Abstract
Nature displays a remarkable ability to carry out site-selective post-translational modification of proteins, therefore enabling a dramatic increase in their functional diversity1. Inspired by this, chemical tools have evolved for the synthetic manipulation of protein structure and function, and have become essential to the continued advancement of chemical biology, molecular biology and medicine. However, the number of chemical transformations suitable for effective protein functionalization is limited because the stringent demands inherent to biological systems preclude the applicability of many potential processes2. Put simply, these chemical transformations often need to be selective at a single site on a protein, proceed with very fast reaction rates, operate under biologically ambient conditions and should provide homogeneous products with near perfect conversion2–7. While many elegant bioconjugation methods exist at cysteine, lysine and tyrosine, we reasoned that a method targeting a less explored amino acid would significantly expand the protein functionalization toolbox. Herein, we report the development of a multifaceted-approach to protein functionalization based on chemoselective labelling at methionine residues. By exploiting the unique electrophilic reactivity of a bespoke hypervalent iodine reagent, one can target the S-Me group in the side-chain of methionine. The bioconjugation reaction is fast, selective, operates at low µM concentrations and is complementary to existing bioconjugation strategies. Moreover, the new reaction produces a protein conjugate that is, itself, a high energy intermediate with reactive properties that can serve as a platform for the development of secondary, visible-light mediated bioorthogonal protein functionalization processes. Taken together, the merger of these approaches provides a versatile platform for the development of distinct transformations that can deliver versatile, information-rich protein conjugates directly from the native biomacromolecules.
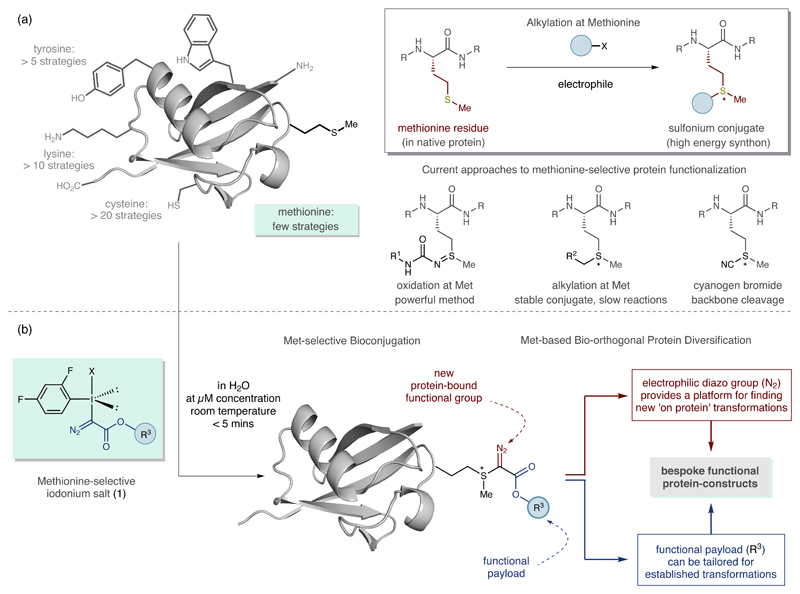
The sheer structural diversity of the proteome in any single organism means that no one protein functionalization method is likely to provide a universal solution for the preparation of protein constructs (Fig. 1a)8–11. Although encoded by the AUG start codon at the beginning of protein synthesis, methionine is often post-translationally excised and thus has a low abundance in proteins (around 2%). It is also frequently used as a replacement for hydrocarbon-containing residues12. The limited function of methionine compared to other residues, being mainly responsible for protection against oxidative stress, means that its functionalization is less likely to impair protein function13. Targeting Met would not only provide a distinct bioconjugation approach but also, using our strategy, the resulting Met conjugates provide exploitable intrinsic reactivity such that they could lead to the rapid synthesis of diverse, functional protein constructs from native proteins.
Figure 1. The evolution of a methionine-selective protein functionalization strategy.
(a) Existing protein functionalization tactics, and the potential for methionine-selective bioconjugation; R = peptide or protein, R1 = various organic groups, R2 = aryl or ester group (b) Functionalized hypervalent iodine reagents enable methionine-selective protein modification, leading to methionine-based bioorthogonal protein diversification. X = leaving group; R3 = functional payload.
To date, the only effective method for bioconjugation at methionine was recently reported by Chang et al, who showed that the oxidation of thioethers with oxaziridine reagents provided the basis for an elegant bio-inspired strategy to form stable protein-bound sulfoximines14. We aimed to target the polarizable thioether on the Met side chain with a suitable electrophile to form a cationic sulfonium species, selectively installing a versatile payload and distinct functionality at a methionine residue, thereby providing a fundamentally different bioconjugation approach (Figure 1a)8–11. Methionine residues react with cyanogen bromide (CNBr), however, such a process cannot function as a bioconjugation method because the reactivity of the resulting unstable cyano-substituted sulfonium cation triggers protein backbone cleavage15. Conversely, methionine residues undergo slow S-alkylation reactions using iodoacetamide or with other benzyl-derived electrophiles to form relatively stable trialkyl sulfonium cations16–19. Striking a balance between the stability of the protein-bound sulfonium cations, compatibility of reaction conditions and the reaction rate of the thioether with suitable electrophiles has, to date, precluded the development of an effective alkylation-based method for bioconjugation at methionine. Guided by these limitations, we posited that a distinct class of electrophile, based on the hypervalent iodine scaffold (λ-3-iodanes)20, could make for a functioning bioconjugation process at methionine. Tailoring the substituents and counteranion on the I(III) atom should allow tuning of the reactivity of the polarizable I(III) nodal center to dovetail with the electron lone pair of the thioether and, also, the stability of the resulting sulfonium conjugate through modulation of the electronic features of the groups directly attached to the cationic sulfur motif. We had noted that a structurally remarkable iodonium salt (1 in Figure 1, R=Et and X=OTf) reacts rapidly with dimethylsulfide (the simplest possible mimic of the thioether motif in methionine) to form a sulfonium adduct21,22. Successful reaction of this iodonium salt with methionine would not only represent a distinct method for bioconjugation, but also deliver a high-energy conjugate equipped with reactive ‘on protein’ groups that could serve as a basis for designing new transformations towards diversely-functionalized protein constructs (Figure 1b).
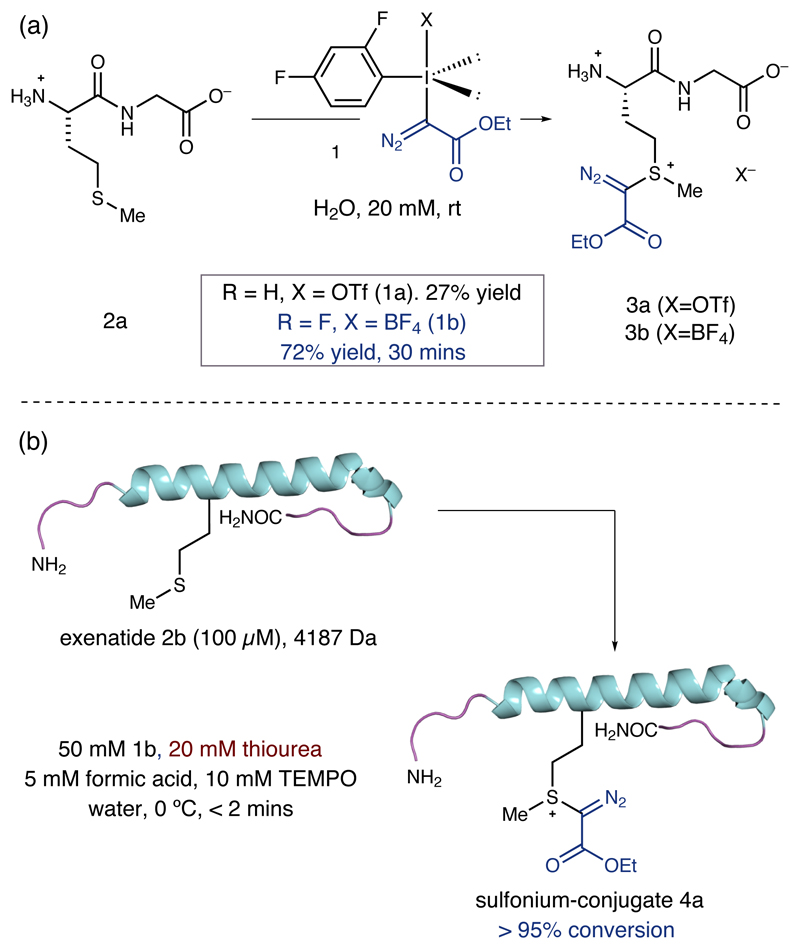
We first examined the reaction between dipeptide 2a and iodonium triflate 1a (Figure 2a). We observed the formation of the desired sulfonium-conjugate 3a, though it was clear from the low yield (27%) that the iodonium salt displayed poor stability in aqueous solution. By tailoring the aryl group of the iodonium salt (to the electron-deficient 2,4-difluorobenzene) and replacing the triflate counteranion with tetrafluoroborate (1b), we found that a readily prepared reagent 1b displayed superior physical properties (half-life in water is > 50 h). Treatment of reagent 1b with dipeptide 2a gave 70% of the desired product 3b accompanied by the corresponding sulfoxide (not shown) after reaction for 30 minutes.
Figure 2. Evolution of a methionine-selective bioconjugation strategy.
(a) Initial results for functionalization at methionine with hypervalent iodine reagents. (b) Optimal process for the thiourea-accelerated methionine selective bioconjugation of exenatide.
Moving to a more complex substrate, the GLP-1 receptor agonist, exenatide (Byetta™, 2b, a 39-residue helical polypeptide containing a single mid-chain methionine, Figure 2b), we found that treatment of a 100 µM aqueous solution with 1b led to decomposition of the polypeptide. A key breakthrough revealed that the addition of a low concentration of thiourea (20 mM) resulted in a dramatic improvement of the reaction, such that sulfonium-conjugate 4a was formed with 68% conversion in less than 2 minutes, accompanied by non-specific oxidation and labelling. Further improvements could be made by running the reaction in the presence of TEMPO (10 mM), which minimized formation of oxidative by-products. Addition of aqueous formic acid solution (5 mM, approximately pH 3) reduced the non-specific labelling by-products to trace levels. Finally, we found that the labelling process proceeded effectively when conducted in distilled water. Routine analysis of ESI-MS and MS-MS fragmentation data confirmed selective reaction at methionine. Although the concentration of thiourea is well below the levels needed for protein denaturation, we were pleased to observe that the CD spectra of exenatide-conjugate 4a confirmed retention of the characteristic helical structure (See the supplementary information S35 for details). While we are not yet certain of the remarkable role of thiourea, it is important to note its presence appears to be fundamental in providing a bioconjugation process at reaction rates needed for transformation on complex biomolecules. With these refined conditions, the thiourea-accelerated bioconjugation was almost instantaneous at exenatide concentrations ranging from 5-500 µM without compromising the conversion, and a larger, milligram-scale reaction enabled purification of conjugate 4a by semi-preparative HPLC to give 79% yield of isolated product. Given the similarity of the exenatide conjugate to the intermediate that would arise from reaction of the polypeptide with cyanogen bromide (leading to peptide cleavage), it is remarkable that 4a has a half-life in water of over 100 hours. In comparison to the corresponding reaction of methionine residues with cyanogen bromide, we believe that the observed stability of 4a is a result of the ethyl diazoacetate motif imparting a lesser electron-withdrawing effect on the sulfonium salt (than the cyano-group), which in turn leads to a species that is less reactive to attack by the proximal carbonyl group and is, hence, a more stable conjugate.
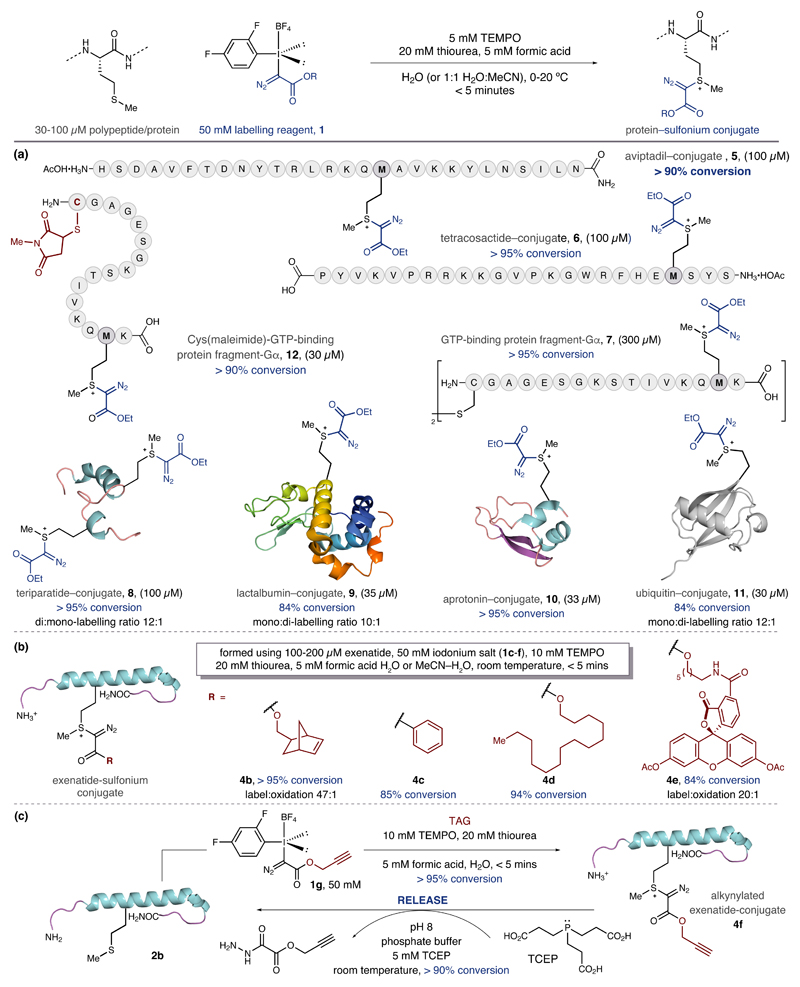
The optimized conditions, using 1b, were used to evaluate the substrate scope (Figure 3a). Random coil polypeptides that contain methionine, such as Aviptadil™ and tetracosactide, are efficiently converted to the sulfonium-conjugates 5 and 6 respectively. GTP-binding protein fragment-Gα, which contains a free sulfhydryl group in a N-terminal cysteine residue, underwent smooth methionine-labelling to 7 with concomitant oxidative disulfide formation. Teriparatide, a polypeptide containing two methionines, was converted to its bis-sulfonium conjugate (8) with good conversions. α-Lactalbumin, a globular protein displaying a readily oxidized methionine residue, undergoes bioconjugation to 9 with minimal competitive oxidation. Aprotinin (Trasylol) also forms the corresponding sulfonium-conjugate 10 in good yield. Particularly significant is the observation that two cysteine-disulfide linkages within the structure of aprotinin (and the labelling to 7) are not affected by 1b, with a single methionine-derived conjugate obtained in high conversion. When the methionine residues are buried within the tertiary structure of the protein, for example with RNA-ase B, the bioconjugation did not occur, highlighting an inherent selectivity for the labeling process between exposed and inaccessible thioether groups. This feature is highlighted by the case of ubiquitin; the N-terminal methionine residue has only moderate surface exposure and could slow down the rate of functionalization to the point where oxidation becomes competitive. Accordingly, reaction under de-oxygenated conditions allowed for efficient conversion to the labeled product (85%), with a 10:1 label:oxidation ratio (11). The methionine-selective bioconjugation strategy effectively functionalizes a range of polypeptides and proteins in high conversion at µM concentrations and in very short reaction times. Given the reactive appearance of 1b – a carbon-electrophile displaying two of the best leaving groups known to organic chemists – it is remarkable that this species selectively engages the moderately nucleophilic methionine residue in the presence of competitively nucleophilic and oxidizable amino acid residues. Bioconjugation strategies that target other amino acids should, therefore, be compatible with, and complementary to, our methionine-functionalization process. To exemplify this, we showed that the GTP-binding protein fragment-Gα could be first labelled at cysteine, using a maleimide derivative,23 and then conjugated at methionine using iodonium salt 1b to form 12. The methionine-selective process does not interfere with the cysteine-maleimide motif, which itself contains a thioether linkage, thereby highlighting possible applications towards multi-site heterolabelling of proteins24.
Figure 3. Scope of the methionine-selective bioconjugation strategy.
(a) Range of polypeptide/protein substrates. (b) Functionalized iodonium reagents compatible with the bioconjugation. (c) A stimuli responsive strategy for reversing methionine bioconjugation.
The modular synthesis of the hypervalent iodine reagents enables facile incorporation of different acyl groups attached to the diazo motif, allowing the transfer of a range of functional payloads to proteins (Figure 3b). Functional groups relevant to other bioorthogonal reactions are readily tolerated in both the reagent synthesis and methionine labelling, smoothly affording sulfonium-conjugates 4b,c with 80-95% conversion to product. Biochemical reporter groups, such as myristyl- and fluorescein-derived esters 4d and 4e, are also readily transferred to exenatide. Interestingly, we found that sulfonium-conjugates of exenatide (such as 4f) underwent reaction with the tertiary phosphine TCEP, a standard biochemical reagent, resulting in the cleavage of the labelling group and return of the parent exenatide 2b in >90% conversion (Figure 3c)25; the cleavage reaction also works for conjugates 5, 6, and 8 in comparable conversions and provides a stimulus-responsive means of reversing the bioconjugation reaction.
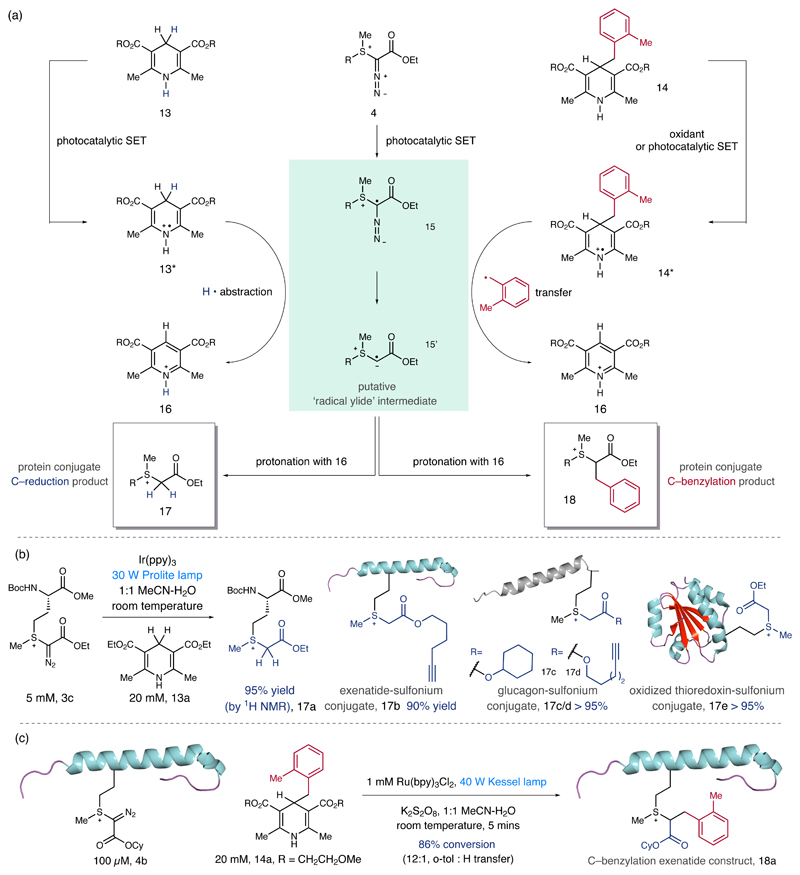
Next, we turned our attention to exploring the multifaceted reactivity that we anticipated would be intrinsic to the high-energy methionine-derived conjugate. The electrophilicity of the diazo sulfonium-conjugate 4 prompted us to investigate the single electron transfer (SET) chemistry of this reactive motif enabled by visible light photocatalysis26,27. Addition of a single electron to the diazo sulfonium conjugate 4 could result in intermediate 15, which upon cleavage of the C–N2 bond would form a putative radical ylide 15’ (Figure 4a). We envisaged two pathways through which we could exploit the reactivity of the previously unexplored radical ylide intermediate. Firstly, combining the radical ylide with photo-activated Hantzsch ester 13* (from 13) may lead to a reduction process resulting in the generation of a trialkylsulfonium motif, which would impart enhanced chemical stability to the protein-conjugate. Furthermore, deployment of a C-4 benzylated Hantzsch ester derivative (14, an established precursor for a benzyl radical)28 to intercept the radical ylide species would lead to a C–benzylation product that could be used to introduce functional diversity to the protein-conjugate. We screened a range of photocatalysts under visible light conditions. When 3c was irradiated with a 30 W lamp in the presence of fac-Ir(ppy)3 and the Hantzsch ester 1329,30, we observed the formation of the reduced trialkylsulfonium product 17a in 95% yield (determined by 1H NMR, Figure 4b)35. Using these conditions, we showed that a range of sulfonium-protein conjugates, including exenatide, glucagon and thioredoxin derivatives are reduced, with excellent conversions, to stable trialkylsulfonium species (17b-e, see the supplementary information S119 for details). Interestingly, reduction of a thioredoxin-derivative31 to its trialkylsulfonium-protein congener 17e proceeds in high conversion without affecting its labile disulfide linkage, serving to highlight the mild nature of this protocol. Additionally, the methionine bioconjugation and photoreduction steps can be carried out in a one pot operation, significantly simplifying the overall process without compromising the yield or purity of the trialkylsulfonium product.
Figure 4. Exploiting the multi-faceted reactivity of the protein-sulfonium conjugate.
(a) A photocatalytic design plan for secondary protein diversification. (b) A system for photo-redox mediated reduction of the sulfonium conjugate and examples of the substrate scope; in the case of 17e, further reduction of C–S bond in 13 was not observed35. Interestingly, the reduction of 13 proceeds in light, without the Ir-photocatalyst, but the yield was greatly diminished. (c). Secondary protein functionalization via photoredox radical cross coupling between diazo sulfonium-protein conjugate and C-4 substituted Hantzsch ester.
In testing the viability of the proposed C–benzylation using a modified Hantzsch ester derivative28, we were delighted to find that treatment of the exenatide-sulfonium conjugate 4b with o-tolyl Hantzsch ester derivative 14a, under slightly modified photocatalytic conditions, led to cross-coupling and the formation of the C-ligation product 18a in high conversion. This unique bioorthogonal protein functionalization reaction not only represents the first example of a synthetic radical-radical cross-coupling using a protein scaffold, but also provides a platform for bioorthogonal protein diversification wherein two distinct functionalities could be introduced sequentially at the same amino acid residue.
In summary, through the merger of methionine-selective bioconjugation and a new visible-light mediated photocatalytic reaction platform, information-rich synthetic constructs can be rapidly assembled by a two-step protocol directly from native proteins. The remarkable reactivity inherent to the methionine conjugate distinguishes the bioconjugation process from other methods. Moreover, the capacity for functional diversification, by tailoring the hypervalent iodine reagent and modified Hantzsch ester derivative, means that highly functional protein conjugates could be made readily available directly from native proteins.
Supplementary Material
Acknowledgments
We thank Dr. Manuel Nappi and Dr. Carine Guerot (AstraZeneca, Cambridge UK) for advice and useful discussions. We thank the Marie Curie Actions program (MTT & MS), AstraZeneca and EPRSC (JEN), and the European Research Council (ERC-SRG-259711), EPSRC (EP/100548X/1) and the Royal Society (Wolfson Merit Award) for fellowships (MJG). We are grateful to Professor Jason Chin, Dr. Nicolas Huguen & Dr. Mark Skehel (Laboratory of Molecular Biology, Cambridge, UK), Dr. Hilary J. Lewis (AstraZeneca, Cambridge UK) and Dr. Matthew J. Edgeworth (MedImmune, Cambridge UK) for assistance with mass spectrometry experiments. Data presented in the paper are included in supplementary materials.
Footnotes
Author contributions
MJG, MTT, JEN and MS conceived the project and designed the experiments. MJG, MTT, JEN and MS performed and analyzed the experiments. MJG, MTT and JEN wrote the paper.
Conflict of interest
The authors declare no conflict of interest
Data availability statement:
The data that support the findings of this study are available within the paper and its supplementary information files. Raw data are available from the corresponding author on reasonable request
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spicer CD, Davis BG. Selective chemical protein modification. Nature Communications. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 4.Koniev O, Wagner A. Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem Soc Rev. 2015;44:5495–5551. doi: 10.1039/c5cs00048c. [DOI] [PubMed] [Google Scholar]
- 5.Dawson PE, Kent SBH. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 6.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 8.Vinodagrova EV, Zhang C, Spokoyny AM, Pentelute BL, Buchwald SL. Organometallic palladium reagents for cysteine bioconjugation. Nature. 2015;526:687–691. doi: 10.1038/nature15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright TH, et al. Posttranslational mutagenesis: a chemical strategy for exploring protein side-chain diversity. Science. 2016;354:597–609. doi: 10.1126/science.aag1465. [DOI] [PubMed] [Google Scholar]
- 10.Yang A, et al. A chemical biology route to site-specific authentic protein modifications. Science. 2016;354:623–626. doi: 10.1126/science.aah4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abegg D, Frei R, Cerato L, Prasad Hari D, Wang C, Waser J, Abidekian A. Proteome-wide profiling of targets of cysteine reactive small molecules by using ethynyl benziodoxolone reagents. Angew Chem Int Ed. 2015;54:10852–10857. doi: 10.1002/anie.201505641. [DOI] [PubMed] [Google Scholar]
- 12.Levine RL, Moskovitz J, Stadtman ER. Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. Life. 2000;50:301–307. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- 13.Cowie DB, Cohen GN, Bolton ET, De Robichon-Szulmajster H. Amino acid analog incorporation into bacterial proteins. Biochem Biophys Acta. 1959;34:39–46. doi: 10.1016/0006-3002(59)90230-6. [DOI] [PubMed] [Google Scholar]
- 14.Lin S, Yang X, Jiu S, Weeks AM, Hornsby M, Lee PS, Nichiporuk RV, Iavarone AT, Wells JA, Toste FD, Chang CJ. Redox-based reagents for chemoselective methionine bioconjugation. Science. 2017;355:597–602. doi: 10.1126/science.aal3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross E, Witkop B. Nonenzymatic cleavage of peptide bonds: The methionine residue in bovine pancreatic ribonuclease. J Biol Chem. 1962;237:1856–1860. [PubMed] [Google Scholar]
- 16.Gundlach HG, Moore S, Stein WH. The nature of the amino acid residues involved in the inactivation of ribonuclase by iodoacetate. J Biological Chem. 1959;234:1754–1760. [PubMed] [Google Scholar]
- 17.Vithayathil PJ, Richards FM. Modification of the Methionine residue in the peptide component of ribonuclease-S. J Biological Chem. 1960;235:2343–2351. [Google Scholar]
- 18.Kramer JR, Deming TJ. Preparation of multifunctional and multireactive polypeptides via methionine alkylation. Biomacromolecules. 2012;13:1719–1723. doi: 10.1021/bm300807b. [DOI] [PubMed] [Google Scholar]
- 19.Kramer JR, Deming TJ. Reversible chemoselective tagging and functionalization of methionine containing peptides. Chem Commun. 2013;49:5144–5146. doi: 10.1039/c3cc42214c. [DOI] [PubMed] [Google Scholar]
- 20.Stang PJ, Zhdankin VV. Organic polyvalent iodine compounds. Chem Rev. 1996;96:1123–1178. doi: 10.1021/cr940424+. [DOI] [PubMed] [Google Scholar]
- 21.Weiss R, Seubert J, Hampel F. α-Aryliodonio diazo compounds: SN reactions at the α-C atom as a novel reaction type for diazo compounds. Angew Chem Int Ed. 1994;33:1952–1953. [Google Scholar]
- 22.Schnaars C, Hennum M, Bonhe-Hansen T. Nucleophilic halogenations of diazo compounds, a complementary principle for the synthesis of halodiazo compounds: experimental and theoretical studies. J Org Chem. 2013;78:7488–7497. doi: 10.1021/jo401050c. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y, Ho SO, Gassman NR, Korlann Y, Landorf EV, Collart FR, Weiss S. Efficient site-specific labelling of proteins via cysteines. Bioconj Chem. 2008;19:786–791. doi: 10.1021/bc7002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mülhberg M, Hösl MG, Kühne C, Dernedde J, Budisa N, Hackenberger CPR. Belstein JOC. 2015;11:784–791. doi: 10.3762/bjoc.11.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staundinger H, Lüscher G. Über darstellung und Reaktionen von Phosphazinen. Helv Chim Acta. 1922;5:75–86. [Google Scholar]
- 26.Prier CK, Rankic DA, Macmillan DWC. Visible light photoredox catalysis with transition metal complexes: applications in organic synthesis. Chem Rev. 2013;113:5322–5363. doi: 10.1021/cr300503r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Kamlet AS, Steinman JB, Liu DR. A biomolecule-compatible visible-light-induced azide reduction from a DNA-encoded reaction-discovery system. Nature Chemistry. 2011;3:146–153. doi: 10.1038/nchem.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Cheng X. Hantzsch esters as multifunctional reagents in visible-light photoredox catalysis. Synlett. 2017;28:148–158. [Google Scholar]
- 29.Fukuzimi S, Hironaka K, Tanaka T. Photoreduction of alkyl halides by an NADH model compound. An electron-transfer chain mechanism. J Am Chem Soc. 1983;105:4722–4727. [Google Scholar]
- 30.Hedstrand DM, Kruizinga WH, Kellog RM. Light induced and dye accelerated reductions of phenacyl onium salts by 1,4-dihydropyridines. Tetrahedron Lett. 1978;19:1255–1258. [Google Scholar]
- 31.Krause G, Lundström J, Barea JL, de la Cuesta CP, Holmgren A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J Biol Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.