Abstract
The linker histone (LH), an auxiliary protein that can bind to chromatin and interact with the linker DNA to form stem motifs, is a key element of chromatin compaction. By affecting the chromatin condensation level, it also plays an active role in gene expression. However, the presence and variable concentration of LH in chromatin fibers with different DNA linker lengths indicate that its folding and condensation are highly adaptable and dependent on the immediate nucleosome environment. Recent experimental studies revealed that the behavior of LH in mononucleosomes markedly differs from that in small nucleosome arrays, but the associated mechanism is unknown. Here we report a structural analysis of the behavior of LH in mononucleosomes and oligonucleosomes (2 to 6 nucleosomes) using mesoscale chromatin simulations. We show that the adapted stem configuration heavily depends on the strength of electrostatic interactions between LH and its parental DNA linkers, and that those interactions tend to be asymmetric in small oligonucleosome systems. Namely, LH in oligonucleosomes dominantly interacts with one DNA linker only, as opposed to mononucleosomes where LH has similar interactions with both linkers and forms a highly stable nucleosome stem. Although we show that the LH condensation depends sensitively on the electrostatic interactions with entering and exiting DNA linkers, other interactions, especially by non-parental cores and non-parental linkers, modulate the structural condensation by softening LH and thus making oligonucleosome more flexible, in comparison to to mono and dinucleosomes. We also find that the overall LH/chromatin interactions sensitively depend on the linker length because the linker length determines the maximal nucleosome stem length. For mononucleosomes with DNA linkers shorter than LH, LH condenses fully, while for DNA linkers comparable or longer than LH, the LH extension in mononucleosomes strongly follows the length of DNA linkers, unhampered by neighboring linker histones. Thus, LH is more condensed for mononucleosomes with short linkers, compared to oligonucleosomes, and its orientation is variable and highly environment dependent. More generally, the work underscores the agility of LH whose folding dynamics critically controls genomic packaging and gene expression.
Graphical Abstract
Introduction
The pivotal discovery of the internal structure of DNA revealed how genetic information is stored in an orderly fashion within the cell nucleus. However, organism complexity could not be matched to the limited number of genes contained in DNA. Therefore, the interactions within the genome and its editing became significant areas of research. The sheer size of DNA (2 meters in every human cell when stretched) requires a highly efficient mechanism of its packing within the micrometer-size cell nucleus. This mechanism must also control the facile access to the DNA when unraveled. To achieve this extraordinary packaging, DNA employs histones, a class of highly conserved proteins, which form globular octamers around which DNA wraps to form nucleosome cores. To accomplish both the high compression of genetic data and its easy release, histone proteins use their tails to interact with neighboring DNA strands, other histones and proteins, and thereby control transcription. Chromatin condensation and decondensation are therefore achieved through those interactions, and through numerous epigenetic markings 1, 2, 3, 4, 5, 6, 7, 8.
The histone tails are not sufficient to account for different packing arrangements of chromatin during the cell cycle. The linker histone (LH) protein, and its sub-variants, also play pivotal roles in the chromatin compaction 9, 10, 11, 12, 13, 14, 15. The LH protein binds the nucleosome core at its dyad axis, close to both exiting and entering DNA strands. In this way it helps form and stabilize “stems” and promotes two-start, or zigzag, arrangement of nucleosomes 11, 12, 16, 17, 18, 19, 20. Similar to the histone tails, LH is a target for reversible posttranslational modifications 21, 22, 23. Because LH belongs to a group of intrinsically disordered proteins (IDP), it achieves the full condensation only when surrounded with DNA and other proteins 2, 24, 25. This property allows it to dynamically regulate chromatin structure.
The detailed structural aspects associated with LH/chromatin interactions are not well understood. Here we focus on the LH condensation patterns in short nucleosome arrays and their possible dependence on the interactions between LH and other chromatin building blocks (DNA linkers, nucleosomes, histone tails and other linker histones), to help interpret recent experimental findings 24. Specifically, we seek to explain why the folding of LH in mononucleosomes appears different from its folding in longer arrays 24. Hayes and c o-workers recently examined the folding of LH in mononucleosomes and oligonucleosomes by Förster resonance energy transfer (FRET) techniques to investigate the dependence of the LH condensation on its environment 24. They confirm that LH, primarily through its C-terminal domain (CTD), while disordered in isolation, is condensed when bound to nucleosome arrays. The observed intensities of the FRET signal indicate that the LH has different conformations when bound to free DNA, mononucleosomes, and oligonucleosomes, suggesting that CTD responds to different environments by adjusting its conformation. However, a structural view of these complex effects is not available.
We apply our established mesoscale chromatin model to examine the LH folding in mononucleosomes as well as in small nucleosome arrays of 2–6 nucleosomes, and dinucleosomes subsaturated with LH (e.g., 1 LH per dinucleosome). The model has successfully addressed the roles of chromatin building blocks on the various aspects of chromatin architecture, such as the role of histone tails 26, 27, 28, and the influence of DNA linker length on fiber architecture and packing density 29, 30. In particular, we showed that the LH-induced fiber condensation is affected by the linker length, since optimal stems form when the DNA linkers are similar in size to the LH length (short to medium linkers) 25, 29, 30, 31, 32. For long linkers, the excess linker DNA forms other interactions with the tails and neighboring units, promoting bending and polymorphic states 32. Previous work has also examined the LH-dependent fiber condensation 30, 31, 33, and the synergistic folding of the LH and the chromatin fiber 25, 34. LH condenses synergistically as the chromatin fiber forms a compact zigzag structure, and its LH condensation depends sensitively on the salt environment. The fiber compaction may lead to asymmetric nucleosome configurations at low monovalent salt (LH interacts with one DNA linker), while more symmetric structures tend to form at high salt (LH interacts with both DNA linkers) 34. Because the LH density controls global chromatin architecture 16, 35, 36, 37, it is important to understand the LH condensation effects for the interpretation of chromatin higher order structures.
Here we explain the enhanced condensation of mononucleosomes compared to small systems of nucleosomes fibers by more intense interactions LH has with parental DNA linkers in mononucleosomes and by the lack of competing electrostatic and steric factors from neighboring fiber units that lead to functionally most optimal stem geometries. We show that the energetically most important interactions with parental DNA linkers are followed by interactions with the parent cores, neighboring LHs, and H3 tails. We also show that the DNA linker length has a strong influence on the LH. In short NRL mononucleosomes, LH is has strong contacts with both parental linkers, while in oligonucleosomes with the same NRL, LH is in contact with only one parental DNA linker and instead interacts strongly with non-parental nucleosomes. Furthermore, LHs in short NRL oligonucleosomes are energetically less stable than in mononucleosomes or in oligonucleosomes with longer NRLs. Our work opens the way to further investigations of LH variants and LH epigenetic marks.
Methods
We analyze the condensation of linker histone in mononucleosomes, oligonucleosomes with 2 to 6 nucleosomes, as well as in dinucleosomes sub-saturated with LH (1 LH per dinucleosome) for a wide range of linker lengths, using our mesoscale model, with our recently refined model of LH (H1) 25. The model was most recently summarized in 38. Essentially, our chromatin model interprets the nucleosome surface with the wrapped DNA, but without the histone tails, via 300 charged beads that robustly reproduce the electrostatic field of the nucleosome at phy siological monovalent salt concentrations 39, 40, 41. The DNA linkers and histone tails are depicted as flexible beads 26, 41. The refined linker histone accounts for the inherently dynamic and disordered nature of LH 15, 36. We use 6 beads for the globular head domain (GH) and 22 beads for the flexible C-terminal domain (CTD) 25, believed to be the main element that binds to the chromatin fiber. The globular head is permanently attached to the nucleosome at the dyad axis. All nucleosome building blocks interact with each other via electrostatic, mechanical (stretching, bending, twisting) and steric van der Waals (VdW) potentials. Equilibrium chromatin array configurations are sampled using efficient Monte-Carlo local and global moves. Full model details are provided in refs. 25, 26, 40, 41, 42. The model is shown in Fig. 1a.
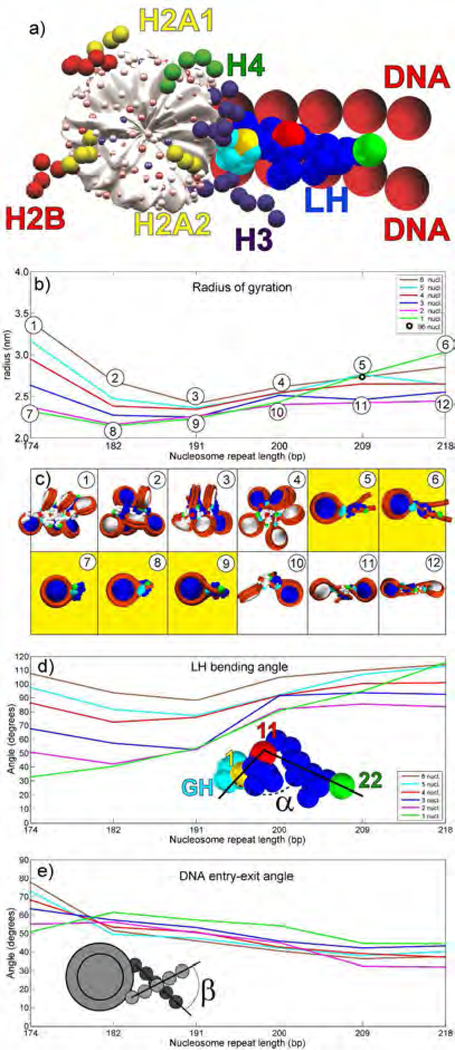
Figure 1.
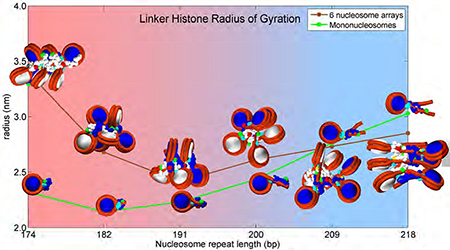
Chromatin dependent condensation of LH. a) Our mesoscale model of nucleosome core with tails, DNA linkers and LH. b) Linker histone radius of gyration as a function of the number of nucleosomes in the array and NRL. Radius of gyration data for 96-core array (NRL = 207 bp) taken from 35. c) Highest LH condensation configurations for each NRL (upper panels), and lowest condensation configurations for each NRL (lower panels). LHs have the same colors as their parent nucleosomes (consecutive nucleosomes are colored white and blue). GH is colored cyan. The first bead in CTD is colored yellow, bead 11 is colored red, and the last bead (22) is green. Mononucleosomes are in yellow boxes. d) CTD bending angle (angle between vectors formed by beads 1–11 and 11–22). e) Angle between DNA linkers entering and exiting the nucleosome as function of the number of cores and NRL.
In each simulated system, every nucleosome has equal entering and exiting DNA linker lengths. The simulated DNA linker lengths of 27, 35, 44, 53, 62, and 71 base-pairs (bp) correspond to nucleosome repeat lengths (NRL) values, usually encountered in nature, NRL = 174, 182, 191, 200, 209, 218 and 226 bp (NRL = 147 bp around the nucleosome plus DNA linker length). Three different intrinsic DNA twist values were sampled (0, ±12º about the mean) to account for small variations naturally present in DNA twist between consecutive nucleosomes. The starting configurations were zigzag geometries, as it is believed that the zigzag organization of nucleosomes is a dominant pattern in chromatin 19. The chromatin fibers were simulated at physiological monovalent salt concentration of 0.15 M and room temperature (293 K), with 1 LH per nucleosome (except for the additional subsaturated dinucleosomes which have 1 LH per dinucleosome). For each 1–6 nucleosome systems considered, every NRL value mentioned above was covered via 12 simulations, using 4 random seeds per each of the three DNA twist angles. Each trajectory was 20 million steps long, sufficient for convergence of these small systems 25, 30. With dinucleosome arrays subsaturated with LH we similarly conducted 12 simulations per each NRL value. In each case, we use the last 10 million steps per trajectory for statistical analysis.
Results and Discussion
As we are interested in the condensation of CTD in short nucleosome arrays, and its dependence on chromatin array elements, we measured the CTD radius of gyration Rgyr and its bending angle, the angle between vectors formed between CTD beads 1–11 and 11–22 (see Fig. 1b-d). Those quantities correspond to FRET intensities measured in 24. Namely, a lower Rgyr and smaller LH bending angle correspond to higher FRET intensities, i.e., decreased distances between the fluorophore-labeled positions. Fig. 1 also depicts the angle between DNA strands entering and leaving nucleosome (see Fig. 1e). We also analyze the influence of various chromatin building blocks (DNA linkers, nucleosome cores, other LHs, histone tails) on the CTD compaction via their contact intensities and energetic contributions (Figs 2 and 3). We also examine the distributions of LH and DNA linker beads as functions of array size and DNA linker length in Figs. 4–6).
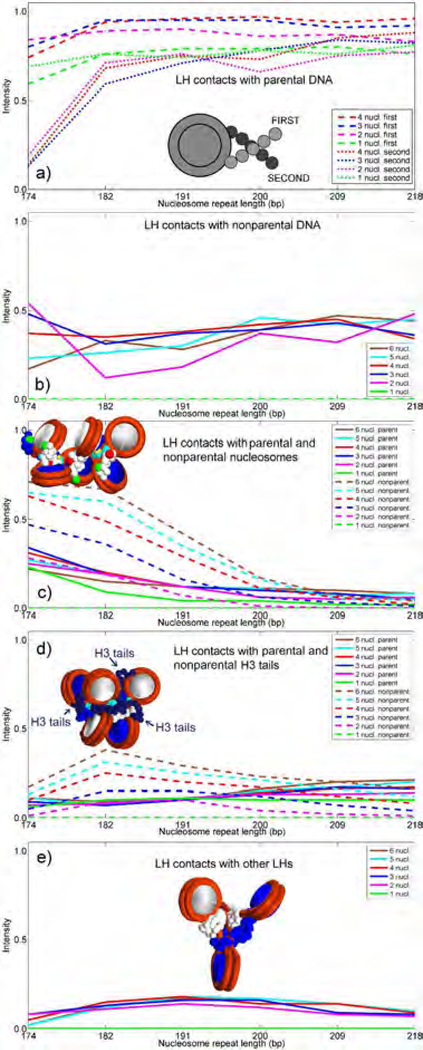
Figure 2.
LH interactions with parental and nonparental DNA linkers, nonparental cores, H3 tails and nonparental LHs. Intensities are counted in a binary manner (on or off) when distances are within half the sum of van der Waals radii of LH (1.8 Å) and other elements (1.8 Å for tails and 2.7 Å for DNA). Total values are normalized. a) LH interactions with first parental (dashed lines) and second parental DNA linkers (dotted lines). b) LH interactions with non-parental DNA. c) LH interactions with parental (full) and non-parental nucleosomes (dashed lines). d) LH interactions with parental (full) and non-parental H3 tails (dashed lines). Only H3 tails are depicted in the chromatin fiber image as chains composed of navy beads, see Fig. 1a. e) LH interactions with non-parental LHs.
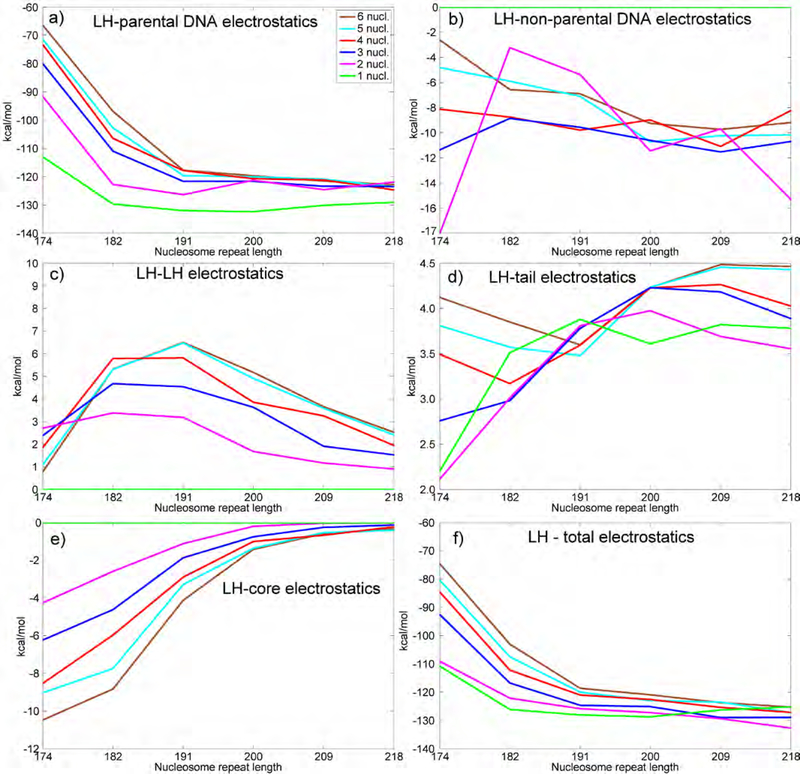
Figure 3.
Array electrostatics. Average electrostatic energies per nucleosome in the chromatin arrays are measured in kcal/mol. a) Electrostatic energy between linker histone and parental DNA strands. b) Electrostatic energy between linker histone and non-parental DNA strands. c) Electrostatic energy among all linker histones. d) Electrostatic energy between linker histone and tails. e) Electrostatic energy between linker histone and nucleosome core(s). f) Total electrostatic energy.
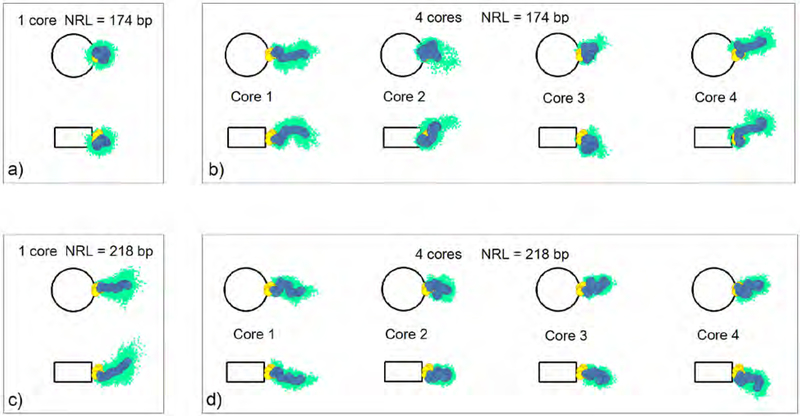
Figure 4.
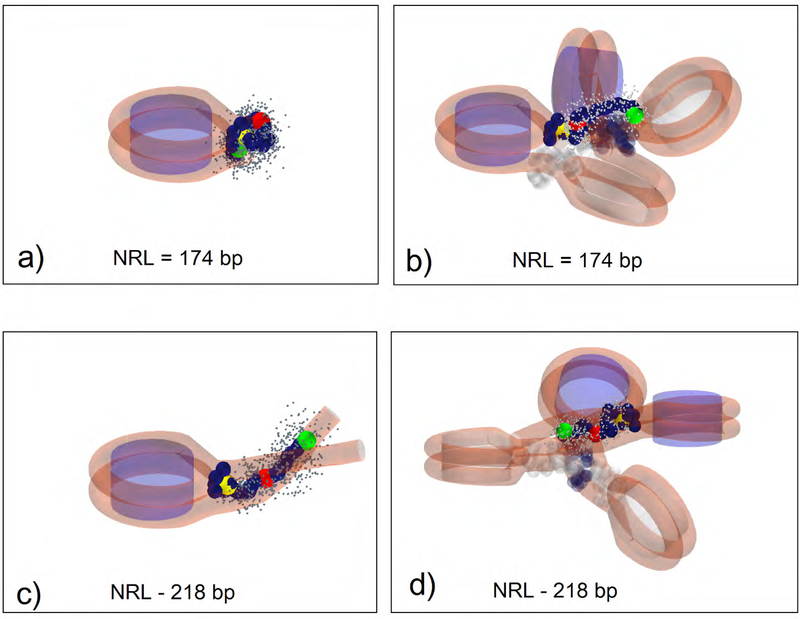
Linker histone distribution for typical fiber configurations in mononucleosomes and 4-nucleosome arrays, for shortest (174 bp) and longest (218 bp) NRLs, projected on the nucleosome and dyad planes. The clouds of light green dots represent linker DNA bead positions in a single trajectory. The average positions of linker histone beads, for a given trajectory, are shown as yellow (GH) and blue (CTD) circles.
Figure 6.
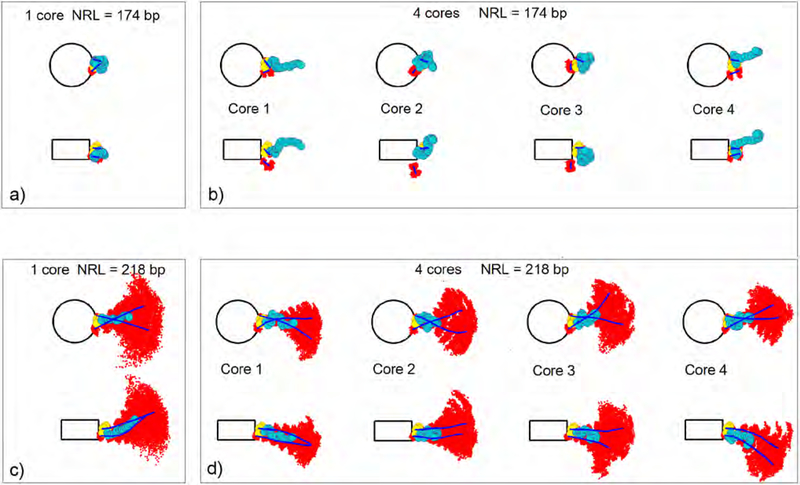
Three-dimensional CTD distributions for typical fiber configurations in mononucleosomes and 4-nucleosome arrays, for shortest (174 bp) and longest (218 bp) NRLs. Each image depicts an average configuration of one LH over the last 5 million steps per a single trajectory. All other LHs are shown as semitransparent and carry the same color as their parental nucleosome (white or navy). Small gray beads are trajectory positions of CTD beads. Analyzed CTDs are opaque, navy colored, with bead 1 in yellow, bead 11 in red, and bead 22 in green. a) Mononucleosome with NRL = 174 bp. b) 4-nucleosome array with NRL = 174 bp; c) Mononucleosome with NRL = 218 bp. d) 4-nucleosome array with NRL = 218 bp.
a. Mononucleosomes Are Most Compact for Short Linkers
From the analysis of Rgyr in Fig. 1b, we see that CTD in arrays with short to medium long linkers (NRL = 174 to 200 bp) follows a pattern observed by Fang et al. 24, namely, CTD achieves highest levels of compaction (lowest radii of gyration and smallest bending angle) in mononucleosomes and dinucleosomes. In a mononucleosome with long linkers (209 to 218 bp), Rgyr is highest. Note that these values reflect averages over the oligonucleosome system; in general peripheral LHs (belonging to the first and last nucleosomes) are less compact than LHs belonging to the internal LHs (data not shown). The images in Fig. 1c depict systems with maximum and minimum Rgyr for each NRL and show that mononucleosomes are most compact for small to medium (typical) NRL values (images 7,8, 9).
The CTD bending angle α (Fig. 1d) confirms these observations. In mononucleosomes up to NRL =209bp, the CTD’s bending toward the nucleosome is much more pronounced than in oligonucleosome arrays (CTD bending angles are smaller in mononucleosomes than in oligonucleosomes). This result is in agreement with the observations by Fang et al. 24 that “the relative distance between residues 101–173 is less than that between 101–195 when H1 is bound to arrays, while the opposite relationship “was found when H1 is bound to mononucleosomes” (see Fig. 3 in 24. With NRL=209 bp and longer, the trend is reversed; in mononucleosomes CTD (LH) is flat, with much higher bending angle. See also Fig S1 for more information on distances within the same LH and between neighboring LHs. In particular, Fig S1c makes clear that the CTD orientations are highly dependent on the NRL. Short-NRL arrays have CTDs pointing toward the array exterior (1–1 distances are much shorter than 22–22 distances), while long-NRL arrays have CTDs pointing toward the fiber interior (1–1 distances are longest). See inset images of 6-nucleosome arrays.
The angle β between DNA strands entering and leaving nucleosomes (Fig. 1e) shows that in arrays with the shortest NRL (174 bp), the entry-exit angle is narrowest in mononucleosome and steadily rises with the number of nucleosomes in the array. In the short linker mononucleosome (174bp), LH is highly compact and strongly bound to DNA, while in a 6-nucleosome array LH makes strong contacts with non-parental nucleosomes and thus forms only a loose stem (see image of the 6-nucleosome array with NRL = 174 bp in Fig. 1c, panel 1, and Fig. 2a and 2c). For all other NRLs, this angle is widest for mononucleosomes. In mononucleosomes with medium to long linkers, DNA has more freedom, while in oligonucleosomes, DNA is sterically restricted due to fiber architecture and interactions with other elements in the array (histone tails, see for example 30.
b. Competition between DNA Linkers and Other Components (Nucleosomes, Tails, Other LHs) Modulate the LH Folding
The analyses of contact patterns between LH and other chromatin building blocks (Fig. 2) and the corresponding energies (Fig. 3) shed light on the puzzling behavior of CTD. We express the LH interaction intensities as the normalized average number of CTD beads (out of 22 beads in CTD) that interact with other elements in the system; an “interaction” is recorded when the distance between them is less than half the sum of their van der Waals radii (1.8 Å for CTD beads). We separately measured the LH interactions with the parental DNA linker entering the nucleosome from the LH interactions with the DNA linker leaving it. In mononucleosomes, the interactions with both parental DNA linkers are similar in intensity. In oligonucleosomes they are always very high for the linker entering the nucleosome (first linker), but vary in strength for the second linker, from weak for short NRL (174 bp) to strong (almost equal in intensity to the LH’s interactions with first linker) for long NRLs (209–218 bp). Thus, that the interactions with parental DNA linkers have a strongest influence on the CTD compaction, in accord with Fang et al. 24, followed by the interactions with nonparental cores and linkers (Fig. 2a-c). The interactions with non-parental DNA linkers (Fig. 2b) are lower but still relatively high, especially for NRLs between 200 and 218 bp. This is because, in oligonucleosomes with medium to long NRLs, interactions with nonparental DNA reduce the CTD extension by pulling CTD away from parental DNA linkers (see Fig. 1c, panels 1–4 and 10–12). In subsaturated dinucleosomes, similar to mononucleosomes with short to medium NRLs, symmetric interactions with both parental DNA linkers increase the electrostatic attraction and also produce low gyration radii (see Fig. S2 for the images of dinucleosomes subsaturated with LH).
The CTD interactions with non-parental nucleosome cores (Fig. 2c) closely mirror interactions with parental DNA, particularly in arrays with short DNA linkers, in agreement with Fang et al. 24. The interactions with nonparent cores are highest in 5 and 6-nucleosome arrays with short NRLs in accord with lower levels of CTD condensation. In short NRL fibers, the presence of neighboring cores with their linkers limits the fiber architecture (particularly entry-exit angle for NRL=174), increases interactions with non-parental cores at the expense of interactions with parental DNA, and allows CTD to escape its parental nucleosome. In mononucleosomes, the LH interactions with parent cores are lower than in oligonucleosomes because LH is tightly bound to DNA and thus has less freedom to explore other fiber constituents.
CTDs also distinctly interact with the H3 tails (Fig. 2d); interactions with other tails are negligible (results not shown). The length of the H3 tails and their positions close to the dyad axis and nucleosome stem (see Fig. 1a) explain their notable interactions with LH, especially interactions between non-parental H3 tails and LH in 5 and 6-nucleosome arrays with short to medium DNA linkers (NRL = 182 to 200 bp).
Interactions with other LHs (Fig. 2e) are less prominent. They are more notable in arrays with medium to long linkers (NRL = 182 to 209 bp), which correlates to reduced 11–11 and 22–22 bead distances (see Fig. S1c). Therefore, in oligonucleosomes with medium to long DNA linkers, steric clashes between CTDs in cooperation with non-parental DNA interactions prevent the full CTD extension.
c. Energetic Analysis Emphasizes Dominance of Linker DNA/LH Interactions
The analysis of the LH electrostatic interaction energies (normalized per LH, Fig. 3) offers quantitative support to the previous observations and shows that chromatin compaction is a complex process that strongly depends on the nucleosome environment. Interactions between positively charged LH and negatively charged parental DNA linkers are highly attractive (Fig. 3a), while interactions with other LHs and histone tails are repulsive (Fig. 3c and 3d). As mentioned above, LH in mononucleosomes strongly interacts with both parental linkers for all NRLs, while in oligonucleosomes, the interaction is more asymmetric, especially for short NRLs. Therefore, in shortto-medium NRL arrays, the parental-DNA electrostatic interactions in mononucleosomes are strongest, while in 6-nucleosome arrays they are weakest. These complex and competitive interactions in oligonucleosomes, with short-to-medium NRLs in particular, reduce the strength of the electrostatic interaction and make the fiber more flexible 25. The electrostatic interactions with non-parental DNA linkers (Fig. 3b) and nucleosome cores (Fig. 3e) are also attractive but less intense compared to primary interactions with parental DNA linkers. Those secondary attractive interactions, however, may help pull LH away from the nucleosome stem, and increase its radius of gyration, when compared to 2-nucleosome arrays. All other LH interaction energies are smaller, but contribute to the overall energetics and help interpret folding behavior, particularly in mononucleosomes.
Our model also offers an explanation on the folding behavior observed by Fang et al. 24. They noticed that the reduction of the free DNA linker length in dinucleosome increases the FRET signal (Fig. 7 in 24. We can explain this trend by an increased and substantially more symmetric contacts of LH with both parental linkers. In our dinucleosome simulations with 1 LH per dinucleosome symmetric interactions with both parental DNA linkers increase the electrostatic attraction and also produce low gyration radii for short to medium NRLs.
Figure 7.
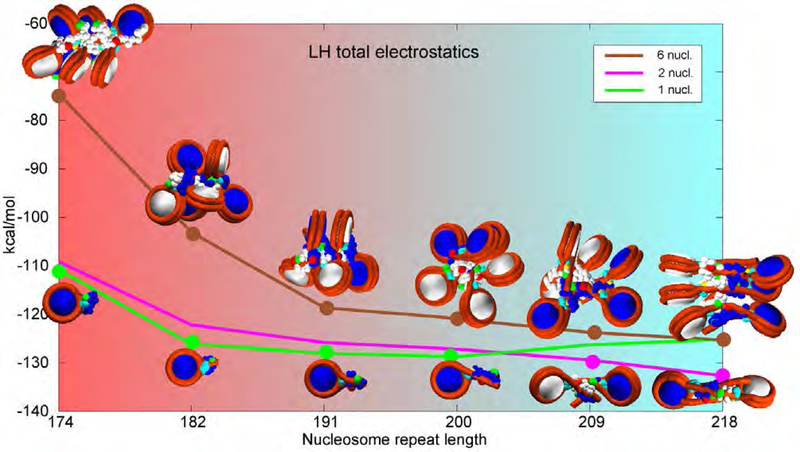
Chromatin structures vs. electrostatic energy for mononucleosomes, dinuclesomes and 6-core arrays over NRLs most commonly encountered in living cells. Representative images correspond to selected points on the energy curves. Mononucleosomes and dinucleosomes are electrostatically most stable nucleosome arrays with strongest electrostatic attractions for a given NRL (green and pink curves respectively). 6-core arrays are closest to chromatin in-vivo. Genetically active cells have loosely bound LHs that interact asymmetrically with one DNA linker only. Mature, transcriptionally inactive cells have LHs tightly bound to parental DNA linkers and thus serve as barriers to transcription. The background gradient represents biological activity of chromatin over different NRLs, from chromatin in genetically active cells with high protein production in red to mature eukaryotic cells with limited protein production in light blue.
d. LH and Linker DNA Flexibility Increases with NRL and Fiber Size
The chromatin fiber is a dynamic entity. Its flexibility stems from the flexibilities of DNA linkers and linker histones and tails. Fig. 4 shows the CTD distributions and average LH bead positions for mononucleosome and tetranucleosome arrays (extracted from single trajectories), for shortest (174 bp) and longest (218 bp) NRLs, projected on the nucleosome and dyad planes. Fig. 5 shows the analogous distributions for the DNA linkers. We see that the CTD and DNA linkers exhibit different dynamic behavior in mononucleosomes versus 4-nucleosome arrays at the same NRL. For NRL = 174 bp (Fig.4a), CTD in the mononucleosome is folded into a compact configuration bent toward the globular head and does not deviate much from the average position. In the 4-nucleosome array with the same NRL (Fig. 4b), CTD is more flexible, evident by the more extended configurations and wider distributions present in all 4 nucleosomes. In long NRL arrays (NRL=218 bp), CTD in mononucleosome (Fig. 4c) is fully extended away from the globular head, with higher mobility. In the 4-nucleosome array with the same NRL (Fig. 4d), the average CTD configurations resemble configurations in the short NRL tetranucleosome, only with tighter distributions. CTDs belonging to terminal (first and fourth) nucleosomes are generally more extended than CTDs in internal (second and third) nucleosomes.
Figure 5.
Linker DNA distribution for typical fiber configurations in mononucleosomes and 4-nucleosome arrays, for shortest (174 bp) and longest (218 bp) NRLs projected on the nucleosome and dyad plane. The clouds of red dots represent linker DNA bead positions. The mean trajectories of the DNA linkers are shown as blue lines. The average positions of linker histone beads are shown as yellow (GH) and turquois (CTD) circles.
From our additional analysis of the CTD radius of gyration distributions in Figure S3, it is also evident that the LH binding to nucleosomes is rather dynamic and NRL dependent. Namely, Figure S3 shows that the distributions of LH radius of gyration in short NRL tetranucleosomes (174 bp) have a rather complex shape, in comparison to mononucleosomes and long-NRL tetranuclesomes (218 bp). This is an indication that LH is only weakly bound to chromatin in short NRL oligonucleosomes (in real world where the LH globular head is not permanently attached to the nucleosome).
From Fig. 5 we see that long NRL arrays have much more flexibility. In the short NRL mononucleosome (NRL = 174 bp), DNA is tightly bound to LH, while in the tetranucleosome with the same NRL, only one parental DNA linker is attached to its LH. In long NRL arrays (NRL = 218 bp), both DNA linkers are bound to their parent linker histone, but also have much more freedom with wider distribution in comparison to their short NRL counterparts. In the long NRL mononucleosome, DNA linkers have wider and more uniform distribution, while in the tetranucleosome, the distribution is more restricted because DNA linkers are limited by the neighboring nucleosomes and their LHs. Fig. 6 provides 3D images in different nucleosome environments to clarify these trends.
Conclusion
Chromatin compaction depends on many factors, including the DNA linker length, the multivalent and divalent ion concentrations, and auxiliary proteins like linker histones. The influence of DNA linker length, histone tails, and LHs had been addressed by mesoscale modeling 25, 27, 29, 30, 32, 33. However, recent experimental data have presented further structural puzzles regarding the different behavior of mononucleosomes compared to oligonucleosome systems 24. Here we examined the CTD compaction as a function of nucleosome environment and linker length. While our model is a simplified coarse-grained interpretation which cannot account for side-chain interactions, DNA sequence, and many other microscopic factors, our results provide a structural explanation to the data and confirm that the LH compaction is strongly influenced by the environment. Namely, the CTD condensation level reflects a compromise between the interactions LH has with the proximal parental DNA linkers as well as parent and non-parent nucleosomes, neighboring LHs and parental and nonparental histone H3 tails. The full stem formation is possible in mononucleosomes with short NRLs, where the CTD is optimally condensed and clustered near the nucleosome core, as well as in mononucleosomes and oligonucleosomes with long NRLs where CTD is maximally extended. In short NRL arrays, LH (CTD primarily) has limited interaction with both linkers (see Fig. 2), and increased interactions with non-parental cores, thus forms suboptimal stems. This trend correlates with the observation that genetically active cells have short NRL fibers sub-saturated with LH while mature cells have longer NRL chromatin fibers fully saturated with LH (see for example Table 1 in30.
Besides showing that CTD achieves maximum compaction in mononucleosomes and dinucleosomes for short to medium DNA linkers, our results show that in oligonucleosomes, CTD exhibits a spectrum of conformations dependent on the DNA linker length, number of nucleosomes, and position within an array (peripheral or internal nucleosomes). The primary source of CTD compaction is an attractive force between LH and the parental and non-parental DNA linkers, and non-parental cores, followed by the repulsions between CTD and neighboring linker histones and H3 tails. The mono and dinucleosomes with short to medium linkers produce electrostatically most stable LH complexes with a highest level of LH-DNA attraction. The introduction of additional nucleosomes reduces the very strong interactions between LH and parental DNA primarily through the attractive interactions with non-parental DNA and repulsive interactions between LH and nonparental LHs and H3 tails. The complex interplay between attractive and repulsive forces softens LH and increases its radius of gyration, as well as its overall flexibility. These results also show that the chromatin exhibits a range of mesoscale configurations dependent on linker length and LH presence, which may influence its behavior over a longer stretches of fiber, and thus control gene expression (see 39. In rapidly growing, genetically active cells, with high protein production, LH, if present, is only loosely bound to chromatin and interacts with one DNA linker only (electrostatic attraction between LH and chromatin is low). In that case, LH has high Rgyr to maximize the surface area in contact with electronegative chromatin constituents (e.g., the nucleosome core surface). In mature, transcriptionally inactive cells, LH is tightly bound to parental DNA linkers, with similar contact intensities to both linkers. In that case, the electrostatic attraction is high, similar to the intensity of electrostatic attraction between LH and chromatin constituents (DNA primarily) in tightly bound mononucleosomes and dinucleosomes. Thus, the strong attraction between LH and DNA represents a barrier to transcription. To maximize electrostatic attraction, LH in oligonucleosomes exhibits a spectrum of configurations, from fully condensed LH in arrays with medium long linkers, to fully extended LH in arrays with very short or very long linkers. This concept is illustrated in Figure 7, which shows a range of configurations for mononucleosomes, dinucleosomes and 6-nucleosome arrays along their electrostatic profiles as functions of NRL. Their varied CTD configurations emphasize the variability in stem and CTD geometries.
Of course it should be emphasized that our coarse-grained model has general limitations as other simplified treatments. The LH and the entire fiber system are modeled only in mesoscopic detail and cannot account for detailed atomistic fluctuations. Energetic approximations are also involved. Furthermore, here we model LH permanently attached to the nucleosome core, as suggested by experimental structures, and do not consider DNA unwrapping. In real systems, LH binds and unbinds dynamically; we have previously modeled the dynamic binding/unbinding of LH to oligonucleosomes and observed fiber softening relative to permanent LH binding 33. Our data here suggest that the stability of LH depends on the fiber size (Fig. S3). Furthermore, the LH type we follow (rat H1) is also longer by about 20 amino acids that the H1ºa system used by Fang et al. 24. Nevertheless, prior studies in agreement with a large array of experimental data 25, 26, 27, 29, 30, 28, 31, 32, 33, 34 suggest that in-silico studies help shed light on chromatin structure and energy trends, especially when conducted on a large set of system25, 26, 27, 29, 30, 28, 31, 32, 33, 34s with systematic parameters variations. Future models that allow DNA unwrapping, dynamic LH binding/unbinding, and other linker histone species can address these limitations.
The results underscore the flexibility and adaptability of the LH whose dynamic interplay with nucleosomes, tails and other proteins determines chromatin architecture. Further studies on reversible LH modifications 39 and LH variants and their influence on the behavior of chromatin constitute interesting avenues for further research.
Supplementary Material
Acknowledgments
Support by NIGMS RO1-055164 and Philip-Morris USA and Philip-Morris International to T.S. is gratefully acknowledged.
Footnotes
Supporting Information Available
The supporting information contains 3 figures (S1 to S3). Figure S1 depicts inter and intranucleosomal bead distances. Figure S2 shows dinucleosome configurations with fully saturated 2-core arrays (upper images) and subsaturated 2-core arrays with 1 LH per dinucleosome (lower images), and Figures S3 shows linker histone radii of gyration histograms for mononucleosomes and 4-nucleosome arrays.
References
- (1).Felsenfeld G Chromatin Unfolds. Cell, 1996, 86, 13–19. [DOI] [PubMed] [Google Scholar]
- (2).Wolffe A Chromatin Structure and Function; Academic Press Inc.: San Diego, CA, third edition, 1998. [Google Scholar]
- (3).Felsenfeld G; Groudine M Controlling the Double Helix. Nature, 2003, 421, 448–453. [DOI] [PubMed] [Google Scholar]
- (4).Kouzarides T Chromatin Modifications and Their Function. Cell, 2007, 128, 693–705. [DOI] [PubMed] [Google Scholar]
- (5).van Holde K; Zlatanova J Chromatin Fiber Structure: Where Is the Problem Now? Sem. Cell Dev. Bio 2007, 18, 651–658. [DOI] [PubMed] [Google Scholar]
- (6).Bartke T; Vermeulen M; Xhemalce B; Robson SC; Mann M; Kouzarides T Nucleosomeinteracting Proteins Regulated by DNA and Histone Methylation. Cell, 2010, 143, 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bannister AJ; Kouzarides T Regulation of Chromatin by Histone Modifications. Cell Res., 2011, 21, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chavez MS; Scorgie JK; Dennehey BK; Noone S; Tyler JK; Churchill MEA The Conformational Flexibility of the C-terminus of Histone H4 Promotes Histone Octamer and Nucleosome Stability and Yeast Viability. Epigenetics & Chromatin, 2012, 5, 1–20, DOI: 10.1186/1756-8935-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pruss D; Bartholomew B; Persinger J; Hayes J; Arents G; Moudrianakis EN; Wolffe AP An Asymmetric Model for the Nucleosome: A Binding Site for Linker Histones Inside the DNA Gyres. Science, 1996, 274, 614–617. [DOI] [PubMed] [Google Scholar]
- (10).McBryant SJ; Adams VH; Hansen JC Chromatin Architectural Proteins. Chromosome Research, 2006, 37, 39–51. [DOI] [PubMed] [Google Scholar]
- (11).Zhou BR; Feng H; Kato H; Dai L; Yang Y; Zhou Y; Bai Y Structural Insights Into the Histone H1nucleosome Complex. Proc. Natl. Acad. Sci. USA, 2013, 110, 19390–19395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhou BR; Jiang J; Feng H; Ghirlando R; Xiao TS; Bai Y Structural Mechanisms of Nucleosome Recognition by Linker Histones. Molecular Cell, 2015, 59, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Robinson P; Fairall L; Huynh V; Rhodes D EM Measurements Define the Dimensions of the “30nm” Chromatin Fiber: Evidence for a Compact, Interdigitated Structure. Proc. Natl. Acad. Sci. USA, 2006, 103, 6506–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Flanagan TW; Files JK; Casano KR; George EM; Brown DT Photobleaching Studies Reveal that a Single Amino Acid Polymorphism is Responsible for the Differential Binding Affinities of Linker Histone Subtypes H1.1 and H1.5. Biology Open, 2016, 5, 372–380. DOI: 10.1242/bio.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pachov GV; Gabdoulline RR; Wade RC On the Structure and Dynamics of the Complex of the Nucleosome and the Linker Histone. Nucleic Acids Res, 2011, 39, 5255–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bednar j.; Horowitz RA; Grigoryev SA; Carruthers LM; Hansen JC; Koster AJ; Woodcock CL Nucleosomes, Linker DNA, and Linker Histone form a Unique Structural Motif that Directs the Higher-Order Folding and Compaction of Chromatin. Proc. Natl. Acad. Sci. USA, 1998, 95, 14173–14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Carruthers LM; Bednar J; Woodcock CL; Hansen JC Linker Histones Stabilize the Intrinsic Salt-Dependent Folding of Nucleosomal Arrays: Mechanistic Ramifications for Higher-Order Chromatin Folding. Biochemistry, 1998, 37, 14776–14787. [DOI] [PubMed] [Google Scholar]
- (18).Lu X; Hansen JC Identification of Specific Functional Subdomains within the Linker Histone H10 Cterminal Domain. Journal of Biological Chemistry, 2004, 279, 8701–8707. [DOI] [PubMed] [Google Scholar]
- (19).Grigoryev S; Arya G; Correll S; Woodcock C; Schlick T Evidence for Heteromorphic Chromatin Fibers from Analysis of Nucleosome Interactions. Proc. Natl. Acad. Sci. USA, 2009, 106, 13317–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lu X; Hamkalo B; Parseghian MH; Hansen JC Chromatin Condensing Functions of the Linker Histone C-terminal Domain are mediated by Specific Amino Acid Composition and Intrinsic Protein Disorder. Biochemistry, 2009, 48, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Dou Y; Mizzen CA; Abrams M; Allis CD; Gorovsky MA Phosphorylation of Linker Histone H1 Regulates Gene Expression in Vivo by Mimicking H1 Removal. Mol. Cell, 1999, 4, 641–647. [DOI] [PubMed] [Google Scholar]
- (22).Wisniewski JR; Zougman A; Krüger S; Mann M Mass Spectrometric Mapping of Linker Histone H1 Variants Reveals Multiple Acetylations, Methylations, and Phosphorylation as Well as Differences between Cell Culture and Tissue. Mol. Cell. Proteomics, 2007, 6, 72–87. [DOI] [PubMed] [Google Scholar]
- (23).Kalashnikova A; Winkler DD; McBryant SJ; Henderson RK; Herman J; DeLuca JG; Luger K; Prenni JE; Hansen JC Linker Histone H1.0 Interacts with an Extensive Network of Proteins Found in the Nucleolus. Nucleic Acids Res, 2013, 41, 4026–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fang H; Wei S; Lee TH; Hayes JJ Chromatin Structure-Dependent Conformations of the H1 CTD. Nucleic Acid. Res, 2016, 44, 9131–9141. DOI: 10.1093/nar/gkw586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Luque A; Collepardo-Guevara R; Grigoryev S; Schlick T Dynamic Condensation of Linker Histone C-terminal Domain Regulates Chromatin Structure. Nucleic Acids Res., 2014, 42, 7553–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Arya G; Schlick T Role of Histone Tails in Chromatin Folding Revealed by a New Mesoscopic Oligonucleosome Model. Proc. Natl. Acad. Sci. USA, 2006, 103, 16236–16241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Arya G; Schlick T A Tale of Tails: How Histone Tails Mediate Chromatin Compaction in Different Salt and Linker Histone Environments. J. Phys. Chem. A, 2009, 113, 4045–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Collepardo-Guevara R; Portella G; Frenkel MD; Schlick T; Orozco M Chromatin Unfolding by Epigenetic Modifications Explained by Dramatic Impairment of Internucleosome Interactions: A Multiscale Computational Study. J. Am. Chem. Soc, 2015, 137, 10205–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Schlick T; Perišić O Mesoscale Simulations of Two Nucleosome-Repeat Length Oligonucleosomes. Phys. Chem. Chem. Phys, 2009, 11, 10729–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Perišić O; Collepardo-Guevara R; Schlick T Modeling Studies of Chromatin Fiber Structure as a Function of DNA Linker Length. J. Mol. Biol, 2010, 403, 777–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Collepardo-Guevara R; Schlick T Crucial Role of Dynamic Linker Histone Binding and Divalent Ions for DNA Accessibility and Gene Regulation Revealed by Mesoscale Modeling of Oligonucleosomes. Nucleic Acids Res, 2012, 40, 8803–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Collepardo-Guevara R; Schlick T Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc. Natl. Acad. Sci. USA, 2014, 111, 8061–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Collepardo-Guevara R; Schlick T The Effect of Linker Histone’s Nucleosome Binding Affinity on Chromatin Unfolding Mechanisms. Biophys. J, 2011, 101, 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Luque A; Ozer G; Schlick T Correlation among DNA Linker Length, Linker Histone Concentration, and Histone Tails in Chromatin. Biophys. J, 2016, 110, 2309–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Grigoryev SA; Bascom G; Buckwalter JM; Schubert MB; Woodcock CL; Schlick T Hierarchical Looping of Zigzag Nucleosome Chains in Metaphase Chromosomes. Proc. Natl. Acad. Sci. USA, 2016, 113, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Caterino TL; Hayes JJ Structure of the H1 C-terminal Domain and Function in Chromatin Condensation. Biochem Cell Biol, 2011, 89, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Bascom GD; Sanbonmatsu KY; Schlick T Mesoscale Modeling Reveals Hierarchical Looping of Chromatin Fibers near Gene Regulatory Elements. J. Phys. Chem. B, 2016, 120, 8642–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bascom G; Schlick T Linking Chromatin Fibers to Gene Folding by Hierarchical Looping. Biophys. J, 2017, 112, 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Perišić O; Schlick T Computational Strategies to Address Chromatin Structure Problems. Phys. Biol, 2016, 13, 035006. [DOI] [PubMed] [Google Scholar]
- (40).Beard D; Schlick T Modeling Salt-Mediated Electrostatics of Macromolecules: The Discrete Surface Charge Optimization Algorithm and Its Application to the Nucleosome. Biopolymers, 2001, 58, 106–115. [DOI] [PubMed] [Google Scholar]
- (41).Zhang Q; Beard D; Schlick T Constructing Irregular Surfaces to Enclose Macromolecular Complexes for Mesoscale Modeling Using the Discrete Surface Charge Optimization (DISCO) Algorithm. J. Comput. Chem, 2003, 24, 2063–2074. [DOI] [PubMed] [Google Scholar]
- (42).Schlick T; Li B; Olson WK The Influence of Salt on the Structure and Energetics of Supercoiled DNA. Biophys. J, 1994, 67, 2146–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.