Abstract
Current re-emergence of Nipah virus (NiV) in India caused 11 deaths so far and many patients were kept in quarantine. A thorough study of previous outbreaks occurred in Malaysia, Bangladesh and India represents cases with high rate of fatality due to acute encephalitis. Our work involves genome analysis of NiV for prediction of miRNAs and their targeted genes in human in order to understand encephalitis origin. Ab-intio program-VMir was used for initial screening of genome, obtained nine pre-miRNAs was analyzed by ViralMir to check for any pseudo pre-miRNAs. Eighteen functional mature miRNAs were extracted from pre-miRNAs by using Mature-Bayes tool, which targets 669 genes in human genome as retrieved by miRDB. Gene ontology terms by PANTHER provide important pathways in which target genes were involved like Axon guidance, T cell activation, and nicotinic acetylcholine receptor signaling. Significant outcome was obtained after NCBI Gene and OMIM database mining and literature search for predicted target genes. TLR3, TJP1, NOTCH2, FHL1, and GRIA3 target genes obtained showed their involvement in host defense, blood brain barrier, neurogenesis, mental retardation and encephalitis. To conclude, we predicted significant genes in human that can be inhibited by miRNAs of NiV and results in etiology of encephalitis.
Key Words: Nipah, miRNA, Encephalitis, Target genes
INTRODUCTION
Nipah virus (NiV) belongs to family Paramyxoviridae and genus Henipavirus was first isolated from a patient in Sungai Nipah (a village in Malaysia) in 1998 [1]. Fruit bats of the genus Pteropus is the natural host of NiV [2]. Being zoonotic in origin, the virus can transmitted from bat to pigs, bat to human, pigs to human, human to human and horse to human. The mode of transmission can be direct contact with the pigs (who consumed infected fruits in farm) and their infectious secretions (respiratory droplets and throat or nasal secretions). The consumption of contaminated date palm or products derived from it also results in spread of virus in human population [3-5]. The first outbreak of the NiV occurred among pig farmers in Malaysia in September 1998, initially the outbreak was suspected to link with Japanese encephalitis [6]. Geographical distribution of NiV is not limited to Malaysia only as evident from outbreaks in other Southeast Asian countries such as Singapore [7], India [8] and Bangladesh [9]. Malaysian outbreak occurred during 1998-99 was the largest claiming 105 deaths with 40 % fatality rate, mostly pig farmers. Bangladesh has greatest number and frequency of outbreaks that occurred from 2001 to 2013 claiming 187 deaths with 80 % fatality rate [10-12]. The first outbreak in India occurred in Siliguri, West Bengal in 2001 claiming 45 deaths with fatality rate of 68 % [13], followed by a second outbreak in 2007 in Nadia district, West Bengal, which borders Bangladesh and resulted in 5 deaths with 100 % fatality rate [14].
Infected patients showed clinical signs similar to flu such as fever, headache, dizziness, myalgia and vomiting. These initial symptoms may be followed by acute encephalitis that is characterized by drowsiness, disorientation, signs of brainstem dysfunction, convulsions, coma and other signs [15-17]. In case of acute encephalitis virus was isolated from CNS, lungs, kidneys, spleen, lymph nodes, and endothelial tissue of the smaller blood vessels [18]. Usually the mean incubation period for virus infection varies from 6-14 days [19-20].
Determination of virus infection can be done by virus isolation, nucleic acid amplification and serological testing but being a biosafety level-4 (BSL-4) pathogen proper physical containment and security measures must be adopted to limit its transmission [21]. Various laboratory techniques used for its diagnosis are RT-PCR, ELISA and in some cases electron microscopy or immunoelectron microscopy can also be used [22, 23]. Currently, there is no approved vaccine available for the treatment of the NiV but antiviral drug ribavirin had some success in reducing the mortality occurred due to acute encephalitis [24, 25]. The recent and third outbreak in India in May 2018 caused 11 deaths and suspected to originate from consuming bat contaminated water. Even the WHO listed NiV in its blueprint list of priority diseases during annual review meeting occurred in Feb-2018. The list comprised diseases that have urgent needs of accelerated research and for which there is no efficacious drugs/vaccine is available.
MicroRNAs (miRNAs) are small non-coding RNAs of size ~21 nucleotides that played role in post-transcriptional gene regulation by binding to complementary sites on mRNA and results either in inhibition of translation or complete cleavage of mRNA [26-27]. In addition to animals, plants and fungi [28-29], miRNAs are also encoded by viruses and involved in penetrating the host defense mechanism, cell differentiation, apoptosis and cell proliferation [30]. Viral miRNAs target specific genes in host that were involved in important pathways (cell growth, axon guidance and cell differentiation), thus helps virus particle to evade host immune system and their continuous proliferation [31-33]. Mostly DNA viruses encode miRNAs but RNA viruses also have potential of coding miRNAs to silent the host target genes [34, 35]. Experimental methods of miRNA identification is relied on expression in specific cell type and time and needs cloning from virus infected cell, therefore the computational approaches are frequently used for prediction of miRNAs and their target genes [36-37].
Many members of RNA virus families such as hepatitis A virus (HAV) [38], Dengue virus (DENV) [39], ZIKA Virus (ZIKV) [40], Ebola virus (EBOV) [41], Japanese Encephalitis virus (JEV) [42] and Kyasanur forest disease virus (KFDV) [43] were predicted to have encoded miRNAs. Recent outbreak in India, concern raised by WHO, no approved vaccine for the NiV and evidences of viral miRNAs targeting host genes encourage us to analyse the genome sequence data of the NiV for possible prediction of miRNAs and their target genes in human.
MATERIALS AND METHODS
Retrieval of NiV Genome sequence data: Complete genome sequence of Nipah virus (Accession number NC_002728.1) was retrieved from NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome/). Genome is single stranded RNA molecule with linear topology and contains 18246 nucleotide base pair.
Precursor miRNAs (pre-miRNAs) identification: An ab-intio pre-miRNAs identification tool, VMir [44] is used for finding self complementary hair pin loop structures in NiV genome. VMir package contains two individual programs: VMir Analyzer and VMir Viewer. Analyzer was used for analyzing sequence for pre-miRNA identification whereas Viewer used for viewing and filtering out results of analyzer. pre-miRNAs were identified by keeping the parameter to default values for analyzer (window count: 500, conformation: linear, orientation: both) and stringent filtering was done by setting min. hairpin size: 70, min. score: 115 and min. window count: 35 in VMir viewer as previously described [45] to select high score candidate pre-miRNAs for further evaluation.
Identification of potential pre-miRNAs : Filtered pre-miRNAs obtained through VMir were subjected to ViralmiR (http://csb.cse.yzu.edu.tw/viralmir/) [46], an online server dedicated to differentiate between potential viral pre-miRNAs from other pseudo pre-miRNAs. It is based on SVM (Support Vector Machine) model and is trained on sequence and structural features of experimentally validated pre-miRNAs data set.
Energy calculation and Secondary structure prediction: The Mfold [47] web server (http://unafold.rna.albany.edu/?q=mfold) with default parameters was used to predict the secondary structure and minimum free energy (MFE) of pre-miRNAs.
Identification of mature miRNAs from pre-miRNAs: Mature miRNAs were identified from pre-miRNAs sequences using Mature Bayes (http://mirna.imbb.forth.gr/MatureBayes. html) [48], an online tool that uses Naive Bayes Classifier (NBC) taking into account sequence as well as structural information of experimental predicted miRNA precursors. All the potential pre-miRNAs identified by ViralmiR was used for analysis.
Prediction of Target genes in human: miRDB (http://mirdb.org/) [49], a web based server was used for prediction of target genes in human. Using custom target prediction, all the mature miRNAs were screened to identify target genes. miRDB uses the seeding approach and scan the 3’ UTR (untranslated regions) of human’s gene for possible hybridization with miRNAs sequence.
GO (Gene Ontology) analysis: Gene ontology analysis of the retrieved target genes was performed using PANTHER (Protein Analysis through Evolutionary Relationships) (http:// www.pantherdb.org) [50] to gain insight in to molecular functional, biological process and cellular component of the target genes products [51]. Gene IDs of target genes were used for this analysis to find GO terms related to gene products.
Screening of target genes and literature data mining: NCBI Gene (https://www.ncbi. nlm.nih.gov/gene) and (https://www.omim.org/) OMIM [52] (Online Mendelian Inheritance in Man) databases was searched for encephalitis disease genes in human and screening was done manually by crosschecking the predicted target genes with database genes. Literature search was performed for the screened target genes to support the evidence.
RESULTS
Computational miRNAs prediction depends on two approaches: ab-intio based and homology based. Evolutionary conservation tracing is the main motive of homology based approach and thus having limitation in finding novel miRNAs. But ab-intio based approach which search for hair-pin loop structure topology in genomic sequence is more of significance in locating novel pre-miRNAs and hence the derivative miRNAs because pre-miRNAs tends to form hair-pin loop structures during their biogenesis [53-55].
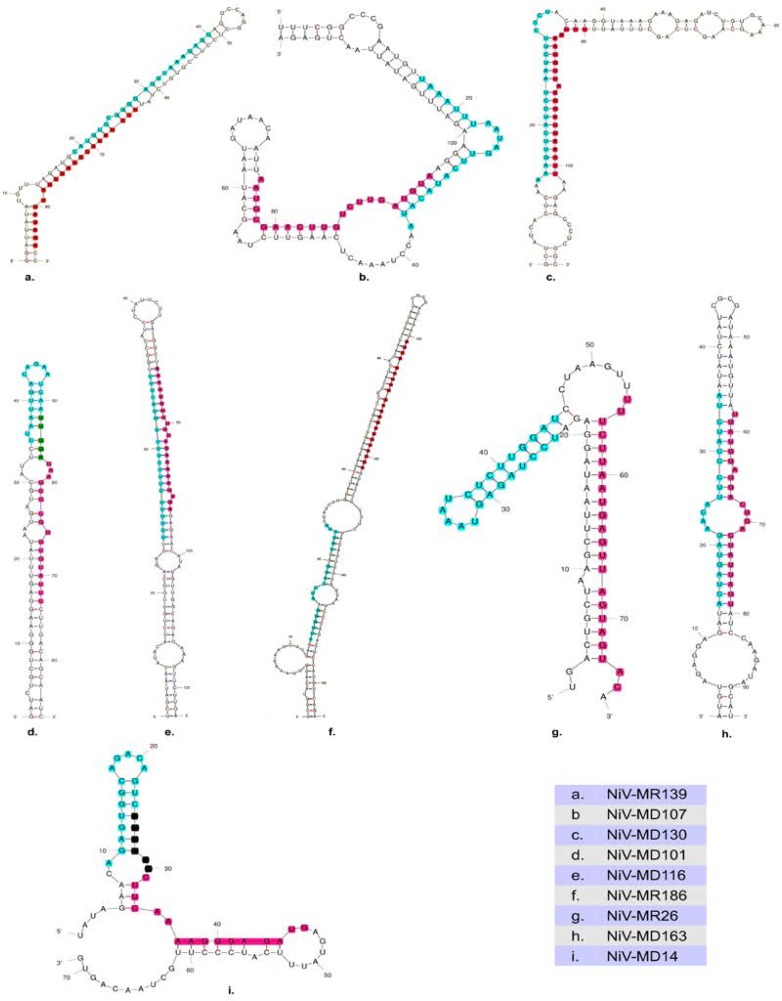
In our study we also used ab-intio based program, VMir for scanning the NiV genome for possible pre-miRNAs prediction. Genome sequence was analysed on both strands (Direct/Reverse) during pre-miRNAs prediction. We got nine pre-miRNAs (Fig. 1) with high score at stringent parameters (min. hairpin size: 70, min. score: 115 and min. window count: 35 in VMir viewer) as previously described [45] to filter out imprecise candidate pre-miRNAs. Six pre-miRNAs predicted were on direct strand and three were on reverse strand. The length of pre-miRNAs was in the range 76-165 nucleotide. The genomic position, Vmir score and rank were shown in Table 1.
Figure 1.
Predicted pre-miRNAs structures. Hairpin loop structures of NiV predicted by m fold, 5' arm of mature miRNAs indicated by cyan, 3' arm by pink whereas dark green show overlapping region
Table 1.
pre-miRNAs predicted by VMir
| Predicted pre-miRNA | Rank | Orientation | Length (nt) | Position on Genome | VMir Score |
|---|---|---|---|---|---|
| NiV-MR139 | 1 | Reverse | 87 | 12613-12699 | 212.3 |
| NiV-MD107 | 2 | Direct | 119 | 11042-11160 | 183.3 |
| NiV-MD130 | 3 | Direct | 114 | 13920-14033 | 180.8 |
| NiV-MD101 | 4 | Direct | 86 | 10445-10530 | 180.1 |
| NiV-MD116 | 5 | Direct | 124 | 12451-12574 | 171.9 |
| NiV-MR186 | 6 | Reverse | 165 | 17226-17390 | 171.6 |
| NiV-MR26 | 7 | Reverse | 76 | 2273-2348 | 155.3 |
| NiV-MD163 | 8 | Direct | 93 | 17254-17346 | 152.6 |
| NiV-MD14 | 9 | Direct | 88 | 1417-1504 | 130.4 |
False-positive pre-miRNAs prediction is common limitation to all ab-intio prediction programs because of selection of the pseudo hair pin loops structures [56, 57], therefore to validate and find reliable results we examined all nine predicted pre-miRNAs by SVM based virus specific tool, ViralmiR. All nine pre-miRNAs were found to be potential in yielding miRNAs. Minimum free energy (MFE) calculation of pre-miRNAs sequence during folding is one of the features that confer stability values to it [58]. We used Mfold web server for MFE calculation (Table 2) and secondary structure prediction of pre-miRNAs.
Table 2.
Potential pre-miRNAs validated by ViralMir and Minimum Free Energy (MFE) calculated by Mfold
| Predicted pre-mi-RNA | Potential/ Non-potential | Minimum Free Energy (MFE) (-∆G. kcal/mol) |
|---|---|---|
| NiV-MR139 | Potential | -30.70 |
| NiV-MD107 | Potential | -24.20 |
| NiV-MD130 | Potential | -34.40 |
| NiV-MD101 | Potential | -27.50 |
| NiV-MD116 | Potential | -35.30 |
| NiV-MR186 | Potential | -48.10 |
| NiV-MR26 | Potential | -25.50 |
| NiV-MD163 | Potential | -25.50 |
| NiV-MD14 | Potential | -27.80 |
After initial identification and validation, pre-miRNAs sequences were subjected to Mature Bayes for retrieving the small mature miRNAs. Large pre-miRNAs sequences were cleaved to short mature miRNAs of 22 nucleotide length. We got eighteen mature miRNAs from nine pre-miRNAs sequences on 5’ and 3’ stem location as shown in Table 3. Because one or both strands can serve as mature miRNA molecule depending on the assembly of RISC complex [59], we kept both for further analysis.
Table 3.
Mature miRNAs sequences predicted by MatureBayes
| Mature miRNAs | Length (nt) | Location | Mature miRNAs sequence |
|---|---|---|---|
| NiV-MR139 5P | 22 | 5’ | CAUGUGCGGGGAGGUAAAGAGG |
| NiV-MR139 3P | 22 | 3’ | CCCUAUACCCAUUUAUUAUAGU |
| NiV-MD107 5P | 22 | 5’ | UCAAGUUCUAAGCAUAAUGAUA |
| NiV-MD107 3P | 22 | 3’ | AAUGCGAACUUGUCUUGAUGUA |
| NiV-MD130 5P | 22 | 5’ | AAAGUUCAUCCUAAUCUUCCCU |
| NiV-MD130 3P | 22 | 3’ | UUGAAAGGUUAAGGAUGAACUU |
| NiV-MD101 5P | 22 | 5’ | CUAAUUGACAGAAUCAAUUGGA |
| NiV-MD101 3P | 22 | 3’ | UUGGAUAAGCGCGGGUGUAUUC |
| NiV-MD116 5P | 22 | 5’ | AUCCUAUUCUUGAGGCUAAAGU |
| NiV-MD116 3P | 22 | 3’ | AAAGUUGCUGCAGAAAAAGUGA |
| NiV-MR186 5P | 22 | 5’ | CUUGAGAUUGGGAAUCCAGGGG |
| NiV-MR186 3P | 22 | 3’ | AUUAGAUGGGAAUGUUCUACUA |
| NiV-MR26 5P | 22 | 5’ | AUCCUAGAGUAAAUCUCUUGGA |
| NiV-MR26 3P | 22 | 3’ | UUUCUUAAUGAGUUAGUAGUAC |
| NiV-MD163 5P | 22 | 5’ | ACUAGUAGAACAUUCCCAUCUA |
| NiV-MD163 3P | 22 | 3’ | AAUGUUAUGAUGGAGGACGGAC |
| NiV-MD14 5P | 22 | 5’ | AGAUGAGUAUUUCAUCCCUUGC |
| NiV-MD14 3P | 22 | 3’ | AUCCCUUGCUAACAGUGUGCCG |
miRNAs exerts their effects by targeting the mRNA of protein coding gene of the cell, thus predicting the target of miRNAs directly revealed their function in the cell [60]. Computational predictions of target depend on the Watson-Crick base pairing between miRNA and mRNA molecule and mostly used seed pairing approach [61, 62]. Target prediction by miRDB for all mature miRNAs gave 769 target genes (supplementary Table) in human genome. The server uses the MirTarget algorithm, which is based on 7-mer seeding approach and custom predict miRNAs targets in human gene’s 3’ UTRs. We selected target genes with miRDB score >80 because a predicted target with prediction score >80 is most likely to be real and not required any other supporting evidence [49].
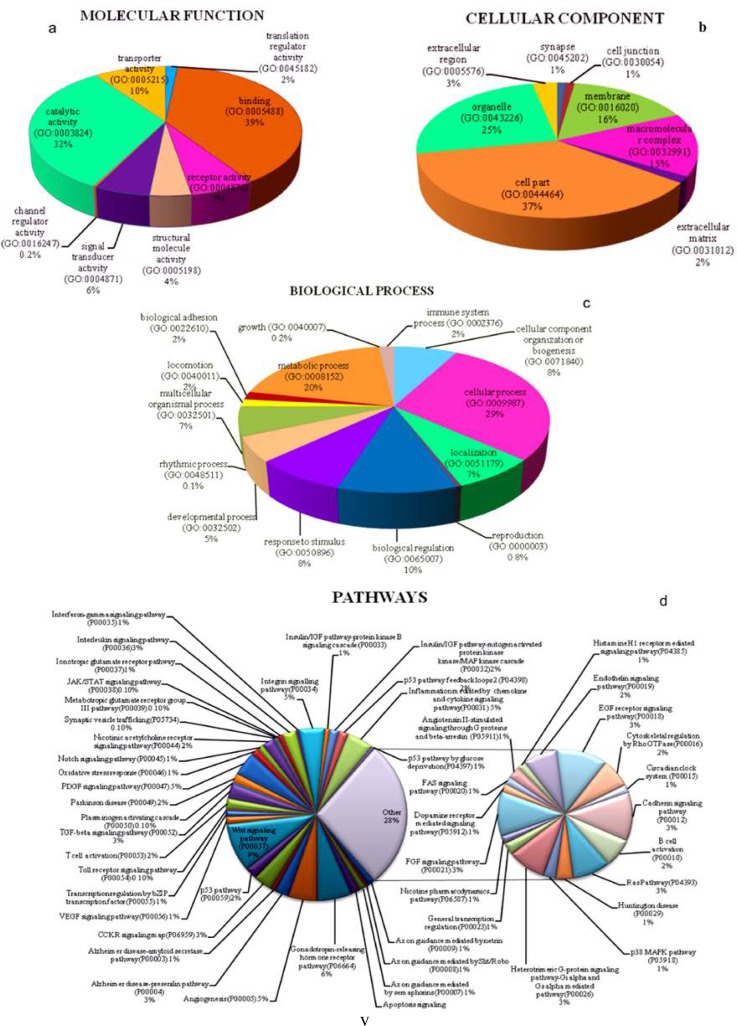
Gene ontology analysis of the target genes by PANTHER revealed their involvement in different clusters of molecular functions, cellular component, biological process and pathway. The clustering approach proved to be significant in determination target gene molecular function (Fig. 2a), cellular component (Fig. 2b), biological process (Fig. 2c) and pathway (Fig. 2d) of large data set at once. In molecular function cluster target genes products were depicted to play role in translation regulator activity (GO:0045182), signal transducer activity (GO:0004871) binding (GO:0005488) and receptor activity (GO:0004872), which shows that inhibition of these gene product might lead to abnormal state in body. Biological process cluster classification have proteins which are involved in immune system response (GO:0002376), response to stimulus (GO:0050896), biological adhesion (GO:0022610) and localization (GO:0051179) etc. Biological processes mentioned are significant for defense against viral infection. Pathway clusters analysis showed pathways that can mediate the disease in human. Interference in pathways like Axon guidance mediated by netrin (P00009), Axon guidance mediated by semaphorins (P00007), T cell activation (P00053), Nicotinic acetylcholine receptor signaling pathway (P00044), Alzheimer disease-amyloid secretase pathway (P00003) and Parkinson disease (P00049) can lead to acute encephalitis. Cellular component classification also suggest the target gene products are part of synapse (GO:0045202), cell junction (GO:0030054), extracellular region (GO:0005576) and organelle (GO:0043226) etc. Significant results obtained through GO analysis prompt us to uncover the encephalitis disease genes among the 769 target genes. Manual screening of target genes with NCBI Gene database and OMIM encephalitis disease genes identified five target genes. Literature mining results (Table 4.) of five target genes discovered their role in normal brain functioning and disorders.
Figure 2.
Go Analysis of target genes: Target genes were found to be involved in molecular function (2a), cellular component (2b), biological process (2c) and pathways (2d)
Table 4.
Screened target genes role and associated disorders
| Mature mi-RNA | Target Gene | Description | Role/Disease | PMID |
|---|---|---|---|---|
| NiV-MR26 5P | TLR3 | Toll Like Receptor 3 | Host defense against viruses | 16877304, 26298326, 15558055 |
| NiV-MR186 5P | TJP1 | Tight Junction Protein 1 | Blood-brain barrier/ Encephalitis | 10595922, 24198423 |
| NiV-MD130 3P | NOTCH2 | Neurogenic Locus Notch Homolog Protein 2 | Neurogenesis | 9720489 |
| NiV-MD130 5P | FHL1 | Four And A Half LIM Domains 1 | Muscular dystrophy | 27765816 |
| NiV-MD116 5P | GRIA3 | Glutamate Ionotropic Receptor AMPA Type Subunit 3 | Neurophysiologic processes/ Rasmussen encephalitis | 16713244, 19338055 |
TLR3 (Toll Like Receptor 3) expressed in human neurons during innate immune response confirmed its role in the host defense against viruses. It works by recognizing the molecular patterns specific to microorganisms [63]. Degradation of TJP1 (Tight Junction Protein 1) during Japanese encephalitis virus and human Immunodeficiency virus-1 infection caused breached in blood-brain barrier (BBB) and hence results in neurological symptoms leads to encephalitis [64, 65]. NOTCH2 (Neurogenic Locus Notch Homolog Protein 2) participates in neurogenesis and is key protein in adult brain, impairment in signaling of NOTCH2 contribute to neurological disease manifestation [66, 67]. The expression evidences of FHL1 (Four And A Half LIM Domains 1) in brain tissue and interaction with PLEKHG2 (Pleckstrin Homology And RhoGEF Domain Containing G2) revealed its role in brain cells [68]. FHL1 mutation lead to muscular dystrophy is main disorder related to this target gene [69]. Lastly, GRIA3 (Glutamate Ionotropic Receptor AMPA Type Subunit 3) are the predominant excitatory neurotransmitter receptors in the mammalian brain and are activated in a variety of normal neurophysiologic processes. Diseases associated with GRIA3 include mental retardation and Rasmussen encephalitis [70, 71].
DISCUSSION
Current outbreak of NiV in India posed serious problem and its transmission to other neighboring countries was suspected. Unavailability of an approved vaccine makes it more fatal due to acute encephalitis it caused in infected patients. Previous studies on NiV dominantly related to its entry mechanism in to host [72, 73] and lack of knowledge about disease manifestation prevent us to tackle this deadly virus. Most of the viruses that caused fatal outbreaks target the brain cells and lead to impairment of normal functioning [74].
Role of human miRNAs interaction with NiV during viral entry was identified [75] but prominent evidences from other viruses [76-78] miRNAs screening and targeting of their host genes were also documented previously and their possible relation to disease mechanism cannot be ruled out. Here in this work, we used computational prediction methods to predict miRNAs in NiV genome and their targeted genes in human. We successfully found eighteen miRNAs from nine pre-miRNAs obtained by genome analysis of NiV. By analyzing the gene ontology terms and screened target genes, we found target gene TLR3 involved in host defense against viruses whereas TJP1 and GRIA3 silencing can lead to encephalitis. NOTCH2 and FHL1 expression are needed for normal neurogenesis and muscle functioning respectively. Pathways obtained through GO analysis also supported the results. Five different miRNAs (NiV-MR26 5P, NiV-MR186 5P, NiV-MD130 3P, NiV-MD130 5P, and NiV-MD116 5P) predicted to code by virus genome can down regulate these genes and results in disease manifestations.
In summary, this is the first paper that predicted miRNAs in NiV genome and their target genes in human. Target genes and pathway analysis gave insight in to underlying disease genes, the predicted miRNAs mimics can be synthesized to check their hybridization with proposed target genes and can become targets for antiviral therapy.
Acknowledgement:
The authors are grateful to University Grants Commission (UGC) for their support.
Conflict of Interest:
The authors declare that they have no conflict of interest.
References
- 1.Lo Presti A, Cella E , Giovanetti M , Lai A , Angeletti S , Zehender G , Ciccozzi M Origin and evolution of Nipah virus. J Med Viro. 2016;88:380–388. doi: 10.1002/jmv.24345. [DOI] [PubMed] [Google Scholar]
- 2.Allocati N, Petrucci AG, Di Giovanni P, Masulli M, Di Ilio C, De Laurenzi V. Bat–man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discov. 2016;2:16048. doi: 10.1038/cddiscovery.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton BA. Nipah virus: transmission of a zoonotic paramyxovirus. Curr Opin Virol. 2017;22:97–104. doi: 10.1016/j.coviro.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Luby SP, Rahman M, Hossain MJ, Blum LS, Husain MM, Gurley E, Khan R, Ahmed BN, Rahman S, Nahar N, Kenah E, Comer JA, Ksiazek TG. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurley ES, Montgomery JM, Hossain JM, Bell M, Azad AK, Islam MR, Molla MAR, Carroll DS, Thomas G, Paul KA, Rota , Lowe L, Comer JA, Rollin P, Czub M, Grolla A, Feldmann H, Stephen P, Jennifer LL, Breiman WRF. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13:1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field H, Young P, Yob JM, Mills J, Hall L, Mackenzie J. The natural history of Hendra and Nipah viruses. Microbes Infect. 2001;3:307–314. doi: 10.1016/s1286-4579(01)01384-3. [DOI] [PubMed] [Google Scholar]
- 7.Chew MH, Arguin PM, Shay DK, Goh KT, Rollin PE, Shieh WJ, Zaki SR, Rota PA, Ling AE, Ksiazek TG, Chew SK, Anderson LJ. Risk factors for Nipah virus infection among abattoir workers in Singapore. J Infect Dis. 2000;181:1760–1763. doi: 10.1086/315443. [DOI] [PubMed] [Google Scholar]
- 8.Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, Ksiazek TG, Mishra CA. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis. 2006;12:235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu VP, Hossain MJ, Parashar UD, Ali MM, Ksiazek TG, Kuzmin I, Niezgoda M, Rupprecht C, Bresee J, Breiman RF. Nipah virus encephalitis rteemergence, Bangladesh. Emerg Infect Dis. 2004;10:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giangaspero M. Nipah virus. Trop Med Surg. 2013;01:2910. [Google Scholar]
- 11.Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001-2007. Emerg Infect Dis. 2009;15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan MZ, Sazzad HMS, Luby SP, Sturm-Ramirez K, Bhuiyan MU, Rahman MZ, Islam MM, Stroher U, Sultana S, Kafi MAH, Daszak P, Rahman M, Gurley ES. Nipah virus contamination of hospital surfaces during outbreaks, Bangladesh, 2013-2014. Emerg Infect Dis. 2018;24:15–21. doi: 10.3201/eid2401.161758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harit AK, Ichhpujani RL, Gupta S, Gill KS, Lal S, Ganguly NK, Agarwal SP. Nipah/Hendra virus outbreak in Siliguri, West Bengal, India in 2001. Indian J Med Res. 2006;123:553–560. [PubMed] [Google Scholar]
- 14.Arankalle VA, Bandyopadhyay BT, Ramdasi AY, Jadi R, Patil DR, Rahman M, Majumdar M, Banerjee PS, Hati AK, Goswami RP, Neogi DK, Mishra AC. Genomic characterization of Nipah virus, West Bengal, India. Emerg Infect Dis. 2011;17:907–909. doi: 10.3201/eid1705.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherrini BA, Chong TT. Nipah encephalitis-an update. Med J Malaysia. 2014;69 Suppl A:103–111. [PubMed] [Google Scholar]
- 16.Ng BY, Lim CC, Yeoh A, Lee WL. Neuropsychiatric sequelae of Nipah virus encephalitis. J Neuropsychiatry Clin Neurosci. 2004;16:500–504. doi: 10.1176/jnp.16.4.500. [DOI] [PubMed] [Google Scholar]
- 17.Goh KJ, Tan CT, Chew NK, Tan PS, Kamarulzaman A, Sarji SA, Wong KT, Abdullah BJ, Chua KB, Lam SK. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342:1229–1235. doi: 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- 18.Wong KT, Shieh WJ, Kumar S, Norain K, Abdullah W, Guarner J, Goldsmith CS, Chua KB, Lam SK, Tan CT, Goh KJ, Chong HT, Jusoh R, Rollin PE, Ksiazek TG, Zaki SR. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol. 2002;161:2153–2167. doi: 10.1016/S0002-9440(10)64493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan CT, Chua KB. Nipah virus encephalitis. Curr Infect Dis Rep. 2008;10:315–320. doi: 10.1007/s11908-008-0051-6. [DOI] [PubMed] [Google Scholar]
- 20.Hossain MJ, Gurley ES, Montgomery JM, Bell M, Carroll DS, Hsu VP, Formenty P, Croisier A, Bertherat E, Faiz MA, Azad AK, Islam R, Molla MA, Ksiazek TG, Rota PA, Comer JA, Rollin PE, Luby SP, Breiman RF. Clinical presentation of nipah virus infection in Bangladesh. Clin Infect Dis. 2008;46:977–984. doi: 10.1086/529147. [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni DD, Tosh C, Venkatesh G, Senthil Kumar D. Nipah virus infection: current scenario. Indian J Virol. 2013;24:398–408. doi: 10.1007/s13337-013-0171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellini WJ, Harcourt BH, Bowden N, Rota PA. Nipah virus: an emergent paramyxovirus causing severe encephalitis in humans. J Neurovirol. 2005;11:481–487. doi: 10.1080/13550280500187435. [DOI] [PubMed] [Google Scholar]
- 23.Daniels P, Ksiazek T, Eaton BT. Laboratory diagnosis of Nipah and Hendra virus infections. Microbes Infect. 2001;3:289–295. doi: 10.1016/s1286-4579(01)01382-x. [DOI] [PubMed] [Google Scholar]
- 24.Epstein JH, Field HE, Luby S, Pulliam JR, Daszak P. Nipah virus: impact, origins, and causes of emergence. Curr Infect Dis Rep. 2006;8:59–65. doi: 10.1007/s11908-006-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong HT, Kamarulzaman A, Tan CT, Goh KJ, Thayaparan T, Kunjapan SR, Chew NK, Chua KB, Lam SK. Treatment of acute Nipah encephalitis with ribavirin. Ann Neurol. 2001;49:810–813. doi: 10.1002/ana.1062. [DOI] [PubMed] [Google Scholar]
- 26.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 27.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 28.Lukasik A, Zielenkiewicz P. Plant microRNAs-novel players in natural medicine. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran Y, Agron M, Praher D, Technau U. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol. 2017;1:27. doi: 10.1038/s41559-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012;8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carl JW, Trgovcich J, Hannenhalli S. Widespread evidence of viral miRNAs targeting host pathways. BMC Bioinformatics. 2013;14(Suppl 2) doi: 10.1186/1471-2105-14-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38 Suppl:S25–30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 33.Cullen BR. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol. 2013;14:205–210. doi: 10.1038/ni.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruscella P, Bottini S, Baudesson C, Pawlotsky JM, Feray C, Trabucchi M. Viruses and miRNAs: More friends than foes. Front Microbiol. 2017;8:824. doi: 10.3389/fmicb.2017.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 37.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi J, Duan Z, Sun J, Wu M, Wang B, Zhang J, Wang H, Hu N, Hu Y. Identification and validation of a novel microRNA-like molecule derived from a cytoplasmic RNA virus antigenome by bioinformatics and experimental approaches. Virol J. 2014;11 doi: 10.1186/1743-422X-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ospina-Bedoya M, Campillo-Pedroza N, Franco-Salazar JP, Gallego-Gomez JC. Computa-tional identification of dengue virus microRNA-like structures and their cellular targets. Bioinform Biol Insights. 2014;8:169–176. doi: 10.4137/BBI.S13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pylro VS, Oliveira FS, Morais DK, Cuadros-Orellana S, Pais FS, Medeiros JD, Geraldo JA, Gilbert J, Volpini AC, Fernandes GR. ZIKV - CDB: A collaborative database to guide research linking SncRNAs and ZIKA virus disease symptoms. PLoS Neg Trop Dis. 2016;10:e0004817. doi: 10.1371/journal.pntd.0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng Y, Wang Y, Zhang X, Liu W, Fan H, Yao H, Lin B, Zhu P, Yuan W, Tong Y, Cao W. Systematic Genome-wide Screening and Prediction of microRNAs in EBOV During the 2014 Ebolavirus Outbreak. Sci Rep. 2015;5 doi: 10.1038/srep09912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxena VL, Dwivedi A. In silico identification of miRNAs and their target prediction from Japanese encephalitis. J Bioinform Seq Anal. 2013;5:25–33. [Google Scholar]
- 43.Saini S, Thakur CJ, Kumar V. Genome wide computational prediction of miRNAs in Kyasanur forest disease virus and their targeted genes in human. Innov Thoug Intern Res J. 2017;5:13–46. [Google Scholar]
- 44.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gammaherpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang KY, Lee TY, Teng YC, Chang TH. ViralmiR: a support-vectormachine method for predicting viral microRNA precursors. BMC Bioinformatics. 2015;16 Suppl 1:S9. doi: 10.1186/1471-2105-16-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gkirtzou K, Tsamardinos I, Tsakalides P, Poirazi P. MatureBayes: a probabilistic algorithm for identifying the mature miRNA within novel precursors. PLoS One. 2010;5:e11843. doi: 10.1371/journal.pone.0011843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2016;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. Gene Ontology ConsortiumThe gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33(Database Issue):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allmer J, Yousef M. Computational methods for ab initio detection of microRNAs. Front Genet. 2012;3:209. doi: 10.3389/fgene.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allmer J. Computational and bioinformatics methods for microRNA gene prediction. Methods Mol Biol. 2014;1107:157–175. doi: 10.1007/978-1-62703-748-8_9. [DOI] [PubMed] [Google Scholar]
- 55.Kumar S, Ansari FA, Scaria V. Prediction of viral microRNA precursors based on human microRNA precursor sequence and structural features. Virol J. 2009;6:129. doi: 10.1186/1743-422X-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomes CP, Cho JH, Hood L, Franco OL, Pereira RW, Wang K. A review of computational tools in microRNA discovery. Front Genet. 2013;4:81. doi: 10.3389/fgene.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saçar Demirci MD, Baumbach J, Allmer J. On the performance of pre-microRNA detection algorithms. Nat Commun. 2017;8:330. doi: 10.1038/s41467-017-00403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopes Ide O, Schliep A, de Carvalho AC. The discriminant power of RNA features for pre-miRNA recognition. BMC Bioinformatics. 2014;15:124. doi: 10.1186/1471-2105-15-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo L, Lu Z. The fate of miRNA strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5:e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene gxpression: An overview of nuclear functions. Int J Mol Sci. 2016;17:1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akbari Moqadam F, Pieters R, den Boer ML. The hunting of targets: challenge in miRNA research. Leukemia. 2013;27:16–23. doi: 10.1038/leu.2012.179. [DOI] [PubMed] [Google Scholar]
- 62.Fan X, Kurgan L. Comprehensive overview and assessment of computational prediction of microRNA targets in animals. Brief Bioinform. 2015;16:780–794. doi: 10.1093/bib/bbu044. [DOI] [PubMed] [Google Scholar]
- 63.Kumar A, Yu F-SX. Toll-Like Receptors and Corneal Innate Immunity. Curr Mol Medi. 2006;6:327–337. doi: 10.2174/156652406776894572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen CJ, Ou YC, Li JR, Chang CY, Pan HC, Lai CY, Liao SL, Raung SL, Chang CJ. Infection of pericytes in vitro by Japanese encephalitis virus disrupts the integrity of the endothelial barrier. J Virol. 2014;88:1150–1161. doi: 10.1128/JVI.02738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dallasta LM, Pisarov LA, Esplen JE, Werley JV, Moses AV, Nelson JA, Achim CL. Blood-brain barrier tight junction disruption in human immunodeficiency virus-1 encephalitis. Am J Pathol. 1999;155:1915–1927. doi: 10.1016/S0002-9440(10)65511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lasky JL, Wu H. Notch signaling, brain development, and human disease. Pediatr Res. 2005;57(5 Pt 2):104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- 67.Higuchi M, Kiyama H, Hayakawa T, Hamada Y, Tsujimoto Y. Differential expression of Notch1 and Notch2 in developing and adult mouse brain. Brain Res Mol Brain Res. 1995;29:263–272. doi: 10.1016/0169-328x(94)00257-f. [DOI] [PubMed] [Google Scholar]
- 68.Sato K, Kimura M, Sugiyama K, Nishikawa M, Okano Y, Nagaoka H, Nagase T, Kitade Y, Ueda H. Four-and-a-half LIM domains 1 (FHL1) protein interacts with the Rho guanine nucleotide exchange factor PLEKHG2/FLJ00018 and regulates cell morphogenesis. J Biol Chem. 2016;291:25227–25238. doi: 10.1074/jbc.M116.759571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang B-Q, Si N, Liu D-F. Identification of a novel four and a half LIM domain 1 mutation in a Chinese male presented with hypertrophic cardiomyopathy and mild skeletal muscle hypertrophy. Chin Med J (Engl) 2015;128:2269–2270. doi: 10.4103/0366-6999.162493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gecz J, Barnett S, Liu J, Hollway G, Donnelly A, Eyre H, Eshkevari HS, Baltazar R, Grunn A, Nagaraja R, Gilliam C, Peltonen L, Sutherland GR, Baron M, Mulley JC. Characterization of the human glutamate receptor subunit 3 gene (GRIA3), a candidate for bipolar disorder and nonspecific X-linked mental retardation. Genomics. 1999;62:356–368. doi: 10.1006/geno.1999.6032. [DOI] [PubMed] [Google Scholar]
- 71.Baranzini SE, Laxer K, Bollen A, Oksenberg JR. Gene expression analysis reveals altered brain transcription of glutamate receptors and inflammatory genes in a patient with chronic focal (Rasmussen's) encephalitis. J Neuroimmunol. 2002;128:9–15. doi: 10.1016/s0165-5728(02)00109-1. [DOI] [PubMed] [Google Scholar]
- 72.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith EC, Popa A, Chang A, Masante C, Dutch RE. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 2009;276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swanson PA, McGavern DB. Viral Diseases of the Central Nervous System. Curr Opin Virol. 2015;11:44–54. doi: 10.1016/j.coviro.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Foo CH, Rootes CL, Cowley K, Marsh GA, Gould CM, Deffrasnes C, Cowled CJ, Klein R, Riddell SJ, Middleton D, Simpson KJ, Wang LF, Bean AG, Stewart CR. Dual microRNA screens reveal that the immune-responsive miR-181 promotes henipavirus entry and cell-cell fusion. PLoS Pathog. 2016;12:e1005974. doi: 10.1371/journal.ppat.1005974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 77.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peng X, Li Y, Walters KA, Rosenzweig ER, Lederer SL, Aicher LD, Proll S, Katze MG. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics. 2009;10:373. doi: 10.1186/1471-2164-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]