Abstract
Extracellular vesicles (EVs) are cell membrane-derived compartments that spontaneously secrete from a wide range of cells and tissues. Extracellular vesicles have shown to be the carriers in delivering drugs and siRNA. Among extracellular vesicles, bacterial outer membrane vesicles (OMVs) recently have gained the interest in vaccine development and targeted drug delivery. In this review, we summarize the current discoveries of OMVs and their functions. In particular, we focus on the biogenesis of OMVs and their functions in bacterial virulence and pathogenesis. Furthermore, we discuss the applications of OMVs in vaccination and targeted drug delivery.
Keywords: OMVs, Secretion pathway, Vaccines, OMVs-based drug delivery
Graphical abstract
Introduction
Despite the advances in engineering of synthetic nanoparticles and their surface bio-conjugation for targeted drug delivery(1–7), reductionist’s bio-functionalization of nanoparticles still remains insufficient in replicating complex intercellular interactions present in nature, thus impossibly avoiding exposure of exogenous features of synthetic nanoparticles to immune systems(8,9). Thus, it is urgent to develop new drug delivery platforms that possess intercellular interaction motifs in nature.
Intercellular communication is critical to maintain the homeostasis in a biological system. Recent studies have shown that cells secrete nanosized membrane vesicles (so called extracellular vesicles (EVs)) to transport signaling cargos between cells in a long distance (10). Many cell types secrete EVs (11–14). There are two types of EVs: exosomes and microvesicles. EVs are spherical membrane structures that are similar with synthetic liposomes, but EVs contain many membrane proteins that facilitate intercellular interactions.
EVs were first discovered in 1983, and it was found that reticulocytes released multi-vesicular bodies to the extracellular space (15). The secretion of EVs is strongly correlated to the pathogenesis of many diseases (such as cancer), therefore EVs have become biomarkers in diagnostics and early cancer detection (16). Interestingly, the recent studies have shown that EVs could serve as novel drug carriers in targeted drug delivery. To translate EVs, it is needed to develop new approaches to solve several issues, such as heterogeneity of EVs in composition and size, low production yield, inefficient drug loading and unlikely scalability. Nitrogen cavitation is exploited to generate cell membrane-derived nanovesicles since the mechanical force produced by nitrogen cavitation rapidly disrupts cells, and subsequently the cell membrane forms nanoscale vesicles (17,18). Their size and membrane composition are similar to those of EVs, thus the nanoscale vesicles become new drug delivery platforms to treat vascular diseases (17,18).
Except EVs derived from eukaryotes, prokaryotes (a unicellular organism that lacks a membrane-bound nucleus) also secret extracellular vesicles (EVs). For example, gram-positive and gram-negative bacteria (13,19–21) have been reported to shed EVs. Gram-positive bacteria have a single lipid membrane surrounded by a cell wall comprised of a thick layer of peptidoglycan and lipoteichoic acid, which is anchored to the cell membrane by diacylglycerol (22). Since gram-positive bacteria don’t have “outer membrane” as compared with gram-negative bacteria, gram-positive bacteria release OMVs from the inner membrane and the released membrane vesicles go through the cell wall and form so-called “OMVs”. Gram-positive bacteria-derived EVs were firstly discovered from Staphylococcus aureus by mass spectrometry (21). The size of EVs from gram-positive bacteria was reported to be ~20-100 nm in diameter, which was similar to EVs derived from gram-negative bacteria (21). Gram-negative bacteria also spontaneously release extracellular vesicles, so-called outer membrane vesicles (OMVs). In this review, we will focus on OMVs derived from gram-negative bacteria.
A gram-negative bacterium possesses a unique membrane structure comprised of two membrane layers (outer membrane and inner membrane), a peptidoglycan layer, and a periplasm (23). Membrane proteins and lipopolysaccharide (LPS) distribute on the outer membrane while phospholipids mainly exist in the inner membrane. Periplasm is an oxidizing environment. A thin and rigid peptidoglycan layer in the periplasm adheres outer and inner membranes via proteins, such as OmpA. Several studies (24,25) have shown that peptidoglycan and outer membrane proteins (such as OmpA) are immunogenic, so they can be used in vaccination.
OMVs are a spherical structure with the size of 20-250nm, and they are comprised of bacterial cytoplasmic components that liberate during bacterial proliferation. OMVs play a vital role in pathogenesis, quorum signaling, nutrient acquisition and horizontal gene transfer (26). For instance, OMVs serve as vehicles to transfer toxins or various enzymes in bacterial infections. OMVs were first discovered in a cell culture medium of Escherichia coli (E.coli), and subsequently were identified by an electron microscope, showing a small spherical structure with a single layer of membrane (27). It is found that other gram-negative bacteria also release similar membrane vesicles, implying an important role of OMVs in biological processes and revolution. Furthermore, OMVs can serve as novel vehicles in immunization and delivering therapeutics in the therapies of cancer and other diseases.
The focus of this review is to discuss the biogenesis of OMVs, and their pathogenesis for bacterial infections. We first review the status on vaccination development and targeted drug delivery using OMVs. In the end, we will summarize the advances in OMVs and discuss the future directions.
Biogenesis of OMVs
It is likely that OMVs are reserved among species. The generation of OMVs is spontaneous without the requirement of ATP. During their formation, OMVs are packed with proteins, lipids and DNAs. There are several steps to generate OMVs. Initially, proteins in a gram-negative bacterial envelop are homogenously distributed, and the outer membrane is linked to peptidoglycan. To start the vesicular formation of outer membrane, the binding between outer membrane and peptidoglycan is lost because the protein linkers detach, therefore the outer membrane liberates from a bacterium to form a vesicle. It was reported that antibiotics or autolysins are able to disrupt peptidoglycan, resulting in the outer membrane release to form OMVs (28). The mechanism of OMVs secretion is categorized based on their compositions and contents (Figure 1). During the formation of OMVs, the linking protein between outer membrane and peptidoglycan could release from the membrane and is incorporated inside OMVs. For example, the studies showed that OmpA, a linking protein was incorporated in OMVs (29–32). In some cases, the location in a bacterial envelope is enriched with vesicular-formation proteins that induce the OMVs generation. In this model, proteins concentrate in the inner membrane. When the adherence of peptidoglycan-outer membrane is removed, the dense proteins may participate in bulging of the bacterial outer membrane, leading to the formation of OMVs (33,34).
Figure 1. Biogenesis of OMVs.
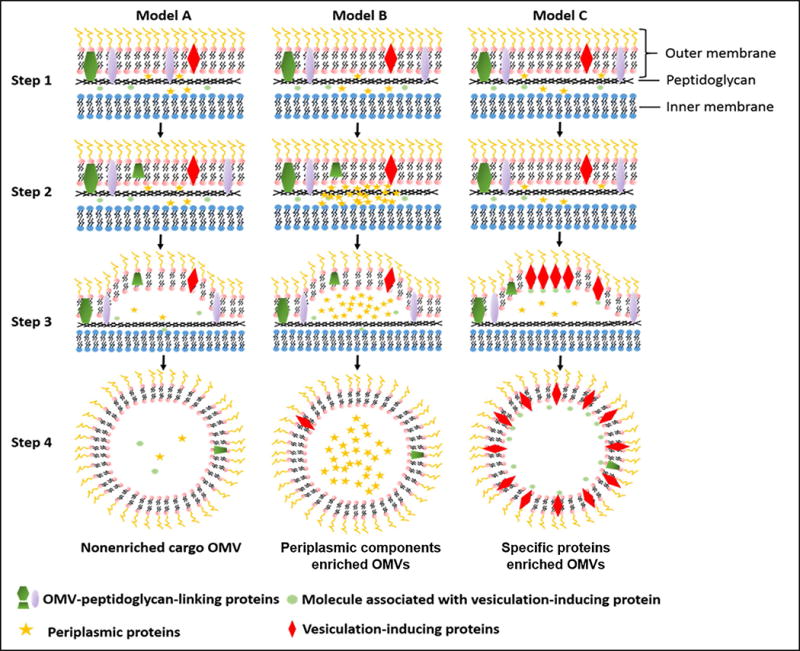
Step1: Gram-negative bacteria cell envelop. In this stage, envelop proteins are homogenously distributed. Outer membrane is linked with peptidoglycan. Step2: Vesiculation initiation. The linking between outer membrane and peptidoglycan is lost through the movement of linking proteins or breaking the connection of outer membrane with peptidoglycan directly. Model A, B and C demonstrate three ways for OMVs production. Model A indicates the basal OMV production. Model B refers to the OMV production with enriched periplasma cargos. Model C shows the formation OMVs is located at specific proteins on the outer surface, and the dense proteins could induce the additional budding of OMV from gram-negative bacteria cell envelop.
It has been an interest to understand how and why gram-negative bacteria secrete OMVs, and whether their secretion is spontaneous or random. For example, under stress, bacteria release OMVs for survival (35). The biogenesis of OMVs is an important topic in the future research.
It is noted that OMVs are complicated and heterogeneous. Analysis on their compositions and protein expression is very important to determine their biofunctions. A. baumannii release OMVs as a mechanism for the horizontal gene transfer, whereby carbapenem resistance genes are delivered to the neighbors (36). Additionally, proteomics is exploited to identify protein features in OMVs. Several studies have been performed to address whether OMVs contain virulence factors of their sources. VacA, a virulence factor, found in Helicobacter pylori OMVs (37), indicates that virulence factors were incorporated into outer membrane vesicles during secretion. The adhesion molecules on OMVs (such as B-type flagellin and pili machinery: (PilA,F,Q,V,Y1)) that can serve as the virulence factors, were also identified in P. aeruginosa (38). Moreover, OMVs also contain outer membrane enzymes (lipases, peptidases, and ribonucleases) that are involved in pathogenesis. They include lipases, PagL (the most abundant protein in OMVs) (39), LipA (40), EstA, peptidases AaaA, PepA, PasP, MucD CtpA, Lon, IcmP and the M23 metaloprotease LasA (41–43). After OMVs were purified via ultracentrifugations, proteomics analysis showed that OMVs lacked the components of inner cell membrane and cytoplasm, but they shared higher similarities with outer membrane, including porins (OprB,C,D,E,H,O,Q), OstA (resistance to organic solvents and antibiotics) and OsmE (associated with the cell envelop integrity) (44,45). However, several studies on OMVs proteomics are not consistent (46–48). The biochemistry shows that protein bands of OMVs using SDS-PAGE are not always identical to those of the outer membrane of bacteria.
As mentioned above in Figure 1, OMVs not only contain the proteins from their source but also concentrate specific proteins compared with their parent bacteria during their formation. Studies on enterotoxigenic Escherichia coli and P. aeruginosa (49,50) show that several proteins are enriched in OMVs, such as LT and aminopeptidase, resulting in the increase of OMVs uptake by epithelial cells.
Proteomics has been applied to analyze the differences between bacteria and their derived vesicles, and to identify the abundance of proteins in OMVs. However, the proteomics results are still complicated. It is possible that the culture media and bacterial growth rates influence the heterogeneity of OMVs, leading to the inconsistence on proteomic analysis results. Another possibility is that proteins and virulence factors might mutate during the formation of OMVs.
Functions of OMVs in bacterial virulence and pathogenesis
OMVs play several roles in physiology and pathogenesis including horizontal gene transfer, quorum sensing, nutrient digestion, toxin secretion, misfolded protein secretion and immunomodulation (Figure 2). Here we focus on the role of OMVs in bacteria virulence and pathogenesis via the secretion of toxins and virulence factors.
Figure 2. Biofunctions of OMVs.
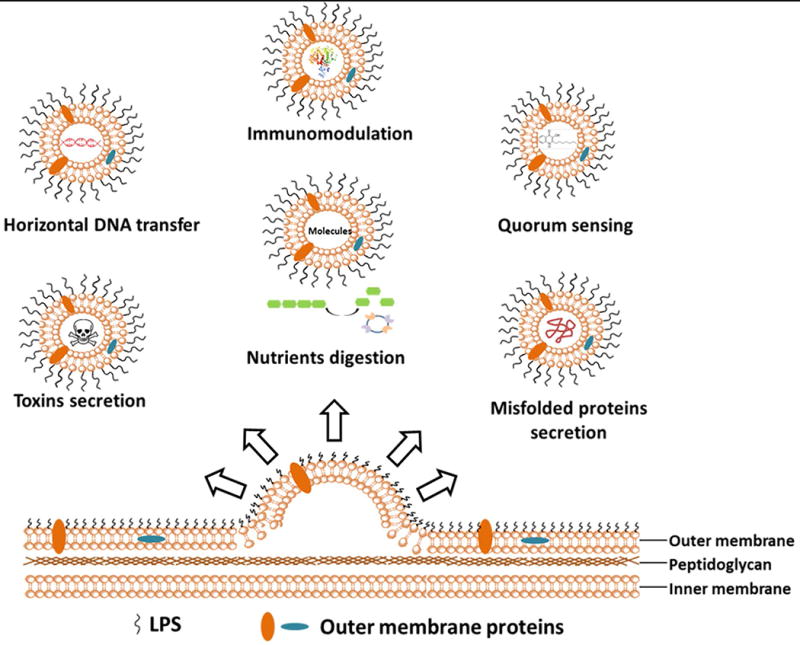
OMVs release from bacteria outer membrane layer and automatically entrap various cellular molecules. Biofunctions of OMVs are categorized and described as above, including horizontal DNA transformation, quorum sensing, toxin secretion, nutrient digestion and misfolded protein secretion.
Several studies have shown that OMVs directly regulate interactions between bacteria and the host via delivering of entrapped toxins, non-toxins, or bacterial virulence into host cells (55). For example, DNA packed in OMVs derived from Neisseria gonorrhoeae and Borrelia burgdorferi mediates the transfer of bacterial virulence to host cells (56). In addition to DNA packing, P. aeruginosa OMVs (38) have been shown to contain FliC, OprF and OprH/OprG and host-bacterium interaction proteins (EstA, FlgE, FlgK, etc) which participate in P. aeruginosa pathogenesis. Particularly, it was reported that EstA, a bacterial virulence factor, can induce nitric oxide and pro-inflammatory cytokines in macrophages. FlgE/K are flagellar proteins that stimulate innate immunity through Toll-like receptor 5 and play a role in the biofilm formation, an important virulence mechanism of P. aeruginosa (57,58). Another example (50) is that enterogenic and uropathogenic E.coli ((ETEC) and (UPEC)) released the heat-labile enterotoxin into OMVs, and the endotoxins were transferred to host cells via OMVs. The mechanism further revealed that endotoxins were likely the ligands that mediated the binding of OMVs to lipid rafts of host cells, thus leading to the uptake of OMVs. Besides toxins and virulence factors, non-toxins including a variety of enzymes and proteins, have been shown to be entrapped in OMVs and delivered to host cells, therefore, affecting bacterium-host cell interactions. For example, P. aeruginosa OMVs released proteins associated with proteolysis, ion transport, and ion binding, which dysregulated host cells (38). The similar study (59) on Treponema denticola OMVs shows that they contain the necessary adhesins and proteolytic arsenal for the adherence to and the damage of eukaryotic cells.
OMVs can also affect bacterium-host cell interactions by entrapping cellular components and directly binding and even destroying host bacterial factors. For example, OMVs produced by H.pylori, presented Lewis antigens on their surface and were able to induce the host immune system activation (60). In this case, the OMVs directly bound to anti-Lewis antibodies in serum to decrease the self-defense ability of host cells, therefore, playing a very important role in H. pylori pathogenesis.
In summary, OMVs play a vital role in pathogenesis of bacterial infections since they contain many toxins from their source, which activate the host defense system. OMVs could be also applied to vaccine development.
OMVs in Vaccination
Infection can cause host immune responses, but sometimes excessive responses may lead to tissue damage, resulting in the death. OMVs play a central role in transporting toxins and virulence factors to host cells, and this transport mediates the host immune response. OMVs could be exploited to train the immune system to combat pathogens if they can be administered to the host in a control manner. Therefore, OMVs are a promising candidate for vaccine development against bacterial infections.
Studies have shown that OMVs could interact with epithelium cells, therefore, inducing the host immune response. The proteomics results from Campylobacter jejuni OMVs revealed that they contained many periplasmic and outer membrane-associated proteins (61). Several molecules are important in survival and pathogenesis, including the cytolethal distending toxin (CDT). Thus, OMVs could be an important alternative for the coordinated delivery of C. jejuni proteins into host cells. This idea was further confirmed by showing that C. jejuni OMVs possessed cytotoxic activity and induced a host immune response in T84 intestinal epithelial cells (IECs) (62). Similar studies in OMVs derived from various mucosal pathogens also showed that OMVs can interact with epithelial cells, resulting in the production of cytokines and chemokines that activated the pro-inflammatory response. For example, OMVs from all strains of P. aeruginosa elicited IL-8 secretion from lung epithelial cells to contribute the inflammation response (63). Moreover, P. aeruginosa-derived OMVs were shown to induce pulmonary inflammation via increasing chemokines and cytokines in the mouse lungs and mouse alveolar macrophages in a rodent model. Interestingly, OMVs could induce inflammatory responses as compared with that of live bacteria (64), indicating that OMVs have the similar ability as live bacteria to induce innate immunity.
Interestingly, several studies in Helicobacter pylori showed that OMVs adhered to the epithelium to cause gastritis, rather than bacteria (65). These OMVs have also been shown to carry CayA and localize in the vicinity of cell-cell contact, therefore they may have an influence on host gene transcriptions, leading to infections and development of cancer (66).
OMVs also interact with various types of immune cells. It is shown that OMVs interact with innate immune cells. A study showed that N. meningitidis OMVs can stimulate human neutrophils, resulting in the production of TNF-α and IL-1β and upregulation of CXCL8, CCL3 and CCL4 (67). L. pneumophila OMVs can generate pro-inflammatory cytokines from macrophages (68). Furthermore, Helicobacter pylori OMVs proteins can induce human eosinophil degranulation (69). Antigen presenting cells (dendritic cells), as a key connection between the innate and adaptive immunity, can be activated by OMVs. For example, OMVs from Salmonella spp induced the expression of CD86 and MHC class II molecules on dendritic cells and the production of TNF-α and IL-12, and promoted the development of protective B cell and T cell response in vivo (70). Similarly, OMVs derived from E.coli showed the increased uptake by dendritic cells and induced IL-6, IL-1β production and antibodies production in vivo (71). OMVs can also interact with other host cells including endothelial cells and platelets cells. OMVs derived from E. coli OMVs up-regulate the expression of endothelial intercellular adhesion molecule-1 (ICAM-1), E-selectin and vascular cell adhesion molecule-1, and enhance the leukocyte binding on human microvascular endothelial cells (72). Moreover, Gingivalis OMVs (73) can enhance the platelet aggregation. In summary, OMVs strongly interact with the host via the activation of the innate and adaptive immune responses, therefore OMVs are an excellent candidate in vaccination.
We have discussed that OMVs are able to interact with a variety of cells. The uptake of OMVs have been investigated (74) and several internalization pathways have been found such as micropinocytosis(75), clathrin-mediated endocytosis (76), non calthrin-mediated endocytosis (lipid raft) (77), and membrane fusion (78). It is needed to address what receptors mediate the internalization of OMVs.
Adjuvants are required in most vaccine formulations to enhance the immune responses (79). Due to the unique features of OMVs, early efforts were focused on utilizing bacterial OMVs as adjuvants that were covalently complexed to antigenic proteins. E. coli-derived OMVs have been combined with malarial proteins in the development of the intranasal vaccine (80). In this work, they confirmed that OMVs can serve as adjuvants which can promote the immune response comparable to the cholera toxin adjuvant. Moreover, the authors suggested that OMVs can be applied as safe adjuvants and replace cholera toxin adjuvant because the cholera toxin adjuvant has the high toxicity and is difficult to be used in human clinical trials.
The studies showed that OMVs could not only be combined with proteins but also with other components. The study showed that meningitides derived OMVs can be complexed with Shigella-specific lipopolysaccharides (LPS) to provide the immunity against Shigella keratoconjuctivitis (81). Another study showed that when combined with inactivated respiratory syncytial virus (iRVS), Neisseria meningitidis OMVs enhanced the protective immunity (82). The main mechanism was elucidated that hexa-acylated Lipid A moiety in the native LPS acted as a stimulator for a TLR4 receptor, therefore activating the innate immunity.
OMVs have been also used as vaccines to prevent bacterial infections. For example, E.coli-derived OMVs efficiently prevented bacterium-induced lethality and OMVs-induced systemic inflammatory response syndrome via Th1 and Th17 cell responses (71,72). In this work, they performed immunization with E. coli-derived OMVs in a rodent model and demonstrated that E. coli-derived OMVs had the high protective effect as shown in Figure 3A. Moreover, they proved that this protective effectiveness can last 42 days after immunization (Figure 3B). It is shown that this protection was dependent on the induction of innate and adaptive immunity, including the production of anti-OMVs specific antibodies and T cell activation (Figure 3C&D). They also demonstrated the passive protection of OMVs via adoptive transfer of serum and splenocytes (data not shown). As mentioned above that immunization of OMVs mainly relies on the activation of innate and adaptive immunity, they also proved that the key antigen presenting cells (dendritic cells), functioning in the presenting of antigens of OMVs to the adaptive immunity-related cells, were fully activated (data not shown therefore). The activation of dendritic cells resulted in the production of immune-modulating cytokines (Th1- and Th17-polarizing cytokines), therefore, inducing the activation of T cells to OMVs. These cytokines included IFN-γand IL-17, the key cytokines produced by Th1 and Th17 cells (data not shown). To understand whether this protective effect was Th-1 and Th-17-dependent and how such IFN-γand IL-17-dependent Th1 and Th 17 cell responses elicited the enhanced vaccine efficacy, IFN-γ, IL-17 and IL-4 knockout mice were used. The results showed that the knockout mice failed the survival compared to the wild-type mice immunized with OMVs (Figure 3E). In summary, they provide a comprehensive and new perspective on the immunological detail regarding OMVs being used alone in vaccination.
Figure 3. Immunization with Escherichia coli outer membrane vesicles protects bacteria-Induced lethality via Th1 and Th17 cell responses.
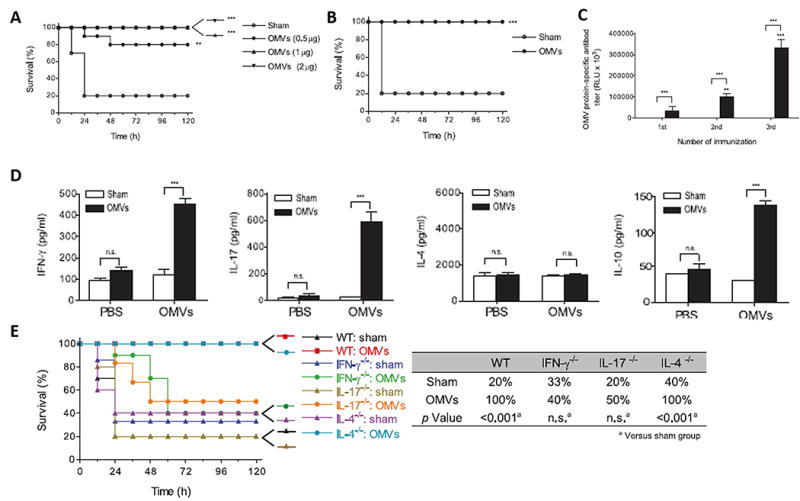
A. Survival rates of OMV- and sham-immunized mice challenged with E.coli; B. Survival rates of OMV- and sham immunized mice challenged with E. coli 42 d after immunization; C. Serum levels of OMV-reactive IgG; D. OMV-specific production of IFN-g, IL-17,IL-4, and IL-10 from splenic T cells; E. Survival rates of wild-type, IFN-g−/−, IL-17−/−, and IL-4−/− mice after the E. coli injection. (Reprinted with permission from Ref. 71. Copyright © 2013 by The American Association of Immunologists, Inc)
Similar studies showed that OMVs can serve as vaccines against Edwardsiellosis because the cytokines and chemokines were significantly increased after administration of OMVs (83). Moreover, B. pertussis-derived OMVs combined with alum adjuvants provided the protection from pertussis in a mouse model and this effect was comparable to the whole-cell formulation of vaccines (84). B. parapertussis derived OMVs also have been shown to have the cross-protection effect against both pertussis and parapertussis (85).
With the promising potential of OMVs in vaccination, OMVs-based vaccines have been tested in clinical trials (86). The Meningitis type B (MenB)-based vaccine has gained much attention. For several serogroups, conjugated vaccines consisting of capsular polysaccharide coupled to a carrier protein have already been in the market. Meningitis type B OMVs have been shown to have the efficacy range in 83%-85% (86,87) and have passed the phase I, phase II and multiple clinical studies (88–91). For instance, in clinical research, they showed that a four-dose schedule (three primary doses and one booster dose) for infants and a two-dose schedule for adolescents of the multi-component Men B vaccine (4CMenB) provided the good result. These vaccines contained three surface-exposed recombinant proteins and New Zealand strain outer membrane vesicles (NZ OMVs) with PorA 1.4 antigenicity. The side effects are mainly associated with the injection site pain/tenderness and fever in infants. They are associated with the injection site pain, malaise and headache in adolescents. Thus, 4CMenB is conservatively estimated to provide 66-91% protection against B group of meningococcal strains worldwide (91). Moreover, Meningococcal-derived OMVs vaccines made of inactivation of lpxL1 gene, were also in the phase I trial (92,93).
With the promising perspectives of OMVs in vaccination, the major concern lies in the stability of OMVs when being administered in vivo. The kinetic biodistribution of OMVs in vivo demonstrated that OMVs were detected at the peak 3 hours after administration, then gradually decreased in most organs within 24 hours post injection (94), indicating that OMVs were likely to be eliminated during circulation. To address this concern, a report demonstrated that gold nanoparticles coated with OMVs can dramatically increase the stability of OMVs, therefore, resulting in higher immune activation as compared with OMVs administration alone (95). In their work, they incorporated gold NPs (AuNPs) into OMVs and proved that the membrane coating can effectively enhance gold NP stability in biological buffers, while gold NP cores stabilized OMVs (Figure 4A). With the higher stability, AuNPs demonstrated better efficacy in inducing B cell and T cell activation via producing higher IgG titer and cytokines (Figure 4B&C). Collectively, the future studies should be focused on increasing the stability of OMVs via membrane coating strategy to enhance OMVs efficacy for vaccination development.
Figure 4. Modulating antibacterial immunity via bacterial membrane-coated nanoparticles.
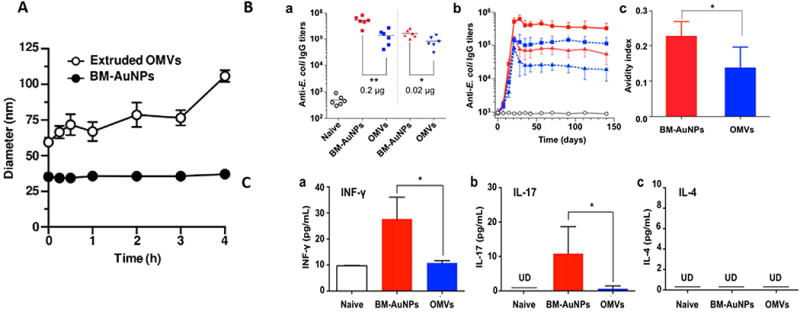
A. Stability of extruded OMVs and BM-AuNPs with time; B. BM-AuNPs eliciting strong bacterium-specific antibody responses in vivo; C. BM-AuNPs inducing pronounced bacterium-specific T cell activation in vivo. (Reprinted with permission from Ref 95. Copyright © 2015 American Chemical Society)
Isolation and purification of OMVs
Purity of OMVs is an essential factor for their applications. Therefore, it is needed to develop novel methods to isolate and purify OMVs.
OMVs are directly obtained from cell suspensions. The most common isolation technique includes several steps of centrifugation. The first step is to separate cell debris through a 0.22 or 0.45 μm filter, followed by the low speed centrifugation (2000-1000× g). To further purify and concentrate OMVs, the centrifugation is combined with the tangential flow microfiltration, such as using a filter of 50-100kDa (96) or a combination of 0.2 and 0.1 μm filters(97). Finally, OMVs are collected after ultracentrifugation at 50,000-200,000× g. The ultra-centrifugation step can also be combined with the density gradient (98). Ammonium sulfate precipitation sometimes is applied as an alternative method for concentration of OMVs (99). In contrast of centrifugation approaches, OMVs are also isolated via detergents, such as DOC (Deoxycholic acid) (100).
However, some contaminants should be cautious during isolation and purification of OMVs. Bacterial components, such as pili, flagella and soluble components, may be mixed with OMVs since they cannot be separated by centrifugation approaches (101).
Bioengineered OMVs in vaccination
OMVs can also be engineered for vaccine development via incorporating of heterologous antigens. These heterologous antigens can be presented with or without surface exposure, attached to the vesicles or non-attached and directly produced by the bacterium or combined in a later production stage. The generation of heterogenous OMVs vaccines mainly includes the following approaches:
1) Recombinant OMVs based on ClyA fusion protein
Antigens can be presented on the surface of OMVs with the exposure to the exterior side of the vesicles via a variety of fusion proteins, such as ClyA fusion (102–104). Cytolysin A (ClyA) is a transmembrane protein (a molecular weight at 34kDa) which is enriched in outer membrane vesicles (105). Genetic modification of OMVs is mainly focused on the fusion of antigens to the C terminus of ClyA which results in production of ClyA-antigen fusion proteins associated with OMVs. Chen’s group (104) was the first to prove the concept via fusing of antigens with ClyA protein and express antigens on the cell surface. Their study showed that green fluorescent protein can be infused with ClyA of E. coli. The in vivo study showed that this recombinant GFP-OMVs can induce a strong production of GFP-specific antibodies without any adjuvants. Inspired by this work, ClyA was fused with antigens to generate anti-bacterial vaccines. Huang’s group (102) fused Omp22 antigens from Acinetobacter baumannii into E. coli DH5α-derived OMVs, as shown in Figure 5, which showed the high protection in a murine sepsis model.
Figure 5. OMVs biomodification in Acinetobacter baumannii vaccines development. Schematic diagram of the construction of recombinant Omp22-OMVs.
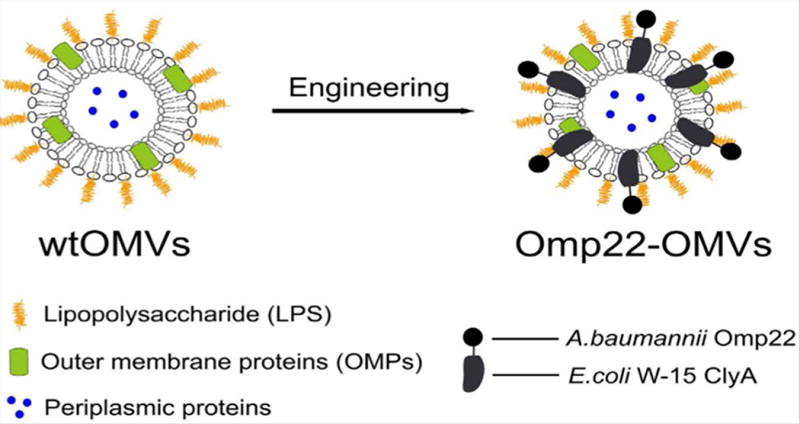
(Reprinted with permission from Ref. 102. Copyright © 2016, Springer Nature)
In this work (102), CytolysinA (ClyA) was successfully infused into E. coli membrane to generate chimeric ClyA fusion proteins, therefore, it can be used for the future engineering of infused Omp22-OMVs. It was confirmed that Omp22 antigen located on the surface of OMVs. This OMV-based vaccine lessened bacterial burdens in various tissues, and antiserum isolated from the mice demonstrated the bactericidal activity. Recombinant OMVs have also shown promising in creating anti-viral vaccines. Rosenthal’s group (103,106) showed that the probiotic E.coli Nissle 1917 strain can be engineered to generate ClyA-GFP OMVs which elicited anti-GFP total IgG titers. The IgG titers were equivalently comparable with that of GFP adjuvanted with alum when the OMVs were administrated into the mice. Furthermore, they proved the protection against influenza infection using OMVs derived from Nissle 1917 strain fused with antigen protein M2e4xHet. Their results showed that bioengineered OMVs could elicit higher anti-M2e IgG2a antibodies which could facilitate the clearance of infected cells. The survival study demonstrated that bioengineered OMVs can save all mice when the mice were challenged with a lethal dose of influenza.
2) Recombinant OMVs based on other carrier fusion protein
Similarly, various fusion proteins are incorporated in OMVs. The fHbp was infused to express the Borrelial surface-exposed lipoprotein OspA on Neisseria meningitidis OMVs surface. The results showed that OMVs could able to elicit antigen specific antibodies as compared with that with luminal expression of OspA inside the cells (107). PspA or Ply fragments (108), ESAT6, Ag85B fragments, Rv2660c (109) and MOMP fragments (110) were able to be fused into fHbp fusion for vaccination.
Antigens can also be expressed in the outer membrane of OMVs. P. aeruginosa A104R antigen can be fused into outer membrane of E.coli-derived OMVs via OprI fusion, and through this way they can protect against African swine fever (111). Ail antigen can also be fused into E.coli-derived OMVs (112). Many studies showed that antigens can be expressed in lumen of OMVs mainly via OmpA fusion. Kesty’s study in 2004 was the first proof to concept to demonstrate the fusing of GFP into E.coli-derived OMVs via Tat signal (112). FLAG tag protein was fused in OMVs lumen (113). Several antigens have been infused with OmpA protein in order to localize them in OMVs lumen to target group A/B Streptococcus disease (109) and Chlamydia (114).
It is interesting to observe that lumen-fused OMVs vaccines only elicited minor specific antibody production. For example, the fusion PspA in Salmonella enteriaca in OMVs lumen showed lower antibody production and decreased protection. OMVs without any antigen fusion or purified fusion antigen showed neither the antibody production nor protective effect (115). One of possibilities is that the expressed antigens may change their conformations. The study observed the higher antibody titer when antigens were in their native structure(109). Additionally, when expressed on the surface of OMVs, bio-engineered OMVs would have higher antibody titers (115). However, the mechanism still remains unclear. It is also not clear whether the non-specific antibody response is dependent on strains. Therefore, more research should be focused on the specificity of OMVs responses.
Currently, most vaccines are generated from acellular organisms and their subunits (116). These vaccines are safer than utilization of live-attenuated or whole inactivated organisms. The vaccines usually don’t contain the whole cell antigens, therefore, lacking of a broad protection compared to a whole inactivated organism. Moreover, the vaccines are also needed to combine with adjuvants to increase the efficacy. OMVs are derived from the bacterial membrane that contains a wide range of antigens required for immunization, thus they are potential to become novel vaccines (19). Since OMVs are nanoscale and can be engineered, they will increase the response of antigen-presenting cells (117,118).
While OMVs demonstrate the potential as a new platform for vaccination, there are several barriers when they are used in clinic. The heterogeneity of OMVs might cause the issues on reproducibility when they are largely scaled up. OMVs secretion strongly depends on the bacterial growth condition, therefore the preparation consistency is not guaranteed. Current techniques cannot scale up the production of OMVs required in clinic. Therefore, it is necessary to develop new approaches to generate not only the high quantity but also high purity of OMVs.
OMVs in drug delivery
Synthetic nanomaterials, such as liposomes, polymers, and metal-based nanoparticles have been broadly studied as drug carriers (119), but the simple bio-conjugation of synthetic nanoparticles is not efficient to replicate intercellular interactions that facilitate nanoparticle trafficking and delivery. Cell membrane derived nanovesicles possess the features of intercellular interactions, thus they are potential to become novel drug delivery platforms.
Similarly, OMVs have been reported to demonstrate a spherical structure which is the size range in 20-250 nm in Figure 6. OMVs contain a broad range of proteins that are derived from their parent cells and can carry diverse cargos, therefore, they can serve as a new platform in targeted drug delivery. (120).
Figure 6. Cryo-TEM visualization OMVs derived from B. pseudomallei outer membrane vesicles (120).
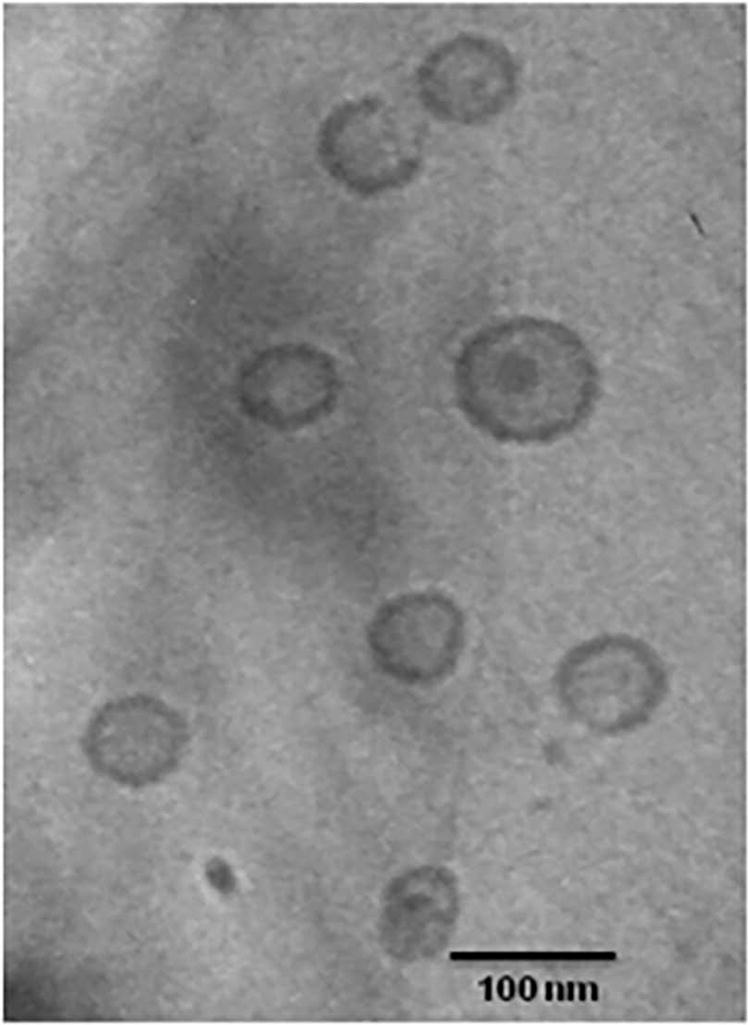
Cryo-transmission electron micrograph of purified OMVs prepared from a late logarithmic culture of B. pseuodomallei strain 1026b. Bar indicates 100 nm.
(Reprinted with permission from Ref.100. Copyright 2010 Plos One).
Enzyme degradation in serum is a problem to effectively deliver, so the incorporation of enzymes in OMVs may resolve this problem. Walper’s group proposed an approach that phosphotriesterase (PTE) (EC 3.1.8.1) from Brevundimonas diminuta, containing a binuclear Zn/Zn active site, was selectively packaged within the OMVs (121,122). In this work, they used OmpA (outer membrane protein A) as an anchor to link PTE as described in Figure 7A. Furthermore, they demonstrated that the PTE-loaded OMVs exhibit native-like enzyme kinetics without changing enzyme activity of PTE (Figure 7B).
Figure 7. Bacterial nanobioreactors–directing enzyme packaging into bacterial outer membrane vesicles.
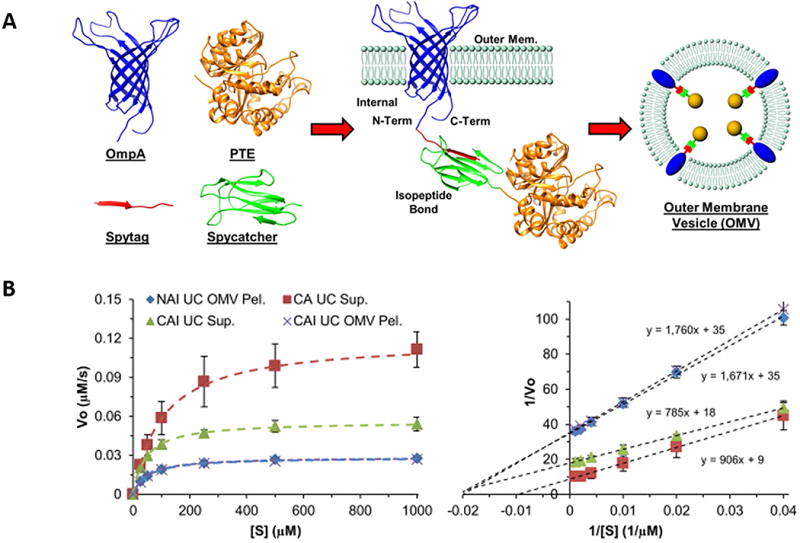
A. Crystal structures for the proteins utilized in the biorthogonal membrane conjugation of PTE for packaging into outer membrane vesicles; B. PTE kinetic data fit to the standard Michaelis–Menten enzyme kinetics equation for NAI and CAI UC pellet, CAI and CA UC supernatant (Left); A Lineweaver–Burk analysis used for determining KM and kcat/KM (Right). (Reprinted with permission from Ref.122 Copyright © 2015 American Chemical Society)
Extracellular vesicles are comprised of the adhesion molecules on vesicle surface and can bind target cells via the ligand-receptor recognition (17,18,123). Similarly, OMVs could target the cells of interest through the genetic and surface modification of OMVs for targeted drug delivery.
A pioneer work developed OMVs using bioengineered bacteria (E.coli) with the fusion of an anti-HER2 affibody to ClyA resulting in OMVs with the affibody displayed on the surface (124,125). OMVs could target and kill cancer cells in a cell-specific manner by electroporation loading with a small interfering RNA (siRNA) therapeutic targeting kinesin spindle protein mRNA (Figure 8 A). Although the loading efficiency was not high (data not shown), the amount of siRNA in the AffiHER2OMVs was still sufficient to exert cytotoxic effects against the HER2-positive tumor cells, because siRNA loaded AffiHER2OMVs were found selectively accumulate in the tumor sites after administration (Figure 8 B). Further study showed that this siRNA delivery strategy exhibited higher ability in tumor growth inhibition due to a significant reduction in KSP protein levels as compared with the free siRNA and non-targeted OMVsiRNA group (Figure 8 C). This genetically engineered OMVs were low toxic, inflammatory and immunological, thus they are considered as a safe platform for cancer therapy.
Figure 8. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy.
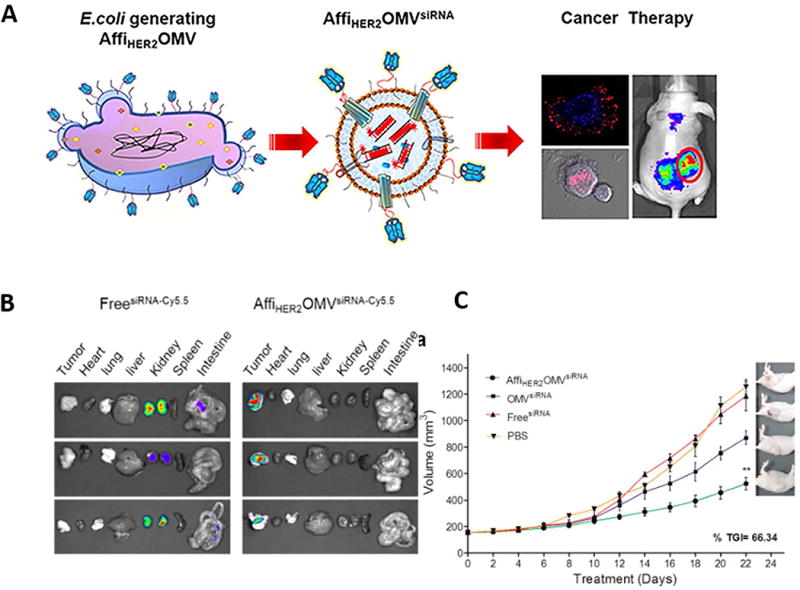
A. Schematic representation of OMVs expressing HER2-specific affibody(AffiHER2OMV) and the application of AffiHER2OMV in cancer therapy; B. Tumor-specific retention and accumulation of delivered siRNA in major vital organs; C.Tumor growth inhibition (TGI) after delivery of siRNA (Reprinted with permission from Ref.125 Copyright © 2014 American Chemical Society)
To enhance their functions, OMVs have been modified using nanotechnologies. The cells were decorated with synthetic nanocarriers to deliver drugs in a more controlled way. Gold nanoshells are nanoparticles which have been shown to successfully treat tumors in mice with tumor remission rate over 90% via being designed and fabricated to allow for the viability of the monocytes/macrophages during recruitment into the tumor (126). Additionally, the study showed that utilizing E.coli OMVs as a drug carrier to coat Au nanoparticles can sufficiently induce the activation and maturation of dendritic cells in the lymph nodes of the vaccinated mice, And these recombinant nanovesicles-induced antibodies production were durable and of higher avidity than OMVs only (95).
Conclusions and Future Perspectives
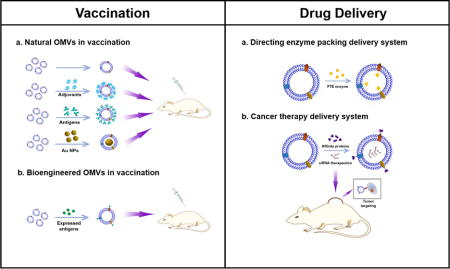
In Figure 9, we have summarized the overview of current research of OMVs. We have discussed the mechanisms of OMVs formation, and addressed how OMVs have been utilized in biomedical applications. We have demonstrated two major applications: vaccines and targeted drug delivery platforms.
Figure 9. OMVs functionality in vaccination and drug delivery.
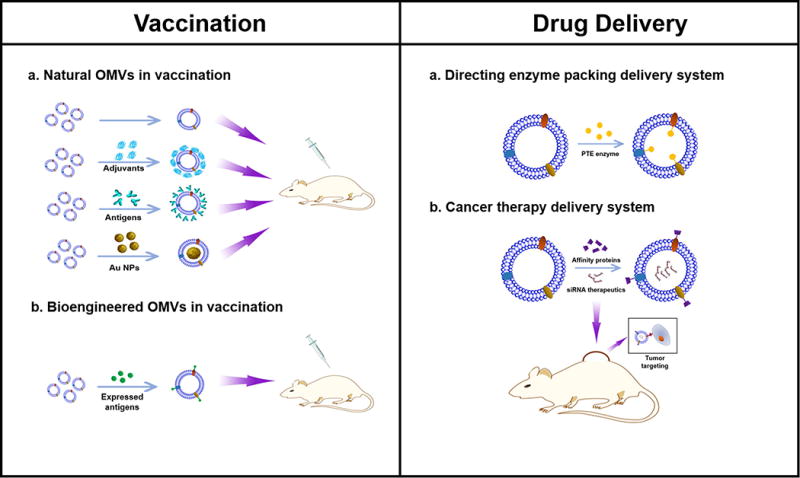
Left: A. Natural derived OMVs vaccine formulas, including OMVs alone, OMVs combined with adjuvants, OMVs combined with bacterial antigens and AuNPs loaded OMVs; B. Bioengineered OMVs vaccine formula. Bacterial antigens are expressed as fusion proteins with outer membrane proteins located on the outer membrane surface of OMVs; Right: A. Directing enzyme packing delivery systems for packing PTE enzymes in OMVs; B. siRNA loaded bioengineered OMVs in cancer therapy.
We have shown that OMVs bulge from bacterium outer membrane via three major mechanisms. During their formation, OMVs can entrap periplasmic proteins. Understanding the mechanism by which OMVs are formed is critical for the medical applications of OMVs, but their biogenesis remains unclear. For example, how and why OMVs are produced? Is the formation of OMVs spontaneous or regulated? If it is regulated, what signaling pathways are involved? If we have a complete picture about their biogenesis, we may resolve the issues on their low production and complex composition.
We believe that we need novel approaches to address these questions. It is shown that secretion of OMVs is a ubiquitous process and OMVs have widely diverse functions than it is currently appreciated. Utilizing proteomics may enable to quantitatively analyze the composition of OMVs and how the composition is associated with their biological functions. Proteomics generates the huge and complex data, so it is needed to develop mathematical approaches to analyze them for the understanding of the OMVs biofunctions. The biogenesis of OMVs is important to develop effective OMVs-based vaccines.
OMVs are exploited as a new drug delivery platform as they are in the nanoscale range. OMVs can be recognized by dendritic cells, thus they may activate the innate and adaptive immune responses. However, the mechanism of OMVs internalization by host cells remains unclear. OMVs have been shown to contain a variety of virulence factors, including LPS and virulent proteins. Therefore, the safety issues should be cautious when OMVs are utilized as drug delivery carriers since LPS could cause innate immune response. This immunotoxicity of OMVs will be an interesting topic in drug delivery applications.
In summary, we have demonstrated that OMVs may be a new drug delivery system used in vaccination and targeted drug delivery. While the research on OMVs is in the early stage, their unique nanosized structure and biofunctions may be a promising platform in nanomedicine.
Table 1.
OMV-associated Virulence proteins
| OMV-associated proteins | Activity | References |
|---|---|---|
| Vacuolating toxins (VacA) | Immunolocalization | (37) |
| Porin proteins (OprF, L,etc) | Membrane proteins, transport small molecules | (44,51,52) |
| OstA | Organic solvent tolerance protein OstA precursor | (44) |
| OsmE | Membrane proteins, cell envelop integrity | (45) |
| FilC | Flagellin type B, motility and attachment | (53) |
| Pili machinery (pilA) | Type 4 fimbrial precursor pilA | (44) |
| PagL | Lipid A 3-O-deacylase | (54) |
| EstA | Secreted factors (enzymes), metabolism | (54) |
| aaaA | Amino acid biogenesis and metabolism | (54) |
Acknowledgments
This work was supported by NIH RO1GM116823 and in part by Environmental Molecular Science Laboratory at Pacific Northwest National Laboratory.
References
- 1.Wang AZ, Langer R, Farokhzad OC. Nanoparticle delivery of cancer drugs. Annu Rev Med. 2012;63:185–198. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 2.Chu D, Gao J, Wang Z. Neutrophil-Mediated Delivery of Therapeutic Nanoparticles across Blood Vessel Barrier for Treatment of Inflammation and Infection. ACS Nano. 2015;9:11800–11811. doi: 10.1021/acsnano.5b05583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conde J, Edelman ER, Artzi N. Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: the jack-of-all-trades in cancer nanotheranostics? Adv Drug Deliv Rev. 2015;81:169–183. doi: 10.1016/j.addr.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Malik AB. Nanoparticles squeezing across the blood-endothelial barrier via caveolae. Ther Deliv. 2013;4:131–133. doi: 10.4155/tde.12.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Tiruppathi C, Cho J, Minshall RD, Malik AB. Delivery of nanoparticle: complexed drugs across the vascular endothelial barrier via caveolae. IUBMB Life. 2011;63:659–667. doi: 10.1002/iub.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Tiruppathi C, Minshall RD, Malik AB. Size and dynamics of caveolae studied using nanoparticles in living endothelial cells. ACS Nano. 2009;3:4110–4116. doi: 10.1021/nn9012274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang CY, Xiong D, Sun Y, Zhao B, Lin WJ, Zhang LJ. Self-assembled micelles based on pH-sensitive PAE-g-MPEG-cholesterol block copolymer for anticancer drug delivery. Int J Nanomedicine. 2014;9:4923–4933. doi: 10.2147/IJN.S69493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng CJ, Tietjen GT, Saucier-Sawyer JK, Saltzman WM. A holistic approach to targeting disease with polymeric nanoparticles. Nat Rev Drug Discov. 2015;14:239–247. doi: 10.1038/nrd4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu CM, Fang RH, Wang KC, Luk BT, Thamphiwatana S, Dehaini D, Nguyen P, Angsantikul P, Wen CH, Kroll AV, Carpenter C, Ramesh M, Qu V, Patel SH, Zhu J, Shi W, Hofman FM, Chen TC, Gao W, Zhang K, Chien S, Zhang L. Nanoparticle biointerfacing by platelet membrane cloaking. Nature. 2015;526:118–121. doi: 10.1038/nature15373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev. 2016;106:148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21:R125–134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 12.Jan AT. Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update. Front Microbiol. 2017;8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 16.Chaput N, Thery C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol. 2011;33:419–440. doi: 10.1007/s00281-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 17.Gao J, Wang S, Wang Z. High yield, scalable and remotely drug-loaded neutrophil-derived extracellular vesicles (EVs) for anti-inflammation therapy. Biomaterials. 2017;135:62–73. doi: 10.1016/j.biomaterials.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Chu D, Wang Z. Cell membrane-formed nanovesicles for disease-targeted delivery. J Control Release. 2016;224:208–216. doi: 10.1016/j.jconrel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, Kim SH, Desiderio DM, Kim YK, Kim KP, Gho YS. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 22.Shockman GD, Barrett JF. Structure, function, and assembly of cell walls of gram-positive bacteria. Annu Rev Microbiol. 1983;37:501–527. doi: 10.1146/annurev.mi.37.100183.002441. [DOI] [PubMed] [Google Scholar]
- 23.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 24.Pore D, Chakrabarti MK. Outer membrane protein A (OmpA) from Shigella flexneri 2a: a promising subunit vaccine candidate. Vaccine. 2013;31:3644–3650. doi: 10.1016/j.vaccine.2013.05.100. [DOI] [PubMed] [Google Scholar]
- 25.Sham LT, Tsui HC, Land AD, Barendt SM, Winkler ME. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr Opin Microbiol. 2012;15:194–203. doi: 10.1016/j.mib.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grenier D, Mayrand D. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun. 1987;55:111–117. doi: 10.1128/iai.55.1.111-117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaron S, Kolling GL, Simon L, Matthews KR. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol. 2000;66:4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: Conceptually new antibiotics. Journal of Bacteriology. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoekstra D, van der Laan JW, de Leij L, Witholt B. Release of outer membrane fragments from normally growing Escherichia coli. Biochim Biophys Acta. 1976;455:889–899. doi: 10.1016/0005-2736(76)90058-4. [DOI] [PubMed] [Google Scholar]
- 30.Loeb MR, Kilner J. Release of a special fraction of the outer membrane from both growing and phage T4-infected Escherichia coli B. Biochim Biophys Acta. 1978;514:117–127. doi: 10.1016/0005-2736(78)90081-0. [DOI] [PubMed] [Google Scholar]
- 31.Mug-Opstelten D, Witholt B. Preferential release of new outer membrane fragments by exponentially growing Escherichia coli. Biochim Biophys Acta. 1978;508:287–295. doi: 10.1016/0005-2736(78)90331-0. [DOI] [PubMed] [Google Scholar]
- 32.Wensink J, Witholt B. Outer-Membrane Vesicles Released by Normally Growing Escherichia-Coli Contain Very Little Lipoprotein. Eur J Biochem. 1981;116:331–335. doi: 10.1111/j.1432-1033.1981.tb05338.x. [DOI] [PubMed] [Google Scholar]
- 33.Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem. 2000;275:12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32:1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 35.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricci V, Chiozzi V, Necchi V, Oldani A, Romano M, Solcia E, Ventura U. Free-soluble and outer membrane vesicle-associated VacA from Helicobacter pylori: Two forms of release, a different activity. Biochem Biophys Res Commun. 2005;337:173–178. doi: 10.1016/j.bbrc.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 38.Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11:3424–3429. doi: 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- 39.Ernst RK, Adams KN, Moskowitz SM, Kraig GM, Kawasaki K, Stead CM, Trent MS, Miller SI. The Pseudomonas aeruginosa lipid A deacylase: selection for expression and loss within the cystic fibrosis airway. J Bacteriol. 2006;188:191–201. doi: 10.1128/JB.188.1.191-201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelzer A, Polen T, Funken H, Rosenau F, Wilhelm S, Bott M, Jaeger KE. Subtilase SprP exerts pleiotropic effects in Pseudomonas aeruginosa. Microbiologyopen. 2014;3:89–103. doi: 10.1002/mbo3.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luckett JC, Darch O, Watters C, Abuoun M, Wright V, Paredes-Osses E, Ward J, Goto H, Heeb S, Pommier S, Rumbaugh KP, Camara M, Hardie KR. A novel virulence strategy for Pseudomonas aeruginosa mediated by an autotransporter with arginine-specific aminopeptidase activity. PLoS Pathog. 2012;8:e1002854. doi: 10.1371/journal.ppat.1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessler E, Safrin M, Gustin JK, Ohman DE. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. Journal of Biological Chemistry. 1998;273:30225–30231. doi: 10.1074/jbc.273.46.30225. [DOI] [PubMed] [Google Scholar]
- 43.Gonzales T, RobertBaudouy J. Bacterial aminopeptidases: Properties and functions. Fems Microbiol Rev. 1996;18:319–344. doi: 10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 44.Maredia R, Devineni N, Lentz P, Dallo SF, Yu J, Guentzel N, Chambers J, Arulanandam B, Haskins WE, Weitao T. Vesiculation from Pseudomonas aeruginosa under SOS. ScientificWorldJournal. 2012;2012:402919. doi: 10.1100/2012/402919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutierrez C, Gordia S, Bonnassie S. Characterization of the osmotically inducible gene osmE of Escherichia coli K-12. Mol Microbiol. 1995;16:553–563. doi: 10.1111/j.1365-2958.1995.tb02418.x. [DOI] [PubMed] [Google Scholar]
- 46.Furuta N, Takeuchi H, Amano A. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect Immun. 2009;77:4761–4770. doi: 10.1128/IAI.00841-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gankema H, Wensink J, Guinee PA, Jansen WH, Witholt B. Some characteristics of the outer membrane material released by growing enterotoxigenic Escherichia coli. Infect Immun. 1980;29:704–713. doi: 10.1128/iai.29.2.704-713.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauman SJ, Kuehn MJ. Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. Bmc Microbiology. 2009;9:26. doi: 10.1186/1471-2180-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23:4538–4549. doi: 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couto N, Schooling SR, Dutcher JR, Barber J. Proteome Profiles of Outer Membrane Vesicles and Extracellular Matrix of Pseudomonas aeruginosa Biofilms. J Proteome Res. 2015;14:4207–4222. doi: 10.1021/acs.jproteome.5b00312. [DOI] [PubMed] [Google Scholar]
- 52.Bauman SJ, Kuehn MJ. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes and Infection. 2006;8:2400–2408. doi: 10.1016/j.micinf.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manabe T, Kato M, Ueno T, Kawasaki K. Flagella proteins contribute to the production of outer membrane vesicles from Escherichia coli W3110. Biochem Biophys Res Commun. 2013;441:151–156. doi: 10.1016/j.bbrc.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 54.Toyofuku M, Roschitzki B, Riedel K, Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J Proteome Res. 2012;11:4906–4915. doi: 10.1021/pr300395j. [DOI] [PubMed] [Google Scholar]
- 55.Kadurugamuwa JL, Beveridge TJ. Virulence Factors Are Released from Pseudomonas-Aeruginosa in Association with Membrane-Vesicles during Normal Growth and Exposure to Gentamicin – a Novel Mechanism of Enzyme-Secretion. Journal of Bacteriology. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorward DW, Garon CF. DNA Is Packaged within Membrane-Derived Vesicles of Gram-Negative but Not Gram-Positive Bacteria. Appl Environ Microbiol. 1990;56:1960–1962. doi: 10.1128/aem.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris AE, Liggitt HD, Hawn TR, Skerrett SJ. Role of Toll-like receptor 5 in the innate immune response to acute P. aeruginosa pneumonia. Am J Physiol Lung Cell Mol Physiol. 2009;297:L1112–1119. doi: 10.1152/ajplung.00155.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deligianni E, Pattison S, Berrar D, Ternan NG, Haylock RW, Moore JE, Elborn SJ, Dooley JS. Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro. Bmc Microbiology. 2010;10:38. doi: 10.1186/1471-2180-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosen G, Naor R, Rahamim E, Yishai R, Sela MN. Proteases of Treponema denticola outer sheath and extracellular vesicles. Infect Immun. 1995;63:3973–3979. doi: 10.1128/iai.63.10.3973-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hynes SO, Keenan JI, Ferris JA, Annuk H, Moran AP. Lewis epitopes on outer membrane vesicles of relevance to Helicobacter pylori pathogenesis. Helicobacter. 2005;10:146–156. doi: 10.1111/j.1523-5378.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 61.Jang KS, Sweredoski MJ, Graham RL, Hess S, Clemons WM., Jr Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni. J Proteomics. 2014;98:90–98. doi: 10.1016/j.jprot.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elmi A, Watson E, Sandu P, Gundogdu O, Mills DC, Inglis NF, Manson E, Imrie L, Bajaj-Elliott M, Wren BW, Smith DG, Dorrell N. Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect Immun. 2012;80:4089–4098. doi: 10.1128/IAI.00161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bauman SJ, Kuehn MJ. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 2006;8:2400–2408. doi: 10.1016/j.micinf.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park KS, Lee J, Jang SC, Kim SR, Jang MH, Lotvall J, Kim YK, Gho YS. Pulmonary inflammation induced by bacteria-free outer membrane vesicles from Pseudomonas aeruginosa. Am J Respir Cell Mol Biol. 2013;49:637–645. doi: 10.1165/rcmb.2012-0370OC. [DOI] [PubMed] [Google Scholar]
- 65.Ismail S, Hampton MB, Keenan JI. Helicobacter pylori outer membrane vesicles modulate proliferation and interleukin-8 production by gastric epithelial cells. Infect Immun. 2003;71:5670–5675. doi: 10.1128/IAI.71.10.5670-5675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turkina MV, Olofsson A, Magnusson KE, Arnqvist A, Vikstrom E. Helicobacter pylori vesicles carrying CagA localize in the vicinity of cell-cell contacts and induce histone H1 binding to ATP in epithelial cells. FEMS Microbiol Lett. 2015;362 doi: 10.1093/femsle/fnv076. [DOI] [PubMed] [Google Scholar]
- 67.Lapinet JA, Scapini P, Calzetti F, Perez O, Cassatella MA. Gene expression and production of tumor necrosis factor alpha, interleukin-1beta (IL-1beta), IL-8, macrophage inflammatory protein 1alpha (MIP-1alpha), MIP-1beta, and gamma interferon-inducible protein 10 by human neutrophils stimulated with group B meningococcal outer membrane vesicles. Infect Immun. 2000;68:6917–6923. doi: 10.1128/iai.68.12.6917-6923.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jager J, Marwitz S, Tiefenau J, Rasch J, Shevchuk O, Kugler C, Goldmann T, Steinert M. Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect Immun. 2014;82:275–285. doi: 10.1128/IAI.00703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ko SH, Jeon JI, Kim YJ, Yoon HJ, Kim H, Kim N, Kim JS, Kim JM. Helicobacter pylori outer membrane vesicle proteins induce human eosinophil degranulation via a beta2 Integrin CD11/CD18- and ICAM-1-dependent mechanism. Mediators Inflamm. 2015;2015:301716. doi: 10.1155/2015/301716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alaniz RC, Deatherage BL, Lara JC, Cookson BT. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo (vol 179, pg 7692, 2007) J Immunol. 2008;180:3612–3612. doi: 10.4049/jimmunol.179.11.7692. [DOI] [PubMed] [Google Scholar]
- 71.Kim OY, Hong BS, Park KS, Yoon YJ, Choi SJ, Lee WH, Roh TY, Lotvall J, Kim YK, Gho YS. Immunization with Escherichia coli outer membrane vesicles protects bacteria-induced lethality via Th1 and Th17 cell responses. J Immunol. 2013;190:4092–4102. doi: 10.4049/jimmunol.1200742. [DOI] [PubMed] [Google Scholar]
- 72.Kim JH, Yoon YJ, Lee J, Choi EJ, Yi N, Park KS, Park J, Lotvall J, Kim YK, Gho YS. Outer Membrane Vesicles Derived from Escherichia coli Up-Regulate Expression of Endothelial Cell Adhesion Molecules In Vitro and In Vivo. Plos One. 2013;8 doi: 10.1371/journal.pone.0059276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharma A, Novak EK, Sojar HT, Swank RT, Kuramitsu HK, Genco RJ. Porphyromonas gingivalis platelet aggregation activity: outer membrane vesicles are potent activators of murine platelets. Oral Microbiol Immunol. 2000;15:393–396. doi: 10.1034/j.1399-302x.2000.150610.x. [DOI] [PubMed] [Google Scholar]
- 74.O’Donoghue EJ, Krachler AM. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol. 2016;18:1508–1517. doi: 10.1111/cmi.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol. 2015;15:375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 76.Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun. 2010;78:5054–5061. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Furuta N, Tsuda K, Omori H, Yoshimori T, Yoshimura F, Amano A. Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect Immun. 2009;77:4187–4196. doi: 10.1128/IAI.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pritsch M, Ben-Khaled N, Chaloupka M, Kobold S, Berens-Riha N, Peter A, Liegl G, Schubert S, Hoelscher M, Loscher T, Wieser A. Comparison of Intranasal Outer Membrane Vesicles with Cholera Toxin and Injected MF59C.1 as Adjuvants for Malaria Transmission Blocking Antigens AnAPN1 and Pfs48/45. J Immunol Res. 2016 doi: 10.1155/2016/3576028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orr N, Robin G, Cohen D, Arnon R, Lowell GH. Immunogenicity and efficacy of oral or intranasal Shigella flexneri 2a and Shigella sonnei proteosome-lipopolysaccharide vaccines in animal models. Infect Immun. 1993;61:2390–2395. doi: 10.1128/iai.61.6.2390-2395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Etchart N, Baaten B, Andersen SR, Hyland L, Wong SY, Hou S. Intranasal immunisation with inactivated RSV and bacterial adjuvants induces mucosal protection and abrogates eosinophilia upon challenge. Eur J Immunol. 2006;36:1136–1144. doi: 10.1002/eji.200535493. [DOI] [PubMed] [Google Scholar]
- 83.Park SB, Jang HB, Nho SW, Cha IS, Hikima J, Ohtani M, Aoki T, Jung TS. Outer membrane vesicles as a candidate vaccine against edwardsiellosis. PLoS One. 2011;6:e17629. doi: 10.1371/journal.pone.0017629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roberts R, Moreno G, Bottero D, Gaillard ME, Fingermann M, Graieb A, Rumbo M, Hozbor D. Outer membrane vesicles as acellular vaccine against pertussis. Vaccine. 2008;26:4639–4646. doi: 10.1016/j.vaccine.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 85.Bottero D, Gaillard ME, Errea A, Moreno G, Zurita E, Pianciola L, Rumbo M, Hozbor D. Outer membrane vesicles derived from Bordetella parapertussis as an acellular vaccine against Bordetella parapertussis and Bordetella pertussis infection. Vaccine. 2013;31:5262–5268. doi: 10.1016/j.vaccine.2013.08.059. [DOI] [PubMed] [Google Scholar]
- 86.Sierra GV, Campa HC, Varcacel NM, Garcia IL, Izquierdo PL, Sotolongo PF, Casanueva GV, Rico CO, Rodriguez CR, Terry MH. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann. 1991;14:195–207. discussion 208-110. [PubMed] [Google Scholar]
- 87.Holst J, Oster P, Arnold R, Tatley MV, Naess LM, Aaberge IS, Galloway Y, McNicholas A, O’Hallahan J, Rosenqvist E, Black S. Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum Vaccin Immunother. 2013;9:1241–1253. doi: 10.4161/hv.24129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Kleijn ED, de Groot R, Labadie J, Lafeber AB, van den Dobbelsteen G, van Alphen L, van Dijken H, Kuipers B, van Omme GW, Wala M, Juttmann R, Rumke HC. Immunogenicity and safety of a hexavalent meningococcal outer-membrane-vesicle vaccine in children of 2-3 and 7-8 years of age. Vaccine. 2000;18:1456–1466. doi: 10.1016/s0264-410x(99)00423-5. [DOI] [PubMed] [Google Scholar]
- 89.de Kleijn E, van Eijndhoven L, Vermont C, Kuipers B, van Dijken H, Rumke H, de Groot R, van Alphen L, van den Dobbelsteen G. Serum bactericidal activity and isotype distribution of antibodies in toddlers and schoolchildren after vaccination with RIVM hexavalent PorA vesicle vaccine. Vaccine. 2001;20:352–358. doi: 10.1016/s0264-410x(01)00371-1. [DOI] [PubMed] [Google Scholar]
- 90.Martin S, Sadler F, Borrow R, Dawson M, Fox A, Cartwright K. IgG antibody subclass responses determined by immunoblot in infants’ sera following vaccination with a meningococcal recombinant hexavalent PorA OMV vaccine. Vaccine. 2001;19:4404–4408. doi: 10.1016/s0264-410x(01)00198-0. [DOI] [PubMed] [Google Scholar]
- 91.O’Ryan M, Stoddard J, Toneatto D, Wassil J, Dull PM. A multi-component meningococcal serogroup B vaccine (4CMenB): the clinical development program. Drugs. 2014;74:15–30. doi: 10.1007/s40265-013-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dowling DJ, Sanders H, Cheng WK, Joshi S, Brightman S, Bergelson I, Pietrasanta C, van Haren SD, van Amsterdam S, Fernandez J, van den Dobbelsteen GP, Levy O. A Meningococcal Outer Membrane Vesicle Vaccine Incorporating Genetically Attenuated Endotoxin Dissociates Inflammation from Immunogenicity. Front Immunol. 2016;7:562. doi: 10.3389/fimmu.2016.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marsay L, Dold C, Green CA, Rollier CS, Norheim G, Sadarangani M, Shanyinde M, Brehony C, Thompson AJ, Sanders H, Chan H, Haworth K, Derrick JP, Feavers IM, Maiden MC, Pollard AJ. A novel meningococcal outer membrane vesicle vaccine with constitutive expression of FetA: A phase I clinical trial. J Infect. 2015;71:326–337. doi: 10.1016/j.jinf.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jang SC, Kim SR, Yoon YJ, Park KS, Kim JH, Lee J, Kim OY, Choi EJ, Kim DK, Choi DS, Kim YK, Park J, Di Vizio D, Gho YS. In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small. 2015;11:456–461. doi: 10.1002/smll.201401803. [DOI] [PubMed] [Google Scholar]
- 95.Gao W, Fang RH, Thamphiwatana S, Luk BT, Li J, Angsantikul P, Zhang Q, Hu CM, Zhang L. Modulating antibacterial immunity via bacterial membrane-coated nanoparticles. Nano Lett. 2015;15:1403–1409. doi: 10.1021/nl504798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ellis TN, Leiman SA, Kuehn MJ. Naturally Produced Outer Membrane Vesicles from Pseudomonas aeruginosa Elicit a Potent Innate Immune Response via Combined Sensing of Both Lipopolysaccharide and Protein Components. Infection and Immunity. 2010;78:3822–3831. doi: 10.1128/IAI.00433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van de Waterbeemd B, Streefland M, van Keulen L, van den IJssel J, de Haan A, Eppink MH, van der Pol LA. Identification and optimization of critical process parameters for the production of NOMV vaccine against Neisseria meningitidis. Vaccine. 2012;30:3683–3690. doi: 10.1016/j.vaccine.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 98.Kim SH, Kim KS, Lee SR, Kim E, Kim MS, Lee EY, Gho YS, Kim JW, Bishop RE, Chang KT. Structural modifications of outer membrane vesicles to refine them as vaccine delivery vehicles. Bba-Biomembranes. 2009;1788:2150–2159. doi: 10.1016/j.bbamem.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, Wolgemuth CW, Limberger RJ, Rowe N. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol. 2009;191:600–607. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Holst J, Martin D, Arnold R, Huergo CC, Oster P, O’Hallahan J, Rosenqvist E. Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine. 2009;27(Suppl 2):B3–12. doi: 10.1016/j.vaccine.2009.04.071. [DOI] [PubMed] [Google Scholar]
- 101.Chutkan H, Macdonald I, Manning A, Kuehn MJ. Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol Biol. 2013;966:259–272. doi: 10.1007/978-1-62703-245-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang W, Wang S, Yao Y, Xia Y, Yang X, Li K, Sun P, Liu C, Sun W, Bai H, Chu X, Li Y, Ma Y. Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci Rep. 2016;6:37242. doi: 10.1038/srep37242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rappazzo CG, Watkins HC, Guarino CM, Chau A, Lopez JL, DeLisa MP, Leifer CA, Whittaker GR, Putnam D. Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine. 2016;34:1252–1258. doi: 10.1016/j.vaccine.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 104.Chen DJ, Osterrieder N, Metzger SM, Buckles E, Doody AM, DeLisa MP, Putnam D. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc Natl Acad Sci U S A. 2010;107:3099–3104. doi: 10.1073/pnas.0805532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 106.El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, Van Rooijen N, Verbeek S, Fiers W, Saelens X. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol. 2011;186:1022–1031. doi: 10.4049/jimmunol.0902147. [DOI] [PubMed] [Google Scholar]
- 107.Salverda ML, Meinderts SM, Hamstra HJ, Wagemakers A, Hovius JW, van der Ark A, Stork M, van der Ley P. Surface display of a borrelial lipoprotein on meningococcal outer membrane vesicles. Vaccine. 2016;34:1025–1033. doi: 10.1016/j.vaccine.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 108.Kuipers K, Daleke-Schermerhorn MH, Jong WS, ten Hagen-Jongman CM, van Opzeeland F, Simonetti E, Luirink J, de Jonge MI. Salmonella outer membrane vesicles displaying high densities of pneumococcal antigen at the surface offer protection against colonization. Vaccine. 2015;33:2022–2029. doi: 10.1016/j.vaccine.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 109.Fantappie L, de Santis M, Chiarot E, Carboni F, Bensi G, Jousson O, Margarit I, Grandi G. Antibody-mediated immunity induced by engineered Escherichia coli OMVs carrying heterologous antigens in their lumen. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daleke-Schermerhorn MH, Felix T, Soprova Z, Ten Hagen-Jongman CM, Vikstrom D, Majlessi L, Beskers J, Follmann F, de Punder K, van der Wel NN, Baumgarten T, Pham TV, Piersma SR, Jimenez CR, van Ulsen P, de Gier JW, Leclerc C, Jong WS, Luirink J. Decoration of outer membrane vesicles with multiple antigens by using an autotransporter approach. Appl Environ Microbiol. 2014;80:5854–5865. doi: 10.1128/AEM.01941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Basto AP, Piedade J, Ramalho R, Alves S, Soares H, Cornelis P, Martins C, Leitao A. A new cloning system based on the OprI lipoprotein for the production of recombinant bacterial cell wall-derived immunogenic formulations. J Biotechnol. 2012;157:50–63. doi: 10.1016/j.jbiotec.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 112.Kesty NC, Kuehn MJ. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J Biol Chem. 2004;279:2069–2076. doi: 10.1074/jbc.M307628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim SH, Kim KS, Lee SR, Kim E, Kim MS, Lee EY, Gho YS, Kim JW, Bishop RE, Chang KT. Structural modifications of outer membrane vesicles to refine them as vaccine delivery vehicles. Biochim Biophys Acta. 2009;1788:2150–2159. doi: 10.1016/j.bbamem.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bartolini E, Ianni E, Frigimelica E, Petracca R, Galli G, Berlanda Scorza F, Norais N, Laera D, Giusti F, Pierleoni A, Donati M, Cevenini R, Finco O, Grandi G, Grifantini R. Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Muralinath M, Kuehn MJ, Roland KL, Curtiss R., 3rd Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect Immun. 2011;79:887–894. doi: 10.1128/IAI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kaminski RW, Oaks EV. Inactivated and subunit vaccines to prevent shigellosis. Expert Rev Vaccines. 2009;8:1693–1704. doi: 10.1586/erv.09.127. [DOI] [PubMed] [Google Scholar]
- 117.Bachmann MF, Jennings GT. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 118.Zepp F. Principles of vaccine design-Lessons from nature. Vaccine. 2010;28(Suppl 3):C14–24. doi: 10.1016/j.vaccine.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 119.Yoo JW, Irvine DJ, Discher DE, Mitragotri S. Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov. 2011;10:521–535. doi: 10.1038/nrd3499. [DOI] [PubMed] [Google Scholar]
- 120.Nieves W, Heang J, Asakrah S, Honer zu Bentrup K, Roy CJ, Morici LA. Immunospecific responses to bacterial elongation factor Tu during Burkholderia infection and immunization. PLoS One. 2010;5:e14361. doi: 10.1371/journal.pone.0014361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alves NJ, Turner KB, Medintz IL, Walper SA. Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles. Sci Rep. 2016;6:24866. doi: 10.1038/srep24866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Alves NJ, Turner KB, Daniele MA, Oh E, Medintz IL, Walper SA. Bacterial Nanobioreactors–Directing Enzyme Packaging into Bacterial Outer Membrane Vesicles. ACS Appl Mater Interfaces. 2015;7:24963–24972. doi: 10.1021/acsami.5b08811. [DOI] [PubMed] [Google Scholar]
- 123.Nolte-’t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113:1977–1981. doi: 10.1182/blood-2008-08-174094. [DOI] [PubMed] [Google Scholar]
- 124.Daniele L, Sapino A. Anti-HER2 treatment and breast cancer: state of the art, recent patents, and new strategies. Recent Pat Anticancer Drug Discov. 2009;4:9–18. doi: 10.2174/157489209787002489. [DOI] [PubMed] [Google Scholar]
- 125.Gujrati V, Kim S, Kim SH, Min JJ, Choy HE, Kim SC, Jon S. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8:1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- 126.O’Neal DP, Hirsch LR, Halas NJ, Payne JD, West JL. Photo-thermal tumor ablation in mice using near infrared-absorbing nanoparticles. Cancer Lett. 2004;209:171–176. doi: 10.1016/j.canlet.2004.02.004. [DOI] [PubMed] [Google Scholar]


