Abstract
Cerebral infarction (CI) is associated with high rates of disability, mortality, and death in China, but its mechanism is unclear. Therefore, early diagnosis of CI and determining its mechanism are very important. Intestinal microecology is thought to be related to cardiovascular and cerebrovascular diseases. We hypothesized that intestinal microecology is also related to CI and that the intestinal microecology in the stool of CI patients differs from that in healthy people.
Fecal samples of healthy subjects and CI patient (all n = 10) and we investigated the intestinal microecology of CI patient and healthy people stool by 16 seconds sequencing and analyzed relative abundance and diversity of microorganisms by unweighted pair-group method with arithmetic mean analysis (UPGMA) and principal co-ordinates analysis (PCoA). We also measured apolipoprotein E (ApoE) levels in the serum by ELISA assay and analyzed the correlation between ApoE and intestinal flora.
We found that the relative structure and diversity of intestinal microecology was significantly different between the stools of CI patients and healthy people. At the class level, Gammaproteobacteria was increased and Bacteroidia was decreased in CI patient stool. We found a correlation between ApoE in the serum and Bacteroidia and Gammaproteobacteria species.
We considered the intestinal flora can be used as an indicator of CI and the up-regulation of ApoE may be the potential mediate for intestinal microecology contribute to CI.
Keywords: apolipoprotein E, cerebral infarction, intestinal microecology
1. Introduction
Although great progress has been made in the diagnosis and treatment of cerebral infarction (CI), disability rates, and mortality remain high.[1] Approximately 15,000,000 people worldwide suffer from stroke each year and 6 million people die from stroke. CI accounts for approximately 70% to80% of all strokes, and most of these incidents result in disability.[2] Therefore, early diagnosis and treatment of CI and its complications are very important.
The structure of the intestinal flora is complex, and a normal flora structure is very important for maintaining human metabolism, immune, endocrine, and other physiological functions. Disruptions of the flora structure can lead to diabetes, obesity, and many other diseases.[3] The intestinal flora is involved in many physiological processes in the host, and recent studies have shown that immune and oxidative stress pathways affect the intestinal microflora and are involved in disease. Improving microecological therapy of the intestinal flora may be useful for preventing and treating some diseases.[4] In this study, we investigated the intestinal flora of patients with CI, explored the relationship between intestinal flora imbalance and CI, and analyzed whether the intestinal microflora can be used as a marker for early diagnosis of CI.
Apolipoprotein E (ApoE) is thought to be closely related to the development of CI and a risk factor for CI.[5,6] This study was designed to investigate the correlation between ApoE and the intestinal flora.
2. Methods
2.1. General information
In this study, 10 patients with CI diagnosed and treated and 10 healthy controls from May 2015 to March 2017 in Tianjin Huanhu Hospital, were selected as subjects. These subjects were 53 to 82 years old and had an average age of 64 years old. There were 10 male and 10 female subjects who had not been treated with antibiotics for 3 months before the study. The healthy control group underwent physical examination and were shown to not to have metabolic disease, cerebrovascular disease, malignant tumors, or other conditions. Approval for the study was granted by the Tianjin Huanhu Hospital medical ethics committee (ethics approval number is 2017-5).
2.2. Genomic DNA extraction and PCR amplification
After sampling, 500 mg of fecal matter was collected and the fecal genome was extracted using an Extraction Kit (Beijing Tiangen Biochemical Technology Co., Ltd., Beijing, China). Total RNA was extracted according to the kit instructions. The samples were diluted to 1 ng/μL with sterile water.
The genomic DNA was diluted and used as a template and specific primers with a barcode were used for PCR amplification using Phusion High-Fidelity PCR Master Mix with GC Buffer (New England Biolabs, Ipswich, MA, USA) and efficient and high-fidelity enzymes.
The 16S and V4 primers (515F and 806R) were used to evaluate bacterial diversity, 18S and V4 primers (528F and 706R) were used to determine the diversity of eukaryotic microorganisms, and ITS1 primers (ITS5-1737F and ITS2-2043R) were used to examine fungal diversity and the 16S V3-V4/16S V4-V5 region, archaeal 16S V4 region, and 18S V9 and ITS2 regions.
2.3. Construction and sequencing of the cDNA library
Using TruSeq DNA PCR-Free Sample Preparation Kit Library Kit (Illumina, San Diego, CA, USA) for library construction, the constructed library was quantified by Qubit and quantitative-PCR using the qualified library. Sequencing was performed using a HiSeq2500 PE250 (Illumina).
2.4. Serum ApoE level
Five milliliters of venous blood were collected from 10 CI patients and healthy volunteer after fasting overnight and centrifuged at 300 × g for 10 minutes. The serum level of ApoE was measured by ELISA kit (R&D Systems, Inc).
2.5. Statistical analysis
SPSS 11.0 software (SPSS, Inc., Chicago, IL, USA) was used for data analysis. Groups were compared using the t test. The Turkey and Wilcox rank sum tests were used to analyze differences in species beta diversity. Differences were considered significant when P < .05. Qiime software (Version, 1.7.0) was used to calculate the Unifrac distance and construct the unweighted pair-group method with arithmetic mean analysis (UPGMA) sample clustering tree. R software (Version 2.15.3) was used to draw and analyze the principal co-ordinates analysis (PCoA) diagram and determine the difference in the beta diversity indices between groups.
3. Results
3.1. Structure differences in intestinal flora in healthy volunteers and patients with CI
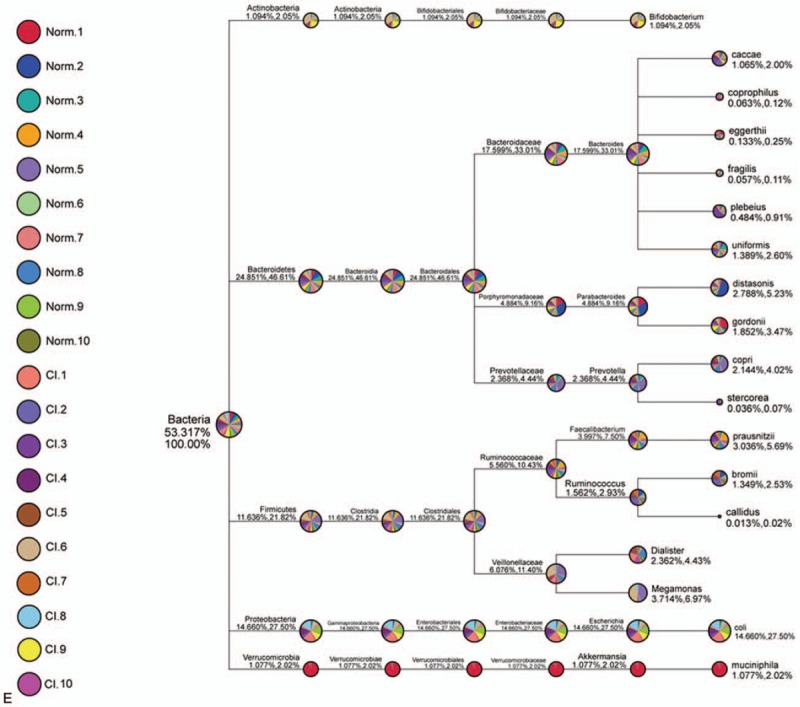
Based on species annotation, we found the maximum abundance of the intestinal flora in healthy volunteers and patients with CI at the class level and ranked these classes as the top 10 (Fig. 1A). A column diagram of the relative abundance of species was generated (Fig. 1B). Species annotation and determination of abundance at the class level were performed for the fecal samples from all healthy volunteers and patients with CI, and the top 35 species were selected for analysis (Fig. 1C). The intestinal flora in the CI patient group significantly differed from that in the healthy control group. An abundance comparison of the top 20 different microorganism is shown in Fig. 1D. We also found that Ruminococcus, Bacteroides, Prevoteella, Parabacteroides, Dialister, Faecalibacterium, Megamonas, Roseburia, and Escherichia were the 9 most abundant genera in CI patients (Fig. 1E).
Figure 1.
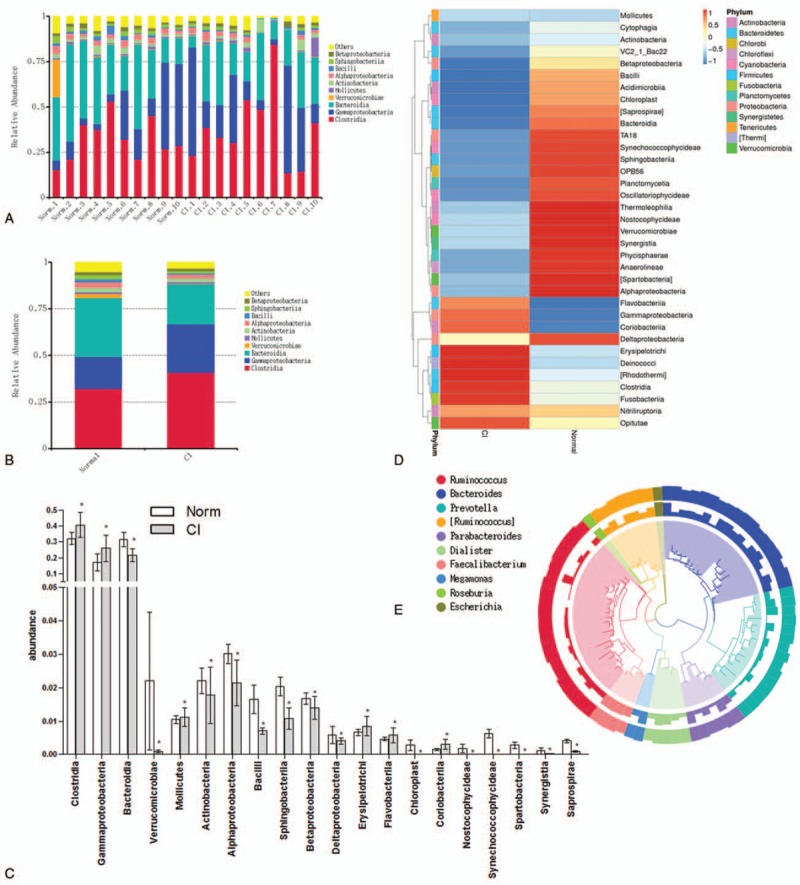
Relative abundance of microorganisms in stool of healthy people and CI patients. (A) Relative abundance of microorganism in stool of healthy people and CI patients. (B) Relative abundance of microorganisms in the stool of the healthy group and CI group. (C) Histogram of relative abundance of microorganism in stool of healthy people and CI patients. ∗, compared to healthy patients, P < .05. (D) Heatmap of microorganisms in stool of healthy people and CI patients. (E) Phylogenetic relationship species annotation of operational taxonomic units of top 9 in the intestinal flora of healthy volunteers and CI patients. CI = cerebral infarction.
3.2. Complexity analysis of intestinal flora in healthy volunteers and patients with CI
The alpha diversity of multiple samples (Fig. 2 A) was compared and analyzed to determine the microbial diversity of each group. We found that microbial diversity in CI patients was lower than that in healthy controls. A beta diversity heatmap was used to analyze the community structure by weighted unifrac and unweighted unifrac (Fig. 2 B). The structure characteristics of each bacteria group were analyzed by PCoA. We found significant differences in the intestinal microflora between healthy volunteers and patients with CI, while the samples in each group showed a similar bacterial structure (Fig. 2 C). UPGMA was conducted to investigate the similarity of each sample (Fig. 2 D). The taxonomic tree of healthy volunteers and CI patients is shown in Fig. 2 E. The abundance percentage of microorganisms by species classification in healthy volunteers and CI patients stool was determined.
Figure 2.
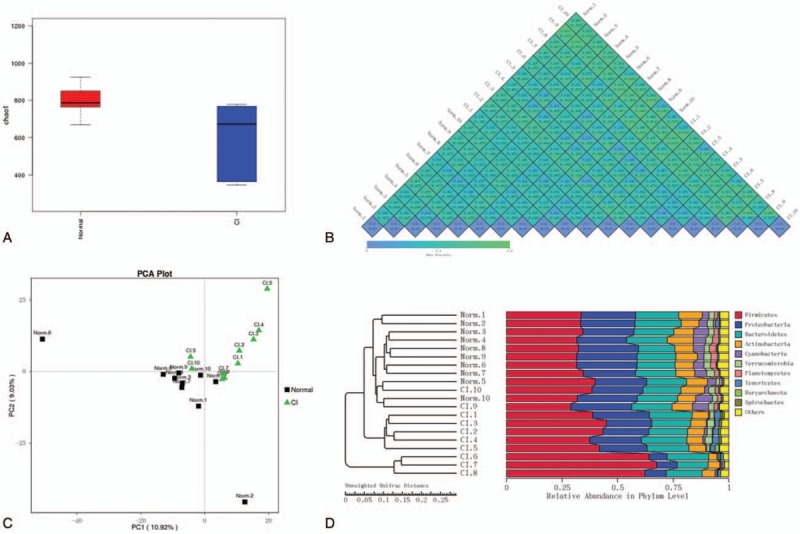
Microorganisms in stool of healthy people and CI patients. (A) Microbial alpha diversity analysis in stool of CI patients and healthy people. (B) Microbial beta diversity analysis in stool of CI patients and healthy people. A lower value represents higher microbial composition consistency. (C) Microbial PCoA analysis in stool of CI patients and healthy people. (D) UPGMA analysis in stool of CI patients and healthy people. (E) Taxtree of stool in CI patients and healthy people. CI = cerebral infarction, PCoA = principal co-ordinates analysis, UPGMA = unweighted pair-group method with arithmetic mean analysis.
3.3. Correlation of ApoE and intestinal flora in healthy volunteers and patients
We examined the correlation of ApoE and Bacteroidia and Gammaproteobacteria in CI patients and healthy volunteers (Fig. 3). The results suggested a correlation with intestinal flora in the CI process.
Figure 2 (Continued).
Microorganisms in stool of healthy people and CI patients. (A) Microbial alpha diversity analysis in stool of CI patients and healthy people. (B) Microbial beta diversity analysis in stool of CI patients and healthy people. A lower value represents higher microbial composition consistency. (C) Microbial PCoA analysis in stool of CI patients and healthy people. (D) UPGMA analysis in stool of CI patients and healthy people. (E) Taxtree of stool in CI patients and healthy people. CI = cerebral infarction, PCoA = principal co-ordinates analysis, UPGMA = unweighted pair-group method with arithmetic mean analysis.
Figure 3.
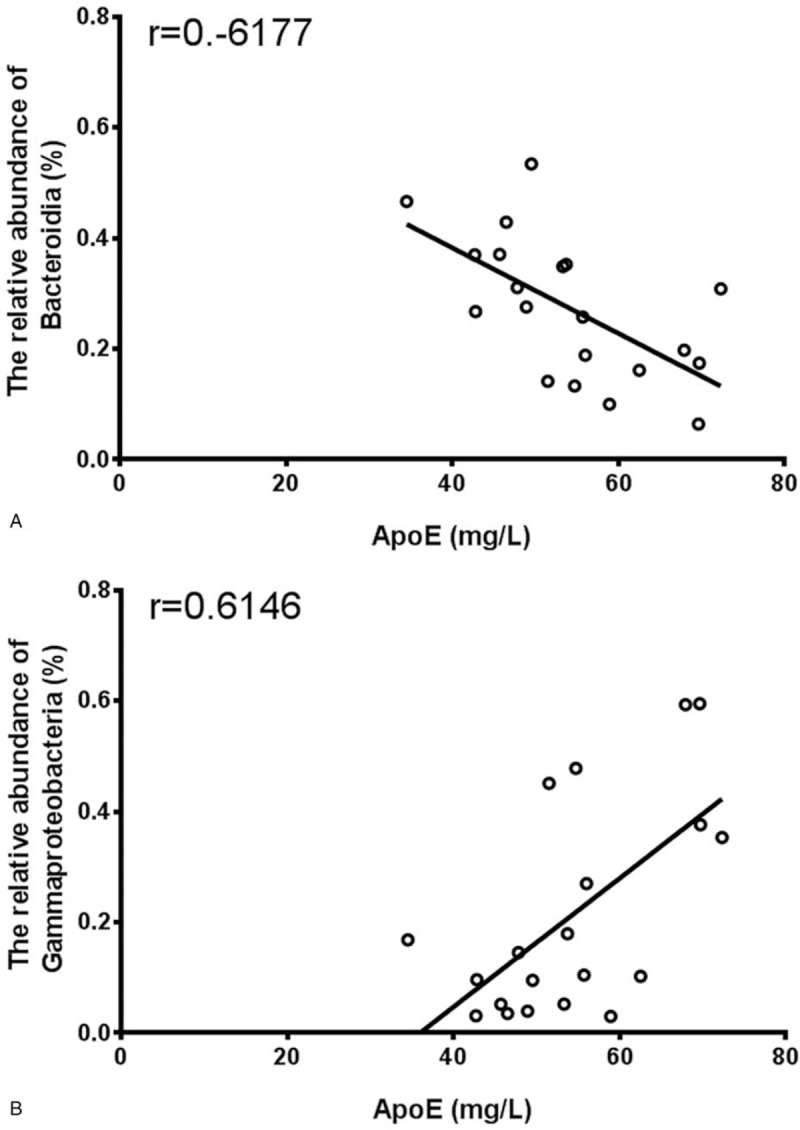
Correlation of serum ApoE and intestinal flora in stool of healthy people and CI patients. Correlative analysis of serum ApoE and Bacteroidia (A) and Gammaproteobacteria (B) in stool of healthy people and CI patients. ApoE = apolipoprotein E, CI = cerebral infarction.
4. Discussion
Intestinal flora can affect an organism by modulating the host metabolic products or via its own metabolic products, and monitoring and controlling the changes in the intestinal flora can help maintain a balance between the micro-ecology and environmental changes.[7] Intestinal flora play an important role in the development of ischemic stroke related risk factors such as obesity, diabetes mellitus, hyperlipidemia, and hypertension.[8]
The brain-gut-microbiome axis was recently proposed and is important in studies of intestinal microflora and the prognosis of stroke.[9] Intestinal microflora can move from the gastrointestinal tract to other organs, inducing systemic immune and inflammatory responses.[10] Immune and inflammatory reactions have been shown to be important in the mechanism of cerebral ischemia injury.[11] This study was conducted to investigate the intestinal flora structure in patients with CI. We found that while the CI and healthy groups had low similarity in intestinal flora, they had similar bacterial flora. In patients with CI, there was a significant increase in fecal bacteria, such as Gammaproteobacteria, while Bacteroidia showed a significant decrease compared with healthy group. The intestinal flora may play an important role in CI. The diagnosis and prognosis of acute CI mainly depend on medical history, physical examination, and imaging examination; thus, characteristic indices showing significant differences in the intestinal microorganisms in patients with CI may be useful for the early diagnosis and prognosis prediction of CI.
ApoE is thought to play a regulatory role in many physiological and pathological processes such as development, metabolic disease, and cardiovascular and cerebrovascular diseases.[12] A previous study showed that ApoE is a multifunctional protein that is important for lipoprotein metabolism and can be used as a biological marker of disease.[13] A recent study found that ApoE was significantly up-regulated in the plasma of patients with CI and is associated with a CI risk in the Asian population,[14,15] suggesting that it plays an important role in CI.
Our previous study showed that ApoE levels in the serum of patients with CI were significantly higher than those in the healthy group.[16,17] Additionally, there was a correlation between ApoE and Bacteroidia and Gammaproteobacteria. Bacteroidia and Gammaproteobacteria are the major microorganisms in the human intestine and play important roles in lipid metabolism. In this study, we found that the abundance of Bacteroidia, which is negatively correlated with ApoE, was decreased, while the abundance of Gammaproteobacteria, which is positively correlated with ApoE, was increased.
In previous reports, there are some ways for gut microbiota affect the lipid metabolism including ApoE, for example, changing food digestive efficiency, producing metabolite such as short-chain fatty acid, secondary bile acids, endotoxin, and so on.[18,19] We considered that ApoE metabolism was impacted due to abnormal abundance of Bacteroidia and Gammaproteobacteria which decreased food digestive efficiency and interfered metabolite of gut microbiota. Therefore, Bacteroidia and Gammaproteobacteria may affect the CI process through ApoE metabolism. There also are some reports described that there was a different abundance of gut microbiota in ApoE knockout rat compared with wide type rat, indicating ApoE metabolism abnormality could change intestinal microecology.[20,21]
5. Conclusion
In sum, we considered the intestinal flora can be used as an indicator of the occurrence and development of CI. We also found observed lipid disorders, for example, ApoE in CI. Therefore, an abnormal intestinal microbiota may induce metabolic disorders involving blood lipids to induce CI. However, there also were some limitations in this study, for example, it need more samples to certified the relationships between the intestinal microbiota and CI, and more penetrating exploration of the mechanism for intestinal microbiota induce the up-regulation of ApoE in further studies.
Acknowledgments
None.
Author contributions
Data curation: Wenyue Wang.
Formal analysis: Xu Li.
Funding acquisition: Yu Zhu, Xiuli Cheng.
Investigation: Xiuhua Yao.
Project administration: Yu Zhu.
Writing – original draft: Xiuli Cheng.
Footnotes
Abbreviations: ApoE = apolipoprotein E, CI = cerebral infarction, PCoA = principal co-ordinates analysis, UPGMA = unweighted pair-group method with arithmetic mean analysis.
WW, XL and XY These authors contributed equally to this work.
Ethics: Approval for the study was granted by the Tianjin Huanhu Hospital medical ethics committee (ethics approval number is 2017-5).
Conflicts of interest: No potential conflicts of interest are disclosed.
Funding: This study was supported by National Natural Science Foundation of China (Grant No.31501159), Tianjin Public Health Key Research Project (Grant No.15KG108), Tianjin Science and Technology Key Project on Chronic Diseases Prevention and Treatment (Grant No. 16ZXMJSY00020), Scientific Research Project on Traditional Chinese Medicine / Integrative Medicine of the Tianjin Health and Family Planning Commission (Grant No.2017084), Tianjin Municipal Special Program of Talents Development for Excellent Youth Scholars, China (Grant No. TJTZJH-QNBJRC-2-9), Tianjin 131 Creative Talents Cultivation Project (1st Class, 2016).
The authors report no conflicts of interest.
References
- [1].Liu J, Xu F, Mohammadtursun N, et al. The analysis of constitutions of traditional Chinese medicine in relation to cerebral infarction in a Chinese sample. J Altern Complement Med 2017;24:458–62. [DOI] [PubMed] [Google Scholar]
- [2].Dong MX, Hu L, Huang YJ, et al. Cerebrovascular risk factors for patients with cerebral watershed infarction: a case-control study based on computed tomography angiography in a population from Southwest China. Medicine (Baltimore) 2017;96:e7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Clavel T, Gomes-Neto JC, Lagkouvardos I, et al. Deciphering interactions between the gut microbiota and the immune system via microbial cultivation and minimal microbiomes. Immunol Rev 2017;279:8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Khanna S, Raffals LE. The microbiome in Crohn's disease: role in pathogenesis and role of microbiome replacement therapies. Gastroenterol Clin North Am 2017;46:481–92. [DOI] [PubMed] [Google Scholar]
- [5].Walker KA, Power MC, Hoogeveen RC, et al. Midlife systemic inflammation, late-life white matter integrity, and cerebral small vessel disease: the atherosclerosis risk in communities study. Stroke 2017;48:3196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rasmussen KL. Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: a review. Atherosclerosis 2016;255:145–55. [DOI] [PubMed] [Google Scholar]
- [7].Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet 2017;18:690–9. [DOI] [PubMed] [Google Scholar]
- [8].Tziatzios G, Giamarellos-Bourboulis EJ, Papanikolaou IS, et al. Is small intestinal bacterial overgrowth involved in the pathogenesis of functional dyspepsia? Med Hypotheses 2017;106:26–32. [DOI] [PubMed] [Google Scholar]
- [9].Allen AP, Dinan TG, Clarke G, et al. A psychology of the human brain-gut-microbiome axis. Soc Personal Psychol Compass 2017;11:e12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mohan M, Chow CT, Ryan CN, et al. Dietary gluten-induced gut dysbiosis is accompanied by selective upregulation of microRNAs with intestinal tight junction and bacteria-binding mMotifs in rhesus macaque model of celiac disease. Nutrients 2016;8: pii: E684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Das S, Kaul S, Jyothy A, et al. Association of APOE (E2, E3 and E4) gene variants and lipid levels in ischemic stroke, its subtypes and hemorrhagic stroke in a South Indian population. Neurosci Lett 2016;628:136–41. [DOI] [PubMed] [Google Scholar]
- [12].Zhong H, Cai Y, Cheng J, et al. Apolipoprotein E epsilon 4 enhances the association between the rs2910164 polymorphism of miR-146a and risk of atherosclerotic cerebral infarction. J Atheroscler Thromb 2016;23:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Woltjer RL, Reese LC, Richardson BE, et al. Pallidal neuronal apolipoprotein E in pantothenate kinase-associated neurodegeneration recapitulates ischemic injury to the globus pallidus. Mol Genet Metab 2015;116:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kumar A, Kumar P, Prasad M, et al. Association between apolipoprotein 4 gene polymorphism and risk of ischemic stroke: a meta-analysis. Ann Neurosci 2016;23:113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang ZD, Xia YF, Zhao Y, et al. Intestinal gutsfeature and role of ApoE and glucose metabolism in cerebral infarction patients. Int J Clin Exp Pathol 2017;10:561–5. [Google Scholar]
- [16].Ji W, Zhu Y, Kan P, et al. Analysis of intestinal microbial communities of cerebral infarction and ischemia patients based on high throughput sequencing technology and glucose and lipid metabolism. Mol Med Rep 2017;16:5413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thanissery R, Winston JA, Theriot CM. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 2017;45:86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nakatani A, Li X, Miyamoto J, et al. Dietary mung bean protein reduces high-fat diet-induced weight gain by modulating host bile acid metabolism in a gut microbiota-dependent manner. Biochem Biophys Res Commun 2018;501:955–61. [DOI] [PubMed] [Google Scholar]
- [19].Liu B, Zhang Y, Wang R, et al. Western diet feeding influences gut microbiota profiles in apoE knockout mice. Lipids Health Dis 2018;17:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bo T, Shao S, Wu D, et al. Relative variations of gut microbiota in disordered cholesterol metabolism caused by high-cholesterol diet and host genetics. Microbiologyopen 2017;6:e491. [DOI] [PMC free article] [PubMed] [Google Scholar]


