Abstract
Altered cannabinoid 1 receptor (CB1R) expression has been reported in the brain of subjects with schizophrenia, a developmental mental illness that usually emerges in late adolescence/early adulthood. However, the developmental period at which changes in the CB1R expression appear in schizophrenia is unknown. To gain insight into this factor, we assessed the postnatal developmental trajectory of CB1R expression in the methylazoxymethanol (MAM) model of schizophrenia. Using in situ hybridization with film and grain analyses, CB1R messenger RNA (mRNA) levels were quantified in multiple brain regions, including the medial prefrontal cortex (mPFC), secondary motor cortex, dorsomedial and dorsolateral striatum, dorsal subregions and ventral subiculum of the hippocampus, of MAM-treated rats and normal controls at three developmental periods [juvenile - postnatal day (PD) 30; adolescence - PD45; and adulthood - PD85]. In all brain regions studied, CB1R mRNA levels were highest in juveniles and then decreased progressively toward adolescent and adult levels in control and MAM-treated rats. However, in MAM-treated rats, CB1R mRNA levels were lower in the mPFC at PD85 and higher in the dorsolateral striatum at PD45 and PD85 relative to controls. Cellular analyses confirmed the changes in CB1R mRNA expression in MAM-treated rats. These findings are in accordance with previous studies showing a decrease in the CB1R mRNA expression from juvenile period to adolescence to adulthood in cortical, striatal, and hippocampal regions. Additionally, similar to most of the schizophrenia-like signs observed in the MAM model, embryonic exposure to MAM leads to schizophrenia-related changes in CB1R mRNA expression that only emerge later in development.
Keywords: cannabinoid, endocannabinoid system, schizophrenia, psychosis, animal model
Introduction
Schizophrenia is a neurodevelopmental disorder that is thought to arise from an interaction between genetic and environmental factors (Kahn et al., 2015). Schizophrenia subjects usually present with the full-blown disorder during late adolescence or early adulthood (Howes and Murray, 2014), when brain circuitry in limbic regions, mainly the prefrontal cortex, undergoes maturation (Bennett, 2009; Lewis et al., 2004).
The pathophysiological features of schizophrenia result from abnormal neurobiological underpinnings that negatively impact synaptic plasticity and synaptic connectivity (Hayashi-Takagi and Sawa, 2010; McGlashan and Hoffman, 2000), including altered synaptic pruning (Feinberg, 1982; McGlashan and Hoffman, 2000), atypical development of GABAergic inhibitory circuits combined with abnormal glutamatergic signaling (Lewis and Moghaddam, 2006; Volk and Lewis, 2002), and increased activity of midbrain dopaminergic pathways (Grace and Gomes, 2018). Additionally, besides the fact that cannabis use, particularly during adolescence, might lead to a higher risk for the later appearance of schizophrenia (Marconi et al., 2016), evidence suggests that alterations in cannabinoid signaling in the brain play a role in the pathophysiology of schizophrenia. For example, measures of the principal cannabinoid receptor in the brain (CB1R) messenger RNA (mRNA) and protein are reported to be lower in the dorsolateral prefrontal cortex of schizophrenia subjects (Eggan et al., 2008; Eggan et al., 2010; Uriguen et al., 2009).
The CB1Rs are selectively expressed in the presynaptic terminal of specific subpopulations of neurons, such as GABAergic and glutamatergic neurons, and their activation inhibits the release of neurotransmitters (Gerdeman and Lovinger, 2001; Wilson and Nicoll, 2001). Also, CB1R-mediated signaling regulates several neurodevelopmental processes, such as synaptic plasticity, axon guidance, and interneuron migration (Berghuis et al., 2005; Berghuis et al., 2007; Mulder et al., 2008). Studies investigating the ontogeny of CB1R expression indicate that its levels change dynamically in a temporally-specific manner throughout development, particularly from adolescence to adulthood (Long et al., 2012). However, when changes in the CB1R expression appear in schizophrenia is unknown.
Here, we assessed whether the postnatal developmental trajectory of CB1R mRNA expression is altered in an animal model of schizophrenia based on neurodevelopmental disruption, named the methylazoxymethanol (MAM) model. In this model, the DNA-methylating agent MAM is administered to pregnant rats at gestational day (GD) 17 which produces many characteristics consistent with schizophrenia-like deficits in the offspring, including neuroanatomical changes (thinning of limbic cortices, loss of parvalbumin-containing GABAergic interneurons), behavioral abnormalities (deficits in prepulse inhibition of startle, behavioral flexibility, and social interaction), enhanced response to amphetamine and phencyclidine, disruption of rhythmic activity in the frontal cortex, and a midbrain hyperdopaminergic state(Modinos et al., 2015; Moore et al., 2006). Interestingly, similar to the onset of the frank disorder in schizophrenia subjects, most of the alterations observed in the MAM model become evident only after puberty, while some behavioral abnormalities, such as social withdrawal and cognitive deficits, are present both before and after puberty (Gomes et al., 2016). Thus, the MAM model seems to be a valuable model for exploring the pathophysiological mechanisms associated with the transition into a schizophrenia-like phenotype in the adult.
Methods and Materials
Animals and methylazoxymethanol (MAM) treatment
Pregnant Sprague–Dawley rats (Envigo, Indianapolis, USA) were obtained on gestational day (GD) 14 and individually housed in ventilated plastic breeding tubs containing only regular bedding and 2–3 sheets of white tissue paper and maintained in a temperature-controlled vivarium on a 12-hour light/dark cycle (lights on at 7 AM; off at 7 PM) with ad libitum access to water and food. MAM (20 mg/kg, i.p., Midwest Research Institute, Kansas City, USA) was administered on GD 17 as previously described (Moore et al., 2006). Control dams received injections of saline (1 mL/kg, i.p.). Male pups were weaned-off on postnatal day (PD) 21 and double or triple housed with littermates, in the same housing conditions described for the pregnant rats, until the time they were killed for tissue processing. Littermates from multiple litters were randomly allocated to the different groups. Experiments were performed in accordance with the policies approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Tissue preparation
CB1R mRNA expression was quantified in different brain regions of saline and MAM-treated rats at three developmental stages corresponding to juvenile period (PD30), adolescence (PD45), and adulthood (PD85) (Spear, 2000). Rats (n = 6–7/group; see supplementary material for details) were killed with CO2, and their brains were removed, fresh frozen on dry ice and then stored at −80°C until cryostat sectioning. Coronal sections from all rat brains were cut on a cryostat at 12μm sections. Five to eight sections evenly spaced at ~72 μm intervals containing the medial prefrontal cortex (mPFC) and secondary motor cortex (+3.2 mm to +2.2 mm from bregma), dorsolateral and dorsomedial striatum (+1.70 mm to +0.20 mm from bregma), dorsal hippocampus (−3.1 mm to −4.1 mm from bregma), and ventral subiculum of hippocampus (−4.8 mm to −6.0 mm from bregma; Supplementary Figure 1) (Paxinos and Watson, 1998) were selected from each rat. Sections were thaw-mounted onto SupraFrost slides (Fisher Scientific, Pittsburgh, PA) and stored at −80°C until use.
In situ Hybridization
A 325-base pair fragment corresponding to bases 1130–1454 of the mouse Cnr1 gene (CB1R) (GenBank Accession Number: NM_007726) was PCR-amplified. Nucleotide sequencing confirmed 100% homology for the amplified fragment to the reported sequence as previously described (Eggan et al., 2012). Sense and antisense riboprobes were produced by in vitro transcription in the presence of 35S-CTP with T7 or SP6 RNA polymerase and then purified. After that, to increase the effectiveness of tissue penetration, riboprobes were reduced to nearly 100 bp by alkaline hydrolysis. Although these riboprobes were designed to quantify levels of CB1R mRNA in the mouse brain as previously described, a pilot study indicated that they also can be used to assess CB1R mRNA expression in the rat brain. It is likely due to the high (94%) homology between mouse and rat for the specific Cnr1 gene sequence targeted by the riboprobe (GenBank; BLAST).
Standard hybridization procedures were performed as previously described (Hashimoto et al., 2003). A detailed descritption of the in situ hybridization is provided in the Supplementary Material. After standard hybridization, for the quantification of CB1R mRNA at the cellular level, slides were then coated with NTB2 emulsion (Kodak) using a mechanical dipper (Auto-dip Emulsion Coater, Ted Pella, Redding, USA), at a constant withdrawal speed and temperature, to ensure the uniformity of emulsion thickness across sections. Slides were exposed at 4°C for 72 hours, developed with D-19 (Kodak), washed, and counterstained with Cresyl violet. The absence of signal with sense probe and the location of grain clusters only over neurons in emulsion-dipped slides confirmed the specificity of the riboprobe,
Quantification
Quantification was carried out blind to age and condition. Trans-illuminated autoradiographic film images of CB1R mRNA were captured using a Microcomputer Imaging Device (MCID) (at 5.1 μm/pixel resolution) and digitized. All images for slides from an experimental run were taken in the same session under identical conditions, including room illumination, gain, black levels, and flatfield correction. CB1R mRNA levels were quantified in the mPFC, secondary motor cortex, dorsolateral and dorsomedial striatum, dorsal hippocampal regions (granular layer of the dentate gyrus and pyramidal layer of the CA1 and C3), and the ventral subiculum of the hippocampus. For each section, optical density (OD) levels of CB1R mRNA were measured bilaterally and expressed as nanocuries per gram of tissue (nCi/g) by reference to carbon-14 standards (ARC Inc., St. Louis, USA) exposed on the same film. All density measures were corrected by subtracting background measured in the white matter. Since the brain regions evaluated are bilateral, measures from each hemisphere were studies and the mean value between them was used as a single measure per section. The data were averaged across at least 3 sections in each animal.
To assess CB1R mRNA at the cellular level, the number of silver grains generated by a 35S-labeled riboprobe in emulsion-dipped Nissl-counterstained sections was counted over neurons using the MCID software and a Nikon microscope with a motorized stage; 54 and 42 sampling boxes (120 × 170 μm) were systematically tiled in both hemispheres of the mPFC and dorsolateral striatum, respectively. Sampling circles with a fixed diameter (12 μm for the dorsolateral striatum and 16 for the mPFC) were placed over each Nissl-stained nucleus that had at least three overlying silver grains. The number of silver grains per neuron from each hemisphere across 1 section was assessed. Background signal was obtained for each tissue section by counting grains in a sampling box placed in the white matter. For the identification of specifically labeled neurons, referred to as CB1R mRNA-positive (+) neurons, a background threshold approach where only cells with 3× background grain density levels was performed before the analysis (Hashimoto et al., 2003). Values were averaged for each rat. Results are presented as the number of CB1R mRNA+ neuron per mm2 area and the number of grains per CB1R mRNA+ neuron.
Statistical analysis
Data were analyzed by two-way analysis of variance (ANOVA) with age (PD30, PD45, and PD85) and condition (saline and MAM-treated rats) as independent factors, followed by Bonferroni post hoc test. Significance was set at p < 0.05, and data are represented as the mean ± SEM.
Results
Changes in CB1R mRNA expression across development
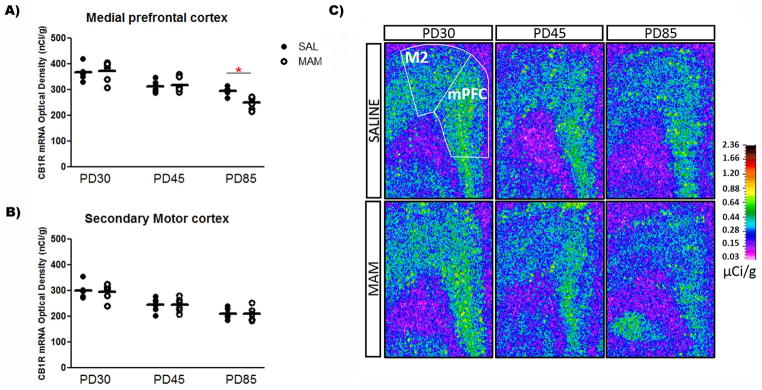
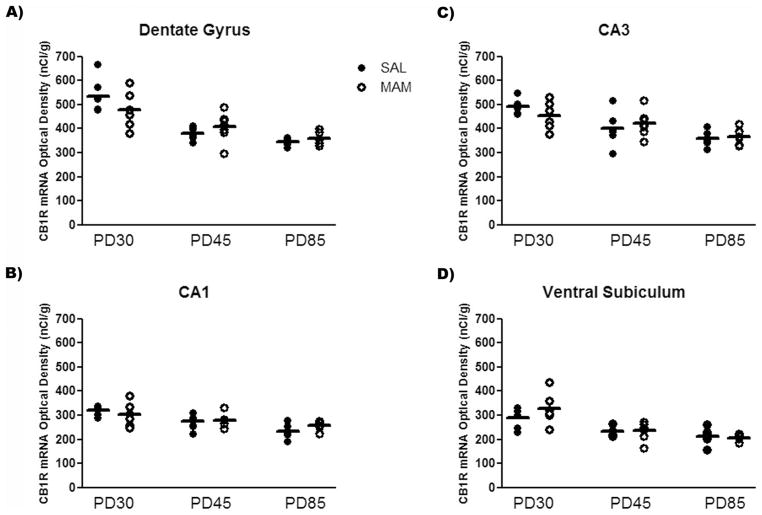
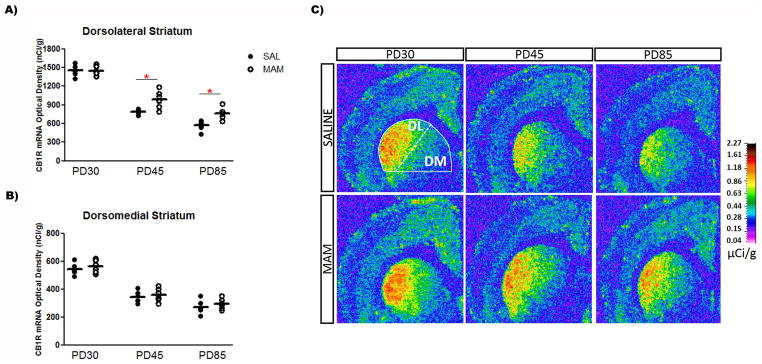
In situ hybridization revealed a significant age-dependent decrease in the expression of CB1R mRNA in all brain regions evaluated in both saline and MAM-treated rats. The two-way ANOVA detected a significant effect of age on the CB1R mRNA levels in the mPFC (F2,30 = 37.1, P < 0.001), secondary motor cortex (F2,30 = 32.2, P < 0.001), dorsomedial striatum (F2,31 = 126.7, P < 0.001) and dorsolateral striatum (F2,31 = 245.3, P < 0.001), dorsal hippocampal regions – dentate gyrus (F2,31 = 26.5, P < 0.001), CA1 (F2,31 = 12.6, P < 0.001), and CA3 (F2,31 = 15.4, P < 0.001) – and ventral subiculum of the hippocampus (F2,31 = 20.9, P < 0.001). The CB1R mRNA levels declined from the juvenile period (PD30) until adolescence (PD45), then declined progressively until adulthood in saline and MAM-treated rats (PD85; Table 1 and Figures 1–3).
Table 1.
Changes (%) in the expression of CB1R mRNA relative to controls at PD30
| Brain area | % of change (relative to control rats at PD30) | ||||
|---|---|---|---|---|---|
| Control rats | MAM-treated rats | ||||
|
|
|
||||
| PD45 | PD85 | PD30 | PD45 | PD85 | |
|
| |||||
| MPFC | −14.6 | −19.5 | +1.5 | −13.1 | −32.2 |
| Secondary motor cortex | −18.4 | −29.9 | −1.7 | −18.8 | −30.1 |
| Dorsomedial striatum | −36.8 | −50.0 | +3.5 | −34.2 | −46.0 |
| Dorsolateral striatum | −45.9 | −60.9 | −0.5 | −32.5 | −48.0 |
| CA1 | −13.9 | −26.7 | −5.5 | −12.3 | −19.1 |
| CA3 | −18.9 | −27.3 | −7.7 | −14.6 | −25.9 |
| Dentate gyrus | −28.5 | −35.3 | −10.5 | −23.6 | −32.8 |
| Ventral subiculum | −19.7 | −26.8 | +12.9 | −18.6 | −29.0 |
Figure 1.
Expression of CB1R mRNA in the (A) mPFC of MAM-treated rats at PD85 is lower than in saline-treated rats, with no change in the (B) secondary motor cortex (n = 6/group). Mean values for saline and MAM-treated rats at each age are indicated by hash marks. *P < 0.05, two-way ANOVA followed by Bonferroni post hoc test. (C) Representative film autoradiograms illustrating the expression of CB1R mRNA. The density of hybridization signal is presented in pseudo-color according to the calibration scale (μCi/g).
Figure 3.
Expression of CB1R mRNA did not differ between saline and MAM-treated rats in different hippocampal subregions [(A) dentate gyrus; (B) CA1; (C) CA3; (D) ventral subiculum] at all ages studied (n = 6–7/group).
Altered CB1 mRNA expression levels in MAM-treated rats
MAM-treated rats showed differential CB1R mRNA expression levels in the mPFC at PD85 and in the dorsolateral striatum at both PD45 and PD85 compared with control rats. In addition to the significant effect of age on the CB1R mRNA levels in all brain regions studied, the two-way ANOVA also showed a significant effect of condition on the mRNA levels in the dorsolateral striatum (F1,31 = 17.9, P < 0.001). There was also significant condition vs. age interaction on the CB1R mRNA levels in the mPFC (F2,30 = 3.5, P < 0.05) and dorsolateral striatum (F2,31 = 4.9, P < 0.01). A preplanned post hoc analysis revealed that the mean expression of CB1R mRNA in the mPFC of MAM-treated rats was 17% lower than in controls at PD85 (Bonferroni post hoc test, P < 0.05; Figure 1). Interestingly, an opposite change was observed in the dorsolateral striatum of MAM-treated rats: the mean expression of CB1R mRNA in the dorsolateral striatum of MAM-treated rats was 25% and 34% higher than controls at PD45 and PD85 (Bonferroni post hoc test, P < 0.01), respectively (Figure 2). No change in the expression of CB1R mRNA in the secondary motor cortex, dorsomedial striatum, dorsal hippocampal regions (dentate gyrus, CA1, and CA3), and ventral subiculum of the hippocampus was observed in MAM-treated rats compared with control rats (Figures 1–3).
Figure 2.
Expression of CB1R mRNA in the (A) dorsolateral striatum of MAM-treated rats at PD45 and PD85 is higher than in saline-treated rats, with no change in the (B) dorsomedial striatum (n = 6–7/group). Mean values for saline and MAM-treated rats at each age are indicated by hash marks. *P < 0.05, two-way ANOVA followed by Bonferroni post hoc test. (C) Representative film autoradiograms illustrating the expression of CB1R mRNA. The density of hybridization signal is presented in pseudo-color according to the calibration scale (μCi/g).
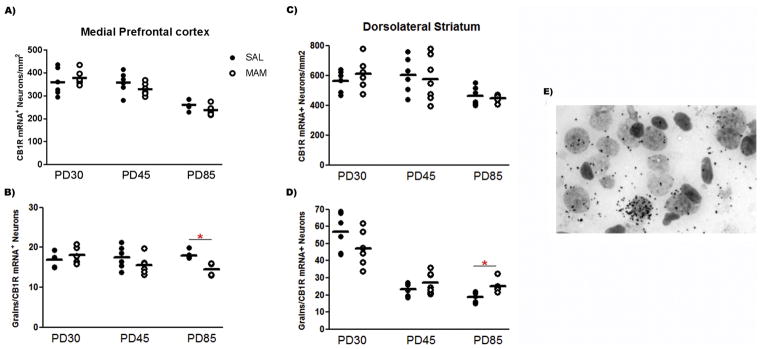
We next sought to determine whether the altered CB1R mRNA expression in the mPFC and dorsolateral striatum of MAM-treat rats was also present at the cellular level. Grain counting analyses revealed an age-dependent decrease in the number of CB1R mRNA+ neurons in both the mPFC (F2,30 = 34.48; P < 0.0001) and dorsolateral striatum (F2,31 = 7.22; P = 0.003). An age-dependent decrease in the number of grains per CB1R mRNA+ neuron was also observed in the dorsolateral striatum (F2,31 = 64.64; P < 0.0001), but not in the mPFC (F2,30 = 0.49; P > 0.05). The two-way ANOVA also detected significant effect of condition on the number of grains per CB1R mRNA+ neuron in the mPFC (F1,30 = 5.51; P = 0.025). There were also significant condition vs. age interaction on the number of grains per CB1R mRNA+ neuron in the mPFC and dorsolateral striatum (F2,31 = 4.44; P = 0.02). Post hoc analysis indicated that, similar to the CB1R mRNA levels by film optical density, MAM-treated rats showed fewer grains per CB1R mRNA+ neuron in the mPFC at PD85 (Bonferroni post hoc test, P < 0.05; Figure 4). An opposite change was found in the dorsolateral striatum of MAM-treated rats at PD85, more grains per CB1R mRNA+ neuron (Bonferroni post hoc test, P < 0.05; Figure 4).
Figure 4.
Cellular grain-counting analysis in the mPFC found (A) no difference in CB1R-labeled neurons/mm2 between saline and MAM-treated rats, but (B) a decrease in the mean number of grains per CB1R-labeled neurons was found in MAM-treated rats at PD85 relative to control rats (n = 6/group). In the dorsolateral striatum (C), no difference in CB1R-labeled neurons/mm2 between saline and MAM-treated rats was found, but (D) an increase in the mean number of grains per CB1R-labeled neurons was found in MAM-treated rats at PD85 relative to control rats (n = 6–7/group). Mean values for each group are indicated by hash marks. *P < 0.05, two-way ANOVA followed by Bonferroni post hoc test. (E) Representative brightfield photomicograph showing CB1R mRNA signals as silver grain clusters over Nissl-staining neurons in the mPFC of a control rat at PD85.
Discussion
The goal of this study was to investigate brain CB1R expression across development in the MAM model of schizophrenia. We found that levels of CB1 mRNA decrease in an age-dependent manner in multiple brain regions, including the mPFC, secondary motor cortex, dorsomedial and dorsolateral striatum, dorsal hippocampal regions and ventral subiculum of the hippocampus, in MAM-treated rats and controls. The expression of CB1R mRNA levels was highest during the juvenile period and exhibited a normal decline through adulthood in all brain regions studied. These findings are consistent with reports of a decrease in the CB1R mRNA expression from juvenile period to adolescence to adulthood in the human prefrontal cortex (Long et al., 2012) and in the mPFC and striatum of rats (Heng et al., 2011; Van Waes et al., 2012), and expand these observations to hippocampal subregions. In these brain regions, although CB1R is present in both inhibitory and excitatory terminals, CB1R mRNA and protein levels are much higher in GABAergic interneurons than in pyramidal cells (Marsicano and Lutz, 1999), and the reduction in CB1R mRNA expression following the juvenile period may be linked to the maturation of GABAergic neurotransmission (Catts et al., 2013; Schmidt and Mirnics, 2015). The dynamic postnatal expression of CB1Rs and the maturation of the GABAergic neurotransmission have been suggested to underlie the higher risk for the development of psychiatric disorders later in life in individuals exposed to adolescent cannabis exposure (Caballero and Tseng, 2012; Cass et al., 2014; Schmidt and Mirnics, 2015).
Although CB1R mRNA expression was unchanged in MAM-treated rats at PD30 and PD45, adult MAM-treated rats (PD85) showed altered CB1R mRNA expression in both the mPFC and dorsolateral striatum relative to age-matched controls. While significantly reduced levels of CB1R mRNA were observed in the mPFC of MAM-treated rats, an increase was observed in the dorsolateral striatum. Altered CB1R mRNA expression in both the mPFC and dorsolateral striatum of adult MAM-treated rats was also observed at the cellular level. These findings appear unlikely to be attributable to a loss of CB1R mRNA expressing neurons as the total number of these neurons was unaltered in the mPFC and dorsolateral striatum of MAM-treated rats relative to controls. Interestingly, the grain counting analysis indicated that the number of labeled neurons in mPFC drops through development, suggesting that some neurons simply stop expressing CB1R altogether across normal development. This does not differ from MAM-treated rats. However, the remaining cells express the same level of CB1R per neuron across development in controls. In contrast, in MAM-treated rats at PD85, the remaining cells are now expressing fewer CB1R. In the dorsolateral striatum, both the number of CB1R mRNA expressing neurons and the number of grains per labeled neuron drop through development. However, MAM-treated rats at PD85 showed more grains per CB1R mRNA-positive neuron in the dorsolateral striatum relative to age-matched controls. Thus, these changes in the grain counting found in MAM-treated rats suggest that having more or less CB1R mRNA per cell may be associated with a loss of phenotype rather than missing/having more CB1R-positive cells. Additionally, mRNA levels for CB1R appear to be unaffected in the secondary motor cortex, dorsomedial striatum, dorsal hippocampal regions (dentate gyrus, CA1, and CA3), and ventral subiculum of the hippocampus of MAM-treated rats. Interestingly, the age at which the changes in the CB1R mRNA expression were observed in the present study is consistent with the appearance of a schizophrenia-like phenotype in MAM-treated rats (Gomes et al., 2016; Moore et al., 2006). These findings suggest that level of CB1R may not be altered until a later stage of development in schizophrenia.
Similar to what has been observed in the dorsolateral prefrontal cortex of subjects with schizophrenia (Eggan et al., 2008; Eggan et al., 2010), a reduced CB1R mRNA expression was present in the mPFC of adult MAM-treated rats. In cortical regions, CB1Rs are heavily localized to inhibitory axon terminals of GABAergic interneurons, primarily those containing the neuropeptide cholecystokinin (CCK), and that target the body and proximal dendrites of pyramidal neurons (Bodor et al., 2005; Marsicano and Lutz, 1999). Activation of CB1Rs suppresses the release of GABA from these terminals and strongly reduces GABA-mediated inhibitory postsynaptic currents in pyramidal neurons (Trettel et al., 2004), playing an important role in regulating network activity patterns by controlling proximal inhibitory input to pyramidal neurons. Deficits in the expression of GABAergic neuron-related mRNAs and proteins, including the enzyme responsible for most GABA synthesis (GAD67), are consistently found in the prefrontal cortex and hippocampus of schizophrenia subjects and are thought to contribute to symptoms in the illness (Akbarian and Huang, 2006; Lewis et al., 2005). Similar to schizophrenia, MAM-treated rats show several abnormalities associated with deficits in GABAergic neurotransmission (Lodge et al., 2009). Interconnected GABA interneurons are crucial for the synchronization of large networks of neurons, including gamma oscillations that increase in the prefrontal cortex with working memory load (Howard et al., 2003), which is disrupted in schizophrenia subjects (Lewis et al., 2005) and in MAM-treated rats (Lodge et al., 2009). As the activation of CB1Rs suppress GABA neurotransmission in the prefrontal cortex, it has been suggested that a lower density of CB1Rs could, by reducing the endocannabinoid-mediated block of GABA release from the terminals of CB1R-containing interneurons, be associated with a compensatory response to lower levels of GAD67-mediated GABA synthesis in those neurons and contribute to a partial, albeit insufficient, normalization of gamma oscillations and working memory function in schizophrenia. In support of this hypothesis, it was found that the extent of alterations in the expression of CB1R mRNA is significantly correlated with that for GAD67 mRNA (Eggan et al., 2008; Hashimoto et al., 2008) and that mice with a GAD67 heterozygous null mutation exhibit lower CB1R mRNA levels in the mPFC than wild-type mice, suggesting that reduced levels of GAD67 mRNA expression drive lower CB1R mRNA expression in the mPFC (Eggan et al., 2012). In contrast, GAD67 mRNA levels were not altered in mice with a CB1R heterozygous or homozygous null mutation, showing that decreased CB1R mRNA expression is unlikely to affect GAD67 mRNA levels (Eggan et al., 2012). Interestingly, similar to the lower levels of CB1R observed in MAM-treated rats only at adulthood, a decreased GAD67 mRNA levels in the mPFC in adult (PD60 and PD70) MAM-treated rats was found without any change at PD30 (Bator et al., 2018). Although MAM-treated rats also show functional deficits in GABAergic interneurons in the hippocampus, the levels of CB1R mRNA in this brain region were found to be similar between control and MAM-treated rats.
In contrast to the mPFC, film analysis showed an increased CB1 mRNA expression in the dorsolateral striatum of MAM-treated rats at both PD45 and PD85. Surprisingly, to the best of our knowledge, there is no study investigating the levels of CB1R mRNA/protein in striatal regions in postmortem tissue of schizophrenia subjects and animal models of schizophrenia. However, a neuroimaging study showed a trend towards an increased CB1R binding in striatal regions (caudate and putamen) was observed in drug-free schizophrenia subjects (Ceccarini et al., 2013). The highest level of CB1R in the brain is found in the striatum and confirming previous studies (Martin et al., 2008; Van Waes et al., 2012), we observed that CB1 mRNA expression in both control and MAM-treated rats at the ages studied exhibited a lateromedial gradient. With a higher expression in the dorsolateral striatum which is more involved in motor and pattern formation and receives input primarily from substantia nigra, and a decrease to a lower expression in the dorsomedial striatum which represents the associative striatum with connections with prelimbic PFC and input from lateral ventral tegmental area (Ikemoto, 2007; Harber, 2016). CB1R, in the striatum, are expressed on terminals of striatal GABAergic, glutamatergic and dopaminergic inputs, on striatal interneurons, and on striatal projection neurons (Hohmann and Herkenham, 2000; Marsicano and Lutz, 1999; Martin et al., 2008). However, it has been suggested that the pattern of density levels of CB1 mRNA as showed by film autoradiography mostly indicates expression on striatal projection neurons (Van Waes et al., 2012), commonly known as medium spiny neurons (MSN). Striatal MSNs receive DA inputs and are divided into two subpopulations, on the basis of the DA postsynaptic receptors that they express, namely D1 positive neurons and D2 positive neurons. Although there is clear evidence for differential distribution of D1 and D2 receptors in dorsal versus ventral striatum (Gagnon et al., 2017), we are not aware of any data suggesting that there are subregion differences on neurons themselves between dorsal striatal subregions. CB1Rs are expressed in both D1 and D2 positive subpopulations and their activation is thought to negatively modulate D1 and D2 receptor-mediated behaviors (Martin et al., 2008). Thus, the increased levels of CB1R mRNA in the dorsolateral striatum could represent a compensatory response to the well documented hyperdopaminergic state found in MAM-treated rats (Modinos et al., 2015; Moore et al., 2006). It is noteworthy that because often mRNA and protein changes do not match, further studies should measure CB1R protein levels as well. This would give a better account of actual changes in function.
In conclusion, these findings demonstrate a trajectory for CB1R mRNA expression in cortical, striatal, and hippocampal regions across postnatal development. MAM-treated rats show a normal development trajectory, except for the mPFC and dorsolateral striatum, as indicated by a decrease and increase in the levels of CB1R mRNA, respectively. These changes seem to appear in adulthood after cellular maturation has been completed, suggesting that levels of CB1R may not be altered early in the course of schizophrenia and, consequently, may not contribute to the schizophrenia-like deficits in MAM-treated rats that emerge before adulthood. Further studies are needed to determine how the altered expression of CB1R mRNA contributes to the schizophrenia-phenotype or to compensatory responses to both the excitatory-inhibitory imbalance and the hyperdopaminergic state present in the MAM model.
Supplementary Material
Acknowledgments
We thank Niki MacMurdo and Christy Smolak for technical assistance. This study was funded by NIH R01MH57440 (to AAG) and NIH R01MH100066 (to DWV).
Footnotes
Contributors
FVG, DWV, and AAG designed the study. FVG performed the experiments and data analyses. JER and DWV helped with the in situ hybridization. FVG wrote the first draft of the manuscript and all authors contributed to and have approved the final version of the manuscript.
Conflict of interest
AAG has received funds from Johnson & Johnson, Lundbeck, Pfizer, GSK, Merck, Takeda, Dainippon Sumitomo, Otsuka, Lilly, Roche, Asubio, Abbott, Autofony, and Janssen. FVG, JRE, and DWV declare no conflict of interest.
Role of the Funding Source
Funding sources did not have any role in the writing of the manuscript or the decision to submit it for publication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev. 2006;52(2):293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Bator E, Latusz J, Wedzony K, Mackowiak M. Adolescent environmental enrichment prevents the emergence of schizophrenia-like abnormalities in a neurodevelopmental model of schizophrenia. Eur Neuropsychopharmacol. 2018;28(1):97–108. doi: 10.1016/j.euroneuro.2017.11.013. [DOI] [PubMed] [Google Scholar]
- Bennett MR. Synapse formation and regression in the cortex during adolescence and in schizophrenia. Med J Aust. 2009;190(4 Suppl):S14–16. doi: 10.5694/j.1326-5377.2009.tb02368.x. [DOI] [PubMed] [Google Scholar]
- Berghuis P, Dobszay MB, Wang X, Spano S, Ledda F, Sousa KM, Schulte G, Ernfors P, Mackie K, Paratcha G, Hurd YL, Harkany T. Endocannabinoids regulate interneuron migration and morphogenesis by transactivating the TrkB receptor. Proc Natl Acad Sci U S A. 2005;102(52):19115–19120. doi: 10.1073/pnas.0509494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghuis P, Rajnicek AM, Morozov YM, Ross RA, Mulder J, Urban GM, Monory K, Marsicano G, Matteoli M, Canty A, Irving AJ, Katona I, Yanagawa Y, Rakic P, Lutz B, Mackie K, Harkany T. Hardwiring the brain: endocannabinoids shape neuronal connectivity. Science. 2007;316(5828):1212–1216. doi: 10.1126/science.1137406. [DOI] [PubMed] [Google Scholar]
- Bodor AL, Katona I, Nyiri G, Mackie K, Ledent C, Hajos N, Freund TF. Endocannabinoid signaling in rat somatosensory cortex: laminar differences and involvement of specific interneuron types. J Neurosci. 2005;25(29):6845–6856. doi: 10.1523/JNEUROSCI.0442-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A, Tseng KY. Association of cannabis use during adolescence, prefrontal CB1 receptor signaling, and schizophrenia. Front Pharmacol. 2012;3:101. doi: 10.3389/fphar.2012.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass DK, Flores-Barrera E, Thomases DR, Vital WF, Caballero A, Tseng KY. CB1 cannabinoid receptor stimulation during adolescence impairs the maturation of GABA function in the adult rat prefrontal cortex. Mol Psychiatry. 2014;19(5):536–543. doi: 10.1038/mp.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catts VS, Fung SJ, Long LE, Joshi D, Vercammen A, Allen KM, Fillman SG, Rothmond DA, Sinclair D, Tiwari Y, Tsai SY, Weickert TW, Shannon Weickert C. Rethinking schizophrenia in the context of normal neurodevelopment. Front Cell Neurosci. 2013;7:60. doi: 10.3389/fncel.2013.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarini J, De Hert M, Van Winkel R, Peuskens J, Bormans G, Kranaster L, Enning F, Koethe D, Leweke FM, Van Laere K. Increased ventral striatal CB1 receptor binding is related to negative symptoms in drug-free patients with schizophrenia. NeuroImage. 2013;79:304–312. doi: 10.1016/j.neuroimage.2013.04.052. [DOI] [PubMed] [Google Scholar]
- Eggan SM, Hashimoto T, Lewis DA. Reduced cortical cannabinoid 1 receptor messenger RNA and protein expression in schizophrenia. Arch Gen Psychiatry. 2008;65(7):772–784. doi: 10.1001/archpsyc.65.7.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan SM, Lazarus MS, Stoyak SR, Volk DW, Glausier JR, Huang ZJ, Lewis DA. Cortical glutamic acid decarboxylase 67 deficiency results in lower cannabinoid 1 receptor messenger RNA expression: implications for schizophrenia. Biol Psychiatry. 2012;71(2):114–119. doi: 10.1016/j.biopsych.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan SM, Stoyak SR, Verrico CD, Lewis DA. Cannabinoid CB1 receptor immunoreactivity in the prefrontal cortex: Comparison of schizophrenia and major depressive disorder. Neuropsychopharmacology. 2010;35(10):2060–2071. doi: 10.1038/npp.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg I. Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res. 1982;17(4):319–334. doi: 10.1016/0022-3956(82)90038-3. [DOI] [PubMed] [Google Scholar]
- Gagnon D, Petryszyn S, Sanchez MG, Bories C, Beaulieu JM, De Koninck Y, Parent A, Parent M. Striatal neurons expressing D1 and D2 receptors are morphologically distinct and differently affected by dopamine denervation in mice. Sci Rep. 2017;7:41432. doi: 10.1038/srep41432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdeman G, Lovinger DM. CB1 cannabinoid receptor inhibits synaptic release of glutamate in rat dorsolateral striatum. J Neurophysiol. 2001;85(1):468–471. doi: 10.1152/jn.2001.85.1.468. [DOI] [PubMed] [Google Scholar]
- Gomes FV, Rincon-Cortes M, Grace AA. Adolescence as a period of vulnerability and intervention in schizophrenia: insights from the MAM model. Neurosci Biobehav Rev. 2016;70:260–270. doi: 10.1016/j.neubiorev.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace AA, Gomes FV. The circuitry of dopamine system regulation and its disruption in schizophrenia: insights into treatment and prevention. Schizophr Bull. 2018 doi: 10.1093/schbul/sbx199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harber SN. Corticostriatal circuitry. Dialogues Clin Neurosci. 2016;18(1):7–21. doi: 10.31887/DCNS.2016.18.1/shaber. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, Mirnics K, Lewis DA. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13(2):147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, Sampson AR, Lewis DA. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23(15):6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Sawa A. Disturbed synaptic connectivity in schizophrenia: convergence of genetic risk factors during neurodevelopment. Brain Res Bull. 2010;83(3–4):140–146. doi: 10.1016/j.brainresbull.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Heng L, Beverley JA, Steiner H, Tseng KY. Differential developmental trajectories for CB1 cannabinoid receptor expression in limbic/associative and sensorimotor cortical areas. Synapse. 2011;65(4):278–286. doi: 10.1002/syn.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann AG, Herkenham M. Localization of cannabinoid CB(1) receptor mRNA in neuronal subpopulations of rat striatum: a double-label in situ hybridization study. Synapse. 2000;37(1):71–80. doi: 10.1002/(SICI)1098-2396(200007)37:1<71::AID-SYN8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Howard MW, Rizzuto DS, Caplan JB, Madsen JR, Lisman J, Aschenbrenner-Scheibe R, Schulze-Bonhage A, Kahana MJ. Gamma oscillations correlate with working memory load in humans. Cer Cortex. 2003;13(12):1369–1374. doi: 10.1093/cercor/bhg084. [DOI] [PubMed] [Google Scholar]
- Howes OD, Murray RM. Schizophrenia: an integrated sociodevelopmental-cognitive model. Lancet. 2014;383(9929):1677–1687. doi: 10.1016/S0140-6736(13)62036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RS, Sommer IE, Murray RM, Meyer-Lindenberg A, Weinberger DR, Cannon TD, O’Donovan M, Correll CU, Kane JM, van Os J, Insel TR. Schizophrenia. Nat Rev Dis Primers. 2015;1:15067. doi: 10.1038/nrdp.2015.67. [DOI] [PubMed] [Google Scholar]
- Ikemoto S. Dopamine reward circuitry: two projection systems from the ventral midbrain to the nucleus accumbens-olfactory tubercle complex. Brain Res Rev. 2007;56:27–78. doi: 10.1016/j.brainresrev.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Cruz D, Eggan S, Erickson S. Postnatal development of prefrontal inhibitory circuits and the pathophysiology of cognitive dysfunction in schizophrenia. Ann N Y Acad Sci. 2004;1021:64–76. doi: 10.1196/annals.1308.008. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6(4):312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Moghaddam B. Cognitive dysfunction in schizophrenia: convergence of gamma-aminobutyric acid and glutamate alterations. Arch Neurol. 2006;63(10):1372–1376. doi: 10.1001/archneur.63.10.1372. [DOI] [PubMed] [Google Scholar]
- Lodge DJ, Behrens MM, Grace AA. A loss of parvalbumin-containing interneurons is associated with diminished oscillatory activity in an animal model of schizophrenia. J Neurosci. 2009;29(8):2344–2354. doi: 10.1523/JNEUROSCI.5419-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long LE, Lind J, Webster M, Weickert CS. Developmental trajectory of the endocannabinoid system in human dorsolateral prefrontal cortex. BMC Neurosci. 2012;13:87. doi: 10.1186/1471-2202-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi A, Di Forti M, Lewis CM, Murray RM, Vassos E. Meta-analysis of the association between the level of cannabis use and risk of psychosis. Schizophr Bull. 2016;42(5):1262–1269. doi: 10.1093/schbul/sbw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsicano G, Lutz B. Expression of the cannabinoid receptor CB1 in distinct neuronal subpopulations in the adult mouse forebrain. Eur J Neurosci. 1999;11(12):4213–4225. doi: 10.1046/j.1460-9568.1999.00847.x. [DOI] [PubMed] [Google Scholar]
- Martin AB, Fernandez-Espejo E, Ferrer B, Gorriti MA, Bilbao A, Navarro M, Rodriguez de Fonseca F, Moratalla R. Expression and function of CB1 receptor in the rat striatum: localization and effects on D1 and D2 dopamine receptor-mediated motor behaviors. Neuropsychopharmacology. 2008;33(7):1667–1679. doi: 10.1038/sj.npp.1301558. [DOI] [PubMed] [Google Scholar]
- McGlashan TH, Hoffman RE. Schizophrenia as a disorder of developmentally reduced synaptic connectivity. Arch Gen Psychiatry. 2000;57(7):637–648. doi: 10.1001/archpsyc.57.7.637. [DOI] [PubMed] [Google Scholar]
- Modinos G, Allen P, Grace AA, McGuire P. Translating the MAM model of psychosis to humans. Trends Neurosci. 2015;38(3):129–138. doi: 10.1016/j.tins.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore H, Jentsch JD, Ghajarnia M, Geyer MA, Grace AA. A neurobehavioral systems analysis of adult rats exposed to methylazoxymethanol acetate on E17: implications for the neuropathology of schizophrenia. Biol Psychiatry. 2006;60(3):253–264. doi: 10.1016/j.biopsych.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J, Aguado T, Keimpema E, Barabas K, Ballester Rosado CJ, Nguyen L, Monory K, Marsicano G, Di Marzo V, Hurd YL, Guillemot F, Mackie K, Lutz B, Guzman M, Lu HC, Galve-Roperh I, Harkany T. Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proc Natl Acad Sci U S A. 2008;105(25):8760–8765. doi: 10.1073/pnas.0803545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4. Academic Press; 1998. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, Mirnics K. Neurodevelopment, GABA system dysfunction, and schizophrenia. Neuropsychopharmacology. 2015;40(1):190–206. doi: 10.1038/npp.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear LP. The adolescent brain and age-related behavioral manifestations. Neurosci Biobehav Rev. 2000;24(4):417–463. doi: 10.1016/s0149-7634(00)00014-2. [DOI] [PubMed] [Google Scholar]
- Trettel J, Fortin DA, Levine ES. Endocannabinoid signalling selectively targets perisomatic inhibitory inputs to pyramidal neurones in juvenile mouse neocortex. J Physiol. 2004;556(Pt 1):95–107. doi: 10.1113/jphysiol.2003.058198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriguen L, Garcia-Fuster MJ, Callado LF, Morentin B, La Harpe R, Casado V, Lluis C, Franco R, Garcia-Sevilla JA, Meana JJ. Immunodensity and mRNA expression of A2A adenosine, D2 dopamine, and CB1 cannabinoid receptors in postmortem frontal cortex of subjects with schizophrenia: effect of antipsychotic treatment. Psychopharmacology. 2009;206(2):313–324. doi: 10.1007/s00213-009-1608-2. [DOI] [PubMed] [Google Scholar]
- Van Waes V, Beverley JA, Siman H, Tseng KY, Steiner H. CB1 cannabinoid receptor expression in the striatum: association with corticostriatal circuits and developmental Regulation. Front Pharmacol. 2012;3:21. doi: 10.3389/fphar.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Lewis DA. Impaired prefrontal inhibition in schizophrenia: relevance for cognitive dysfunction. Physiol Behav. 2002;77(4–5):501–505. doi: 10.1016/s0031-9384(02)00936-8. [DOI] [PubMed] [Google Scholar]
- Wilson RI, Nicoll RA. Endogenous cannabinoids mediate retrograde signalling at hippocampal synapses. Nature. 2001;410(6828):588–592. doi: 10.1038/35069076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.