Abstract
Background
Protozoan parasites such as Giardia duodenalis, Cryptosporidium spp., Cyclospora cayetanensis, Toxoplasma gondii and Entamoeba histolytica represent a great challenge to the systems producing water for human consumption because their cystic forms are persistent in the environment and resist to the disinfection methods conventionally used for their control. In this study, we investigated the presence of these protozoan pathogens in both raw and treated water samples used for the production of drinking water in Nariño Department, southwest Colombia. We collected 110 water samples (10 lof each sample) and analyzed them with real-time PCR (qPCR). qPCR-positive samples were genotyped with PCR and DNA sequencing.
Results
Giardia duodenalis was detected in 35/110 (31.8%) of the samples and Cryptosporidium spp. in 9/110 (8.2%) of the samples; no sample was positive for T. gondii, E. histolytica or C. cayetanensis. Giardia duodenalis was detected in samples of both raw water (Drinking Water Treatment Plants (DWTP): 47.83%;Drinking Water Rural Plants (DWRP): 18.42%) and water collected either after conventional physicochemical treatment (26.09%) or after disinfection by chlorine (50%), whereas Cryptosporidium spp. were only detected in raw waters (DWTP: 17.39%; DWRP: 13.16%). The two pathogens were detected in both types of treatment plants supplying water to urban areas and to rural zones. Analysis of gdh and tpi markers identified assemblages AI, AII and H of G. duodenalis, while analysis of the small subunit rRNA and gp60 markers of Cryptosporidium-positive samples identified C. parvum (Subtype IIcA5G3c), C. galli, C. molnari, Cryptosporidium sp. genotype II of bats and Cryptosporidium sp. genotype VIII of birds.
Conclusions
The results obtained demonstrate the presence of protozoan parasites in the water of the study region, and the need to improve the surveillance systems for these pathogens and identify the corresponding sources of contamination.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-3147-3) contains supplementary material, which is available to authorized users.
Keywords: Protozoan parasites, Raw water, Treated water, PCR, Sequencing analysis
Background
Water is an essential resource for life and thus access to safe water is currently considered a fundamental human right [1]. However, more than a billion people currently lack access to drinking water worldwide, presenting a risk for public health in affected regions [2]. In Colombia, on average, 78% of the population has access to drinking water; however, there are large differences in coverage between urban and rural areas, and it has been reported that around 1300 children die each year from diarrheal diseases caused by unsafe quality water consumption [3]. Nariño is a department located in southwest Colombia that has a risk index of water quality for human consumption (IRCA) of 50.27, which places it as a department at high risk within the country. Among the health problems related to water in Nariño, acute diarrheal disease has an incidence rate of 65.8 cases per 1000 inhabitants and a mortality rate of 18.2 cases per 1,000,000 inhabitants [4]. Additionally, in this department, an outbreak was recorded in 2017 with 2560 cases of acute diarrheal disease, in which it is presumed that water was the main vehicle of transmission of the causal agent (not yet identified) [5].
The most common and widespread risk associated with water is contamination by pathogenic microorganisms, such as viruses, bacteria and helminth and protozoan parasites. Giardia and Cryptosporidium, the causative agents of giardiasis and cryptosporidiosis, respectively, are the protozoan parasites most commonly associated with transmission by water. These pathogens affect not only humans, but a wide range of domestic and wild animals. Similarly, protozoan parasites such as Entamoeba histolytica, Toxoplasma gondii and Cyclospora cayetanensis, responsible for amebiasis, toxoplasmosis and cyclosporiasis, respectively, may also be transmitted by contaminated water sources and affect global health [6]. These protozoan parasites have been responsible for large numbers of outbreaks worldwide, in the period 2011–2016 at least 381 outbreaks caused by the transmission of water-borne parasitic protozoa were reported [7–9]. Protozoan parasites also represent a challenge to the production of water suitable for human consumption because their transmissible forms (cysts and oocysts) are highly stable and persistent in the environment, they can cross the physical barriers used to remove contaminants, and are resistant to several conventional disinfectants widely used in the treatment system of drinking water, such as chlorine and chloramines [10–12]. Finally, it must be emphasized that these pathogens can cause infection at rather low concentrations [13].
Current regulations in Colombia recently included the monitoring of Giardia spp. and Cryptosporidium spp. in the parameters for the microbiological control of water quality in the treatment plants responsible for the distribution of drinking water [14]. In Colombia, the presence of Giardia cysts and Cryptosporidium oocysts is regularly checked with immunofluorescence microscopy, according to the method validated by the United States Environmental Protection Agency in municipal drinking water (USEPA 1623). However, this method has several disadvantages and limitations, described by several authors as extensive experience in the microscopic differentiation of cystic forms and rigorous laboratory staff, high costs, subject to interference resulting from the presence of ions in the sample (manganese, iron and calcium) and it is not capable to identify species or genotypes, which is important for the determination of public health significance [15, 16]. As far as we know, to date there are no validated methodologies for the detection of T. gondii, E. histolytica and C. cayetanensis in water samples. Molecular biological techniques offer a methodological alternative in the study of protozoan parasites because their sensitivity and specificity are greater than those of traditional methods [10, 17]. For this reason, various studies have used these techniques to detect protozoan parasites such as Giardia [11, 18], Cryptosporidium [11, 18–21], C. cayetanensis [20, 22, 23] and T. gondii [20, 24, 25].
One of the great advantages of molecular biological techniques is that they allow the discrimination of microorganisms at the species and genotype levels, information that may be relevant in evaluating the sources of infection in humans and in the study of the potential risks posed by protozoan parasites [26]. For example, several assemblages of G. duodenalis (A-H) and about 37 species of Cryptosporidium have been described as associated with different hosts. Of these, particular genotypes of C. parvum, C. hominis, C. andersoni, C. meleagridis, C. ubiquitum, C. cuniculus, C. suis and G. duodenalis assemblages A and B are of special interest because they have been reported in water sources and may also present zoonotic potential [27, 28]. The identification of the different genotypes of these protozoan parasites in water sources can be useful to determine the possible sources of contamination through their association with the type of host they parasitize.
In this context, the aim of this study was to investigate the presence of protozoan parasites such as G. duodenalis, Cryptosporidium spp., C. cayetanensis, E. histolytica and T. gondii in samples of raw and treated water from water treatment plants that supply water for human consumption to urban areas and rural areas in the department of Nariño (southwest Colombia), using real-time PCR. We also identified the Cryptosporidium species and G. duodenalis assemblages with PCR and DNA sequencing.
Methods
Study area
This study was performed in three Drinking Water Treatment Plants (DWTP) currently supplying drinking water to urban areas and 11 Rural Plants currently supplying water to rural areas (DWRP), in the municipalities of Pasto (1°12'52"N, 77°16'41"W; altitude: 2527 meters above sea level (masl); average temperature: 12 °C), Ipiales (0°49'44"N, 77°38'26"W; altitude: 2900 masl; average temperature: 12 °C), Túquerres (1°05'14"N, 77°37'08W; altitude: 3104 masl; average temperature: 11 °C), and Tumaco (1°48'24"N, 78°45'53W; altitude: 1 masl; average temperature: 26 °C), all located in the Department of Nariño, southwestern Colombia (Fig. 1). The three DWTPs collect the surface waters from rivers as their supply source and use a conventional physicochemical treatment to produce drinking water, including the steps of coagulation, flocculation, sedimentation, filtration and disinfection by the addition of chlorine. The DWRPs of the municipality of Ipiales use surface waters of streams as their sources of supply (DWRP-IA, DWRP-IB, DWRP-IC, DWRP-ID and DWRP-IE) and the DWRPs of the municipality of Tumaco use the surface waters of rivers (DWRP-TE) and water from underground wells (DWRP-TA, DWRP-TB, DWRP-TC, DWRP-TD and DWRP-TF) as their sources of supply. In general, DWRPs undertake minimum water treatment before consumption, consisting mainly of the addition of chlorine. However, during the sampling period, only one of the 11 DWRPs (DWRP-TF) in the study applied some type of treatment.
Fig. 1.
Geographical location of the sampling municipalities, all located in the Department of Nariño, southwestern Colombia
Sampling
In total 117 water samples of 10 leach sample were collected (Additional file 1: Table S1). Of these, 72 water samples were collected in DWTPs at three different points: (i) 24 samples of raw water at the inlets to the plants; (ii) 24 samples after the physicochemical treatment; and (iii) 24 samples after disinfection with chlorine. The remaining 45 DWRP samples collected consisted of only raw water, because these plants did not apply any type of treatment at the times of sampling. The samples were collected on two occasions in 2016, one in March (rainy season) and the other in August-September (dry season). The sampling months were selected for convenience, based on the average monthly precipitation in the Department of Nariño, provided by the Institute of Hydrology, Meteorology, and Environmental Studies of Colombia.
Sample processing
Recovery of protozoan parasites
The raw water samples were filtered through cellulose membranes with 3 μm pore size (47 mm in diameter). The water samples taken after the physicochemical treatment and after the disinfection process were also filtered through cellulose ester membranes with 1.2 μm pore size (47 mm in diameter). The membranes used in the filtration processes were washed twice with 5 ml of elution solution (0.01% Tween 80 and antifoam). To concentrate the samples, the volumes recovered from the washes were transferred to sterile polystyrene tubes and centrifuged at 1500× g for 15 min. Afterwards, the supernatant was discarded, leaving 1 ml of it on the pellet, which was transferred to a new sterile tube.
DNA extraction
The DNA was extracted from 300 μl of the previously obtained cell suspensions, using the FastDNA® SPIN Kit for Soil (MP Biomedicals, Santa Ana, USA), according to the manufacturer’s instructions. This was preceded by two cycles in a Mini-BeatBeater cell disruptor (California, USA) for 3 min and incubation on ice for 2 min to preferentially break the cystic forms of the protozoans. The DNA was eluted in 50 ml of elution buffer and stored at -20 °C until analysis. All water samples were spiked with a known concentration of a recombinant plasmid containing a sequence of an aquaporin of Arabidopsis thaliana as the internal control of amplification (IAC). This allowed the detection of PCR inhibitors in the samples [29].
Detection of parasites using real-time PCR
The DNA of G. duodenalis, Cryptosporidium spp., T. gondii and E. histolytica was detected with real-time PCR (qPCR) with endpoint detection, using primers and TaqMan probes previously reported for each of the protozoans [17, 19, 25]. The sequences of the primers and probes used are listed in Table 1. The qPCR assays were performed independently for each parasite in 96-well MicroAmp plates (Applied Biosystems, Foster City, USA), with a final reaction volume of 9 μl containing 3.5 μl of FastStart Universal Probe Master (Rox) (Roche, Basel, Switzerland), 1.0 μl of each forward and reverse primer (10 μM) and 0.2 μl of TaqMan probe (5 μM) specific for each parasite or for the internal control (IC), 0.3 μl of water, and 2.0 μl of DNA. We used the sequence of an aquaporin of Arabidopsis thaliana cloned into a plasmid as the internal control, as reported elsewhere [29, 30]. To detect Cryptosporidium spp., the final reaction volume was 10 μl because 3.0 μl of DNA was included. The samples were processed in duplicate in the Applied Biosystems 7500 system, using the default parameters and 40 cycles of amplification, except for Cryptosporidium spp., for which 50 cycles were used. The results of qPCR were considered negative if the cycle threshold (Ct) values were > 38. This cycle threshold was determined in a previous study by our research group. The qPCR results were considered negative if the cycle threshold values (Ct) were > 38. To corroborate the Ct value, we conducted experiments to establish the dynamic range of our assay using standards from 10,000 fg/μl to 1 fg/μl. For quantification, plasmids containing the target sequences were cloned into the pGEM-T Easy Vector System I (Promega, Madison, USA), according to the manufacturer’s instructions, and transformed into XL1-Blue Escherichia coli (Agilent Technologies, California, USA). The transformed colonies containing the plasmids were extracted by using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). The purified plasmid DNA was quantified by using a Nanodrop and diluted to have a concentration range of 10,000 fg/ μl to 1 fg/μl. The dynamic range established that the limit of detection was the proposed by Sanchez et al. [29]. We used DNA from G. duodenalis, Cryptosporidium spp., T. gondii and E. histolytica provided by the Parasitology Laboratory of the National University of Colombia as the positive controls in the corresponding reactions, and type I water as the negative control. We detected C. cayetanensis in collaboration with the Division of Parasitic Diseases and Malaria of the Centers for Disease Control and Prevention (Atlanta, USA), using a previously described method [31].
Table 1.
Primers and probes used for the molecular detection of the protozoan parasites under study. In bold the fluorophores and quenchers
| Parasite | Primer | Sequence (5'-3') | Target | Reference |
|---|---|---|---|---|
| Cryptosporidium spp. | CF | GTTTTCATTAATCAAGAACGAAAGTTAGG | 18S rRNA | [19] |
| CR | GAGTAAGGAACAACCTCCAATCTCTAG | |||
| CP | 6FAM/TCAGATACCGTCGTAGTCTTAACCATAAACTATGCC/TAMRA | |||
| SSUrRNAF | AGTGACAAGAAATAACAATACAGG | 18S rRNA | [34] | |
| SSUrRNAR | CCTGCTTTAAGCACTCTAATTTTC | |||
| GpF | GCCGTTCCACTCAGAGGAAC | gp60 | [34] | |
| GpR | CCACATTACAAATGAAGTGCCGC | |||
| Giardia duodenalis | GdF | CATGCATGCCCGCTCA | 18S rRNA | [17] |
| GdR | AGCGGTGTCCGGCTAGC | |||
| GdP | 6FAM/AGGACAACGGTTGCAC/MGB | |||
| GDHeF | TCAACGTYAAYCGYGGYTTCCGT | gdh | [33] | |
| GDHiF | CAGTACAACTCYGCTCTCGG | |||
| GDHiR | GTTRTCCTTGCACATCTCC | |||
| AL3543 | AAATIATGCCTGCTCGTCG | tpi | [32] | |
| AL3546 | CAACATTITCCGCAAACC | |||
| AL3544 | CCCCTTCATCGGIGGTAACTT | |||
| AL3545 | GTGGCCACCACICCCGTGCC | |||
| Entamoeba histolytica | EhF | GTTTGTATTAGTACAAAATGGCCAATTC | 18S rRNA | [17] |
| EhR | TCGTGGCATCCTAACTCACTTAGA | |||
| EhP | 6FAM/CAATGAATTGAGAAATGACA/MGB | |||
| Toxoplasma gondii | TgF | TCCCCTCTGCTGGCGAAAAGT | B1 gene | [25] |
| TgR | AGCGTTCGTGGTCAACTATCGATTG | |||
| TgP | 6FAM/TCTGTGCAACTTTGGTGTATTCGCAG/TAMRA | |||
| Cyclospora cayetanensis | CcF | TAGTAACCGAACGGATCGCATT | 18S rRNA | [31] |
| CcR | AAT GCC ACG GTA GGC CAA TA | |||
| CcP | HEX/CCGGCGATAGATCATTCAAGTTTCTGACC/DABCYL | |||
| Internal control (Aquaglyceroporine) | CiF | ACCGTCATGGAACAGCACGTA | AQGP | [30] |
| CiR | CTCCCGCAACAAACCCTATAAA | |||
| CiP | VIC/AGCATCTGTTCTTGAAGGT/NFQ-MGB |
Identification of G. duodenalis assemblages and Cryptosporidium species
Samples positive on qPCR for G. duodenalis and Cryptosporidium spp. were genotyped with conventional PCR and DNA sequencing. To identify the G. duodenalis assemblages, we used the following molecular markers: gdh (glutamate dehydrogenase) amplified with the primers GDHeF, GDHiF and GDHiR and tpi (triose phosphate isomerase) using the primers AL3543, AL3546, AL3544 and AL3545, as previously described [32, 33]. To identify the Cryptosporidium species the small subunit (SSU) rRNA marker was used, using the primers SSUrRNAF and SSUrRNAR, and subtyping of C. parvum was based on sequence analysis of gp60 genes as previously reported [29, 34]. The sequences of the primers are listed in Table 1. The samples were processed in a MultiGene OptiMax Thermal Cycler (Labnet, California, USA). The amplification products were verified on 2% agarose gels stained with the SYBR® Safe Stain (California, USA). The gel was visualized in an E-Gel Imager (Life Technologies, Carlsband, USA).
The PCR products were sequenced by the dideoxy-terminal method in an automated capillary sequencer (Applied Biosystems, Foster City, USA). The sequences were verified and edited in the MEGA 6.0 program [35] and then aligned with sequences from the GenBank database using the NCBI BLAST tool [36] and with reference sequences in the program ClustalW v.1.8. Phylogenetic reconstruction was performed using a maximum likelihood analysis in MEGA v6.0, using the Tamura 3-parameter method with 1000 bootstrap replicates [35]. The reference sequences considered for the gdh marker of G. duodenalis were: assemblage AI (M84604.1), assemblage AII (AY178737.1), assemblage BIII (AF069059.1), assemblage BIV (AY178739.1), assemblage C (U60982.2), assemblage D (U60986.2), assemblage E (AY178741.1), assemblage F (AB569384.1), assemblage G (AF069058.2) and assemblage H (GU176089.1). The sequence of G. ardeae (AF069060.2) was used as the outgroup. For the G. duodenalis tpi marker, the following reference sequences were used: assemblage AI (AF069556.1), assemblage AII (AF069557.1), assemblage BIII (AF069561.1), assemblage BIV (AF069560.1), assemblage C (AF069563.1), assemblage E (AY228645.1) and assemblage F (AF069558.1). The sequence of G. microti (AY228649.1) was used as the outgroup. For Cryptosporidium, the reference sequences used were: C. andersoni (AF093496.1), C. baileyi (L19068.1), C. bovis (AY741305.1), C. canis (AF112576.1), C. fayeri (AF159112.1), C. felis (AF108862.1), C. fragile (EU162751.1), C. galli (AF316624.1), C. hominis (AF108865.1), C. macropodum (AF513227.2), C. meleagridis (AF112574.1), C. cf. molnari (AY524773.1), C. muris (AB089284.1), C. parvum (AF112571.1), C. ryanae (AY587166.1), C. cf. scophthalmi (KR340588.1), C. serpentis (AF151376.2), C. suis (AF115377.1), C. varanii (AF112573.1) and C. wrairi (AF115378.1). The sequences obtained were deposited in GenBank under the accession numbers MH730625–MH730644.
Data analysis
Descriptive statistics was used to describe the main events of interest; the results are reported as percentages and frequencies. Statistically significant associations between the detection of the parasites examined and variables, such as the place of sampling, type of water and time of sampling, were determined by logistic regression analysis, using the statistical program Statgraphics Centurion XVII (Royal Technologies, Bogota, Colombia). Differences were considered significant at P < 0.05.
Results
Detection of protozoan parasites in water samples with qPCR
A total of 117 water samples were collected, 110 of which were analyzed and seven were excluded from the study because the internal control was not amplified in any of the qPCRs. The seven samples excluded were four water samples collected in DWTPs and three water samples taken from DWRPs. Of the 110 samples analyzed, 31.82% (35/110) were positive for G. duodenalis and 8.18% (9/110) for Cryptosporidium spp. We detected both protozoan parasites G. duodenalis and Cryptosporidium together in 2.73% (3/110) of the water samples analyzed. None of the processed samples were positive for T. gondii (0/110), E. histolytica (0/110) or C. cayetanensis (0/110).
Giardia duodenalis was detected in the three DWTPs studied and in five of the 11 DWRPs (DWRP-IA, DWRP-IC, DWRP-ID, DWRP-TA and DWRP-TB). Most positive samples were found in the DWTP in Ipiales, followed by the DWTPs in Túquerres and Pasto, indicating contamination was prevalent in water from plants supplying urban areas. Cryptosporidium was detected in two of the three DWTPs and in four of the 11 DWRPs (DWRP-IE, DWRP-TA, DWRP-TE and DWRP-TF). We detected the greatest numbers of positive samples in the DWRPs of Tumaco, followed by the DWTP of Pasto; Cryptosporidium was most frequently found in treatment plants supplying rural areas. However, we found no statistically significant correlation between the sampling site and the detection of G. duodenalis (logistic regression, P = 0.1017) or Cryptosporidium (logistic regression, P = 0.4780) (Table 2).
Table 2.
Number and percentage of positive samples for G. duodenalis and Cryptosporidium spp. for each sampling site
| Site (n) | G. duodenalis (%) | Cryptosporidium spp. (%) |
|---|---|---|
| DWTP Pasto (n = 21) | 6 (28.6) | 3 (14.3) |
| DWTP Ipiales (n = 23) | 13 (56.5) | 1 (4.3) |
| DWTP Túquerres (n = 24) | 9 (37.5) | – |
| DWRP Ipiales (n = 17) | 3 (17.6) | 1 (5.9) |
| DWRP Tumaco (n = 25) | 4 (16.0) | 4 (16.0) |
Abbreviations: DWTP Drinking Water Treatment Plants, DWRP Drinking Water Rural Plants, n number of samples
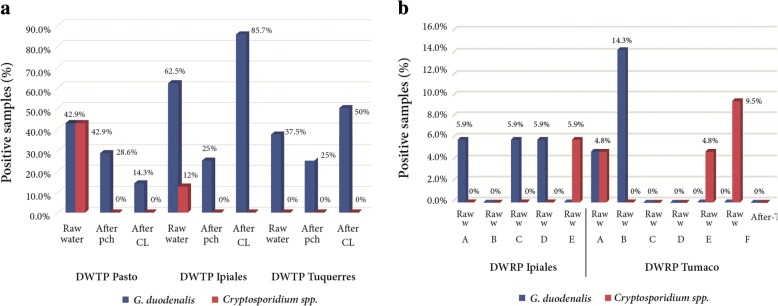
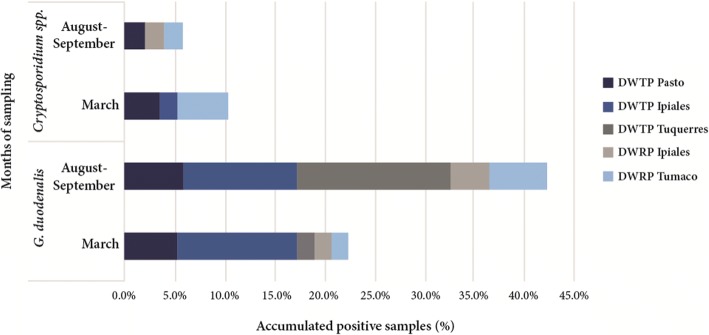
Of the two parasites detected, G. duodenalis was found in both raw and treated water samples from DWTPs (raw water: 11/23, 47.83%; after physicochemical treatment: 6/23, 26.09%; and after chlorination: 11/22, 50%) and in raw water samples from DWRPs (7/38, 18.42%). However, we detected no statistically significant relationship between the type of water and presence of this microorganism (logistic regression, P > 0.6835). In the DWTP of the municipality of Pasto, G. duodenalis was detected with greater frequency in raw water (3/7, 42.9%) than in either water collected after physicochemical treatment (2/7, 28.6%) or after chlorination (1/7, 14.3%). In the DWTPs of the municipalities of Ipiales and Túquerres, this protozoan was more frequent in the water collected after chlorination (Ipiales DWTP 6/7, 85.7%; Túquerres DWTP 4/8, 50%) than in the raw water (Ipiales DWTP 5/8, 62.5%; Túquerres DWTP 3/8, 37.5%). The fewest positive samples of G. duodenalis were detected in water collected after the physicochemical treatment (Ipiales DWTP 2/8, 25%; Túquerres DWTP 2/8, 25%). Cryptosporidium spp. were only found in raw water samples in both the DWTPs (raw water: 4/23, 17.39%) and DWRPs (5/38, 13.16%), and there was a statistically significant relationship between the type of water evaluated and this microorganism (P < 0.0097) (Fig. 2). Giardia duodenalis was detected more frequently in samples collected in the dry months of August-September (22/52, 42.31%) than in the rainy month of March (13/58, 22.41 %; P < 0.0078). However, the detection of Cryptosporidium did not vary significantly between the samples collected in August-September (3/52, 5.77%) and March (6/58, 10.34%; P > 0.3768) (Fig. 3).
Fig. 2.
Molecular detection of G. duodenalis and Cryptosporidium spp. a In Drinking Water Treatment Plants currently supplying drinking water to urban areas from the municipalities of Pasto, Ipiales and Túquerres. Abbreviations: Raw water, water samples collected before treatment; After pch, water samples collected after the physicochemical treatment; After Cl, water samples collected after disinfection with chlorine. b In Rural Plants currently supplying water to rural zones from the municipalities of Ipiales and Tumaco. Abbreviations: Raw w, water samples collected before treatment; After T, water samples collected after treatment. DWRP Ipiales: A (Yaramal), B (La Orejuela), C (Charandu), D (Loma de Zuras) and E (Chaguaipe). DWRP Tumaco: A (Km 36), B (Cajapí), C (El Ceibito), D (Inguapí el Carmen), E (Bajo Jagua) and F (Pueblo Nuevo)
Fig. 3.
Detection of G. duodenalis and Cryptosporidium spp. in the two seasons of sampling: March (rainy season) and August-September (dry season). Abbreviations: DWTP, Drinking Water Treatment Plant; DWRP, Rural Plant
Identification of G. duodenalis assemblages and Cryptosporidium species
To identify the assemblages of G. duodenalis from the qPCR-positive samples (35/110, 31.82%), we determined the nucleotide sequences of 17 PCR products with the gdh marker and 28 PCR products with tpi marker using Sanger sequencing (the remaining samples showed only faint bands or no bands and could not be sequenced). Nucleotide sequences were determined satisfactorily for 13 samples (five with the gdh marker and eight with the tpi marker), and identified assemblages AI (1/5, 20%), AII (1/5, 20%) and H (1/5, 20%) (particularly in Tumaco) with the gdh marker and AI (1/8, 12.5%) and AII (4/8, 50%) with the tpi marker. The other samples had multiple nucleotide sequences in the same sample and therefore could not be analyzed. The same results were obtained when the samples generating these aberrant sequences were re-amplified and the PCR products sequenced again.
Cryptosporidium species present in the qPCR-positive water samples (9/110, 8.18%) were identified by sequencing 9 PCR products with the SSU rRNA marker. The nucleotide sequences were determined satisfactorily for seven samples, and detected C. parvum (1/9, 11.1%), C. galli (1/9, 11.1%), C. molnari (1/9, 11.1%), Cryptosporidium sp. genotype II of bats (1/9, 11.1%) and Cryptosporidium sp. genotype VIII of birds (3/9, 33.3%). The result of subtyping the only C. parvum sample showed the presence of genotype IIcA5G3c. The sequences of the two remaining samples (2/9, 22.2%) only allowed their identification to the genus level.
Discussion
The methods used in this study allowed the recovery and detection of protozoan parasites from water samples. In the case of membrane filtration, this methodology has been used in some Latin American studies, mainly in countries such as Brazil for the recovery of protozoan parasites from various environmental samples [37]. Among its advantages are its low cost, shorter processing time in the laboratory and allowing the processing of samples with complex physicochemical composition (oils, fats, organic matter). However, a high turbidity can limit the use of this methodology due to the obstruction of the filter pores, making the use of several membranes necessary for the filtration of a single sample [15]. One limitation of our study was the use of membrane filtration. Further studies must consider the cartridge filters used in Method 1623 [1].
Regarding the qPCR, this is a technique with great potential in the detection of protozoan parasites due to its high sensitivity, a feature of great importance in environmental samples in which this type of microorganism can occur at low concentrations [11, 38]. Nevertheless, this molecular technique also has some limitations, due to the presence of PCR inhibitors that can be found in water samples and their dependence on the quality of the DNA obtained [6, 39]. In order to avoid false negatives due to the presence of inhibitors in the sample, an internal control was used in this study; this internal control identified samples that did not amplify the target DNA during the qPCR, due to the presence of substances that can act as PCR inhibitors, and therefore should be excluded from the study. In future studies it would also be important to consider the use of PCR enhancers, which could also improve the results.
With respect to the results obtained, the detection of G. duodenalis and Cryptosporidium by qPCR in water samples in this study was consistent with reports worldwide [12, 40]. Giardia duodenalis and Cryptosporidium spp. can be detected in water samples because they are widely distributed in the environment and affect both humans and a wide range of domestic and wild animals [41, 42], facilitating their transmission to water resources that are exposed to contamination [13, 18, 43]. In the DWTPs that supply water to urban areas in Nariño Department, the sources of supply are the surface waters of rivers, which are exposed to the impact of anthropogenic activities before their capture by the treatment plants. Contaminants mainly include discharges of domestic and agricultural origin, industrial effluents, runoff from waste, or the products of livestock, agriculture and human activities [44, 45].
In the DWTP of Pasto, reductions in the numbers of positive samples of G. duodenalis and Cryptosporidium were observed as the treatment of the drinking water progressed. However, this trend was not observed in the DWTPs of Ipiales and Túquerres. Giardia duodenalis was detected in water samples taken after the physicochemical treatment and disinfection with chlorine, at considerably higher frequencies than those in the untreated water, indicating that the procedures used in these plants must be reviewed; these findings may indicate, e.g. poor maintenance of the disinfection units, accumulation of biofilms within the pipelines or lack of optimization in these treatment facilities. One of the key factors that should be reviewed in the three DWTPs examined in this study is the physicochemical treatment (coagulation, flocculation, sedimentation and filtration), because the elimination of protozoan parasites correlates strongly with the adequate operation of each stage of this process [46]. Special attention should be paid to the steps of coagulation and flocculation [47].
In the case of G. duodenalis, its detection in post-treatment water samples may also be related to the resistance of the cystic forms to oxidative conventional disinfectants, such as chlorine [43, 46], which is the method applied in the plants studied. Likewise, it could be due to post-treatment contamination by cysts of this protozoan parasite during the production process of drinking water, caused by infiltrations in the treatment system through leaks, open or crossed connections, manipulation of system elements, repair of pipes, or the formation of biofilms of Giardia in the pipes, representing a possible source of secondary contamination of water, due to the accumulation and concentration of cysts that may occur during periods of low flow. Studies conducted in Colombia by other authors also report the presence of Giardia in drinking water [6, 48, 49]. However, it is important to note that in this study there was no information on the viability or infectivity of the protozoan parasites detected and therefore no conjecture could be made about the risk associated with the detection of G. duodenalis in the post-treatment water.
In the DWRPs of the municipalities of Ipiales and Tumaco, both G. duodenalis and Cryptosporidium spp. were detected, indicating the contamination of the natural water sources captured for human consumption in rural areas. In the DWRPs of Ipiales, the water sources used for supply could be exposed to domestic discharges, residues from crops, and the care or breeding of animals, because different anthropogenic activities are undertaken in this area. In Tumaco’s DWRPs, the groundwater wells could be contaminated because they are directly exposed to the environment and the passage of domestic and wild animals, and they are inadequately maintained. The sizes of both Giardia cysts and Cryptosporidium oocysts could also allow them to leach into the groundwater, which has previously been reported as a threat to groundwater [50, 51]. In the treatment plants that take water from rivers, the sources of contamination could be the various activities frequently performed in the rivers, including recreation, personal hygiene and laundry, and the general residual wastes of domestic origin.
Regarding the G. duodenalis assemblages in the qPCR-positive samples, we identified assemblage A in the DWTPs of Pasto and Ipiales. This assemblage has been reported previously in humans, livestock, companion animals, some species of marine animals and wild mammals; it is essential to consider its zoonotic potential [33, 52]. In the DWTP of Pasto, the sub-assemblage AI was identified, and in the DWTP of Ipiales, the sub-assemblages AI and AII were identified. Of these, the sub-assemblage AI has been reported predominantly in livestock and pets, and AII in humans [53]. Therefore, the sources of contamination are probably associated with these hosts. We also detected a sample containing assemblage H, so far only reported in seals and gulls [27, 52]. This represents the first description of assemblage H in Colombia or South America. Currently, the animal reservoirs of assemblage H remain undefined. Interestingly, this sample was detected in Tumaco, located at the pacific coast of the country. One plausible explanation might be that the Giardia assemblage H came from a seabird or marine vertebrates. However, this premise is too speculative and future studies to determine the exact frequency of this assemblage in the country and the region, as well as its hosts, are needed. Unfortunately, it was only possible to sequence a limited number of samples to identify the G. duodenalis assemblages, but similar results have been obtained in other studies [13, 39, 54]. The failure to amplify all qPCR-positive samples of G. duodenalis with nested PCR may be attributable to that the molecular marker used in the qPCR (18S rRNA) has a higher number of copies than the gdh and tpi molecular markers used in the nested PCR. However, it is also possible that some qPCR results were false positive results, due to the complexity of the environmental samples and the presence of other DNA that could cross-prime with the qPCR assays of Cryptosporidium spp. and G. duodenalis in the water samples collected and possibly with the excess of PCR cycles reported for this assay [39].
Of the Cryptosporidium species detected in the DWTPs, we identified C. parvum, C. molnari, C. galli and Cryptosporidium sp. genotype II of bats. Cryptosporidium parvum is considered one of the most widely distributed Cryptosporidium species, reported in more than 150 mammalian hosts, including humans, mice, cows, horses, sheep, goats, pigs and deer, and is one of the main causal agents of outbreaks of water-borne cryptosporidiosis [34, 55]. Particularly, we found the genotype IIcA5G3c, which is considered zoonotic and has been previously reported in Colombia [29]. The other species identified are considered to be host specific; C. molnari has mainly been described in fish, C. galli in birds such as chickens and finches [41], and Cryptosporidium sp. genotype II corresponds to the sequence of an isolate obtained from bats. It is important to remember that within the genus Cryptosporidium, there are several species and genotypes for which a definitive classification is still pending, so the genus is in continuous review [19, 56].
Little information is available on the relationship between the protozoan parasites detected in water resources across seasons in Latin America, where countries experience only rainy and dry seasons [16, 57]. In the present study, the number of samples positive for G. duodenalis was greater in August-September, which are dry months. However, there was no significant variation in the detection of Cryptosporidium in the dry and rainy months sampled. To better analyze this issue at a regional level, we recommend that future studies monitor these types of microorganisms over longer periods, taking into account the effects of phenomena such as El Niño and La Niña, which can alter the climatic patterns considered typical or normal for a specific region.
Conclusions
We used molecular methodologies to determine the presence of protozoan parasites, such as G. duodenalis and Cryptosporidium, in water samples collected from the Department of Nariño. Our results provide important insights into the transmission dynamics of these protozoans in water resources. They emphasize the need for continued research and monitoring for the presence of these types of microorganisms in the water sources for human consumption, and the measures and precautions that must be considered to mitigate water contamination before its arrival at treatment plants.
Additional file
Table S1. Metadata information of the water samples collected and submitted to molecular detection of Giardia duodenalis, Cryptosporidium, Entamoeba histolytica, Toxoplasma gondii and Cyclospora cayetanensis. (DOCX 24 kb)
Acknowledgements
We thank Janine Miller, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Funding
The authors are grateful for the funding received from Agua Potable Nariño - CT&I Fund of the SGR research project, conceded to Nariño Department, Colombia (BPIN 2014000100020). CS is also grateful for the scholarship granted by the Government of Nariño through CEIBA Foundation (2015) to undertake her MSc study. The authors thank the Dirección de Investigación e Innovación de la Universidad del Rosario and Universidad de Nariño.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its Additional file.
Abbreviations
- DWRP
Rural plants currently supplying water to rural areas
- DWTP
Drinking Water Treatment Plants currently supplying drinking water to urban areas
- qPCR
Real-time PCR
- SSU rRNA
Small subunit ribosomal ribonucleic acid
- USEPA
United States Environmental Protection Agency
- gdh
Glutamate dehydrogenase
- tpi
Triose phosphate isomerase
- masl
Meters above sea level
Authors' contributions
CS wrote the manuscript, analyzed the data and carried out the sampling and experimental tests of molecular biology. YQ and KH performed the detection of Cyclospora cayetanensis. AG, MCL and JDR analyzed the data, reviewed the final version of the manuscript and supervised the project. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Claudia Sánchez, Email: clasanchezor@unal.edu.co.
Myriam Consuelo López, Email: mclopezp@unal.edu.co.
Luis Alejandro Galeano, Email: alejandrogaleano@udenar.edu.co.
Yvonne Qvarnstrom, Email: bvp2@cdc.gov.
Katelyn Houghton, Email: bvp3@cdc.gov.
Juan David Ramírez, Email: juand.ramirez@urosario.edu.co.
References
- 1.UN-Water . Annual Report 2014. Switzerland: Genève; 2014. UN-Water Technical Advisory Unit. [Google Scholar]
- 2.WHO. Guidelines for Drinking-Water Quality . Volume 1. 3. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 3.Pérez A, Lozada P, Cruz C. Water safety plans. Fundamentals and prospects for implementing them in Colombia. Ing Inv. 2009;29:79–85. [Google Scholar]
- 4.Ministerio de Salud y Protección Social. National Report on the Quality of Water for Human Consumption Based on the IRCA. Bogotá D.C. Colombia; 2016. https://www.minsalud.gov.co/sites/rid/Lists/BibliotecaDigital/RIDE/VS/PP/SA/inca-2015_reducido.pdf. Accessed 08 Apr 2018.
- 5.Instituto Departamental de Salud de Nariño. Epidemiological Bulletin. Ipiales, Colombia. 2017. p. 1–27. http://www.idsn.gov.co/images/documentos/epidemiologia/boletines/bepidemiologia.pdf. Accessed 08 Apr 2018.
- 6.Triviño-Valencia J, Lora F, Zuluaga JD, Gomez-Marin JE. Detection by PCR of pathogenic protozoa in raw and drinkable water samples in Colombia. Parasitol Res. 2016;115:1789–1797. doi: 10.1007/s00436-016-4917-5. [DOI] [PubMed] [Google Scholar]
- 7.Efstratiou Artemis, Ongerth Jerry E., Karanis Panagiotis. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks - An update 2011–2016. Water Research. 2017;114:14–22. doi: 10.1016/j.watres.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks - an update 2004–2010. Water Res. 2011;45:6603–6614. doi: 10.1016/j.watres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Karanis P, Kourenti C, Smith H. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J Water Health. 2007;5:1–38. doi: 10.2166/wh.2006.002. [DOI] [PubMed] [Google Scholar]
- 10.Ali MA, Al-Herrawy AZ, El-Hawaary SE. Detection of enteric viruses, Giardia and Cryptosporidium in two different types of drinking water treatment facilities. Water Res. 2004;38:3931–3939. doi: 10.1016/j.watres.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Kumar T, Majid MAA, Onichandran S, Jaturas N, Andiappan H, Salibay CC, et al. Presence of Cryptosporidium parvum and Giardia lamblia in water samples from Southeast Asia: towards an integrated water detection system. Infect Dis Poverty. 2016;5:3. doi: 10.1186/s40249-016-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro-Hermida JA, González-Warleta M, Mezo M. Cryptosporidium spp. and Giardia duodenalis as pathogenic contaminants of water in Galicia, Spain: the need for safe drinking water. Int J Hyg Environ Health. 2015;218:132–138. doi: 10.1016/j.ijheh.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Castro-Hermida JA, García-Presedo I, Almeida A, González-Warleta M, Correia Da Costa JM, Mezo M. Detection of Cryptosporidium spp. and Giardia duodenalis in surface water: a health risk for humans and animals. Water Res. 2009;43:4133–4142. doi: 10.1016/j.watres.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Ministerio de la Protección Social. Resolution 2115: The resolution indicates the characteristics, basic instruments and frequencies of the control and monitoring system for the quality of water for human consumption. Colombia; 2007. p. 1–23. http://www.minambiente.gov.co/images/GestionIntegraldelRecursoHidrico/pdf/Legislaci%C3%B3n_del_agua/Resoluci%C3%B3n_2115.pdf. Accessed 08 Apr 2018.
- 15.Bueno R, Hachich E, Zanolli M, Naveira R, Silva E, Campos M, et al. Performance evaluation of different methodologies for detection of Cryptosporidium spp. and Giardia spp. in water for human consumption to meet the demands of the Environmental Health Surveillance in Brazil. Epidemiol Serv Saude. 2012;21:233–242. doi: 10.5123/S1679-49742012000200006. [DOI] [Google Scholar]
- 16.Rosado-García FM, Guerrero-Flórez M, Karanis G, Hinojosa MDC, Karanis P. Water-borne protozoa parasites: the Latin American perspective. Int J Hyg Environ Health. 2017;220:783–798. doi: 10.1016/j.ijheh.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Mejia R, Vicuña Y, Broncano N, Sandoval C, Vaca M, Chico M, et al. A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Am J Trop Med Hyg. 2013;88:1041–1047. doi: 10.4269/ajtmh.12-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmi K, Skraber S, Burnet JB, Leblanc L, Hoffmann L, Cauchie HM. Two-year monitoring of Cryptosporidium parvum and Giardia lamblia occurrence in a recreational and drinking water reservoir using standard microscopic and molecular biology techniques. Environ Monit Assess. 2011;179:163–175. doi: 10.1007/s10661-010-1726-7. [DOI] [PubMed] [Google Scholar]
- 19.Burnet JB, Ogorzaly L, Tissier A, Penny C, Cauchie HM. Novel quantitative TaqMan real-time PCR assays for detection of Cryptosporidium at the genus level and genotyping of major human and cattle-infecting species. J Appl Microbiol. 2013;114:1211–1222. doi: 10.1111/jam.12103. [DOI] [PubMed] [Google Scholar]
- 20.Lalonde LF, Gajadhar AA. Detection and differentiation of coccidian oocysts by real-time PCR and melting curve analysis. J Parasitol. 2011;97:725–730. doi: 10.1645/GE-2706.1. [DOI] [PubMed] [Google Scholar]
- 21.Li N, Neumann NF, Ruecker N, Alderisio KA, Sturbaum GD, Villegas EN, et al. Development and evaluation of three real-time PCR assays for genotyping and source tracking Cryptosporidium spp. in water. Appl Environ Microbiol. 2015;81:5845–5854. doi: 10.1128/AEM.01699-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitajima M, Haramoto E, Iker BC, Gerba CP. Occurrence of Cryptosporidium, Giardia, and Cyclospora in influent and effluent water at wastewater treatment plants in Arizona. Sci Total Environ. 2014;484:129–136. doi: 10.1016/j.scitotenv.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Varma M, Hester JD, Schaefer FW, Ware MW, Lindquist HDA. Detection of Cyclospora cayetanensis using a quantitative real-time PCR assay. J Microbiol Methods. 2003;53:27–36. doi: 10.1016/S0167-7012(02)00209-9. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro K, Mazet JAK, Schriewer A, Wuertz S, Fritz H, Miller WA, et al. Detection of Toxoplasma gondii oocysts and surrogate microspheres in water using ultrafiltration and capsule filtration. Water Res. 2010;44:893–903. doi: 10.1016/j.watres.2009.09.061. [DOI] [PubMed] [Google Scholar]
- 25.Lin MH, Chen TC, Kuo TT, Tseng CC, Tseng CP. Real-time PCR for quantitative detection of Toxoplasma gondii. J Clin Microbiol. 2000;38:4121–4125. doi: 10.1128/jcm.38.11.4121-4125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao L, Feng Y. Food and waterborne parasitology molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monis P, Caccio S, Thompson A. Variation in Giardia: towards a taxonomic revision of the genus. Trends Parasitol. 2009;25:93–100. doi: 10.1016/j.pt.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Zahedi A, Gofton AW, Greay T, Monis P, Oskam C, Ball A, et al. Profiling the diversity of Cryptosporidium species and genotypes in wastewater treatment plants in Australia using next generation sequencing. Sci Total Environ. 2018;644:635–648. doi: 10.1016/j.scitotenv.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Sánchez A, Muñoz M, Gómez N, Tabares J, Segura L, Salazar Á, et al. Molecular epidemiology of Giardia, Blastocystis and Cryptosporidium among Indigenous children from the Colombian Amazon basin. Front Microbiol. 2017;8:248. doi: 10.3389/fmicb.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duffy Tomas, Cura Carolina I., Ramirez Juan C., Abate Teresa, Cayo Nelly M., Parrado Rudy, Bello Zoraida Diaz, Velazquez Elsa, Muñoz-Calderon Arturo, Juiz Natalia A., Basile Joaquín, Garcia Lineth, Riarte Adelina, Nasser Julio R., Ocampo Susana B., Yadon Zaida E., Torrico Faustino, de Noya Belkisyole Alarcón, Ribeiro Isabela, Schijman Alejandro G. Analytical Performance of a Multiplex Real-Time PCR Assay Using TaqMan Probes for Quantification of Trypanosoma cruzi Satellite DNA in Blood Samples. PLoS Neglected Tropical Diseases. 2013;7(1):e2000. doi: 10.1371/journal.pntd.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qvarnstrom Y, Benedict T, Marcet PL, Wiegand RE, Herwaldt BL, da Silva AJ. Molecular detection of Cyclospora cayetanensis in human stool specimens using UNEX-based DNA extraction and real-time PCR. Parasitology. 2018;145:865–870. doi: 10.1017/S0031182017001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read CM, Monis PT, Thompson RCA. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Hunter PR, Hadfield SJ, Wilkinson D, Lake IR, Harrison FCD, Chalmers RM. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerg Infect Dis. 2007;13:82–88. doi: 10.3201/eid1301.060481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura Koichiro, Stecher Glen, Peterson Daniel, Filipski Alan, Kumar Sudhir. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Bueno M, Branco N, Guiguet A. Methods of concentration and detection of Cryptosporidium spp. and Giardia spp. in water samples. Rev Patol Trop. 2012;41:119–135. [Google Scholar]
- 38.Carey C, Lee H, Trevors J. Biology, persistence and detection of Cryptosporidium parvum and Cryptosporidium hominis oocyst. Water Res. 2004;38:818–862. doi: 10.1016/j.watres.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Plutzer J, Karanis P, Domokos K, Törökné A, Márialigeti K. Detection and characterisation of Giardia and Cryptosporidium in Hungarian raw, surface and sewage water samples by IFT, PCR and sequence analysis of the SSUrRNA and gdh genes. Int J Hyg Environ Health. 2008;211:524–533. doi: 10.1016/j.ijheh.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Sato MIZ, Galvani AT, Padula JA, Nardocci AC, Lauretto M de S, Razzolini MTP, et al. Assessing the infection risk of Giardia and Cryptosporidium in public drinking water delivered by surface water systems in Sao Paulo State, Brazil. Sci Total Environ. 2013;442:389–396. doi: 10.1016/j.scitotenv.2012.09.077. [DOI] [PubMed] [Google Scholar]
- 41.Fayer Ronald. Cryptosporidium: a water-borne zoonotic parasite. Veterinary Parasitology. 2004;126(1-2):37–56. doi: 10.1016/j.vetpar.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Plutzer J, Ongerth J, Karanis P. Giardia taxonomy, phylogeny and epidemiology: facts and open questions. Int J Hyg Environ Health. 2010;213:321–333. doi: 10.1016/j.ijheh.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Castro-Hermida JA, García-Presedo I, Almeida A, González-Warleta M, Correia Da Costa JM, Mezo M. Presence of Cryptosporidium spp. and Giardia duodenalis through drinking water. Sci Total Environ. 2008;405:45–53. doi: 10.1016/j.scitotenv.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 44.CORPONARIÑO. Plan for the Regulation of the Main Stream of the Rio Pasto. Pasto, Nariño; 2011. http://corponarino.gov.co/expedientes/descontaminacion/porhriopasto.pdf. Accessed 08 Apr 2018.
- 45.CORPONARIÑO.Plan for the Regulation and Management of the Hydrographic Basin of the Tio Blanco. Ipiales, Nariño; 2011. http://corponarino.gov.co/expedientes/descontaminacion/porhrioblanco.pdf. Accessed 08 Apr 2018.
- 46.Schoenen D. Role of disinfection in suppressing the spread of pathogens with drinking water: possibilities and limitations. Water Res. 2002;36:3874–3888. doi: 10.1016/S0043-1354(02)00076-3. [DOI] [PubMed] [Google Scholar]
- 47.Betancourt WQ, Rose JB. Drinking water treatment processes for removal of Cryptosporidium and Giardia. Vet Parasitol. 2004;126:219–234. doi: 10.1016/j.vetpar.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Alarcón M, Beltrán M, Cárdenas M, Campos M. Presence and viability of Giardia spp. and Cryptosporidium spp. in drinking water and wastewater in the high basin of Bogotá river. Biomédica. 2005;25:353–365. doi: 10.7705/biomedica.v25i3.1360. [DOI] [PubMed] [Google Scholar]
- 49.Lora-Suarez F, Rivera R, Triviño-Valencia J, Gomez-Marin JE. Detection of protozoa in water samples by formalin/ether concentration method. Water Res. 2016;100:377–381. doi: 10.1016/j.watres.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 50.Elfadaly HA, Hassanain NA, Hassanain MA, Barakat AM, Shaapan RM. Evaluation of primitive ground water supplies as a risk factor for the development of major waterborne zoonosis in Egyptian children living in rural areas. J Infect Public Health. 2018;11:203–208. doi: 10.1016/j.jiph.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 51.Slifko TR, Smith HV, Rose JB. Emerging parasite zoonoses associated with water and food. Int J Parasitol. 2000;30:1379–1393. doi: 10.1016/S0020-7519(00)00128-4. [DOI] [PubMed] [Google Scholar]
- 52.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan Una, Paparini Andrea, Monis Paul, Hijjawi Nawal. It's official – Cryptosporidium is a gregarine: What are the implications for the water industry? Water Research. 2016;105:305–313. doi: 10.1016/j.watres.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen Tram Thuy, Traub Rebecca, Pham Phuc Duc, Nguyen Hung Viet, Nguyen Khuong Cong, Phung Cam Dac, Dalsgaard Anders. Prevalence and molecular characterization of Cryptosporidum spp. and Giardia spp. in environmental samples in Hanam province, Vietnam. Food and Waterborne Parasitology. 2016;3:13–20. doi: 10.1016/j.fawpar.2016.03.003. [DOI] [Google Scholar]
- 55.Abeywardena H, Jex AR, Nolan MJ, Haydon SR, Stevens MA, McAnulty RW, et al. Genetic characterisation of Cryptosporidium and Giardia from dairy calves: discovery of species/genotypes consistent with those found in humans. Infect Genet Evol. 2012;12:1984–1993. doi: 10.1016/j.meegid.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Navarro-I-Martinez L, Del Águila C, Bornay-Llinares FJ. Cryptosporidium: a genus in revision. The situation in Spain. Enferm Infecc Microbiol Clin. 2011;29:135–43 (In Spanish). [DOI] [PubMed]
- 57.Neto RC, Dos Santos LU, Sato MIZ, Franco RMB. Cryptosporidium spp. and Giardia spp. in surface water supply of Campinas, Southeast Brazil. Water Sci Technol. 2010;62:217–222. doi: 10.2166/wst.2010.312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Metadata information of the water samples collected and submitted to molecular detection of Giardia duodenalis, Cryptosporidium, Entamoeba histolytica, Toxoplasma gondii and Cyclospora cayetanensis. (DOCX 24 kb)
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its Additional file.