INTRODUCTION
Understanding the intricate relationship between macromolecular structure and function represents a central goal of undergraduate biology education (1–3). In teaching complex three-dimensional (3D) concepts, instructors typically depend on static two-dimensional (2D) textbook images or computer-based visualization software, which can lead to unintended misconceptions (4–6). While chemical and molecular kits exist, these models cannot handle the size and detail of macromolecules. Consequently, students may graduate in the life sciences without understanding how structure underlies function or acquiring skills to translate between 2D and 3D molecular models (5, 7).
Building on recent technological advances, 3D printing (3DP) potentiates an era in which students learn through direct interaction with dynamic 3D structural models. With 3DP, instructors have the opportunity to use tailor-made models of virtually any size molecule. For example, protein models can be designed to relate enzyme active site structures to kinetic activity. Furthermore, instructors can use diverse printing materials and accessories to demonstrate molecular properties, dynamics, and interactions (Fig. 1). In this article and supplemental guide, we present an example of how to incorporate a 3D model-based lesson on DNA supercoiling in an undergraduate biochemistry classroom and best practices for designing and printing 3D models.
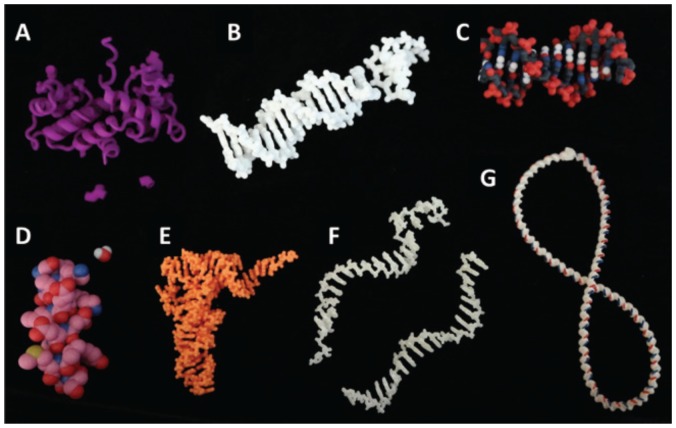
FIGURE 1.
A selection of 3D models with interactive features. A) The Enterobacteria phage λ transcription factor has interchangeable amino acids that allow investigation of point mutations on DNA binding. B) A DNA helix with LEGO-style replaceable base pairs allows investigation of DNA mutations. Models in A and B can be used together to allow investigation of compensatory mutations. Multicolored, detailed models of a DNA helix (C) or protein α-helix and water molecule (D) allow investigation of chemical details, for example, the size of the major and minor grooves or the diameter of the inside of a helix. Flexible models of Phe-tRNA (E), single-stranded RNA and DNA (F), and a long DNA duplex with magnetic ends (G) allow students to engage with the molecular dynamics, investigating folding of complex structures and demonstrating chemical attack, base stacking, or DNA supercoiling.
PROCEDURE
Classroom integration
To address learning goals related to DNA structure and function, we designed and printed flexible plastic models with magnetic ends to mimic DNA supercoiling (Appendices 1 and 2). We selected this model material so students could feel DNA relaxation and witness contortions resulting from twists in DNA. We developed a Qualtrics-based interactive activity to help students use the models to classify supercoiled DNA, predict the effects of DNA wrapping around nucleosomes, and differentiate between topoisomerase activities (Fig. 2). We divided an upper-level undergraduate biochemistry lecture class into groups of two to three students to foster peer-learning, and we provided each group with one model set. The models were also made available at our library resource center. Interactive questions required students to measure and explore physical aspects of the models. It took students roughly 50 minutes to complete the activity during class, and it was interspersed with lecture and demonstration via a digital overhead. Alternative deployments and tips for effective model implementation in a variety of course formats are outlined in Figure 3. Students reported in interviews that models were valuable for their learning because “physically seeing it makes something abstract very real.” In a survey, 60 to 70 percent of students agreed that physical models made it easier to learn the material being taught.
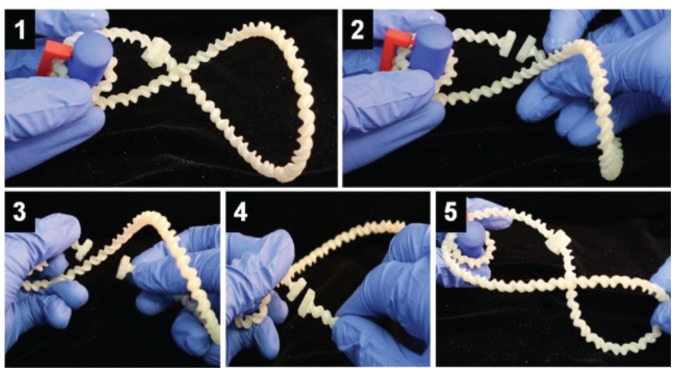
FIGURE 2.
Investigating DNA supercoiling. In step 1, students wrapped the DNA model (white) around a nucleosome model (blue) and characterized the resulting supercoil. In steps 2–4, students mimicked Topoisomerase II by cleaving the DNA and passing the intact strand of DNA through the cleaved site before re-adhering the ends. In step 5, students characterized the resulting supercoil and evaluated Topoisomerase II activity.
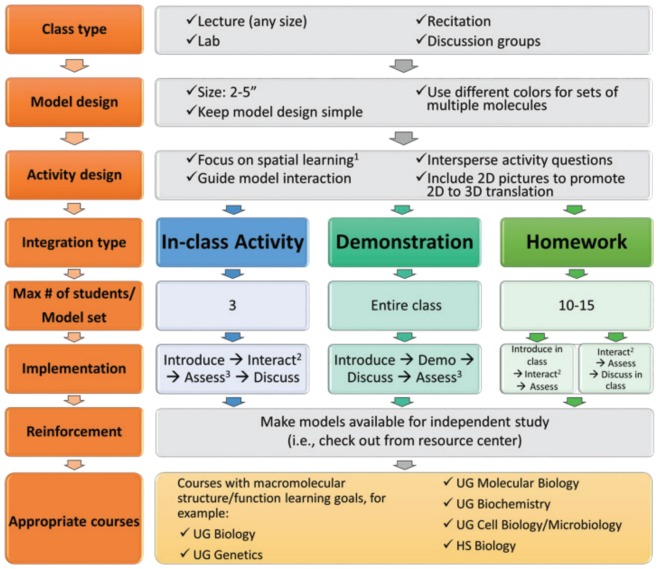
FIGURE 3.
Course integration of 3D instructional models. Integration tips are outlined for in-class activities (blue sequence), in-class demonstrations (teal sequence), and out-of-class homework (green sequence). 1 Based on cognitive theory of multimedia learning (11), instructors are recommended to include 2D and 3D models in addition to lecture. 2 For small classes, a one-on-one interaction with the instructor is preferred; for large classes or homework assignments, an adaptive response-guided activity can be substituted. 3 Formative assessments can provide instant feedback if they include in-class questions. UG = undergraduate; HS = high school.
Making 3D models
We designed our 3DP models around student misconceptions. Misconceptions regarding chemical structure were addressed with precise molecular replicas, and misconceptions about molecular interactions were addressed with simplified models that could replicate movement. A step-by-step video and text guide for our design of a flexible DNA model are provided as an example (Appendices 1 and 2 and https://digitalcommons.unl.edu/structuralmodels/22).
Numerous free online software programs and tutorials exist to facilitate designing 3D models. Molecular coordinates for thousands of macromolecules are housed in the Protein Data Bank and Nucleic Acid Data Bank. To print all or part of a macromolecule, start by downloading and opening the molecular coordinates in a molecular visualization software program, such as PyMOL or Chimera, then adjust the molecule’s size, color, thickness, and representation (e.g., ball-and-stick, ribbon, space-filling). Finally, export the structure to a graphics program, such as Blender or MeshLab, to add LEGO-style attachments (Figs. 1A and B), holes for magnets (Fig. 2), or other design elements. Alternatively, a simplified model can be built de novo using these same graphics programs and piecing together various shapes, as we did for the DNA supercoiling model (Fig. 2).
Following the design process, the object coordinates must be exported in a file format that can be read by a 3D printer, such as .stl or .x3d (8). Because most macromolecules lack a broad base that completely supports the structure, a type of 3DP called selective laser sintering (SLS) produces the best results. It utilizes a growing bed of powder to support otherwise unsupported parts of the model being printed. For each layer, a CO2 laser beam fuses the powder in a specific 2D pattern according to the design file. This process repeats across the entire model as each layer is successively fused to the previous layer; the unbound powder is then blown away, leaving the fused product (9). While SLS printers are cost-prohibitive, cost-effective SLS printing is accessible through various online printing services, such as Shapeways (http://shapeways.com), where our typical models cost between $5 and $30 (Figs. 1 and 2). Three-dimensional printing services offer numerous printing substrates, including a variety of plastics, metals, and sandstone. While plastics are durable and can be flexible, they are one color. Sandstone is a low-cost multicolor material, but resulting models are brittle and need to be clear-coated to increase their strength. Some print services offer a coated sandstone option, or clear epoxy can be applied after printing. Metals are stronger than plastic but are rigid and more expensive.
Most personal 3D printers extrude plastic from a movable source, slowly building a 3D model in a process referred to as fused deposition modeling (FDM). Although more time consuming, FDM can produce accurate 3D macromolecules if a support matrix can be printed that can be dissolved upon print completion. Personal 3D printers are usually highly cost effective, especially if the machine and expertise are already available, though materials are limited by the printer’s capabilities. Note that all 3D printers have potential environmental and health risks, including toxic nanoparticle and chemical vapor emissions, as well as heat, electrical, and mechanical risks, so users should consider the health risks of the materials used and consult their owner’s manual for safe operating procedures (10).
Instructors can freely download the printer files and print any of the custom 3D macromolecular models we developed and tested with students (Fig. 1; https://digitalcommons.unl.edu/structuralmodels/ or https://3dprint.nih.gov/discover) or inexpensively purchase (at no profit to ourselves) the corresponding models at www.shapeways.com/shops/macromolecules. We encourage use or adaptation of these models as needed. Please review these repositories for future developments and contact us with questions.
CONCLUSION
Three-dimensional printing represents an emerging technology with significant potential to advance life-science education by allowing students to physically explore macromolecular structure-function relationships and observe molecular dynamics and interactions. As this technology develops, the cost, resolution, strength, material options, and convenience of 3DP will improve, making 3D models an even more accessible teaching tool. While instructors who wish to design their own models potentially face a learning curve, this report and accompanying guide lay out the basic steps and considerations needed to start designing and printing 3D models.
SUPPLEMENTAL MATERIALS
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grant # NSF DUE-1625804. The authors declare that there are no conflicts of interest.
Footnotes
Supplemental materials available at http://asmscience.org/jmbe
REFERENCES
- 1.American Association for the Advancement of Science. Vision and change in undergraduate biology education: a call to action: a summary of recommendations made at a national conference organized by the American Association for the Advancement of Science; July 15–17, 2009; Washington, DC. 2011. [Google Scholar]
- 2.ASBMB. Biochemistry/molecular biology and liberal education: a report to the Teagle Foundation. Vol. 95. The American Society for Biochemistry and Molecular Biology; Washington, DC: 2008. [Google Scholar]
- 3.Tansey JT, Baird T, Jr, Cox MM, Fox KM, Knight J, Sears D, Bell E. Foundational concepts and underlying theories for majors in “biochemistry and molecular biology”. Biochem Mol Biol Educ. 2013;41:289–296. doi: 10.1002/bmb.20727. [DOI] [PubMed] [Google Scholar]
- 4.Roberts JR, Hagedorn E, Dillenburg P, Patrick M, Herman T. Physical models enhance molecular three-dimensional literacy in an introductory biochemistry course. Biochem Mol Biol Educ. 2005;33:105–110. doi: 10.1002/bmb.2005.494033022426. [DOI] [PubMed] [Google Scholar]
- 5.Jittivadhna K, Ruenwongsa P, Panijpan B. Beyond textbook illustrations: hand-held models of ordered DNA and protein structures as 3D supplements to enhance student learning of helical biopolymers. Biochem Mol Biol Educ. 2010;38:359–364. doi: 10.1002/bmb.20427. [DOI] [PubMed] [Google Scholar]
- 6.Canning DR, Cox JR. Teaching the structural nature of biological molecules: molecular visualization in the classroom and in the hands of students. Chem Educ Res Pract Eur. 2001;2:109–122. doi: 10.1039/B1RP90013G. [DOI] [Google Scholar]
- 7.Gilbert JK. Models and modelling: routes to more authentic science education. Int J Sci Math Educ. 2004;2:115–130. doi: 10.1007/s10763-004-3186-4. [DOI] [Google Scholar]
- 8.Gross BC, Erkal JL, Lockwood SY, Chen C, Spence DM. Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences. Analyt Chem. 2014;86:3240–3253. doi: 10.1021/ac403397r. [DOI] [PubMed] [Google Scholar]
- 9.Chia HN, Wu BM. Recent advances in 3D printing of biomaterials. J Biol Eng. 2015;9:4. doi: 10.1186/s13036-015-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azimi P, Zhao D, Pouzet C, Crain NE, Stephens B. Emissions of ultrafine particles and volatile organic compounds from commercially available desktop three-dimensional printers with multiple filaments. Environ Sci Technol. 2016;50:1260–1268. doi: 10.1021/acs.est.5b04983. [DOI] [PubMed] [Google Scholar]
- 11.Mayer RE. Multimedia learning. 2nd ed. Cambridge University Press; Cambridge, MA: 2001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.