Abstract
Although several studies have evaluated the role of p16INK4a as a diagnostic marker of cervical intraepithelial neoplasia (CIN) and its association with disease progression, studies regarding the role of p16INK4a in human immunodeficiency virus (HIV)-infected patients remain scarce. The present study was designed to determine the potential utility of p16INK4a as a diagnostic marker for CIN and invasive cervical cancer in HIV-positive and negative cervical specimens. An immunohistochemical analysis of p16INK4a was performed in 326 cervical tissue microarray specimens. Performance indicators were calculated and compared using receiving operating characteristics curve (ROC)/area under the curve. In HIV-1-negative women, the percentage of cells that was positive for p16INK4a expression was significantly correlated with the severity of CIN (p < 0.0001). A ROC curve with a cut-off value of 55.28% resulted in a sensitivity of 89%, a specificity of 81%, a positive predictive value of 91% and a negative predictive value of 78%. HIV-seropositive women exhibited decreased expression of p16INK4a in CIN2–3 specimens compared with HIV-negative specimens (p = 0.031). The ROC data underscore the potential utility of p16INK4a under defined conditions as a diagnostic marker for CIN 2–3 staging and invasive cervical cancer. HIV-1 infection, however, is associated with relatively reduced p16INK4a expression in CIN 2–3.
Keywords: CIN, p16INK4a, HPV, HIV, TMA, ROC
P16INK4a is a cyclin-dependent kinase inhibitor that regulates the transition from the G1 to S phase and negatively influences cell proliferation in conjunction with other tumour suppressor proteins, such as the retinoblastoma gene (pRb) (Tringler et al. 2004). As pRb is functionally inactivated by the high-risk human papilloma virus (HPV) oncoprotein E7, there is a concomitant overexpression of p16INK4a. New data suggest that p16 overexpression in E7-expressing cells does not appear to result from pRB degradation, but from the induction of histone demethylases by HPV E7 (McLaughlin-Drubin et al. 2011). Although p16INK4a immunostaining has been correlated with the severity of cytological and histological abnormalities in cervical lesions, variations in interpretation and a lack of standardised methodologies has resulted in uncertainty regarding the most appropriate cut-offs for the analysis of P16INK4a levels (Branca et al. 2004, Wentzensen et al. 2005, Valasoulis et al. 2011). This dilemma is underscored by the fact that P16INK4a expression can be upregulated in non-dysplastic cervical lesions, including common squamous metaplasia. More-over, published studies for the diagnostic performance of P16INK4a in human immunodeficiency virus (HIV)-infected patients remain limited (Queiroz et al. 2006, Kreuter et al. 2010). In the present study, we assessed the expression of p16INK4a in normal and abnormal cervical epithelium in HIV-positive and negative women and determined the diagnostic performance [receiving operating characteristics curve (ROC), sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV)] of P16INK4a for detecting cervical intraepithelial neoplasia (CIN) and invasive cervical cancer in tissue microarray (TMA) samples in each group.
SUBJECTS, MATERIALS AND METHODS
TMA and patient samples
A total of 326 TMA specimens of formalin-fixed, paraffin-embedded cervical tissues (from individual patients) were immunohistochemically analysed. One hundred sixty-nine specimens were obtained from the archive files of the Department of Pathology at the Fernandes Figueira Institute, Oswaldo Cruz Foundation (Fiocruz), Rio de Janeiro, Brazil, between March-December 2009. Among these specimens, 31 were cervical samples from HIV/HPV coinfected patients. The TMA blocks were constructed as previously described by Pires et al. (2006). The punches were 1 mm in diameter and consisted of two cores with full thickness of the cervical epithelium. Clinical information was obtained from the patients’ medical records. Another set of four TMA slides (a total of 142 specimens) was obtained from US Biomax-USA (CR 804, CIN 481, BC 10021 and CR 2081) and all of the specimens tested negative for HIV. In addition, 15 cases of CIN were obtained from the Department of Pathology at Ohio State University, USA. The cases were chosen at random from those available in each diagnostic category. The specimens were identified by a final diagnosis on the histopathology report. The present study was approved by the Fiocruz Institutional Ethical Review Board.
Immunohistochemical analysis
Five micron sections were cut onto silane-coated slides (Sigma, St. Louis, MO, USA) and processed for immunohistochemistry as previously described (Nicol et al. 2008a). We used mouse monoclonal primary antibodies against p16INK4a (CINtec Cytology kit, MTM Lab). Briefly, tissue slides were deparaffinised and antigen retrieval was performed by treating the sections with target retrieval solution, pH 6.0 (S1699, DAKO, Copenhagen, Denmark). Primary antibody (100 μL) was applied in a humidified chamber at 4°C overnight. The LSAB system HRP (Dakocy-tomation, Carpinteria, CA, USA) method was adapted for immunolabelling with the universal biotinylated link antibody and the slides were incubated with the streptavidin-HRP conjugate for 30 min. The slides were washed three times in Tris (pH 7.6) between each incubation step. Antibody binding was visualised with 3,3’-diaminobenzidine (Sigma Chemical Co, St. Louis, MO, USA) and 85 µL of 0.3% hydrogen peroxide. Finally, the slides were counterstained with haematoxylin, dehydrated and mounted in a resinous mounting medium (Merck, Darmstadt, Germany). Negative controls were generated for all of the tissues by omitting the primary antibody.
Immunohistochemical evaluation
The microscopic analysis of the slides was independently performed by two investigators. Digitalised photographs were taken with a Nikon Coolpix camera DP12 and the images were stored in a computer-based software program for documentation. Quantitative results were expressed as the percentage of positive cells per field on total cell count. Only cells within the cervical epithelium were counted. All of the section slides were assessed at 400X magnification and separately evaluated by two observers. At least 200 nuclei were assessed in each case. The counts were performed manually and the percentage of posi-tively stained cells in representative microscopic fields was recorded. The TMA was reviewed by an experienced molecular pathologist (GJ Nuovo). The reaction was considered positive for p16INK4a when a dark brown colour was observed in the nuclei and/or cytoplasmic compartments. The results were compared with the HIV serostatus of the patients.
In situ hybridisation and co-expression analyses
We performed HPV in situ hybridisation on selected cases using a previously published protocol (Nicol et al. 2008a). The HPV in situ hybridisation was performed to analyse the levels of HPV and the expression levels of P16INK4a in the same sections to assess whether HPV infection influenced P16INK4a expression. This analysis was performed with the Nuance system as previously described by Nuovo (2010). Briefly, the Nuance system isolates the blue and brown spectra of the HPV DNA and P16INK4a protein, respectively, converts them to fluorescence-based signals and combines the two to determine if a given cell is producing none, one, or both of the targets.
Statistical analysis
Data analysis was conducted using SAS version 9.1 and R 2.11.1. The variables of p16INK4a expression are presented as medians and interquartile ranges. Correlations with different CIN grades were determined by counting cells (% stained cells/field). Mann-Whitney U, Kruskal-Wallis and Dunn’s tests were applied to compare the means of the positive cells in the epithelium of all of the cases [controls, low-grade CIN, high-grade (HGCIN) and invasive cancer]. A p value < 0.05 indicated statistical significance. The accuracy of p16INK4a to diagnose HGCIN and invasive tumours was evaluated using receiver ROC analyses according to standard protocols (Eng 2005, Fan et al. 2006). The ROC curve was determined to obtain the best cut-off value and the highest sensibility and specificity point corresponding to the highest deviation to the left curve or the nearest of the two arms.
RESULTS
HIV-seropositive subjects
A total of 31 HIV-infected cervical specimens were analysed. Fourteen were CIN 1 and seventeen were CIN 2–3. The mean age of the HIV-seropositive subjects was 36 (the range was 31–42).
HIV-seronegative subjects
A total of 295 HIV-negative cervical specimens were included in the present study (67 normal controls, 24 CIN 1, 20 CIN 2–3 and 184 invasive tumours). The mean age of the HIV-seronega-tive patients was 44 (the range was 39–50).
Immunohistochemical data
Among the 67 histo-logically normal samples (controls) that were used in the present study, only one case exhibited a weak positive reaction for P16INK4a. P16INK4a expression increased in the basal, middle and superficial layers (all compartments) during the progression from CIN 1 to CIN 2–3 and invasive tumour stages (Fig. 1, Table I). Dunn’s test revealed statistically significant differences in the number of positively stained cells in the cervical epithelium between the control and CIN 1–3 specimens, the control and invasive cancer specimens and the invasive cancer and CIN 1 specimens (p < 0.0001).
Fig. 1:
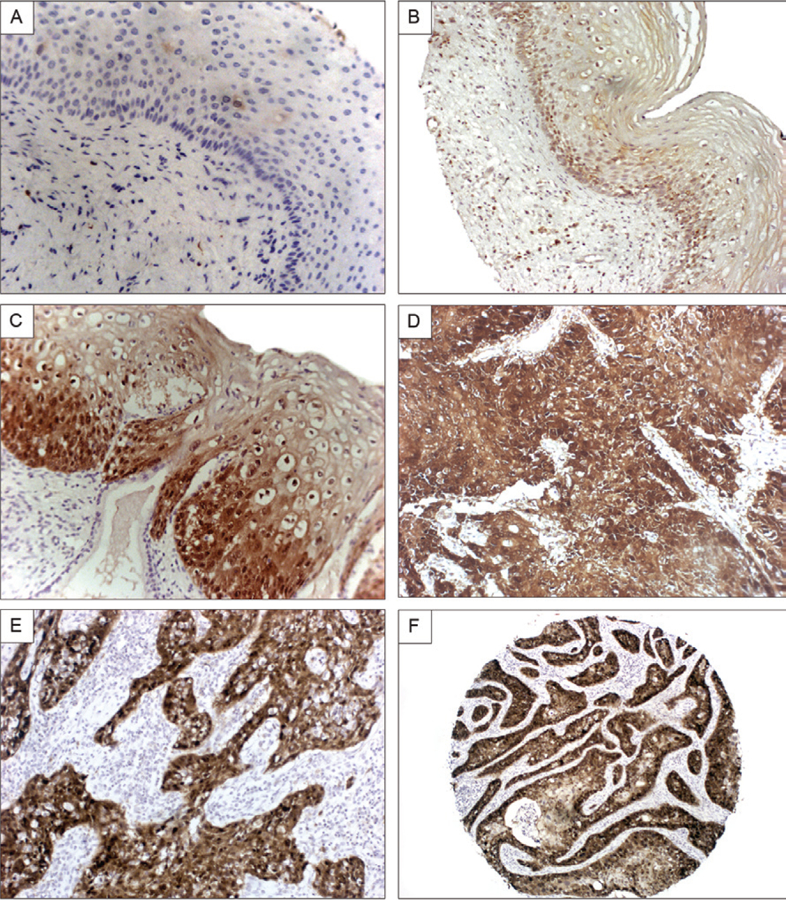
correlation of p16INK4a expression with histologic features of cervical lesions. A shows the lack of expression of p16INK4a in normal cervical epithelia. B [human immunodeficiency virus (HIV) positive cervice, low magnification] and C (HIV negative cervice, high magnification) illustrate the increased basal expression of the protein in low-grade cervical intraepithelial neoplasia lesions. In comparison, a much greater percentage of the epithelial cells are shown to express p16INK4a in the invasive cervical cancers (D, E). F shows a very low magnification of a cervical cancer that underscores that the signal is present in the cancer cells and not the surrounding stromal cells.
TABLE I.
Crude table showing the total of stained cells in the cervical epithelium
| p16INK4a | Total (%) |
|---|---|
| Normal controls | |
| Mean (EP) | 0.1 (0.1) |
| Median (IQR) | 0 (0–0) |
| n | 67 |
| LGCIN | |
| Mean (SD) | 15.1 (4.7) |
| Median (IQR) | 0.5 (0–11.7) |
| n | 38 |
| HGCIN | |
| Mean (SD) | 31.8 (6.8) |
| Median (IQR) | 0.7 (0.6–76.2) |
| n | 37 |
| Invasive cancer | |
| Mean (SD) | 41.7 (2.9) |
| Median (IQR) | 53.2 (0.8–77.8) |
| n | 184 |
| p | < 0.0001 |
Dunn’s test for multiple comparisons revealed significant differences among normal controls compared with low-grade cervical intraepithelial neoplasia (LGCIN), high-grade (HGCIN) and invasive tumour and between LGCIN compared to invasive tumour; b: one case was positive in control. The values are expressed in standard deviation (SD) and interquartile range (IQR).
Table I depicts the total percentage of positive cells for p16INK4a according to the histological diagnosis in all of the analysed samples. Decreased expression of p16INK4a was observed in the CIN 2–3 HIV-positive cervices compared with the corresponding HIV-negative CIN 2–3 lesions (p = 0.031) (Table II).
TABLE II.
Comparison of p16INK4a expression between human immunodeficiency virus (HIV) positive and negative cervices
| p16INK4a |
|||
|---|---|---|---|
| Crude stained cells/field (%) |
|||
| Histological diagnosis |
|||
| HIV negative | HIV positive | p | |
| LGCINa | |||
| Mean (SD) | 23.6 (6.9) | 0.5 (0.5) | 1.000 |
| Median (IQR) | 2.7 (0–49.7) | 0.5 (0.3–0.6) | - |
| n | 24 | 14 | - |
| HGCIN | |||
| Mean (SD) | 58.4 (9.1) | 0.6 (0.6) | 0.031 |
| Median (IQR) | 76.1 (0–91.2) | 0.6 (0.6–0.7) | - |
| n | 20 | 17 | - |
Mann-Whitney test; HGCIN: high-grade cervical intraepithelial neoplasia; IQR: interquartile range; LGCIN: low-grade cervical intraepithelial neoplasia; SD: standard deviation.
No staining was observed in the cervical epithelia of the control specimens. CIN 1 specimens exhibited a diffuse and intense staining pattern in the dysplastic areas that was typically limited to the lower one-third (basal layer) of the epithelium. CIN 2–3 specimens exhibited intense staining in all of the epithelial layers. The invasive tumour specimens exhibited an intense and diffuse staining with a high frequency of P16INK4a positivity in the neoplastic cells (Fig. 2).
Fig. 2:
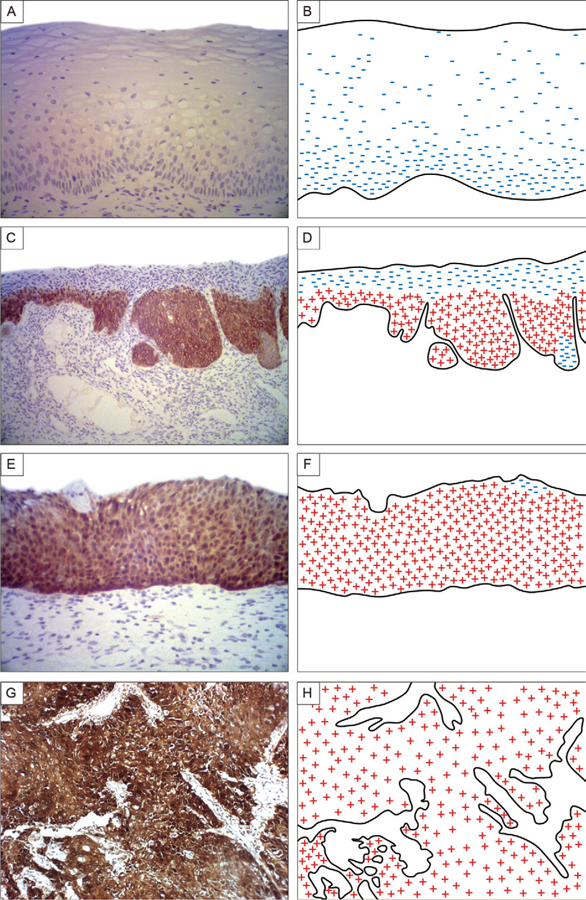
a simplified schematic representation of the different expression patterns in the cervical epithelium. No staining was observed in the cervical epithelium of control specimens (A, B). Low-grade cervical intraepithelial neoplasia (CIN) showed a diffuse and intense staining pattern in the displasic area limited to the one-third basal layer of the epithelium (C, D). High-grade CIN showed an intense staining in all epithelium layers (E, F). The invasive tumour specimens showed an intense and diffuse staining with high frequency of p16INK4a positivity in neoplastic cells (G, H).
We investigated whether the p16INK4a-positive cells were contributing to HPV infection because CIN lesions are invariably associated with HPV infection. Using the Nuance system to analyse the co-labelling of HPV 16 DNA in situ and the p16INK4a, we found few cells with detectable HPV 16 DNA that exhibited co-expression of p16INK4a. Interestingly, the cells that co-expressed HPV 16 DNA and p16INK4a were invariably located towards the surface in the lesion. Towards the base of the CIN, p16INK4a-positive cells were not found with detectable HPV 16 DNA, which reflected the generally low copy of HPV DNA that is typical of the basal cells of CIN lesions (Fig. 3). Thus, our findings show that the p16INK4a-positive cells lack the cytological features of productive HPV infection (i.e., the koilocyte) in most cases and a poor correlation between in situ detection of HPV 16 DNA and p16INK4a protein co-expression.
Fig. 3:
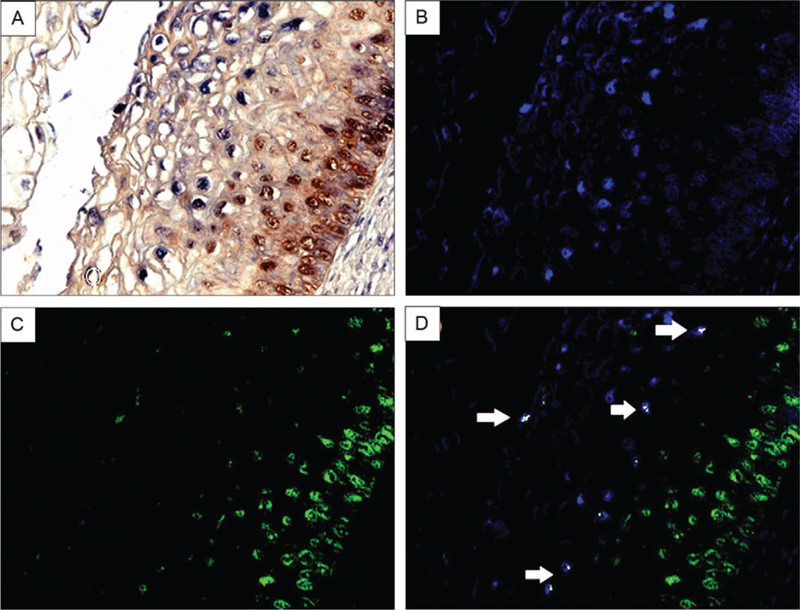
co-expression analysis of p16 INK4a and human papilloma virus (HPV) DNA 16 in low-grade cervical intraepithelial neoplasia (CIN). Co-expression analysis of HPV DNA 16 and p16INK4a in a CIN1 lesion. HPV DNA 16 was detected by in situ hybridization (blue signal), followed by co-expression analysis of p16 (brown signal) in the same tissue (A). The image was then analyzed by the computer-based NUANCE system which converted the HPV DNA signal to fluorescent blue (B) and p16 to fluorescent green (C). These latter two images are merged where cells with detectable HPV 16 and p16 would be fluorescent yellow (D); note the absence of any co-expression of these two targets where p16 dominates in the basal layer and HPV DNA is most abundant towards the cells at the surface of the CIN lesion.
Evaluation of the sensitivity and specificity of p16INK4a as a diagnostic indicator of HGCIN and invasive tumours
Performance indicators were calculated and compared using the ROC curve with a cut-off of 55.28% and an area under the curve (AUC) of 0.8884%. A total of 326 specimens were used to construct the ROC curve. Under these conditions, the ability of p16INK4a expression to diagnose either CIN 2–3 or invasive cervical cancers had a relatively high sensitivity (89.14%), specificity (81.90%), PPV (91.20%) and NPV (78.18%). The results of the ROC curve, sensitivity and specificity calculations are shown in Fig. 4.
Fig. 4:

diagnostic performance of p16INK4a under receiving operating characteristics/area under the (ROC/AUC) curves. The performance of p16INK4a under ROC/AUC curve to predict high-grade cervical intraepithelial neoplasia and invasive tumour after applying the cut off of 55.28% in all analyzed specimens that resulted in relatively high sensitivity (89.14%), specificity (81.90%), positive predictive value (PPV), (91.20%) and negative predictive value (NPV) (78.18%). CI: confidence interval.
The ability of P16INK4a under the ROC/AUC curve to predict HGCIN and invasive tumours after applying the cut-off of 55.28% to all of the analysed specimens resulted in relatively high sensitivity (89.14%), specificity (81.90%), PPV (91.20%) and NPV (78.18%).
DISCUSSION
There are several reports on the prognostic value of p16INK4a in diagnosing CIN (Branca et al. 2004, Tringler et al. 2004, Queiroz et al. 2006). However, the sensitivity and specificity estimates are confounded by a lack of definitive standards that differentiate between background noise and signal. Background noise not only refers to nonspecific staining of epithelial tissue, but also to the well-documented observation that p16INK4a protein can be detected in cervical cells that are clearly not dysplastic. In the present study, we found that p16INK4a expression increased from normal tissue to progressively invasive cervical cancer as defined by the percentage of positive squamous cells per category. We employed an ROC curve to test the predictive power of p16INK4a as a diagnostic marker for CIN 2–3 and invasive cervical cancer. ROC analysis addresses the variance of sensitivity and specificity and the AUC is the most commonly used index of performance associated with ROC analysis (Eng 2005). A cut-off value of 55.28% was established as described a: Dunn’s test for multiple comparisons revealed significant differences among normal controls compared with low-grade cervical intraepithelial neoplasia (LGCIN), high-grade (HG-CIN) and invasive tumour and between LGCIN compared to invasive tumour; b: one case was positive in control. The values are expressed in standard deviation (SD) and interquartile range (IQR). above and a relatively high sensitivity (89%), specificity (81%), PPV (91%) and NPV (78%) were obtained, which reinforced that p16INK4a might be a useful adjunct marker to diagnosis and distinguish CIN from other similar tumours and establish the risk of CIN 2–3. These findings are consistent with previous studies in terms of the potential utility of p16INK4a as a predictive CIN marker (Branca et al. 2004, Tringler et al. 2004, Wang et al. 2004, Queiroz et al. 2006). A previous study found similar nuclear staining for p16INK4a, but reported variation in cytoplasmic intensity according to the CIN grade, which suggests that the overexpression of p16INK4a in the cytoplasm in higher grade lesions might reflect the increased synthesis of p16INK4a (Queiroz et al. 2006). The present study found a continuous staining pattern from the basement membrane that extended upward in proportion to the lesion grade, which was consistent with a previous report (Galgano et al. 2010). The present observations suggest that the staining pattern might be the more important variable in the interpretation of p16INK4a (i.e., rather than the signal intensity in any given cell). Discrepant results have been reported regarding sensitivity, specificity, PPV and NPV for p16INK4a in different clinical and pathologic scenarios in CIN (Branca et al. 2004, Tringler et al. 2004, Queiroz et al. 2006). These disparate results could be attributed in part to the variations in the interpretation of positive immunostaining and by the lack of standardised methodology. One large cohort study found that the PPV varied from 2.5–20.4% and the NPV varied from 99.7–100% across various studies (Wang et al. 2004).
In agreement with the results of the present study, a previous Brazilian clinical trial that diagnosed 90 CIN2 and high-risk human papillomavirus infections concluded that p16INK4a expression could be useful in the diagnosis of CIN2; however, the trial failed to predict a: Mann-Whitney test; HGCIN: high-grade cervical intraepithelial neoplasia; IQR: interquartile range; LGCIN: low-grade cervical intraepithelial neoplasia; SD: standard deviation. the outcome of CIN2. Importantly, no ROC curve was constructed to assess the accuracy of each immunostaining (Guedes et al. 2007).
Few reports are available regarding the diagnostic performance of p16NK4a in HIV-infected patients (Queiroz et al. 2006, Kreuter et al. 2010). One previous report (Kreuter et al. 2010) evaluated p16NK4a performance in HIV anal intraepithelial neoplasia (AIN) samples and found 100% sensitivity and 100% specificity to diagnose high-grade AIN. However these reports only presented the sensitivity and specificity of p16NK4a. We were able to calculate the PPV, NPV and ROC curves to establish the best cut-off point for p16NK4a. Previously, we demonstrated significantly altered expression of regulatory and cell cycle proteins in the cervix between HIV-positive and HIV-negative cervices (Nicol et al. 2008b). In the present study, we observed a decreasing percentage of p16INK4a-positive stained cells in the HIV-positive cervices compared with the HIV-negative cervices in both CIN 1 and CIN 2–3. This result appears contradictory to the well-established notion that HIV-1 infection increases the risk of transition from CIN 1 to CIN 2–3 and invasive cancer progression in women. Nevertheless, the increased risk of cervical cancer in HIV-1-infected women likely reflects the inability of the patients to clear the HPV from the cervix due to their immunocompromised state (Nuovo & Pedemonte 1990); however, the basis for this observation warrants further study. Importantly, studies have established that HIV-1 rarely infects CIN cells (Nicol et al. 2005); thus, the reduced expression of p16INK4a in HIV-1-positive CIN epithelia cannot reflect a direct reduction of p16INK4a by HIV-1. In agree-ment with the results of the present study, co-infection of HPV and HIV-1 likely results in a markedly altered cytokine profile in the lamina propria of the cervix, including factors such as interleukin (IL)-6, which could result in decreased p16INK4a expression in CIN cells.
Limitations of the present study included a relatively small number of HIV-positive specimens, which limited adequate subgroup analyses and comparisons amongst HIV-infected cervices. Recent studies have suggested that testing the performance of p16INK4a in combination with other biomarkers could improve its performance (Galgano et al. 2010, Gupta et al. 2010), but we were unable to assess combinations in this study. Another limitation is that the ROC/AUC values were obtained with a convenience sample, which limits the reproducibility of our findings. However, this is the typical procedure employed by similar studies (Pinto et al. 2008, Galgano et al. 2010).
The present data revealed effective sensitivity and specificity of p16INK4a and confirm the potential utility of p16INK4a as a diagnostic marker for CIN, particularly in CIN 2–3 and invasive cervical cancer lesions. In addition, the present study found significantly decreased expression of p16INK4a from CIN 1 to CIN 2–3 in HIV-1-positive cervices (p = 0.031), which underscored another reason for caution when interpreting the immunohistochemical profile of p16INK4a in cervical lesions.
ACKNOWLEDGEMENTS
To Margareth Nuovo, Rodrigo Mexas and Heloisa MN Diniz, for figures.
Financial support: LIPMED-IOC-FIOCRUZ, Lewis Foundation AFN and SMAF are scholars in IARTP and are supported by the FIC, NIH Research (2 D 43 TW000010–23-AITRP), NO and WM are scholars of FAPERJ and SMAF from CAPES.
REFERENCES
- Branca M, Ciotti M, Santini D, Di Bonito L, Giorgi C, Benedetto A, Paba P, Favalli C, Costa S, Agarossi A, Alderisio M, Syrjänen K 2004. p16(INK4A) expression is related to grade of cin and high-risk human papillomavirus but does not predict virus clearance after conization or disease outcome. Int J Gynecol Pathol 23: 354–365. [DOI] [PubMed] [Google Scholar]
- Eng J 2005. Receiver operating characteristic analysis: a primer. Acad Radiol 12: 909–916. [DOI] [PubMed] [Google Scholar]
- Fan J, Upadhye S, Worster A 2006. Understanding receiver operating characteristic (ROC) curves. CJEM 8: 19–20. [DOI] [PubMed] [Google Scholar]
- Galgano MT, Castle PE, Atkins KA, Brix WK, Nassau SR, Stoler MH 2010. Using biomarkers as objective standards in the diagnosis of cervical biopsies. Am J Surg Pathol 34: 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes AC, Brenna SM, Coelho SA, Martinez EZ, Syrjanen KJ, Zeferino LC 2007. p16(INK4a) expression does not predict the outcome of cervical intraepithelial neoplasia grade 2. Int J Gynecol Cancer 17: 1099–1103. [DOI] [PubMed] [Google Scholar]
- Gupta N, Srinivasan R, Rajwanshi A 2010. Functional biomarkers in cervical precancer: an overview. Diagn Cytopathol 38: 618–623. [DOI] [PubMed] [Google Scholar]
- Kreuter A, Jesse M, Potthoff A, Brockmeyer NH, Gambichler T, Stucker M, Bechara FG, Pfister H, Wieland U 2010. Expression of proliferative biomarkers in anal intraepithelial neoplasia of HIV-positive men. J Am Acad Dermatol 63: 490–498. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Crum CP, Munger K 2011. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA 108: 2130–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol AF, Fernandes AT, Grinsztejn B, Russomano F, Tristao A, Perez M de A, Nuovo GJ, Martinez-Maza O, Bonecini-Almeida M da G 2005. Distribution of immune cell subsets and cytokine-producing cells in the uterine cervix of human papillomavirus (HPV)-infected women: influence of HIV-1 coinfection. Diagn Mol Pathol 14: 39–47. [DOI] [PubMed] [Google Scholar]
- Nicol AF, Nuovo GJ, Salomao-Estevez A, Grinsztejn B, Tristao A, Russomano F, Lapa ESJR, Oliveira MP, Pirmez C 2008a. Immune factors involved in the cervical immune response in the HIV/HPV co-infection. J Clin Pathol 61: 84–88. [DOI] [PubMed] [Google Scholar]
- Nicol AF, Pires AR, de Souza SR, Nuovo GJ, Grinsztejn B, Tristao A, Russomano FB, Velasque L, Lapa e Silva JR, Pirmez C 2008b. Cell-cycle and suppressor proteins expression in uterine cervix in HIV/HPV co-infection: comparative study by tissue micro-array (TMA). BMC Cancer 8: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo GJ 2010. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods 52: 307–315. [DOI] [PubMed] [Google Scholar]
- Nuovo GJ, Pedemonte BM 1990. Human papillomavirus types and recurrent genital warts. JAMA 263: 1223–1226. [PubMed] [Google Scholar]
- Pinto AP, Schlecht NF, Woo TY, Crum CP, Cibas ES 2008. Bio-marker (ProEx C, p16(INK4A) and MiB-1) distinction of high-grade squamous intraepithelial lesion from its mimics. Mod Pathol 21: 1067–1074. [DOI] [PubMed] [Google Scholar]
- Pires AR, Andreiuolo F da M, de Souza SR 2006. TMA for all: a new method for the construction of tissue microarrays without recipient paraffin block using custom-built needles. Diagn Pathol 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz C, Silva TC, Alves VA, Villa LL, Costa MC, Travassos AG, Filho JB, Studart E, Cheto T, de Freitas LA 2006. P16(INK4a) expression as a potential prognostic marker in cervical pre-neo-plastic and neoplastic lesions. Pathol Res Pract 202: 77–83. [DOI] [PubMed] [Google Scholar]
- Tringler B, Gup CJ, Singh M, Groshong S, Shroyer AL, Heinz DE, Shroyer KR 2004. Evaluation of p16INK4a and pRb expression in cervical squamous and glandular neoplasia. Hum Pathol 35: 689–696. [DOI] [PubMed] [Google Scholar]
- Valasoulis G, Tsoumpou I, Founta C, Kyrgiou M, Dalkalitsis N, Na-sioutziki M, Kassanos D, Paraskevaidis E, Karakitsos P 2011. The role of p16(INK4a) immunostaining in the risk assessment of women with LSIL cytology: a prospective pragmatic study. Eur J Gynaecol Oncol 32: 150–152. [PubMed] [Google Scholar]
- Wang SS, Trunk M, Schiffman M, Herrero R, Sherman ME, Burk RD, Hildesheim A, Bratti MC, Wright T, Rodriguez AC, Chen S, Reichert A, von Knebel Doeberitz C, Ridder R, von Knebel Doeberitz M 2004. Validation of p16INK4a as a marker of oncogenic human papillomavirus infection in cervical biopsies from population-based cohort in Costa Rica. Cancer Epidemiol Bio-markers Prev 13: 1355–1360. [PubMed] [Google Scholar]
- Wentzensen N, Bergeron C, Cas F, Eschenbach D, Vinokurova S, von Knebel Doeberitz M 2005. Evaluation of a nuclear score for p16INK4a-stained cervical squamous cells in liquid-based cytology samples. Cancer 105: 461–467. [DOI] [PubMed] [Google Scholar]


