YA (2002). The neurotrophin receptor p75(NTR): Novel functions and implications for diseases of the nervous system. Nature Neuroscience, 5(11), 1131–1136. 10.1038/nn1102-1131. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002). Unraveling hot spots in binding interfaces: Progress and challenges. Current Opinion in Structural Biology, 12(1), 14–20. [DOI] [PubMed] [Google Scholar]
- de Mol NJ, & Fischer MJ (2010). Surface plasmon resonance: A general introduction. Methods in Molecular Biology, 627, 1–14. 10.1007/978-1-60761-670-2_1. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Vassar R, & Golde T (2010). The secretases: Enzymes with therapeutic potential in Alzheimer disease. Nature Reviews. Neurology, 6(2), 99–107. 10.1038/nrneurol.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilek N, Poirier N, Hulin P, Coulon F, Mary C, Ville S, et al. (2013). Targeting CD28, CTLA-4 and PD-L1 costimulation differentially controls immune synapses and function of human regulatory and conventional T-cells. PLoS One, 8(12), e83139 10.1371/journal.pone.0083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson LF, Boomer JS, Deppong CM, Shah DD, Sim J, Bricker TL, et al. (2009). Targeted knock-in mice expressing mutations of CD28 reveal an essential pathway for costimulation. Molecular and Cellular Biology, 29(13), 3710–3721. 10.1128/MCB.01869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan DE, & Gupta S (2014). PD-1 pathway inhibitors: Changing the landscape of cancer immunotherapy. Cancer Control, 21(3), 231–238. [DOI] [PubMed] [Google Scholar]
- Dunbar AY, Kamada Y, Jenkins GJ, Lowe ER, Billecke SS, & Osawa Y (2004). Ubiquitination and degradation of neuronal nitric-oxide synthase in vitro: Dimer stabilization protects the enzyme from proteolysis. Molecular Pharmacology, 66(4), 964–969. 10.1124/mol.104.000125. [DOI] [PubMed] [Google Scholar]
- Eggert S, Midthune B, Cottrell B, & Koo EH (2009). Induced dimerization of the amyloid precursor protein leads to decreased amyloid-beta protein production. The Journal of Biological Chemistry, 284(42), 28943–28952. 10.1074/jbc.M109.038646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, et al. (2013). Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell, 152(3), 543–556. 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson DA (2006). Fragment-based lead discovery: A chemical update. Current Opinion in Biotechnology, 17(6), 643–652. 10.1016/j.copbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Ferguson KM (2004). Active and inactive conformations of the epidermal growth factor receptor. Biochemical Society Transactions, 32(Pt. 5), 742–745. 10.1042/BST0320742. [DOI] [PubMed] [Google Scholar]
- Ferguson KM (2008). Structure-based view of epidermal growth factor receptor regulation. Annual Review of Biophysics, 37, 353–373. 10.1146/annurev.biophys.37.032807.125829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, & Lemmon MA (2003). EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Molecular Cell, 11(2), 507–517. [DOI] [PubMed] [Google Scholar]
- Fichter CD, Timme S, Braun JA, Gudernatsch V, Schopflin A, Bogatyreva L, et al. (2014). EGFR, HER2 and HER3 dimerization patterns guide targeted inhibition in two histotypes of esophageal cancer. International Journal of Cancer, 135(7), 1517–1530. 10.1002/ijc.28771. [DOI] [PubMed] [Google Scholar]
- Ford ML, Adams AB, & Pearson TC (2014). Targeting co-stimulatory pathways: Transplantation and autoimmunity. Nature Reviews. Nephrology, 10(1), 14–24. 10.1038/nrneph.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox S, Farr-Jones S, Sopchak L, Boggs A, Nicely HW, Khoury R, et al. (2006). High-throughput screening: Update on practices and success. Journal of Biomolecular Screening, 11(7), 864–869. 10.1177/1087057106292473. [DOI] [PubMed] [Google Scholar]
- Franco R, Casado V, Cortes A, Ferrada C, Mallol J, Woods A, et al. (2007). Basic concepts in G-protein-coupled receptor homo- and heterodimerization. ScientificWorldJournal, 7, 48–57. 10.1100/tsw.2007.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Martinez-Pinilla E, Lanciego JL, & Navarro G (2016). Basic pharmacological and structural evidence for class A G-protein-coupled receptor heteromerization. Frontiers in Pharmacology, 7, 76 10.3389/fphar.2016.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, et al. (2002). Protein detection using proximity-dependent DNA ligation assays. Nature Bio-technology, 20(5), 473–477. 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, & Schioth HB (2003). The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Molecular Pharmacology, 63(6), 1256–1272. 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Freeman GJ (2008). Structures of PD-1 with its ligands: Sideways and dancing cheek to cheek. Proceedings of the National Academy of Sciences of the United States of America, 105(30), 10275–10276. 10.1073/pnas.0805459105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend LD, Shah DD, Deppong C, Lin J, Bricker TL, Juehne TI, et al. (2006). A dose-dependent requirement for the proline motif of CD28 in cellular and humoral immunity revealed by a targeted knockin mutant. The Journal of Experimental Medicine, 203(9), 2121–2133. 10.1084/jem.20052230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry D, Huang KS, Di Lello P, Mohr P, Muller K, So SS, et al. (2013). Design of libraries targeting protein-protein interfaces. ChemMedChem, 8(5), 726–732. 10.1002/cmdc.201200540. [DOI] [PubMed] [Google Scholar]
- Gardner D, Jeffery LE, & Sansom DM (2014). Understanding the CD28/CTLA-4 (CD152) pathway and its implications for costimulatory blockade. American Journal of Transplantation, 14(9), 1985–1991. 10.1111/ajt.12834. [DOI] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, et al. (2002). Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell, 110(6), 763–773. [DOI] [PubMed] [Google Scholar]
- Gentry JJ, Rutkoski NJ, Burke TL, & Carter BD (2004). A functional interaction between the p75 neurotrophin receptor interacting factors, TRAF6 and NRIF. The Journal of Biological Chemistry, 279(16), 16646–16656. 10.1074/jbc.M309209200. [DOI] [PubMed] [Google Scholar]
- Gokhale A, Kanthala S, Latendresse J, Taneja V, & Satyanarayanajois S (2013). Immunosuppression by co-stimulatory molecules: Inhibition of CD2-CD48/CD58 interaction by peptides from CD2 to suppress progression of collagen-induced arthritis in mice. Chemical Biology & Drug Design, 82(1), 106–118. 10.1111/cbdd.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale AS, & Satyanarayanajois S (2014). Peptides and peptidomimetics as immuno-modulators. Immunotherapy, 6(6), 755–774. 10.2217/imt.14.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Weldeghiorghis TK, Taneja V, & Satyanarayanajois SD (2011). Conformationally constrained peptides from CD2 to modulate protein-protein interactions between CD2 and CD58. Journal of Medicinal Chemistry, 54(15), 5307–5319. 10.1021/jm200004e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, & Olson AJ (2000). Structural symmetry and protein function. Annual Review of Biophysics and Biomolecular Structure, 29, 105–153. 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- Graham J, Muhsin M, & Kirkpatrick P (2004). Cetuximab. Nature Reviews. Drug Discovery, 3(7), 549–550. 10.1038/nrd1445. [DOI] [PubMed] [Google Scholar]
- Greenwald RJ, Freeman GJ, & Sharpe AH (2005). The B7 family revisited. Annual Review of Immunology, 23, 515–548. 10.1146/annurev.immunol.23021704.115611. [DOI] [PubMed] [Google Scholar]
- Guo W, Wisniewski JA, & Ji H (2014). Hot spot-based design of small-molecule inhibitors for protein-protein interactions. Bioorganic & Medicinal Chemistry Letters, 24(11), 2546–2554. 10.1016/j.bmcl.2014.03.095. [DOI] [PubMed] [Google Scholar]
- Hall RA (2005). Co-immunoprecipitation as a strategy to evaluate receptor-receptor or receptor-protein interactions. G Protein-Coupled Receptor-Protein Interactions, 9 165À178. [Google Scholar]
- Hanold LE, Oruganty K, Ton NT, Beedle AM, Kannan N, & Kennedy EJ (2015). Inhibiting EGFR dimerization using triazolyl-bridged dimerization arm mimics. PLoS One, 10(3), e0118796 10.1371/journal.pone.0118796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel TT, Kropshofer H, Singer T, Mitchell JA, & George AJ (2010). The safety and side effects of monoclonal antibodies. Nature Reviews. Drug Discovery, 9(4), 325–338. 10.1038/nrd3003. [DOI] [PubMed] [Google Scholar]
- Hikita H, Takehara T, Shimizu S, Kodama T, Shigekawa M, Iwase K, et al. (2010). The Bcl-xL inhibitor, ABT-737, efficiently induces apoptosis and suppresses growth of hepatoma cells in combination with sorafenib. Hepatology, 52(4), 1310–1321. 10.1002/hep.23836. [DOI] [PubMed] [Google Scholar]
- Hiller C, Kuhhorn J, & Gmeiner P (2013). Class A G-protein-coupled receptor (GPCR) dimers and bivalent ligands. Journal of Medicinal Chemistry, 56(17), 6542–6559. 10.1021/jm4004335. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Jotte RM, Berry CA, Mencia WA, Stowell SA, & Gardner AJ (2014). Quality of care of patients with non-small-cell lung cancer: A report of a performance improvement initiative. Cancer Control: Journal of the Moffitt Cancer Center, 21(1), 90–97. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M, & Cappuzzo F (2009). Predictive value of EGFR and HER2 overexpression in advanced non-small-cell lung cancer. Oncogene, 28(Suppl. 1), S32–37. 10.1038/onc.2009.199. [DOI] [PubMed] [Google Scholar]
- Hunig T (2007). Manipulation of regulatory T-cell number and function with CD28-specific monoclonal antibodies. Advances in Immunology, 95, 111–148. 10.1016/S0065-2776(07)95004-X. [DOI] [PubMed] [Google Scholar]
- Ianelli CJ, Edson CM, & Thorley-Lawson DA (1997). A ligand for human CD48 on epithelial cells. Journal of Immunology, 159(8), 3910–3920. [PubMed] [Google Scholar]
- Ishida Y, Agata Y, Shibahara K, & Honjo T (1992). Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. The EMBO Journal, 11(11), 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ispolatov I, Yuryev A, Mazo I, & Maslov S (2005). Binding properties and evolution of homodimers in protein-protein interaction networks. Nucleic Acids Research, 33(11), 3629–3635. 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VV (2016). A review of stapled peptides and small molecules to inhibit protein-protein interactions in cancer. Current Medicinal Chemistry, 23(27), 3025–3043. [DOI] [PubMed] [Google Scholar]
- Jacoby E, Bouhelal R, Gerspacher M, & Seuwen K (2006). The 7 TM G-protein-coupled receptor target family. ChemMedChem, 1(8), 761–782. 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- James JR, Oliveira MI, Carmo AM, Iaboni A, & Davis SJ (2006). A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nature Methods, 3(12), 1001–1006. 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- Jencks WP (1981). On the attribution and additivity of binding energies. Proceedings of the National Academy of Sciences of the United States of America, 78(7), 4046–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Wang W, & Fang G (2014). Targeting protein-protein interaction by small molecules. Annual Review of Pharmacology and Toxicology, 54, 435–456. 10.1146/annurev-pharmtox-011613-140028. [DOI] [PubMed] [Google Scholar]
- Jones CD, Jevnikar MG, Pike AJ, Peters MK, Black LJ, Thompson AR, et al. (1984). Antiestrogens. 2. Structure-activity studies in a series of 3-aroyl-2-arylbenzo[b] thiophene derivatives leading to [6-hydroxy-2-(4-hydroxyphenyl)benzo[b]thien-3-yl] [4-[2-(1-piperidinyl)ethoxy]-phenyl]methanone hydrochloride (LY156758), a remarkably effective estrogen antagonist with only minimal intrinsic estrogenicity. Journal of Medicinal Chemistry, 27(8), 1057–1066. [DOI] [PubMed] [Google Scholar]
- Jubb H, Higueruelo AP, Winter A, & Blundell TL (2012). Structural biology and drug discovery for protein-protein interactions. Trends in Pharmacological Sciences, 33(5), 241–248. 10.1016/j.tips.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Jung JI, Ran Y, Cruz PE, Rosario AM, Ladd TB, Kukar TL, et al. (2014). Complex relationships between substrate sequence and sensitivity to alterations in gamma-secretase processivity induced by gamma-secretase modulators. Biochemistry, 53(12), 1947–1957. 10.1021/bi401521t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaake RM, Wang X, Burke A, Yu C, Kandur W, Yang Y, et al. (2014). A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Molecular & Cellular Proteomics, 13(12), 3533–3543. 10.1074/mcp.M114.042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadmiel M, & Cidlowski JA (2013). Glucocorticoid receptor signaling in health and disease. Trends in Pharmacological Sciences, 34(9), 518–530. 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthala S, Banappagari S, Gokhale A, Liu YY, Xin G, Zhao Y, et al. (2015). Novel peptidomimetics for inhibition of HER2:HER3 heterodimerization in HER2-positive breast cancer. Chemical Biology & Drug Design, 85, 702–714. 10.1111/cbdd.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthala S, Gauthier T, & Satyanarayanajois S (2014). Structure-activity relationships of peptidomimetics that inhibit PPI of HER2-HER3. Biopolymers, 101(6), 693–702. 10.1002/bip.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova T, & McNally JG (2006). Detecting protein-protein interactions with CFPYFP FRET by acceptor photobleaching. Current Protocols in Cytometry. Chapter 12, Unit12.7 10.1002/0471142956.cy1207s35. [DOI] [PubMed] [Google Scholar]
- Khalifa NB, Van Hees J, Tasiaux B, Huysseune S, Smith SO, Constantinescu SN, et al. (2010). What is the role of amyloid precursor protein dimerization? Cell Adhesion & Migration, 4(2), 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Sun ZY, Byron O, Campbell G, Wagner G, Wang J, et al. (2001). Molecular dissection of the CD2-CD58 counter-receptor interface identifies CD2 Tyr86 and CD58 Lys34 residues as the functional “hot spot”. Journal of Molecular Biology, 312(4), 711–720. 10.1006/jmbi.2001.4980. [DOI] [PubMed] [Google Scholar]
- Kocan M, & Pfleger KD (2011). Study of GPCR-protein interactions by BRET. Methods in Molecular Biology, 746, 357–371. 10.1007/978-1-61779-126-0_20. [DOI] [PubMed] [Google Scholar]
- Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, et al. (2015).The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nature Protocols, 10(5), 733–755. 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna SS, & Aravind L (2010). The bridge-region of the Ku superfamily is an atypical zinc ribbon domain. Journal of Structural Biology, 172(3), 294–299. 10.1016/j.jsb.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CP, Pearson TC, Adams AB, Tso P, Shirasugi N, Strobert E, et al. (2005). Rational development of LEA29Y (belatacept), a high-affinity variant of CTLA4-Ig with potent immunosuppressive properties. American Journal of Transplantation, 5(3), 443–453. 10.1111/j.1600-6143.2005.00749.x. [DOI] [PubMed] [Google Scholar]
- Lazar-Molnar E, Yan Q, Cao E, Ramagopal U, Nathenson SG, & Almo SC (2008). Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proceedings of the National Academy of Sciences of the United States of America, 105(30), 10483–10488. 10.1073/pnas.0804453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP, et al. (2008). A central role for HER3 in HER2-amplified breast cancer: Implications for targeted therapy. Cancer Research, 68(14), 5878–5887. 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- Leitner J, Kuschei W, Grabmeier-Pfistershammer K, Woitek R, Kriehuber E, Majdic O, et al. (2010). T cell stimulator cells, an efficient and versatile cellular system to assess the role of costimulatory ligands in the activation of human T cells. Journal of Immunological Methods, 362(1–2), 131–141. 10.1016/j.jim.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, et al. (2016). Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment. Proteins, 84(Suppl. 1), 323–348. 10.1002/prot.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED, Pereira-Leal JB, Chothia C, & Teichmann SA (2006). 3D complex: A structural classification of protein complexes. PLoS Computational Biology, 2(11), e155 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xiao H, Lin L, Jou D, Kumari V, Lin J, et al. (2014). Drug design targeting protein-protein interactions (PPIs) using multiple ligand simultaneous docking (MLSD) and drug repositioning: Discovery of raloxifene and bazedoxifene as novel inhibitors of IL-6/GP130 interface. Journal of Medicinal Chemistry, 57(3), 632–641. 10.1021/jm401144z. [DOI] [PubMed] [Google Scholar]
- Lin DY, Tanaka Y, Iwasaki M, Gittis AG, Su HP, Mikami B, et al. (2008). The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proceedings of the National Academy of Sciences of the United States of America, 105(8), 3011–3016. 10.1073/pnas.0712278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Tann JY, Goh ET, Kelly C, Lim KB, Gao JF, et al. (2015). Structural basis of death domain signaling in the p75 neurotrophin receptor. eLife, 4, e11692 10.7554/eLife.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, & Nadler SG (2009). The clinical utility of inhibiting CD28-mediated costimulation. Immunological Reviews, 229(1), 307–321. 10.1111/j.1600-065X.2009.00780.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ross JF, Bodine PV, & Billiard J (2007). Homodimerization of Ror2 tyrosine kinase receptor induces 14–3–3(beta) phosphorylation and promotes osteoblast differentiation and bone formation. Molecular Endocrinology, 21(12), 3050–3061. 10.1210/me.2007-0323. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, & Janin J (1999). The atomic structure of protein-protein recognition sites. Journal of Molecular Biology, 285(5), 2177–2198. [DOI] [PubMed] [Google Scholar]
- London N, Raveh B, & Schueler-Furman O (2013). Druggable protein-protein interactions—From hot spots to hot segments. Current Opinion in Chemical Biology, 17(6), 952–959. 10.1016/j.cbpa.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Long EO (2011). ICAM-1: Getting a grip on leukocyte adhesion. Journal of Immunology, 186(9), 5021–5023. 10.4049/jimmunol.1100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, & Nogales E (2001). Refined structure of alpha beta-tubulin at 3.5 A resolution. Journal of Molecular Biology, 313(5), 1045–1057. 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Lu C, Mi LZ, Grey MJ, Zhu J, Graef E, Yokoyama S, et al. (2010). Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Molecular and Cellular Biology, 30(22), 5432–5443. 10.1128/MCB.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM (2006). Transmembrane signaling by G protein-coupled receptors. Methods in Molecular Biology, 332, 3–49. 10.1385/1-59745-048-0:1. [DOI] [PubMed] [Google Scholar]
- Madauss KP, Bledsoe RK, McLay I, Stewart EL, Uings IJ, Weingarten G, et al. (2008). The first X-ray crystal structure of the glucocorticoid receptor bound to a nonsteroidal agonist. Bioorganic & Medicinal Chemistry Letters, 18(23), 6097–6099. 10.1016/j.bmcl.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Marianayagam NJ, Sunde M, & Matthews JM (2004). The power of two: Protein dimerization in biology. Trends in Biochemical Sciences, 29(11), 618–625. 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, & Beyreuther K (1985). Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proceedings of the National Academy of Sciences of the United States of America, 82(12), 4245–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr LM, & Fuerst P (2008). The future of high-throughput screening. Journal of Bio-molecular Screening, 13(6), 443–448. 10.1177/1087057108319644. [DOI] [PubMed] [Google Scholar]
- Medina PJ, & Adams VR (2016). PD-1 pathway inhibitors: Immuno-oncology agents for restoring antitumor immune responses. Pharmacotherapy, 36(3), 317–334. 10.1002/phar.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi LZ, Grey MJ, Nishida N, Walz T, Lu C, & Springer TA (2008). Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid nanodiscs. Biochemistry, 47(39), 10314–10323. 10.1021/bi801006s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi LZ, Lu C, Li Z, Nishida N, Walz T, & Springer TA (2011). Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nature Structural & Molecular Biology, 18(9), 984–989. 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineev KS, Bocharov EV, Pustovalova YE, Bocharova OV, Chupin VV, & Arseniev AS (2010). Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. Journal of Molecular Biology, 400(2), 231–243. 10.1016/j.jmb.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Moreira IS, Fernandes PA, & Ramos MJ (2007). Hot spots—A review of the protein-protein interface determinant amino-acid residues. Proteins, 68(4), 803–812. 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- Morelli X, Bourgeas R, & Roche P (2011). Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I). Current Opinion in Chemical Biology, 15(4), 475–481. 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Mullard A (2012). Protein-protein interaction inhibitors get into the groove. Nature Reviews. Drug Discovery, 11(3), 173–175. 10.1038/nrd3680. [DOI] [PubMed] [Google Scholar]
- Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, et al. (2007). GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. The EMBO Journal, 26(6), 1702–1712. 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadezhdin KD, Garcia-Carpio I, Goncharuk SA, Mineev KS, Arseniev AS, & Vilar M (2016). Structural basis of p75 transmembrane domain dimerization. The Journal of Biological Chemistry, 291(23), 12346–12357. 10.1074/jbc.M116.723585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, & Goodrich JA (2006). Protein-protein interaction assays: Eliminating false positive interactions. Nature Methods, 3(2), 135–139. 10.1038/nmeth0206-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE, & Petersen CM (2005). p75NTR—Live or let die. Current Opinion in Neurobiology, 15(1), 49–57. 10.1016/j.conb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Oakley RH, & Cidlowski JA (2011). Cellular processing of the glucocorticoid receptor gene and protein: New mechanisms for generating tissue-specific actions of glucocorticoids. The Journal of Biological Chemistry, 286(5), 3177–3184. 10.1074/jbc.R110.179325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oasa S, Sasaki A, Yamamoto J, Mikuni S, & Kinjo M (2015). Homodimerization of glucocorticoid receptor from single cells investigated using fluorescence correlation spectroscopy and microwells. FEBS Letters, 589(17), 2171–2178. 10.1016/j.febslet.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, et al. (2002). Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell, 110(6), 775–787. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. (2005). An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature, 435(7042), 677–681. 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Oxnard GR, Arcila ME, Chmielecki J, Ladanyi M, Miller VA, & Pao W (2011). New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 17(17), 5530–5537. 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsaki M, & Costanzo A (2013). Treatment of psoriasis and psoriatic arthritis. BioDrugs, 27(Suppl. 1), 3–12. 10.1007/BF03325637. [DOI] [PubMed] [Google Scholar]
- Papp KA, Dekoven J, Parsons L, Pirzada S, Robern M, Robertson L, et al. (2012). Biologic therapy in psoriasis: Perspectives on associated risks and patient management. Journal of Cutaneous Medicine and Surgery, 16(3), 153–168. 10.1177/120347541201600305. [DOI] [PubMed] [Google Scholar]
- Petta I, Lievens S, Libert C, Tavernier J, & De Bosscher K (2016). Modulation of protein-protein interactions for the development of novel therapeutics. Molecular Therapy, 24(4), 707–718. 10.1038/mt.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. (2005). Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. The New England Journal of Medicine, 353(16), 1659–1672. 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- Poger D, & Mark AE (2014). Activation of the epidermal growth factor receptor: A series of twists and turns. Biochemistry, 53(16), 2710–2721. 10.1021/bi401632z. [DOI] [PubMed] [Google Scholar]
- Poirier N, Azimzadeh AM, Zhang T, Dilek N, Mary C, Nguyen B, et al. (2010). Inducing CTLA-4-dependent immune regulation by selective CD28 blockade promotes regulatory T cells in organ transplantation. Science Translational Medicine, 2(17), 17ra10 10.1126/scitranslmed.3000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier N, Blancho G, & Vanhove B (2011). A more selective costimulatory blockade of the CD28-B7 pathway. Transplant International, 24(1), 2–11. 10.1111/j.1432-2277.2010.01176.x. [DOI] [PubMed] [Google Scholar]
- Poirier N, Mary C, Dilek N, Hervouet J, Minault D, Blancho G, et al. (2012). Pre-clinical efficacy and immunological safety of FR104, an antagonist anti-CD28 monova-lent Fab’ antibody. American Journal of Transplantation, 12(10), 2630–2640. 10.1111/j.1600-6143.2012.04164.x. [DOI] [PubMed] [Google Scholar]
- Poth AG, Chan LY, & Craik DJ (2013). Cyclotides as grafting frameworks for protein engineering and drug design applications. Biopolymers, 100(5), 480–491. 10.1002/bip.22284. [DOI] [PubMed] [Google Scholar]
- Reichmann D, Rahat O, Cohen M, Neuvirth H, & Schreiber G (2007). The molecular architecture of protein-protein binding sites. Current Opinion in Structural Biology, 17(1), 67–76. 10.1016/j.sbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Richter L, Munter LM, Ness J, Hildebrand PW, Dasari M, Unterreitmeier S, et al. (2010). Amyloid beta 42 peptide (Abeta42)-lowering compounds directly bind to Abeta and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proceedings of the National Academy of Sciences of the United States of America, 107(33), 14597–14602. 10.1073/pnas.1003026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roarty K, Shore AN, Creighton CJ, & Rosen JM (2015). Ror2 regulates branching, differentiation, and actin-cytoskeletal dynamics within the mammary epithelium. The Journal of Cell Biology, 208(3), 351–366. 10.1083/jcb.201408058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R Jr. (2014). The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacological Research, 79, 34–74. 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Rudd CE, Taylor A, & Schneider H (2009). CD28 and CTLA-4 coreceptor expression and signal transduction. Immunological Reviews, 229(1), 12–26. 10.1111/j.1600-065X.2009.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable R, Durek T, Taneja V, Craik DJ, Pallerla S, Gauthier T, et al. (2016). Constrained cyclic peptides as immunomodulatory inhibitors of the CD2:CD58 protein-protein interaction. ACS Chemical Biology, 11(8), 2366–2374. 10.1021/acschembio.6b00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable R, & Jois S (2015). Surfing the protein-protein interaction surface using docking methods: Application to the design of PPI inhibitors. Molecules, 20(6), 11569–11603. 10.3390/molecules200611569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh AZ, Greenman KL, Billings S, Van Vranken DL, & Krolewski JJ (2005). Binding of madindoline A to the extracellular domain of gp130. Biochemistry, 44(32), 10822–10827. 10.1021/bi050439+. [DOI] [PubMed] [Google Scholar]
- Sato T, Kienlen-Campard P, Ahmed M, Liu W, Li H, Elliott JI, et al. (2006). Inhibitors of amyloid toxicity based on beta-sheet packing of Abeta40 and Abeta42. Biochemistry, 45(17), 5503–5516. 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Tang TC, Reubins G, Fei JZ, Fujimoto T, Kienlen-Campard P, et al. (2009). A helix-to-coil transition at the epsilon-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proceedings of the National Academy of Sciences of the United States of America, 106(5), 1421–1426. 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayanajois S, Villalba S, Jianchao L, & Lin GM (2009). Design, synthesis, and docking studies of peptidomimetics based on HER2-herceptin binding site with potential antiproliferative activity against breast cancer cell lines. Chemical Biology & Drug Design, 74(3), 246–257. 10.1111/j.1747-0285.2009.00855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann S, Hambsch B, Hesse L, Stumm J, Schmidt C, Beher D, et al. (2001). Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer’s disease. The Journal of Biological Chemistry, 276(36), 33923–33929. 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- Schwartz RH (2003). T cell anergy. Annual Review of Immunology, 21, 305–334. 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- Scott DE, Coyne AG, Hudson SA, & Abell C (2012). Fragment-based approaches in drug discovery and chemical biology. Biochemistry, 51(25), 4990–5003. 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- Shankaran H, Wiley HS, & Resat H (2006). Modeling the effects of HER/ErbB1–3 coexpression on receptor dimerization and biological response. Biophysical Journal, 90(11), 3993–4009. 10.1529/biophysj.105.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe AH (2009). Mechanisms of costimulation. Immunological Reviews, 229(1), 5–11. 10.1111/j.1600-065X.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe AH, Wherry EJ, Ahmed R, & Freeman GJ (2007). The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nature Immunology, 8(3), 239–245. 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- Sheppard JD, Torkildsen GL, Lonsdale JD, D’Ambrosio FA Jr., McLaurin EB, Eiferman RA, et al. (2014). Lifitegrast ophthalmic solution 5.0% for treatment of dry eye disease: Results of the OPUS-1 phase 3 study. Ophthalmology, 121(2), 475–483. 10.1016/j.ophtha.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, & Fesik SW (1996). Discovering high-affinity ligands for proteins: SAR by NMR. Science, 274(5292), 1531–1534. [DOI] [PubMed] [Google Scholar]
- Skwarczynska M, & Ottmann C (2015). Protein-protein interactions as drug targets. Future Medicinal Chemistry, 7(16), 2195–2219. 10.4155/fmc.15.138. [DOI] [PubMed] [Google Scholar]
- Soba P, Eggert S, Wagner K, Zentgraf H, Siehl K, Kreger S, et al. (2005). Homo- and heterodimerization of APP family members promotes intercellular adhesion. The EMBO Journal, 24(20), 3624–3634. 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowmya G, Breen EJ, & Ranganathan S (2015). Linking structural features of protein complexes and biological function. Protein Science, 24(9), 1486–1494. 10.1002/pro.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl B, Seifert MH, & Berg T (2009). Natural product inhibitors of protein-protein interactions mediated by Src-family SH2 domains. Bioorganic & Medicinal Chemistry Letters, 19(12), 3305–3309. 10.1016/j.bmcl.2009.04.083. [DOI] [PubMed] [Google Scholar]
- Stamos J, Sliwkowski MX, & Eigenbrot C (2002). Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. The Journal of Biological Chemistry, 277(48), 46265–46272. 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, et al. (2008). Estimating the size of the human interactome. Proceedings of the National Academy of Sciences of the United States of America, 105(19), 6959–6964. 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swapna LS, Srikeerthana K, & Srinivasan N (2012). Extent of structural asymmetry in homodimeric proteins: Prevalence and relevance. PLoS One, 7(5), e36688 10.1371/journal.pone.0036688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szidonya L, Cserzo M, & Hunyady L (2008). Dimerization and oligomerization of G-protein-coupled receptors: Debated structures with established and emerging functions. The Journal of Endocrinology, 196(3), 435–453. 10.1677/JOE-07-0573. [DOI] [PubMed] [Google Scholar]
- Tampellini D, Magrane J, Takahashi RH, Li F, Lin MT, Almeida CG, et al. (2007). Internalized antibodies to the Abeta domain of APP reduce neuronal Abeta and protect against synaptic alterations. The Journal of Biological Chemistry, 282(26), 18895–18906. 10.1074/jbc.M700373200. [DOI] [PubMed] [Google Scholar]
- Tang PA, & Heng DY (2013). Programmed death 1 pathway inhibition in metastatic renal cell cancer and prostate cancer. Current Oncology Reports, 15(2), 98–104. 10.1007/s11912-012-0284-2. [DOI] [PubMed] [Google Scholar]
- Tao RH, & Maruyama IN (2008). All EGF(ErbB) receptors have preformed homo- and heterodimeric structures in living cells. Journal of Cell Science, 121(Pt. 19), 3207–3217. 10.1242/jcs.033399. [DOI] [PubMed] [Google Scholar]
- Tezuka K, Tsuji T, Hirano D, Tamatani T, Sakamaki K, Kobayashi Y, et al. (2000). Identification and characterization of rat AILIM/ICOS, a novel T-cell costimulatory molecule, related to the CD28/CTLA4 family. Biochemical and Biophysical Research Communications, 276(1), 335–345. 10.1006/bbrc.2000.3466. [DOI] [PubMed] [Google Scholar]
- Tilley JW, Chen L, Fry DC, Emerson SD, Powers GD, Biondi D, et al. (1997). Identification of a small molecule inhibitor of the IL-2/IL-2Rα receptor interaction which binds to IL-2. Journal of the American Chemical Society, 119(32), 7589–7590. [Google Scholar]
- Trifilieff P, Rives ML, Urizar E, Piskorowski RA, Vishwasrao HD, Castrillon J, et al. (2011). Detection of antigen interactions ex vivo by proximity ligation assay: Endogenous dopamine D2-adenosine A2A receptor complexes in the striatum. BioTechniques, 51(2), 111–118. 10.2144/000113719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndall JD, & Sandilya R (2005). GPCR agonists and antagonists in the clinic. Medicinal Chemistry, 1(4), 405–421. [DOI] [PubMed] [Google Scholar]
- Uvebrant K, da Graca Thrige D, Rosen A, Akesson M, Berg H, Walse B, et al. (2007). Discovery of selective small-molecule CD80 inhibitors. Journal of Biomolecular Screening, 12(4), 464–472. 10.1177/1087057107300464. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, & Davis SJ (2003). Molecular interactions mediating T cell antigen recognition. Annual Review of Immunology, 21, 659–684. 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, et al. (1999). Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science, 286(5440), 735–741. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science, 303(5659), 844–848. 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Velazquez-Campoy A, Leavitt SA, & Freire E (2004). Characterization of protein-protein interactions by isothermal titration calorimetry. Methods in Molecular Biology, 261, 35–54. 10.1385/1-59259-762-9:035. [DOI] [PubMed] [Google Scholar]
- Velikovsky CA, Deng L, Chlewicki LK, Fernandez MM, Kumar V, & Mariuzza RA (2007). Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family. Immunity, 27(4), 572–584. 10.1016/j.immuni.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinde CL, Rudenko G, & Hol WG (1992). In search of new lead compounds for trypanosomiasis drug design: A protein structure-based linked-fragment approach. Journal of Computer-Aided Molecular Design, 6(2), 131–147. [DOI] [PubMed] [Google Scholar]
- Vilar M, Sung TC, Chen Z, Garcia-Carpio I, Fernandez EM, Xu J, et al. (2014). Heterodimerization of p45-p75 modulates p75 signaling: Structural basis and mechanism of action. PLoS Biology, 12(8), e1001918 10.1371/journal.pbio.1001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilasco M, Communal L, Mourra N, Courtin A, Forgez P, & Gompel A (2011). Glucocorticoid receptor and breast cancer. Breast Cancer Research and Treatment, 130(1), 1–10. 10.1007/s10549-011-1689-6. [DOI] [PubMed] [Google Scholar]
- Vlieghe P, Lisowski V, Martinez J, & Khrestchatisky M (2010). Synthetic therapeutic peptides: Science and market. Drug Discovery Today, 15(1–2), 40–56. 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Walker LS (2013). Treg and CTLA-4: Two intertwining pathways to immune tolerance. Journal of Autoimmunity, 45, 49–57. 10.1016/j.jaut.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Smolyar A, Tan K, Liu JH, Kim M, Sun ZY, et al. (1999). Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) count-erreceptors. Cell, 97(6), 791–803. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, et al. (2003). BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nature Immunology, 4(7), 670–679. 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- Webber A, Hirose R, & Vincenti F (2011). Novel strategies in immunosuppression: Issues in perspective. Transplantation, 91(10), 1057–1064. 10.1097/TP.0b013e3182145306. [DOI] [PubMed] [Google Scholar]
- Wells JA, & McClendon CL (2007). Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature, 450(7172), 1001–1009. 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Wilson WD (2002). Tech.Sight. Analyzing biomolecular interactions. Science, 295(5562),2103–2105. 10.1126/science.295.5562.2103. [DOI] [PubMed] [Google Scholar]
- Wilson AJ (2009). Inhibition of protein-protein interactions using designed molecules. Chemical Society Reviews, 38(12), 3289–3300. 10.1039/b807197g. [DOI] [PubMed] [Google Scholar]
- Xu L, Josan JS, Vagner J, Caplan MR, Hruby VJ, Mash EA, et al. (2012). Heterobivalent ligands target cell-surface receptor combinations in vivo. Proceedings of the National Academy of Sciences of the United States of America, 109(52), 21295–21300. 10.1073/pnas.1211762109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CM, Ji S, Li Y, Fu LY, Jiang T, & Meng FD (2017). Ror2, a developmentally regulated kinase, is associated with tumor growth, apoptosis, migration, and invasion in renal cell carcinoma. Oncology Research, 25(2), 195–205. 10.3727/096504016X14732772150424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RY, Yang KS, Pike LJ, & Marshall GR (2010). Targeting the dimerization of epidermal growth factor receptors with small-molecule inhibitors. Chemical Biology & Drug Design, 76(1), 1–9. 10.1111/j.1747-0285.2010.00986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Chu A, Li W, Wang B, Shelton F, Otero F, et al. (2009). Lipid G protein-coupled receptor ligand identification using beta-arrestin PathHunter assay. The Journal of Biological Chemistry, 284(18), 12328–12338. 10.1074/jbc.M806516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X, & Allison JP (2007). The B7 family and cancer therapy: Costimulation and coinhibition. Clinical Cancer Research, 13(18 Pt 1), 5271–5279. 10.1158/1078-0432.CCR-07-1030. [DOI] [PubMed] [Google Scholar]
- Zhang W, Ben-David M, & Sidhu SS (2016). Engineering cell signaling modulators from native protein-protein interactions. Current Opinion in Structural Biology, 45, 25–35. 10.1016/j.sbi.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang T, Fresnay S, Welty E, Sangrampurkar N, Rybak E, Zhou H, et al. (2011). Selective CD28 blockade attenuates acute and chronic rejection of murine cardiac allo-grafts in a CTLA-4-dependent manner. American Journal of Transplantation, 11(8), 1599–1609. 10.1111/j.1600-6143.2011.03624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Schwartz JC, Guo X, Bhatia S, Cao E, Lorenz M, et al. (2004). Structural and functional analysis of the costimulatory receptor programmed death-1. Immunity, 20(3), 337–347. [DOI] [PubMed] [Google Scholar]
- Zhanhua C, Gan JG, Lei L, Sakharkar MK, & Kangueane P (2005). Protein subunit interfaces: Heterodimers versus homodimers. Bioinformation, 1(2), 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L, & Kroemer G (2012). Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology, 1(8), 1223–1225. 10.4161/onci.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]