Abstract
White adipose tissue (WAT) is the primary energy storage organ and its excess contributes to obesity, while brown adipose tissue (BAT) and inducible thermogenic (beige/brite) adipocytes in WAT dissipate energy via Ucp1 to maintain body temperature. BAT and subcutaneous WAT develop perinatally while visceral WAT forms after birth from precursors expressing distinct markers, such as Myf5, Pref-1, Wt1, and Prx1, depending on the anatomical location. In addition to the embryonic adipose precursors, a pool of endothelial cells or mural cells expressing Pparγ, Pdgfrβ, Sma and Zfp423 may become adipocytes during WAT expansion in adults. Several markers, such as Cd29, Cd34, Sca1, Cd24, Pdgfrα and Pref-1 are detected in adult WAT SVF cells that can be differentiated into adipocytes. However, potential heterogeneity and differences in developmental stage of these cells are not clear. Beige cells form in a depot- and condition-specific manner by de novo differentiation of precursors or by transdifferentiation. Thermogenic gene activation in brown and beige adipocytes relies on common transcriptional machinery that includes Prdm16, Zfp516, Pgc1α and Ebf2. Moreover, through changing the chromatin landscape, histone methyltransferases, such as Mll3/4 and Ehmt1, as well as demethylases, such as Lsd1, play an important role in regulating the thermogenic gene program. With the presence of BAT and beige/brite cells in human adults, increasing thermogenic activity of BAT and BAT-like tissues may help promote energy expenditure to combat obesity.
Introduction
Adipose tissue plays a crucial role in mammalian metabolism. White adipose tissue (WAT) stores excess energy as triglycerides (TAGs) in a unilocular lipid droplet within adipocytes. WAT is also considered an endocrine organ that secretes adipokines to affect various processes including food intake and insulin sensitivity 1. In contrast, brown adipose tissue (BAT) serves mostly as an oxidative tissue to regulate body temperature but also is beneficial to glucose and lipid homeostasis 2,3. Brown adipocytes contain multilocular lipid droplets and abundant mitochondria with the unique protein Ucp1, which uncouples substrate oxidation from ATP synthesis to generate heat. In rodents, BAT is located primarily in the interscapular region, whereas WAT depots are found in various but specific regions in the body. More recently, “thermogenic” Ucp1 positive adipocytes, so called “beige” or “brite” cells, have been found in mainly subcutaneous WAT, following cold exposure or stimulation by β3-adrenergic agonists, drawing much attention due to their potential benefit in weight-loss 4–6. This review will focus on the developmental origin of adipocytes, highlighting transcriptional and epigenetic control of brown and beige adipogenesis.
Developmental origin of WAT
Researchers have long puzzled over the origin of adipose tissue as well as its development. WAT is categorized into subcutaneous and visceral WAT. Subcutaneous WAT is found in inguinal (posterior) and intrascapular (anterior) regions, whereas visceral WAT is found in perigonadal (often referred as epididymal WAT in males), perirenal, epicardial, retroperitoneal, mesenteric and omental regions (Shown in Figure 1A). Subcutaneous and visceral WAT are believed to have distinct response mechanisms as well as health consequence upon high-fat diet-induced expansion 7–10. In addition, subcutaneous as opposed to visceral WAT, is considered to be the major site of “browning” during cold exposure (will be discussed in detail in a later section). Moreover, although both subcutaneous and visceral WAT are believed to be of mesodermal origin, it has been unclear whether these WAT depots have the same origin or development.
Figure 1.
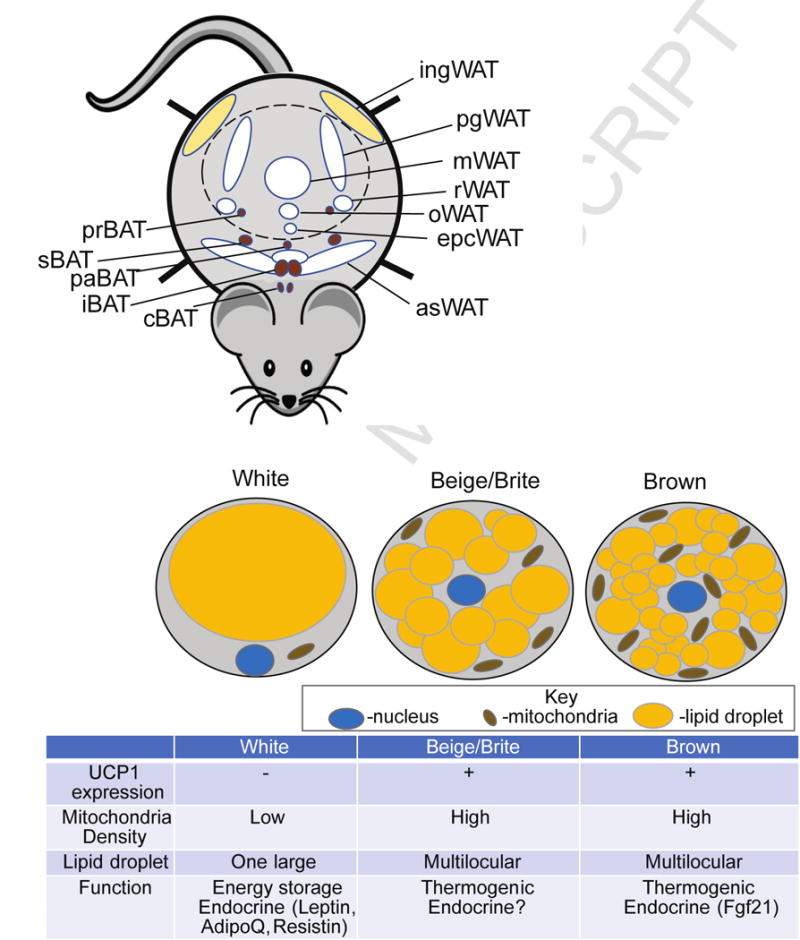
Anatomical location of murine adipose tissue and three types of adipocytes. A. Anatomical location of mouse white adipose tissue (W AT) and brown adipose tissue (BAT). ingWAT-inguinal WAT also known as posterior subcutaneous, pgW AT-perigonadal WAT, rWAT-retroperitoneal WAT, mWAT-mesenteric WAT, oWAT-omental WAT, epcWAT- epicardial WAT, asWAT- anterior subcutaneous WAT. pgWAT, rWAT, mWAT, oWAT, epcWAT are collectively called visceral WAT, while ingWAT and asWAT are combined into subcutaneous WAT. prBAT- perirenal BAT, sBAT-subscapular BAT, paBAT-periaortal BAT, iBAT-interscapular BAT, cBAT-cervical BAT.
The peritoneum is depicted as a dotted line. B. Functional and morphological differences between 3 types of adipocyte: white, beige/brite and brown.
Role of Pref-1 cells in adipose tissue development
To address the temporal and developmental origin of adipose tissue, Hudak et al. utilized lineage tracing with an inducible Preadipocyte factor 1 (Pref-1 or Dlk1) promoter coupled with two fluorescent reporters- H2BGFP for transient labeling and Rosa26-flox-stop-flox-tdTomato for permanent labeling in mice 11. Pref-1 represents a useful tool for studying adipose tissue development since its expression is detected only in adipose precursors and is absent from differentiated adipocytes 12. From transient labeling studies, it became apparent that Pref-1+ cells first started appearing at E10.5 in mouse embryogenesis in the dorsal mesenteric region at the presumptive inguinal and dorsal subcutaneous depots. By E13.5 these cells formed a line at the dorsal edge of the embryo under the skin, were proliferative, but did not yet contain lipids. At E17.5, these precursor cells differentiated into lipid-containing adipocytes forming subcutaneous WAT. By E19.5, the number of lipid-filled cells more than doubled in this region, indicating hyperplasia as a mechanism for WAT expansion during embryogenesis. This temporal aspect of adipogenesis that occurs prenatally was confirmed using permanent labeling that showed that the lipid filled tdTomato positive cells in the subcutaneous region at E19.5 were descendants of Pref-1 cells. In contrast, no Pref-1 marked cells or lipid containing adipocytes were detected in the presumptive visceral WAT during embryogenesis in this study. It is not until P6 that Pref-1 cells were detected as lipid-laden adipocytes in the visceral WAT. This temporal difference in subcutaneous and visceral WAT development provided the first solid evidence that subcutaneous WAT starts its development perinatally, while visceral fat development takes place after birth. These findings are in agreement with studies utilizing the AdipoChaser model, an inducible system for permanent labeling with LacZ, relying on the adipocyte specific activity of the AdipoQ promoter 13. Regardless, in humans, since WAT is estimated to account for 16% of body weight of newborns, at least some of the adipose depots must develop prenatally 14. Although genetic approaches cannot be used in humans, light microscopy examination showed the first traces of a fat organ between the 14th and 16th weeks of prenatal life 15.
Development of subcutaneous vs. visceral WAT
Given the temporal difference in the formation of subcutaneous versus visceral WAT, it is unclear whether they arise from the same or distinct precursors. However, a study reported that formation of six different visceral WAT depots, but not subcutaneous WAT or BAT, occurred from cells expressing Wilms Tumor 1 (Wt1) in late gestation 16. This work also suggested that WAT depots associated with visceral organs have a mesothelial layer that serves as a source of adipocyte precursors, while, the subcutaneous depot was derived from cells marked by Prx1, a homeobox transcription factor expressed in embryonic limb and bud mesenchyme 17,18. By lineage tracing using membrane fluorescent reporter, mT/mG, Myf5-Cre, which was thought to be active in muscle and interscapular BAT as described below, also labeled most or all cells of anterior subcutaneous and renal WAT, but not inguinal or perigonadal WAT 19. Moreover, by utilizing Sox10-Cre and R26-YFP to trace neural crest cells, Billon et al. found in adult mice that cephalic WAT around salivary glands, but not subcutaneous, gonadal, perirenal or interscapular WAT, was labelled by Sox10-YFP+ cells that co-expressed Perilipin, indicating that the craniofacial adipocytes arise from neural crest cells 20. These studies, overall, highlight that a simplistic division of WAT into visceral and subcutaneous may need to be reconsidered. It is probable that different WAT depots may have different origins and even cells within the same adipose tissue may be heterogeneous in origin.
Adipose tissue expansion in adults
Numerous studies also investigated the origin of white adipocytes during adipose expansion in adults. Mural cells (pericytes and vascular smooth muscle cells) are mainly derived from mesodermal lineages and, within WAT, were shown first to be a likely pool of adipose progenitors 21–24.The evidence came from Tang et al. who utilized Pparγ locus driving GFP or LacZ expression that detected Pparγ+ cells within blood vessels in adult WAT but not in other tissues 24. These Pparγ+ cells also closely resembled mural cells based on the expression of Pdgfrβ, neural/glial antigen 2 (Ng2) and smooth muscle actin (Sma). In a later study, the same group reported that Pparγ marked both developmental and adult WAT progenitors that derived from distinct lineages, but only adult adipose progenitors came from Sma+ mural lineage 22. However, Pparγ is known to be expressed in macrophages and dendritic cells 25,26 in addition to being master a regulator of adipogenesis 27. Therefore, Pparγ lineage tracing gives some information about adipocyte origin, but potential contribution from macrophages and dendritic cells cannot be completely ruled out. Another study that followed cells expressing Zfp423, a zinc-finger transcription factor that regulates Pparγ expression, reported that adult adipose tissue was derived from not only mural cells but also from endothelial cells 28,29. In a later study, Zfp423+; Pdgfrβ+ mural cells were shown to contribute to WAT hyperplasia in diet-induced obesity in a sex- and depot-dependent manner 30. Additionally, two distinct pools of Perilipin+ preadipocytes for both developmental and adult adipose expansion were identified but the majority of embryonic preadipocytes were not derived from the Pdgfrβ+ mural compartment 31, highlighting the importance of mural precursors for adult adipose tissue expansion. Recently, cells expressing Vstm2a, which is secreted from preadipocytes 32, were also associated closely with the vasculature, but interestingly, they did not express endothelial or mural markers. In addition, neural crest-derived adipose precursors were detected in subcutaneous WAT as well 33.
In addition to mural cells, other mesenchymal precursors have been shown to play a role in adult WAT expansion. Ablation of Pref-1+ cells using diphtheria toxin in early embryogenesis resulted in a severe lack of adipose tissue postnatally, further indicating the importance of Pref-1 expressing mesenchymal precursors in embryonic and postnatal adipogenesis. Upon high-fat diet feeding, Pref-1+ cells proliferated and contributed to WAT expansion through hyperplasia. However, Pref-1+ adipose precursors did not express hematopoietic markers (Cd45) and were not of endothelial (Cd31−, Ve-cadherin−) or pericyte origin (Sma−, Pdgfrβ− or Cd146−) 11. Permanent dual labeling with Tie2-GFP and Pref-1-tdTomato also showed that those cells labeled by Pref-1 were not co-stained with Tie2-GFP that labels vasculature, further excluding an endothelial origin of Pref-1+ cells. This study showed that Pref-1 labels a population of early mesenchymal adipose precursors that do not coincide with precursor/preadipocyte populations associated with the vasculature. Similarly, Pdgfrα+ adipose precursor cells that give rise to all adult WAT depots and proliferate in visceral WAT upon high-fat diet 34,35, albeit in close proximity to capillaries, were negative not only for Pparγ but also for Sma, Pdgfrβ, and the endothelial marker, isolectin Ib4 34. These data indicate that the Pdgfrα+ progenitor cell population may reside outside of the mural compartment. These studies, overall, suggest that very early precursors expressing Pref-1 may migrate closer to the vasculature upon loss of Pref-1 and acquire mural or endothelial markers and then become Pdgfrα expressing cells. Alternatively, the mural cells or those cells in the vasculature may represent a different progenitor pool from Pref-1 or Pdgfrα positive pools. However, the exact relationship between adipose cell populations traced by various markers and their relative contribution to each WAT depot in development and in adults in various conditions still need further investigation.
Isolation of adipose precursors in SVF of adult WAT
In adipose tissue, in addition to adipocytes, there are multiple other types of cells, such as preadipocytes or adipose precursors, stem cells, fibroblasts, endothelial cells, macrophages, and leukocytes, often collectively called SVF based on the separation from lipid-containing adipocytes. Heterogeneity within SVF has long been an obstacle in isolating and characterizing pure precursor populations. Fluorescence activated cell sorting (FACS) using several stem cell markers allowed researchers to enrich SVF for the precursor/progenitor population and, thus, sorting for Lin− (Cd31−, Cd45−, Ter119−) eliminated the majority of endothelial, hematopoietic cells and erythrocytes 36. However, it is not clear whether these markers may label cells at different stages of differentiation, such as stem cells, committed preadipocytes, or those cells at early stage of differentiation.
Rodeheffer et al. isolated a Lin−: Cd29+: Cd34+: Sca1+: Cd24+ population of proliferating adipose precursors that gave rise to Cd24− cells in vivo during adipogenesis 36. In a later study, Cd24− cells of the precursor population were shown to represent preadipocytes that express Pparγ and C/ebpα, key adipogenic transcription factors 35. Since Pdgfrα+ labeled both Cd24+ and Cd24− precursor populations in WAT, isolation of Lin−: Pdgfrα+ cells may represent a strategy to enrich for adipocyte precursors in adipose tissue 37,38. In an attempt to characterize Pref-1+ cells in the context of the adipogenic lineage, Hudak et al. performed gene expression and immunostaining analysis of Pref-1+ cells and showed their mesenchymal origin (Sox9+, Cd29+, Sca1+, Cd105+ and Cd34+) 11. The transiently labeled Pref-1+ cells did not yet express the adipogenic transcription factor Pparγ or adipose commitment factor Zfp423, but were proliferative precursors based on expression of Cd24 and Ki-67 and incorporation of BrdU. Some of the permanently labelled Pref-1 cells had lipid laden morphology and expressed Zfp423, Pparγ and C/ebpα indicating that Pref-1 cells indeed are adipose precursors. Additionally, when SVF from Pref-1-tdTomato mice was injected into SCID mice, after 2 weeks these cells differentiated into adipocytes with a characteristic gene expressing signature. These findings suggest that Pref-1 precursors represent a very early stem cell-like population and highlight the usefulness of Pref-1 as a marker for FACS or conditional ablation from adipose precursors at early stages of adipogenesis. Further research will shed light on when and under which physiological and molecular cues Pref-1 expression is lost in order for adipogenesis in vivo to proceed. Overall, these studies suggest that adipose SVF contains a hierarchy of progenitor populations with different degree of progression from adipose commitment to differentiation. Additionally, obese individuals show excessive fibrosis in WAT 39,40. It has recently been reported that myofibroblasts arising from diet-induced obesity came from highly proliferative Cd9high, Pdgfrα+ cells with high Pref-1 expression. In contract, Cd9low, Pdgfrα+ population was enriched for Pparγ and C/ebpα, with low Pref-1 expression, having low proliferative capacity but high adipogenic potential 41. Overall, the relationship between cell populations identified based on the expression of various markers needs further investigation. Identification and characterization of stage specific markers may help to isolate and define various precursor populations and their relationship during WAT development.
Developmental origin of BAT
Given the morphological and functional differences between brown and white adipocytes, these two types of adipocytes may have different developmental origins. Indeed, interscapular BAT (iBAT) formation in mice starts earlier than WAT during embryogenesis and BAT is fully thermogenically-competent at birth, providing a defense mechanism against cold stress in newborns. As early as at E9.5, cells expressing engrailed 1 (En1), a homeobox domain containing gene that marks the central dermomyotome, were shown to give rise to iBAT 42. Moreover, cells that express En1 at E10.5-11.5 gave rise to iBAT as well as dermis and epaxial muscle, indicating a very early specification of cells to BAT and that BAT and muscle cells may share the same progenitors in early development. The next evidence came from a study by Timmons et al. where, by microarray analysis of primary preadipocytes, they found that brown fat preadipocytes had a myogenic transcription signature (expressing transcription factors for muscle differentiation, such as MyoD and Myf5), differing from immortalized adipogenic cell lines 43. Seale and colleagues then utilized mice harboring Myf5-Cre coupled with R26R3-YFP and found that Myf5 expressing cells gave rise to not only muscle but also iBAT and perirenal BAT 4. Later, a study with Myf5-Cre coupled with mT/mG, showed that Myf5+ cells also gave rise to some WAT depots in the anterior subcutaneous, interscapular subcutaneous, and retroperitoneal regions. However, not all BAT depots may come from Myf5+ cells, with only interscapular and subscapular BAT being fully labeled by Myf5-Cre, and only a population of cells in cervical BAT and none from periaortal or perirenal BAT 19. Additionally, a study using a CreERT2 knock-in allele at the paired-homeodomain transcription factor- Pax7 locus combined with a LacZ reporter, showed that somatic Pax7 expressing cells marked at E9.5 gave rise to dorsal dermis, BAT, trunk muscle, and diaphragm muscle. After E12.5 these cells become restricted to the muscle lineage 44. Moreover, although Pref-1 is required for WAT development and expansion, only a few cells labeled by Pref-1 were detected in BAT, further indicating an early divergence of BAT precursors in development prior to Pref-1 expression 11. Overall, it can be viewed that interscapular and subscapular BAT and muscle originate from common Myf5+, Pax7+ progenitors, while brown adipocytes from periaortic and perirenal BAT as well as the most well studied WAT depots, such as inguinal and epididymal do not share the same developmental origin.
“Browning” of WAT
Typical adipocytes in WAT have a unilocular lipid droplet morphology and few mitochondria (Summarized in Figure 1B). Upon cold exposure or β-adrenergic stimulation, some cells in WAT acquire Ucp1 expression and have multilocular lipid droplets and abundant mitochondria. Thus, WAT may undergo “browning” with the appearance of thermogenic “beige” or “brite” adipocytes that share some similar features with brown adipocytes 4–6. Historically, the first evidence of existence of these multilocular adipocytes in WAT came from a study in 1984, in which cold acclimation caused the appearance of brown fat like cells in the parametrial WAT depot of female BALB/c mice judged by morphology in EM and presence of Ucp1 45. Later in 1992, a report showed Ucp1 expression in various WAT depots of rats, that was induced by cold exposure 46. Furthermore, mouse subcutaneous inguinal WAT depot is much more susceptible to “browning” even with mild stimulation compared to visceral epididymal WAT. Additionally, capacity to “brown” depends on the mouse strain: the A/J strain showed a marked induction in Ucp1 in WAT depots following β3-adrenergic stimulation compared to C57BL/6J 47.
Transdifferentiation as a potential mechanism for “browning”
A study by Himms-Hagen et al. showed that multilocular brown-like adipocytes in retroperitoneal WAT following a 7-day treatment with β3-adrenergic agonist- CL-316,243, did not incorporate BrdU 48. The authors concluded that these brown-like adipocytes did not derive from actively proliferating cells, and rather came from conversion of white adipocytes. Consistently, 95% of brown-like multilocular cells with numerous mitochondria also did not incorporate BrdU in retroperitoneal WAT in rats following β3-adrenergic stimulation 49. More recently, Rosenwald et al. showed the appearance of Ucp1+ cells in ingWAT after one week cold exposure 50 by using two mouse models, Ucp1-GFP for transient labeling of Ucp1 expressing cells and Ucp1-CreER-ROSA-tdRFP for permanent labeling. They also reported that beige/brite adipocytes were not eliminated by apoptosis and reverted to a white adipocyte unilocular morphology with characteristic gene expression after 5 weeks of warm adaptation. Furthermore, if subjected to a one-week cold exposure again, these cells could convert back to beige/brite cells. However, after restimulation, only half of the beige/brite adipocytes detected were from warm–adapted white, but previously beige adipocytes. These data suggested that in addition to transdifferentiation some of the beige adipocytes may have formed from de novo differentiation of precursor cells. In line with this evidence for transdifferentiation, another study showed that most if not all beige/brite adipocytes in ingWAT, but not abdominal WAT, following cold exposure did not arise from recruitment or proliferation precursors and rather came from unilocular adipocytes 51. All together, these studies showed transdifferentiation to be a mechanism for “browning” of WAT.
De novo differentiation of beige/brite adipocytes
In contrast to the concept of transdifferentiation, various researchers have reported that, upon cold exposure or β3-adrenergic stimulation, a subset of SVF cells isolated from WAT by using different markers, such as Pdgfrα and Ebf2, proliferate and become Ucp1+ beige or brown-like cells 34,52. The use of the AdipoChaser model also revealed that most of the “browning” in subcutaneous WAT following cold exposure or β3-adrenergic stimulation occurred from de novo adipogenesis 13. Overall, these studies indicate the presence of precursors cells in WAT that upon cold or β3-adrenergic stimulation may undergo browning and acquire Ucp1 expression. Are beige precursors distinct from typical white or brown adipocyte precursors? Unlike brown adipocytes, the Ucp1+ cells in WAT were originally believed to come from a Myf5− lineage 4. However, a more recent study using Myf5-Cre coupled with fluorescent reporters (R26R3-YFP, R26R-LacZ or mT/mG) showed that beige/brite adipocytes can come from Myf5+ or Myf5− lineages depending on the WAT depot and the type of stimulation 4,19. To understand the molecular identity of beige precursor cells, Wu et al. isolated clonal lines from ingWAT SVF to compare gene expression signature of adipogenic clones. This analysis revealed the existence of a subset of cells in ingWAT that showed gene expression patterns more similar to bona fide brown fat cell lines than other ingWAT cell lines, suggesting the presence of a distinct pool of progenitors that generate “beige” cells in ingWAT that are more similar to classical BAT progenitors. These “beige” progenitors in basal conditions had low expression of thermogenic genes. However, upon stimulation with cAMP, they responded by an increase in Ucp1 expression to levels similar to brown fat cells, having enhanced respiration 53. Additionally, Pdgfrα+ cells from abdominal WAT, which represent white adipose precursors, can also become Ucp1+ cells in response to β3-adrenergic stimulation but only in abdominal WAT 34. Early B-cell factor 2 (Ebf2), a transcription factor critical for BAT development, was shown also to mark SVF cells in ingWAT capable of acquiring Ucp1 upon differentiation in culture 52. The number of these Ebf2+ cells in ingWAT increased upon cold-exposure contributing to de novo beige adipogenesis. In another study, beige/brite adipocytes were shown to share a molecular signature with smooth-muscle cells not observed in classical brown adipocytes 54. A fate-mapping approach based on the myosin heavy chain 11 (Myh11) promoter active in smooth muscle or smooth muscle-like cells marked some of the beige adipocytes in ingWAT following 2-weeks of cold exposure 54. Gene expression analysis from Myh11+ cells of WAT revealed that these cells were not enriched in pericyte, endothelial or hematopoietic markers, but they expressed some precursor/preadipocyte genes (Sca1, Pdgfrα, Zfp423). The authors concluded that the smooth muscle lineage may overlap with previously described preadipocyte populations. Another study showed that cells expressing the mural marker Pdgfrβ which has been demonstrated to play a role in WAT adipogenesis were also recruited to become beige adipocytes after prolonged cold exposure 30. Interestingly, after a short-term cold exposure (1 week), however, very few beige cells came from Pdgfrβ+ cells, indicating that distinct “browning” mechanisms may exist in short versus long term cold exposure. However, it remains unclear whether they represent a distinct pool or share precursors with classical WAT or BAT cells. Interestingly, it has also been reported that, following cold exposure, eWAT exhibited an increase in adipogenesis; however, the newly formed adipocytes in this depot appeared to be Ucp1− further proving evidence that “browning” of WAT is depot- dependent 13. Together these studies suggest the presence of a subpopulation within WAT precursors that is capable of differentiating into Ucp1+ adipocytes. Further investigation is needed in order to establish markers that distinguish this cell population predisposed to “browning” to affect energy metabolism in vivo.
How can such a discrepancy in the origin of beige/brite adipocytes be explained? Potentially, different degrees of innervation exist between subcutaneous and visceral WAT and may play a role in different mechanisms governing “browning” between the two depots. Indeed, Granneman and coworkers reported that Pdgfrα+ precursors from abdominal WAT proliferated to become Ucp1+ cells, while brown-like adipocytes emerged in subcutaneous WAT (ingWAT) upon stimulation were from transdifferentiation of unilocular white adipocytes 34,51. Regardless, results explaining either transdifferentiation or de novo adipogenesis for “browning” may partly arise due to technical limitations such as caveats associated with long lasting effect of tamoxifen when using Cre-ER or difficulty in substrate penetrance for β–galactosidase when using LacZ reporter mice 5,51. Overall, all these studies point to the emerging complexity in the origin of brite or beige adipocytes. The mechanism of their formation may be highly dependent on the mouse strain and sex, the specific WAT depot analyzed, as well as the specific stimulation applied. Better studies using lineage- tracing and co-labeling with multiple precursor markers may help determine the relative contribution of each mechanism.
Human BAT: beige or brown?
Is human BAT considered beige or classical brown? While BAT as an organ was originally described in 1551 55, it was not described to be present in all mammals until the 20th century. The presence in humans was discovered almost 40 years ago but was limited to outdoor workers, skid row alcoholics, and those with pro-brown adipogenic tumors 56,57. Indeed, a breakthrough in 2009 showed that BAT in humans is either widespread or universal under cold stimulus and could be metabolically relevant58–63. In infancy, human BAT is localized to interscapular and perirenal depots, the molecular signature of which closely resembles classical rodent iBAT 60. Recent studies suggest that in adult humans UCP1+ adipocytes can be found in various depots around the body that are typically heterogeneous, also containing UCP1− adipocytes. Brown-like adipocytes have been detected in the supraclavicular region, being the most enriched for UCP1+ adipocytes, and also around the aorta, carotid artery and subscapular region, as well as others 2,6. Whether these UCP1+ adipocytes in human adults represent beige or true brown adipocytes remains controversial. On the gene expression level, supraclavicular UCP1+ adipocytes more closely resembled mouse beige adipocytes. Lee et al. showed that these adipocytes expressed some common BAT markers such as UCP1, PGC1α, PRDM16 and DIO2, but not MPZL2 which is thought to only be expressed in classical iBAT of rodents 64. On the other hand, these cells expressed beige fat-specific genes such as Tmem26 and Hoxc9. Another study confirmed the “beige-like” molecular signature of human UCP1+ adipocytes and discovered novel markers K3 (KCNK3) and mitochondrial tumor suppressor 1 (MTUS1) to be enriched in human beige cells versus white adipocytes 65. However UCP1+ adipocytes from other human depots such as cervical and perirenal regions, showed expression of classical BAT markers, ZIC1 and LHX8 66,67. All these studies point out heterogeneous composition of human BAT as well as an emerging need for novel markers that will allow researchers to clearly distinguish classical BAT vs beige cells.
Transcriptional regulation of BAT development and “browning” of WAT
Since BAT mass is inversely correlated with BMI in humans, increasing BAT activity could be a promising strategy for weight-loss and management of obesity-associated diseases 2,68. With greatly higher mass of WAT in comparison to BAT, increasing WAT “browning” may improve insulin sensitivity and reduce weight gain under high fat diet as shown in mice 3,69–73. This section summarizes the transcriptional regulation involved in BAT development and “browning” of WAT. The well-established transcriptional program for white adipocyte differentiation has been extensively reviewed elsewhere (Rosen and Spiegelman, 2014; Farmer, 2006).
General adipogenic transcription factors, Prdm16, and Zfp516
Early work describing the transcriptional regulation of BAT centered on the norepinephrine-β3-adrenergic receptor-cAMP- cyclic AMP response element binding protein (CREB)/ p38 MAP kinase axis central to the response to cold. Thus, several target genes of this signaling pathway have been described such as Pparγ-coactivator 1α (Pgc1α), CCAAT-enhancer binding protein β (C/ebpβ), diodinase 2 (Dio2) as well as Ucp1 itself 74–76. Further work showed that BAT gene regulation requires general adipogenic machinery including peroxisome proliferator-activated receptor gamma (Pparγ), RXR, and the aforementioned C/ebpβ 77. Importantly, by RT-qPCR analysis of WAT and BAT to identify transcription-related genes enriched in BAT, Seale et al. first identified Prdm16 as a BAT-enriched coregulator of the BAT gene program 78. Prdm16 interacts with a wide variety of transcription factors and cofactors including C/ebpβ, C-terminal binding protein 1 and 2 (Ctbp1, Ctbp2), histone deacetylase 1 and 2 (Hdac1/2), mediator complex subunit 1 (Med1), Pgc1α, Pparγ, and Zfp516 as well as epigenetic regulators-euchromatic histone lysine methyltransferase 1 (Ehmt1) and lysine specific demethylase 1 (Lsd1) 71,78–84. Interestingly, Harms et al. showed that while Prdm16 is required for maintenance of brown adipocyte identity, Prdm16 is not required for BAT development. While there may be some compensatory activity of Prdm3, prenatal development was not affected by Prdm3/Prdm16 double knockout 85. Med1 that interacts with Prdm16 may mediate the active chromatin structure at BAT specific genes bringing distal enhancers to proximity to the transcription start site facilitating transcriptional activation 86. However, markers of active transcription such as H3K27ac were only mildly affected by Prdm16 knockout. Interestingly, Prdm16 is not regulated by cold and must rely on interacting factors to facilitate Prdm16-mediated BAT gene induction. One such factor is Zfp516, which is induced upon cold exposure. Zfp516 binds and activates the Ucp1 promoter in response to cold to upregulate Ucp171. In this regard, Zfp516 KO embryos show dramatically reduced BAT mass, while mice overexpressing Zfp516 in adipose tissue demonstrate enhanced “browning” of WAT even at room temperature. Although Zfp516 directly binds to Prdm16 and may be responsible for cold-inducible Prdm16-mediated activity, Dempersmier et al. showed that Zfp516 can induce Ucp1 expression in the absence of Prdm16. Perhaps in the absence of Prdm16, Zfp516 binding to a Prdm16 related protein, such as Prdm3, or a yet to be identified factor, may drive BAT gene expression.
Other factors involved in the cold-inducible regulation of Ucp1 include Pgc1α, a cofactor known to regulate mitochondrial biogenesis 87. However, Pgc1α-deficient adipose tissue shows only a mild thermogenic defect 88. Another cold inducible factor, Irf4 interacts with Pgc1α to drive expression of Ucp1, and Irf4-deficient mice have a greater thermogenic deficiency 89. However, Irf4 has been shown to be important for general adipogenesis and thus, the BAT specific role is unclear 90. Recent work identified a critical role for histone deacetylase 3 (Hdac3) in basal and cold-inducible activation of thermogenic gene promoters including Ucp1. While normally working as a transcriptional repressor, Hdac3 was shown to deacetylate Pgc1α resulting in activation of thermogenic gene transcription even in the absence of thermogenic stimuli 91. Another factor recently identified to play a role in regulation of thermogenic gene expression through inducing Pgc1α in response to acute cold exposure is the sirtuin, Sirt6. Yao et al. found that Sirt6 was induced by cold where it interacts with phosphorylated activating transcription factor 2 (Atf2) to drive Pgc1α promoter activity. Adipocyte specific Sirt6 knockout mice had significantly reduced Pgc1α levels as well as Pgc1α binding to the Ucp1 promoter. Interestingly, Sirt6 ablation did not result in decreased Pgc1α acetylation, which has been shown to be important for Pgc1α transcriptional coactivity, and the authors did not examine the role of Sirt6 deacetylase activity in regulating Pgc1α promoter activity92.
Ebf2 as a pioneer factor for the BAT gene program
While analyzing differential binding of Pparγ in WAT and BAT, Rajakumari et al. found that BAT-specific Pparγ response elements (PPRE) coincide commonly with early B-cell factor cis-elements and thus identified a critical role of Ebf2 recruiting Pparγ to the Prdm16 promoter for Prdm16 transcription 93. Further work identified Dpf3 as an Ebf2-interacting histone reader, which identifies genes of the BAT program, leading to Ebf2-mediated recruitment of the chromatin remodeler, Brg1, resulting in opening of chromatin for transcription 94. Thus, Ebf2 appears to be working as a pioneer factor during adipogenesis leading to activation of the BAT gene program.
Transcription factors specifically regulating “browning”
While most of the transcriptional regulators mentioned above are common for brown and beige adipogenesis in order to induce thermogenic genes such as Ucp1, several genes were reported to be specifically important for induction or repression of “browning”. For example, Myocardin-related transcription factor A (Mrtfa) was shown to inhibit beige adipocyte differentiation, and Mrtfa KO mice show an increased number of beige adipocytes in WAT. These mice were protected from diet induced obesity and insulin resistance 95. Additionally, Smad3, a key mediator of Tgfβ signaling was shown to inhibit beige adipocyte differentiation 96,97. Klf11 was reported to activate selective beige gene expression by cooperation with Pparγ at superenhancers 98. Interestingly, transcription factor Hes1, which is activated by Notch signaling, repressed Prdm16 and Pgc1α transcription during beige but not classical brown adipogenesis 99. These studies describe common as well as distinct transcriptional mechanisms involved in BAT and beige adipogenesis. Given that not all brown and beige cells have the same origin, it is tempting to speculate that tissue-specific early developmental transcription factors may exist to establish brown versus beige cell differentiation and development.
Epigenetic control of the BAT gene program
As in most biological processes, interactions between genes and the environment, such as temperature or diet 100,101, may influence BAT gene expression and thermogenesis, by involving epigenetic events, i.e., heritable changes in traits without changes in DNA sequence. The broad umbrella of epigenetics research includes both DNA and histone modifications as well as microRNA and long noncoding RNA (lncRNA) either inhibiting or enhancing transcription. In this regard, DNA is wrapped around histone proteins- the building block of the nucleosome (the histone octamer core contains 2 copies of H2A, H2B, H3 and H4) the structure of which is critical for regulation of transcription 102,103. Histone modifications, such as methylation and acetylation, affect transcription by altering nucleosome compaction, changing the chromatin landscape, and thus DNA accessibility of transcription factors and co-regulators 104,105. For example, H3K4me3 is a well-recognized hallmark of transcription activation, whereas H3K9me3 represents a repressive mark, all at the tail region of H3 106,107. The field of BAT epigenetics has been extensively reviewed of late 108–112 and this section focuses on writers and erasers of histone methylation marks, specifically, methyltransferases and demethylases, which are critical components of epigenetic regulation.
Mll3/4
Lysine Methyltransferase 2C (KMT2C, Mll3/4), a H3K4 methyl transferase, has been shown to be involved in adipogenesis, and mice with catalytic dead mutations show reduced adiposity. However, recent studies into the role of Mll3/4 in immortalized brown adipocytes found that Mll3/4 identifies critical super enhancers for BAT and general adipogenic genes and recruits Cbp/p300 to poised enhancers to drive transcription 113,114. Interestingly, this study showed a broad spectrum of histone modifications in both brown preadipocytes and brown adipocytes, with differential patterning of H3K4me1/2, H3K9me2, H3K27me3, H3K27ac, and H3K36me3 implicating many yet to be identified histone methyltransferases in this process.
Ehmt1
Ehmt1, identified as a binding partner for Prdm16, catalyzes the methylation of H3K9 di- or tri-methylation (H3K9me2/3). Loss of Ehmt1 resulted in loss of BAT gene expression and decreased BAT tissue mass 81. However, the proposed mechanism of action was not due to the methylation activity, but due to stabilization of Prdm16 protein, a mechanism proposed for the Pparγ agonist and “browning” agent, rosiglitazone 115. Thus, the relative importance of Prdm16 stabilization versus the potential Ehmt1 demethylase activity in the BAT gene program remains to be studied.
Lsd1
Another H3K9 demethylase, that has been shown to regulate the BAT gene program by histone modification, is lysine-specific demethylase 1 (Lsd1) which catalyzes the demethylation of mono- and dimethylated H3K9. Through direct interaction with Zfp516, Lsd1 is recruited to BAT gene promoters to promote transcription in vivo 82. Indeed, BAT of Lsd1 KO mice using Ucp1-Cre showed reduced Ucp1 expression, which accompanied increased H3K9 mono and demethylation at the proximal Ucp1 promoter, a site where both Lsd1 and Zfp516 were bound. Therefore, BAT-specific Lsd1 ablation compromised BAT gene expression as well as development of BAT, with the tissue resembling WAT with reduced thermogenic activity. Thus, Ucp1-driven Lsd1 ablation resulted in obesity with impaired glucose tolerance upon high fat feeding. Interestingly, later work by Zeng et al. identified an LSD1 complex containing Zfp516 and Prdm16 as well as Ctbp1/2, Hdac1/2 and others 84. This complex was found to suppress WAT-specific genes by colocalizing in regions of H3K4me1 demethylation. Another group identified an Lsd1 interaction with Nrf1 to drive mitochondriogenesis and thermogenic transcription in brown adipocytes maintaining their BAT identity 116.
Jhdm2a
Jumanji-C domain containing histone demethylase 2A (Jhdm2a, also known as Jmjd1a), a mono-and di-methyl H3K9 demethylase, was originally identified as a regulator of thermogenic gene expression when a global knockout, generated to identify the role of Jhdm2a in spermatogenesis, developed an obese phenotype. A closer examination of these mice showed that Jhdm2a was recruited to the Ucp1 promoter during cold and isoproterenol treatment to remove H3K9 methylation resulting in promoter activation 117,118. Interestingly, later work showed that the H3K9 demethylation activity of Jhdm2a may actually be a secondary activity and that Jhdm2a primarily functions as a PKA-mediated, phosphorylation-dependent scaffold protein that interacts with SWI/SNF complex members Arid1a, Brg1, and Baf60b as well as Pparγ in multiple locations of target promoters. Interestingly, non-phosphorylatable Jhdm2a mutants were unable to be recruited to target promoters resulting in decreased UCP1 expression and activity in vitro and in vivo 119. The authors hypothesized that this may be facilitating long-range chromatin interaction, but further work using chromosome conformation capture (3C) needs to be done to validate these claims.
Demethylation of trimethylated H3K27: potential players
Several recent papers have identified the importance of the demethylation of trimethylated H3K27. Interestingly, two different enzymes have been attributed to be responsible for this activity, Jmjd3 and ubiquitously transcribed tetratricopeptide repeat on chromosome X (Utx). In cells, knockout of Jmjd3 as well as chemical inhibition of Jmjd3 enzymatic activity results in a decrease in Ucp1 mRNA and protein. However, in vivo, transgenic expression or chemical inhibition of Jmjd3 only affected Ucp1 expression in aged mice 120. Similarly, Utx, which increases during brown adipogenesis and cold exposure, was found to decrease H3K27me3 at the Ucp1 enhancer and transcription start site (TSS). It is proposed that Utx then recruits Cbp to acetylate H3K27 thereby promoting BAT gene transcription 121. This pro-thermogenic program of Utx is antagonized by the activity of Hdac1, which deacetylates H3K27 and recruits Ezh2 and Suz12 to facilitate H3K27 methylation while preventing Utx binding 122. However, both studies into the function of Utx were performed in vitro so the relative contributions of Jmjd3 and Utx in vivo have yet to be discerned. Given the overlap in functionality, the lack of a strong phenotype in the Jmjd3 knockout mice could be due to compensatory activity of Utx. Further studies are needed to verify this hypothesis. These studies, overall, indicate a critical role for both transcriptional and epigenetic regulation of thermogenic gene expression. Further studies will provide molecular details of the chromatin modifications required for the thermogenic gene program (See figure 2).
Figure 2.
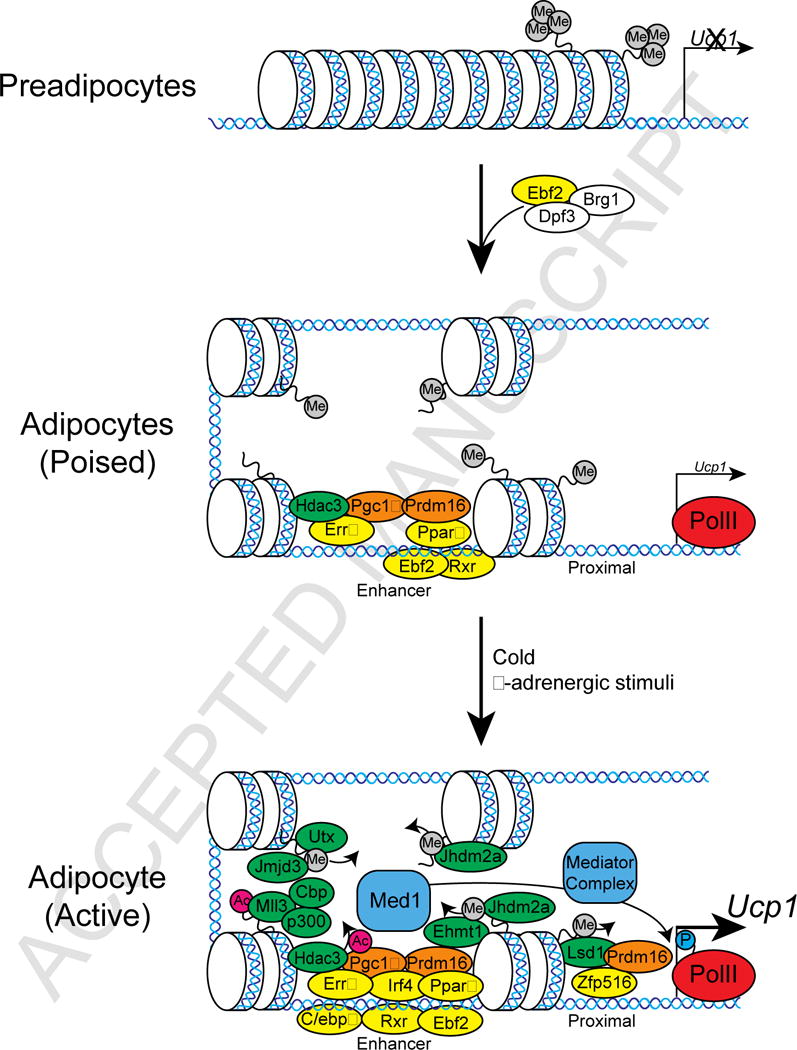
Graphical Representation of Transcriptional and Epigenetic Regulators binding to the UCP1 promoter in beige/brown adipocytes. The UCP1 promoter is silent in preadipocytes. During differentiation, chromatin remodeling occurs and basal transcriptional machinery is recruited to the UCP1 promoter (poised). During a cold challenge, further transcriptional activators are recruited for the activation of the UCP1 promoter (active).
Future directions
Understanding WAT and BAT development and the underlying mechanism to promote “browning” of WAT may provide targets for combating and preventing obesity and associated diseases. For some of the markers of adipose precursors recently identified, further investigation is needed to establish their contribution in embryogenic versus postnatal adipogenesis. Better FACS using multiple markers coupled with immunostaining and lineage tracing approaches will be needed. Moreover, single cell level genomic studies would allow the study of the possible mosaic developmental origin of WAT as well as potential mechanisms for “browning” of WAT. Additionally, many of the studies have focused on a specific depot or condition, e.g. adipose expansion during high fat diet or “browning” after cold exposure. Given that different WAT depots may have different developmental origins it would be beneficial to examine those markers in various depots and different conditions. In addition, as various “browning” mechanisms may exist depending on the type of stimulus applied, the relative contribution of transdifferentiation versus de novo differentiation need to be assessed under different conditions. Moreover, the relationship among transcription factors involved in transitioning from completely “closed” silent chromatin to open/poised chromatin in brown/beige adipocytes needs further investigation. With striking differences in histone modifications during brown/beige adipose development, the enzymes catalyzing these modifications and their contribution to thermogenic gene regulation remains to be elucidated.
Table 1.
Genes involved in adipose tissue expansion and development
| Marker | Embryonic origin | Tissue marked | Expression in adult | Ref | ||
|---|---|---|---|---|---|---|
| Precursor | Preadipocyte | Adipocyte | ||||
| Cd24 | Mesoderm | WAT | + | − | − | 35,36 |
| En1, Pax7 | Dermomyotome | iBAT and muscle | + | ? | − | 42,44 |
| Myf5 | Dermomyotome | iBAT, muscle, asWAT | + | ? | − | 4,19,43,12,3 |
| Ng2 | Dermomyotome | Subcutaneous WAT | ? | + | − | 24 |
| Pdgfrα | Mesoderm | Subcutaneous and visceral WAT, BAT? | +? | + | − | 34,35,51 |
| Pdgfrβ | Mesoderm | ingWAT, visceral WAT, retroperitoneal WAT | ? | + | − | 24,30,31 |
| Perilipin | Mesoderm | WAT? | − | + | + | 31 |
| Pref-1 | Mesoderm | Subcutaneous and visceral WAT | + | + | − | 11 |
| Pparγ | Mesoderm | All WAT and BAT | − | + | ++ | 24 |
| Prx1 | Embryonic limb and bud mesenchyme | Subcutaneous WAT | + | ? | − | 17 |
| Sma | Dermomyotome | Subcutaneous and visceral WAT | ? | + | − | 22,24 |
| Vstm2a | Mesoderm | ? | − | + | − | 32 |
| Wt1 | Mesothelium | Visceral WAT | + | ++ | − | 16 |
| Zfp423 | Mesoderm | ? | − | + | + | 28–30 |
Table 2.
Genes involved in the genetic and epigenetic regulation of thermogenic transcription
| Gene | Activity | Promoter State | Reference(s) |
|---|---|---|---|
| Brg1 | Chromatin remodeler | Poised/Active | 94 |
| C/EBPβ | DNA binding | Active | 74–78 |
| Creb | DNA binding | Active | 74–76 |
| Ctbp1 | DNA binding | Poised/Active | 79,84 |
| Ctbp2 | DNA binding | Poised/Active | 79,84 |
| Ebf2 | DNA binding | Poised/Active | 93,94 |
| Ehmt1 | H3K9 methyl transferase | Active | 81,115 |
| Errα | DNA binding | Poised/Active | 91 |
| Hdac1 | H3K27 deacetylase | Poised/Active (?) | 84,122 |
| Hdac3 | Pgc1a Deacetylase | Poised/Active | 91 |
| Hes1 | DNA binding | Poised/Active | 99 |
| Irf4 | DNA binding | Active | 89,90 |
| Jhdm2a | H3K9 demethylase | Active | 117–119 |
| Jmjd3 | H3K27 demethylase | Active | 120 |
| Klf11 | DNA binding | Poised/Active (?) | 98 |
| Lsd1 | H3K9 demethylase | Active | 82,84,116 |
| Med1 | Cofactor | Active | 86 |
| Mll3/4 | H3K4 methyl transferase | Active | 113,114 |
| Mrtfa | DNA binding | Poised/Active | 95 |
| Pgc1α | Cofactor | Poised/Active | 64,71,74–76 |
| Pparγ | DNA binding | Poised/Active | 71,77–84 |
| Prdm16 | Cofactor | Poised/Active | 71,78–86 |
| Rxr | DNA binding | Poised/Active | 77 |
| Sirt6 | Cofactor | Active | 92 |
| Smad3 | DNA binding | Poised/Active | 96,97 |
| Utx | H3K27 demethylase | Active | 121,122 |
| Zfp516 | DNA binding | Active | 70,71 |
Acknowledgments
The research programs of the authors have been supported in part by an NIDDK grant to H.S.S.
List of abbreviations
- WAT
white adipose tissue
- BAT
brown adipose tissue
- Ucp1
uncoupling protein 1
- Pref-1
preadipocyte factor 1
- TAG
triglycerides
- H2BGFP
histone2B fused green fluorescent protein
- EM
electron microscopy
- BMI
body mass index
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kajimura S, Spiegelman BM, Seale P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Seale P. Control of brown and beige fat development. Nat Rev Mol Cell Biol. 2016;17:691–702. doi: 10.1038/nrm.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-Gurmaches J, Hung CM, Guertin DA. Emerging Complexities in Adipocyte Origins and Identity. Trends Cell Biol. 2016;26:313–326. doi: 10.1016/j.tcb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeffery E, Church CD, Holtrup B, Colman L, Rodeheffer MS. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat Cell Biol. 2015;17:376–385. doi: 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffery E, et al. The Adipose Tissue Microenvironment Regulates Depot-Specific Adipogenesis in Obesity. Cell Metab. 2016;24:142–150. doi: 10.1016/j.cmet.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SM, et al. Loss of White Adipose Hyperplastic Potential Is Associated with Enhanced Susceptibility to Insulin Resistance. Cell Metab. 2014;20:1049–1058. doi: 10.1016/j.cmet.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macotela Y, et al. Intrinsic Differences in Adipocyte Precursor Cells From Different White Fat Depots. Diabetes. 2012;61:1691–1699. doi: 10.2337/db11-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudak CS, et al. Pref-1 Marks Very Early Mesenchymal Precursors Required for Adipose Tissue Development and Expansion. Cell Rep. 2014;8:678–687. doi: 10.1016/j.celrep.2014.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 13.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poissonnet CM, Burdi AR, Garn SM. The chronology of adipose tissue appearance and distribution in the human fetus. Early Hum Dev. 1984;10:1–11. doi: 10.1016/0378-3782(84)90106-3. [DOI] [PubMed] [Google Scholar]
- 15.Ailhaud G, Hauner H. Development of White Adipose Tissue. Handbook of obesity. :483. [Google Scholar]
- 16.Chau YY, et al. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol. 2014;16:367–375. doi: 10.1038/ncb2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krueger KC, Costa MJ, Du H, Feldman BJ. Characterization of Cre Recombinase Activity for In Vivo Targeting of Adipocyte Precursor Cells. Stem Cell Rep. 2014;3:1147–1158. doi: 10.1016/j.stemcr.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Gurmaches J, Hsiao WY, Guertin DA. Highly Selective In Vivo Labeling of Subcutaneous White Adipocyte Precursors with Prx1-Cre. Stem Cell Rep. 2015;4:541–550. doi: 10.1016/j.stemcr.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Gurmaches J, Guertin DA. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat Commun. 2014;5:4099. doi: 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Billon N, et al. The generation of adipocytes by the neural crest. Dev Camb Engl. 2007;134:2283–2292. doi: 10.1242/dev.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armulik A, Genové G, Betsholtz C. Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises. Dev Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Berry DC, Tang W, Graff JM. Independent Stem Cell Lineages Regulate Adipose Organogenesis and Adipose Homeostasis. Cell Rep. 2014;9:1007–1022. doi: 10.1016/j.celrep.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinha S, Iyer D, Granata A. Embryonic origins of human vascular smooth muscle cells: implications for in vitro modeling and clinical application. Cell Mol Life Sci. 2014;71:2271–2288. doi: 10.1007/s00018-013-1554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang W, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 26.Szatmari I, et al. PPARgamma regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood. 2007;110:3271–3280. doi: 10.1182/blood-2007-06-096222. [DOI] [PubMed] [Google Scholar]
- 27.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 28.Gupta RK, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta RK, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vishvanath L, et al. Pdgfrβ+ Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell Metab. 2016;23:350–359. doi: 10.1016/j.cmet.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong KY, et al. Perilipin+ embryonic preadipocytes actively proliferate along growing vasculatures for adipose expansion. Dev Camb Engl. 2015;142:2623–2632. doi: 10.1242/dev.125336. [DOI] [PubMed] [Google Scholar]
- 32.Secco B, et al. Amplification of Adipogenic Commitment by VSTM2A. Cell Rep. 2017;18:93–106. doi: 10.1016/j.celrep.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sowa Y, et al. Adipose Stromal Cells Contain Phenotypically Distinct Adipogenic Progenitors Derived from Neural Crest. PLOS ONE. 2013;8:e84206. doi: 10.1371/journal.pone.0084206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YH, Petkova AP, Mottillo EP, Granneman JG. In Vivo Identification of Bipotential Adipocyte Progenitors Recruited by β3-Adrenoceptor Activation and High-Fat Feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 37.Festa E, et al. Adipocyte Lineage Cells Contribute to the Skin Stem Cell Niche to Drive Hair Cycling. Cell. 2011;146:761–771. doi: 10.1016/j.cell.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uezumi A, Fukada S, Yamamoto N, Takeda S’ichi, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 39.Iwayama T, et al. PDGFRα signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity. Genes Dev. 2015;29:1106–1119. doi: 10.1101/gad.260554.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joe AWB, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcelin G, et al. A PDGFRα-Mediated Switch toward CD9(high) Adipocyte Progenitors Controls Obesity-Induced Adipose Tissue Fibrosis. Cell Metab. 2017;25:673–685. doi: 10.1016/j.cmet.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 42.Atit R, et al. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 43.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genes N Y N 2000. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett. 1984;167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- 46.Cousin B, et al. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103:931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- 47.Collins S, Daniel KW, Petro AE, Surwit RS. Strain-specific response to beta 3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology. 1997;138:405–413. doi: 10.1210/endo.138.1.4829. [DOI] [PubMed] [Google Scholar]
- 48.Himms-Hagen J, et al. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am J Physiol Cell Physiol. 2000;279:C670–681. doi: 10.1152/ajpcell.2000.279.3.C670. [DOI] [PubMed] [Google Scholar]
- 49.Cinti S. Adipocyte differentiation and transdifferentiation: Plasticity of the adipose organ. J Endocrinol Invest. 2002;25:823–835. doi: 10.1007/BF03344046. [DOI] [PubMed] [Google Scholar]
- 50.Rosenwald M, Perdikari A, Rülicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013;15:659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 51.Lee YH, Petkova AP, Konkar AA, Granneman JG. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J Off Publ Fed Am Soc Exp Biol. 2015;29:286–299. doi: 10.1096/fj.14-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W, et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A. 2014;111:14466–14471. doi: 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J, et al. Beige Adipocytes are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long JZ, et al. Ribosomal Profiling Provides Evidence for a Smooth Muscle-Like Origin of Beige Adipocytes. Cell Metab. 2014;19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gessner C. Conradi Gesneri medici Tigurini Historiæ animalium Lib I de quadrupedibus uiuiparis : opus philosophis, medicis, grammaticis, philologis, poëtis, & omnibus rerum linguarumq´; uariarum studiosis, utilissimum simul iucundissimumq´; futurum. (Apud Christ. Froschouerum, 1551) [Google Scholar]
- 56.Huttunen P, Hirvonen J, Kinnula V. The occurrence of brown adipose tissue in outdoor workers. Eur J Appl Physiol. 1981;46:339–345. doi: 10.1007/BF00422121. [DOI] [PubMed] [Google Scholar]
- 57.Ricquier D, Mory G. 4 - Factors affecting brown adipose tissue activity in animals and man. Clin Endocrinol Metab. 1984;13:501–520. doi: 10.1016/s0300-595x(84)80035-3. [DOI] [PubMed] [Google Scholar]
- 58.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cypess AM, et al. Cold but not sympathomimetics activates human brown adipose tissue in vivo. Proc Natl Acad Sci U S A. 2012;109:10001–10005. doi: 10.1073/pnas.1207911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lidell ME, et al. Evidence for two types of brown adipose tissue in humans. Nat Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 61.Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 62.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 63.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 64.Lee P, Werner CD, Kebebew E, Celi FS. Functional thermogenic beige adipogenesis is inducible in human neck fat. Int J Obes Lond. 2014;38:170–6. doi: 10.1038/ijo.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shinoda K, et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015;21:389–394. doi: 10.1038/nm.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue R, et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat Med. 2015;21:760–768. doi: 10.1038/nm.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kir S, et al. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature. 2014;513:100–104. doi: 10.1038/nature13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brestoff JR, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 2015;519:242–246. doi: 10.1038/nature14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dempersmier J, Sul HS. Shades of brown: a model for thermogenic fat. Front Endocrinol. 2015;6:71. doi: 10.3389/fendo.2015.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dempersmier J, et al. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol Cell. 2015;57:235–246. doi: 10.1016/j.molcel.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rao RR, et al. Meteorin-like Is a Hormone that Regulates Immune-Adipose Interactions to Increase Beige Fat Thermogenesis. Cell. 2014;157:1279–1291. doi: 10.1016/j.cell.2014.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shao M, et al. Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab. 2016;23:1167–1184. doi: 10.1016/j.cmet.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao W, Medvedev AV, Daniel KW, Collins S. β-Adrenergic Activation of p38 MAP Kinase in Adipocytes cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 map kinase. J Biol Chem. 2001;276:27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 75.Puigserver P, et al. A Cold-Inducible Coactivator of Nuclear Receptors Linked to Adaptive Thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 76.Xue B, Coulter A, Rim JS, Koza RA, Kozak LP. Transcriptional Synergy and the Regulation of Ucp1 during Brown Adipocyte Induction in White Fat Depots. Mol Cell Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barak Y, et al. PPARγ Is Required for Placental, Cardiac, and Adipose Tissue Development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 78.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kajimura S, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sambeat A, et al. LSD1 interacts with Zfp516 to promote UCP1 Transcription and Brown Fat Program. Cell Rep. 2016;15:2536–2549. doi: 10.1016/j.celrep.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng X, et al. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev. 2016;30:1822–1836. doi: 10.1101/gad.285312.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harms MJ, et al. Prdm16 Is Required for the Maintenance of Brown Adipocyte Identity and Function in Adult Mice. Cell Metab. 2014;19:593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harms MJ, et al. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 2015;29:298–307. doi: 10.1101/gad.252734.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arany Z, et al. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 88.Kleiner S, et al. Development of insulin resistance in mice lacking PGC-1α in adipose tissues. Proc Natl Acad Sci. 2012;109:9635–9640. doi: 10.1073/pnas.1207287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kong X, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell. 2014;158:69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eguchi J, et al. Interferon-regulatory factors (IRFs) are transcriptional regulators of adipogenesis. Cell Metab. 2008;7:86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Emmett MJ, et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017;546:544–548. doi: 10.1038/nature22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yao L, et al. Cold-Inducible SIRT6 Regulates Thermogenesis of Brown and Beige Fat. Cell Rep. 2017;20:641–654. doi: 10.1016/j.celrep.2017.06.069. [DOI] [PubMed] [Google Scholar]
- 93.Rajakumari S, et al. EBF2 Determines and Maintains Brown Adipocyte Identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shapira SN, et al. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev. 2017;31:660–673. doi: 10.1101/gad.294405.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McDonald ME, et al. Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell. 2015;160:105–118. doi: 10.1016/j.cell.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koncarevic A, et al. A novel therapeutic approach to treating obesity through modulation of TGFβ signaling. Endocrinology. 2012;153:3133–3146. doi: 10.1210/en.2012-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yadav H, et al. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab. 2011;14:67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loft A, et al. Browning of human adipocytes requires KLF11 and reprogramming of PPARγ superenhancers. Genes Dev. 2014;114 doi: 10.1101/gad.250829.114. gad.250829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bi P, et al. Notch signaling regulates adipose browning and energy metabolism. Nat Med. 2014;20:911–918. doi: 10.1038/nm.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mazzio EA, Soliman KFA. Epigenetics and nutritional environmental signals. Integr Comp Biol. 2014;54:21–30. doi: 10.1093/icb/icu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang G, Sun Q, Liu C. Influencing Factors of Thermogenic Adipose Tissue Activity. Front Physiol. 2016;7:29. doi: 10.3389/fphys.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Patel DJ, Wang Z. Readout of epigenetic modifications. Annu Rev Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Petty E, Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet TIG. 2013;29:621–629. doi: 10.1016/j.tig.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 105.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 106.Kebede AF, Schneider R, Daujat S. Novel types and sites of histone modifications emerge as players in the transcriptional regulation contest. FEBS J. 2015;282:1658–1674. doi: 10.1111/febs.13047. [DOI] [PubMed] [Google Scholar]
- 107.Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 108.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sambeat A, Gulyaeva O, Dempersmier J, Sul HS. Epigenetic Regulation of the Thermogenic Adipose Program. Trends Endocrinol Metab. 2017;28:19–31. doi: 10.1016/j.tem.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shamsi F, Zhang H, Tseng Y-H. MicroRNA Regulation of Brown Adipogenesis and Thermogenic Energy Expenditure. Front Endocrinol. 2017;8 doi: 10.3389/fendo.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Trajkovski M, Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol Metab. 2013;24:442–450. doi: 10.1016/j.tem.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu S, Chen P, Sun L. Regulatory networks of non-coding RNAs in brown/beige adipogenesis. Biosci Rep. 2015;35 doi: 10.1042/BSR20150155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lai B, et al. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res. 2017;45:6388–6403. doi: 10.1093/nar/gkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duteil D, et al. Lsd1 Ablation Triggers Metabolic Reprogramming of Brown Adipose Tissue. Cell Rep. 2016;17:1008–1021. doi: 10.1016/j.celrep.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Inagaki T, et al. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells Devoted Mol Cell Mech. 2009;14:991–1001. doi: 10.1111/j.1365-2443.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 118.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abe Y, et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat Commun. 2015;6 doi: 10.1038/ncomms8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pan D, et al. Jmjd3-Mediated H3K27me3 Dynamics Orchestrate Brown Fat Development and Regulate White Fat Plasticity. Dev Cell. 2015;35:568–583. doi: 10.1016/j.devcel.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zha L, et al. The Histone Demethylase UTX Promotes Brown Adipocyte Thermogenic Program Via Coordinated Regulation of H3K27 Demethylation and Acetylation. J Biol Chem. 2015;290:25151–25163. doi: 10.1074/jbc.M115.662650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li F, et al. Histone Deacetylase 1 (HDAC1) Negatively Regulates Thermogenic Program in Brown Adipocytes via Coordinated Regulation of Histone H3 Lysine 27 (H3K27) Deacetylation and Methylation. J Biol Chem. 2016;291:4523–4536. doi: 10.1074/jbc.M115.677930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanchez-Gurmaches J, et al. PTEN Loss in the Myf5 Lineage Redistributes Body Fat and Reveals Subsets of White Adipocytes that Arise from Myf5 Precursors. Cell Metab. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


