Abstract
Objective
This study determined the clinical utility of an fMRI classification algorithm predicting medication‐class of response in patients with challenging mood diagnoses.
Methods
Ninety‐nine 16–27‐year‐olds underwent resting state fMRI scans in three groups—BD, MDD and healthy controls. A predictive algorithm was trained and cross‐validated on the known‐diagnosis patients using maximally spatially independent components (ICs), constructing a similarity matrix among subjects, partitioning the matrix in kernel space and optimizing support vector machine classifiers and IC combinations. This classifier was also applied to each of 12 new individual patients with unclear mood disorder diagnoses.
Results
Classification within the known‐diagnosis group was approximately 92.4% accurate. The five maximally contributory ICs were identified. Applied to the complicated patients, the algorithm diagnosis was consistent with optimal medication‐class of response to sustained recovery in 11 of 12 cases (i.e., almost 92% accuracy).
Conclusion
This classification algorithm performed well for the know‐diagnosis but also predicted medication‐class of response in difficult‐to‐diagnose patients. Further research can enhance this approach and extend these findings to be more clinically accessible.
Keywords: mood disorders, bipolar disorder, functional neuroimaging, machine learning, differential diagnosis
Significant outcomes.
The machine learning algorithm described here was robust for classifying the adolescent/young adult patients in the known major depressive disorder, bipolar disorder, and healthy control groups it was trained on, with 92.4% accuracy.
The classification algorithm was then utilized to assess its accuracy for treatment response in a new sample of 12 complex patients with unknown diagnoses and mapped accurately onto predicting the medication class of response 91.7% of the time.
The classification algorithm utilized five independent components for classification, which included components of classic functional networks. These comprised regions of the default mode network, salience network, dorsal attention network, and frontoparietal central executive network. The caudate body, thalamus, and parahippocampal gyrus were also involved in the optimal classification.
Limitations.
In five cases of 12 the medication‐class of response in the complex patient group was consistent with medication used at the time of the scan, leading to questions about whether the algorithm was detecting this medication itself rather than a trait feature of the patient.
The scanning, algorithm creation, and algorithm application were all highly technical, time‐consuming, and expensive. Further research is needed to extend these findings to develop an accessible neuro‐functional biomarker for the diagnosis of mood and other psychiatric disorders that answers the important question: “What medication‐class is most likely to help my patient attain sustained recovery?”
Introduction
Differentiating bipolar disorder (BD) from major depressive disorder (MDD) using the current “gold standard”, the Diagnostic and Statistical Manual (DSM), is challenging in patients without obvious mania. Since patients with BD generally spend more time in depressive than manic states 1, BD is often misdiagnosed as MDD. BD patients go 6–10 years without a proper diagnosis 2. And while the adoption of a spectrum approach to mood disorder diagnosis is appealing 3, the treatment response of patients to the different medication classes, antidepressants (AD) vs. mood stabilizers (MS), suggests that there is a fundamental difference between MDD and BD in many cases. MSs often do not effectively treat MDD while ADs may make BD type I worse 4. Inadequately treated BD is costly to the medical system and adds to human suffering 5. Thus, obtaining the correct mood diagnosis, or more importantly the correct medication‐class to best support recovery of the patient, is imperative.
Numerous efforts have successfully distinguished DSM‐diagnosed BD from MDD using fMRI 6, 7, 8, including those of our own group 9, 10, 11, 12. However, these studies involved patients with diagnoses made according to the DSM or related criteria, with patients not meeting such criteria excluded from study. The imaging algorithms were then created to classify those patients according to this DSM “gold standard”. The lack of complexity in the patients included in these studies leads to questions about the utility of such algorithms as a facilitative strategy in real clinical work.
This same concern about complexity in the “real world” of clinical care also applies to randomized clinical trials evaluating medication (and other) treatment responses within specific patient populations. The careful selection of research participants with clear‐cut DSM diagnoses leads to challenges in implementing treatment recommendations in the clinical reality of multiple comorbid diagnoses, fluctuating time‐courses of illness, uncertain reporting of personal and family histories, and unclear responses to past medication trials. In clinical work, diagnoses are often not as well defined as in research protocols.
Because our understanding of the brain is such that psychiatric diagnoses are not based on pathophysiology, diagnosis in complex patients is often only imperfectly predictive of the medication‐class of response. The situations most demanding of a biologically based classification algorithm (biomarker) are, in fact, those in which the DSM classification is unclear and the clinical impression is confusing. Practicing clinicians need to prescribe for such complicated patients. Additionally, in emerging adults without a long personal history of psychiatric illness clinical presentation is often not classical 13. Yet emerging adults with mood disorders often face a long future of treatment responsiveness or refractoriness, making it imperative to choose the right medication‐class to optimize recovery. The science of psychiatric diagnosis must move beyond the DSM when the DSM is insufficient.
We aimed to use two patient groups, those with “known” BD and MDD as well as healthy controls (HC) to create a classification algorithm and then use that algorithm to retrospectively predict medication‐class of response in individuals from a new patient sample whose diagnoses were complicated and unclear. Our hypothesis was that the classification algorithm created using known DSM‐based MDD vs. BD, type I patients would illuminate neurophysiological differences that could then be used to predict AD vs. MS response, respectively, in complex patients whose diagnoses were unclear by the DSM. This approach could reveal biomarkers of medication‐class of response within mood disorders in the clinical realities of psychiatric care, an important step beyond the DSM when that classification is insufficient.
Material and methods
Participants
Data collection was approved by the University of Western Ontario Research Ethics Board. All participants were provided with a written description of the study and had the opportunity to ask questions. Written, informed consent was obtained. When signing consent participants indicated up to five family members they allowed for contact to collect corroborative information regarding family mental health. Consent to participate by family members was taken verbally over the phone and verified by completion of the interview.
Participants used in algorithm creation were aged 16–27, in four categories: 32 with BD, type I (BD); 34 with MDD; and 33 HCs. BD and MDD patients were recruited from the First Episode Mood and Anxiety Program (FEMAP), and other psychiatric services at London Health Sciences Centre, London, Ontario, Canada, who had presented with a primary mood disorder. HCs were recruited from the general community. The patients had been treated to euthymia according to standard psychiatric practice based on their clinical presentations and were then recruited as either MDD or BD participants. Diagnoses were made using the Structured Clinical Interview for DSM disorders‐IV, (SCID‐IV) 14 or the Diagnostic Interview for Genetic Studies (DIGS) 15 and were confirmed by the clinical psychiatric diagnostic assessment (patient groups only). Agreement between clinical diagnosis and SCID/DIGS diagnosis of MDD or BD‐type I, in full remission, was required for these patient groups.
No participant was included in the MDD group if they had a family history of BD by the Family Interview for Genetic Studies (FIGS) 16, or if the patient met screening criteria for BD as per the CIDI‐based BPD screening scale. Participants in the HC group were excluded if they had a family history of mood disorders.
Inability to meet the inclusion criteria above led to creation of an “unknown” (UNK) group of 12 participants. Reasons for classification as UNK were prospective: disagreement between DIGS (no UNK participant underwent the SCID‐IV) and psychiatrist diagnosis, first degree relative with mental illness (recruited as HC), or uncertain diagnosis on psychiatrist evaluation. Or retrospective: change in diagnosis over the course of treatment or response to a medication‐class inconsistent with DSM diagnosis.
Chart review was used to determine the medication‐class used to help each patient in the UNK group attain sustained euthymia, as evaluated by the treating clinician, lasting at least 6 months. Medication‐class was simplified to either an AD or MS (lithium, lamotrigine, carbamazepine, divalproex sodium). Three participants were taking an atypical antipsychotic (AP) and this was noted.
Medications were unchanged for 3 weeks prior to scanning. All subjects’ moods were euthymic at the time of the scan as evaluated with the Montgomery‐Asberg Depression Rating Scale and Young Mania Rating Scale. Some individuals in the UNK group had relapses following the scan and underwent a medication change to result in sustained (over 3 months) euthymia, which was then considered the medication‐class of response.
Imaging data and processing
MRI images were collected using a 3.0 T Siemens Verio MRI scanner at the Lawson Health Research Institute using a 32‐channel phased‐array head coil. A T1‐weighted, 3D magnetization‐prepared rapid gradient echo sequence was used for anatomical images. Acquisition parameters were: repetition time (TR) = 3000 ms, echo time (TE) = 2.98 ms, flip angle = 9°, field of view (FOV) = 256 ×256 mm, matrix size = 256 × 256, 176 sagittal slices, voxel size = 1 × 1 × 1 mm. Functional scans were gradient‐echo, echo‐planar scans with TR = 2000 ms, TE = 30 ms, flip angle = 90°, FOV = 240 × 240 mm, matrix size = 80 × 80, 40 axial slices and thickness = 3 mm, with no parallel acceleration. Scans covered whole brain with an isotropic spatial resolution of 3 mm for a total time of approximately 8 min (164 brain volumes). No participant reported falling asleep during the scan when asked immediately after scanning.
The fMRI images were preprocessed using statistical parametric mapping software (SPM12, http://www.fil.ion.ucl.ac.uk/spm/). To allow for magnetization equilibrium, the first 10 images were discarded. The remaining 154 images were first corrected for the acquisition time delay among different slices, and were realigned to the first volume for head‐motion correction. fMRI images were spatially normalized to Montreal Neurological Institute (MNI) space by diffeomorphic anatomical registration using exponentiated Lie algebra (DARTEL) and spatially smoothed with a 6‐mm FWHM Gaussian kernel.
Statistical analysis
We extended a previous classification method 17, 18, which our main framework was based on, consisting of four parts: estimating maximally spatially independent components (ICs) from fMRI data 18, constructing a similarity matrix between subjects which was combined with sigmoid kernel to build the final kernel function for classification 18, partitioning the matrix in kernel space, and conducting nested cross‐validation of the support vector machine (SVM) classifiers and IC combinations (Fig. 1).
Figure 1.
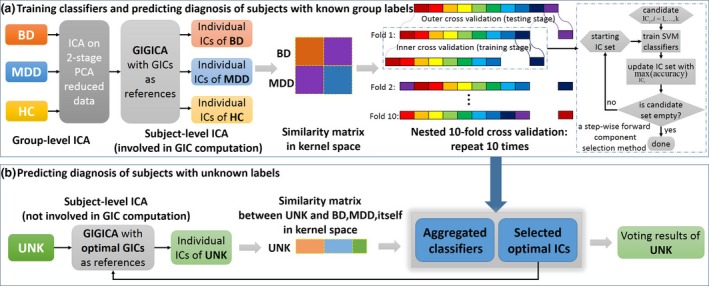
Flowchart of method. (a) Training classifiers and predicting diagnosis of subjects with known group labels. The training stage is composed of four parts as mentioned. Group independent component analysis (ICA) is computed on temporally concatenated fMRI data of bipolar disorder (BD) patients, major depressive disorder (MDD) patients, and healthy controls (HCs) resulting in individual subject maps computed by GIG‐ICA 17. Note that the UNK subjects were not involved in the computation of the group‐level ICA. For each cross‐validation loop, similarity matrices for BD, MDD were computed and classified via a kernel support vector machine (SVM) from the hold‐out data using 10‐fold cross‐validation. Namely, inner loop (dotted line frame) generated 9 SVM models, and a whole nested 10‐fold cross‐validation generated 90 SVM models. After repeating 10 times, 900 SVM classifiers were generated for the majority voting in prediction. (b) Predicting diagnosis of subjects with unknown labels. Following group ICA and individual subject map calculation, a similarity matrix between the UNK and the BD and MDD individuals was computed. Diagnosis of the UNK group was based on a majority voting mechanism using an ensemble classifier with a hypothesis supposing each UNK individual were either BD or MDD due to total blind diagnosis prediction of our binary classification method. [Colour figure can be viewed at wileyonlinelibrary.com]
This approach made use of multiple fMRI ICs to build linear subspaces that were calculated by adopting a principal angle‐based Riemannian distance for each individual subject, which facilitated a comprehensive characterization of fMRI data for individuals with known group labels including BD and MDD. Then, in conjunction with a SVM classifier, a forward component selection technique selected optimal ICs for constructing the most group‐discriminative combinations and determined the final diagnosis of UNK by majority voting. Note that the (cross‐validated) training of the known BD/MDD/HC and the prediction of the UNK individuals were totally separated from each another. The prediction of the UNK individuals used the final group IC‐based classifier extracted from the trained BD/MDD/HC stage. As shown in Fig. 1, four steps were included in the classification framework. This methodology is more fully explained in the supporting materials.
Individual‐specific spatial component extraction
Figure 1 shows the schematic flowchart of group information guided ICA (GIG‐ICA) to identify subject‐specific component maps. As an extension of regular group ICA 19, 20, which incorporates an initial ICA to identify group spatial maps, GIG‐ICA used the group component map as the reference to back‐reconstruct individual subject components using spatially constrained ICA. The advantage of GIG‐ICA is that the estimated individual IC maps are more accurate and robust 21, 22, and it is able to estimate corresponding‐matched components from new data sets, the unknown subjects in this case, for classification purposes. GIG‐ICA was performed for HC, BD, and MDD, respectively, to build group‐specific individual ICs, while the UNK subjects were not included, to avoid potential bias. We chose 20 ICs to reduce computational complexity, similar to typical ICA based fMRI studies 17, 18, 23.
Subspace similarity between individuals
Because we aimed for the optimal IC combination, and thereby the highest classification accuracy, a subspace spanned by a combination of discriminative ICs was used. This subspace representation of ICs conveyed richer information in a concise manner, compared with treating ICs as voxel‐wise spatial maps 18, 24. A Riemannian similarity/distance measure 25 was computed based on principal angles between spatial component maps of different individuals, representing subspace similarities between individuals spanning a subset of ICs 26 (see supporting materials). As shown in Fig. 1a, the subspace similarity was represented by a symmetric matrix, which was computed from ICs of BD and MDD individuals using the method mentioned.
Forward component selection using SVM classifiers
The classification process involved the training and testing stage of known BD and MDD patients in a 10‐fold nested 10‐fold cross‐validation (Fig. 1a), and the testing stage (Fig. 1b) for the additional UNK individuals. We adopted a step‐wise forward selection method 27 such that, in each training step, a component in the candidate set was added to the optimizing component set and used to build a new component set. We then performed SVM classification with sigmoid kernel on test sets of known BD and MDD individuals using LIBSVM 28, and selected the component with the highest accuracy to add into the optimization set. This process continued until the candidate set was empty (Fig. 1). The classification performance was estimated by averaging the classifiers’ performance for all testing individuals. Comparing the classification performance of IC combinations with different IC numbers used in training identified the optimal group‐discriminative IC combinations. Finally, we constructed an ensemble classifier of 900 voters (see supporting materials) based on all known BD and MDD patients whose base classifier was built with the 100 optimal IC combinations (10 × 10) from the training sets.
Classifying subjects with unknown labels
Imaging analysts were blind to the medication‐class of response in the UNK patients. The aggregated classifiers were used to vote for the labels of each UNK patient for individual subject classification. We predicted group membership for each new subject by calculating their spatial components via GIG‐ICA and entering each independently into the subspace representation. Note the UNK information was not use in either the training or testing stages, ensuring an unbiased prediction (Fig. 1b). Again, the training stage of known BD & MDD and the predicting stage of UNK were totally separated from each other. The predicting stage relied on the group ICs extracted from BD&MDD&HC and classifier models and optimal IC combinations trained from BD&MDD of the training stage. Thus, each patient classification represented an individual test of the classification algorithm at the single subject level. Classification vote was for either BD or MDD, as per the training set in the algorithm, and this was compared with medication‐class of response.
Results
A summary of known BD, MDD and HC participants’ demographic information is presented in Table 1. There were no significant sex differences across groups; though the BD subjects were significantly older than the MDD patients at the time of scanning. Nevertheless, P‐values from a MANCOVA showed no difference related to either sex or age among groups. We ran a cross‐validation after regressing out demographic covariates of each IC, and the results were the same as those without this regression.
Table 1.
Demographic and clinical data of participant scans
| Group | BD | MDD | HC | P value (BD/MDD/HC) | P value (BD/MDD) | P value MANCOVA |
|---|---|---|---|---|---|---|
| Number | 32 | 34 | 33 | – | – | – |
| Age (mean ± SD)Range (years) | 21.3 ± 2.916–27 | 19.7 ± 2.616–25 | 20.2 ± 2.017–24 | 0.05a | 0.03b | 0.85e |
| Sex (M/F) | 16/16 | 10/24 | 13/20 | 0.23c | 0.09d | 0.96e |
The P value was obtained by analysis of variance (ANOVA) of BD&MDD&HC.
The P value was obtained by two‐sample two‐tailed t‐test of BD&MDD.
The P value was obtained by cross‐tabulation of BD&MDD&HC.
The P value was obtained by cross‐tabulation of BD&MDD.
The P values of age and sex were obtained by a multivariate analysis of covariance (MANCOVA) (Allen et al. 33).
To confirm the limited role of demographics, we also ran a classification of BD and MDD based on only gender and age using SVM with the same parameters as with the imaging data. The accuracy was 49.32%, with sensitivity of 89.19%, and specificity of 8.33%. The classification rates in this model were all approximately 50%.
Classification of known HC, BD, & MDD
As shown in Fig. 2a, classification accuracies between known BD and MDD were all above 84% with different numbers of ICs used. The best classification rate, of 92.4% (sensitivity 87.5%, specificity 97.1%), was obtained using five ICs. Using these five ICs classification accuracy between BD vs. HC was 91.4% and between MDD vs. HC was 94.2%. This was an improvement over our previous approach 10 both in accuracy and time complexity, taking only 39.8 h for one 10‐fold cross‐validation vs. 143.1 h on a CentOS Linux computer cluster with 19 servers and 188 Intel Xeon CPUs.
Figure 2.
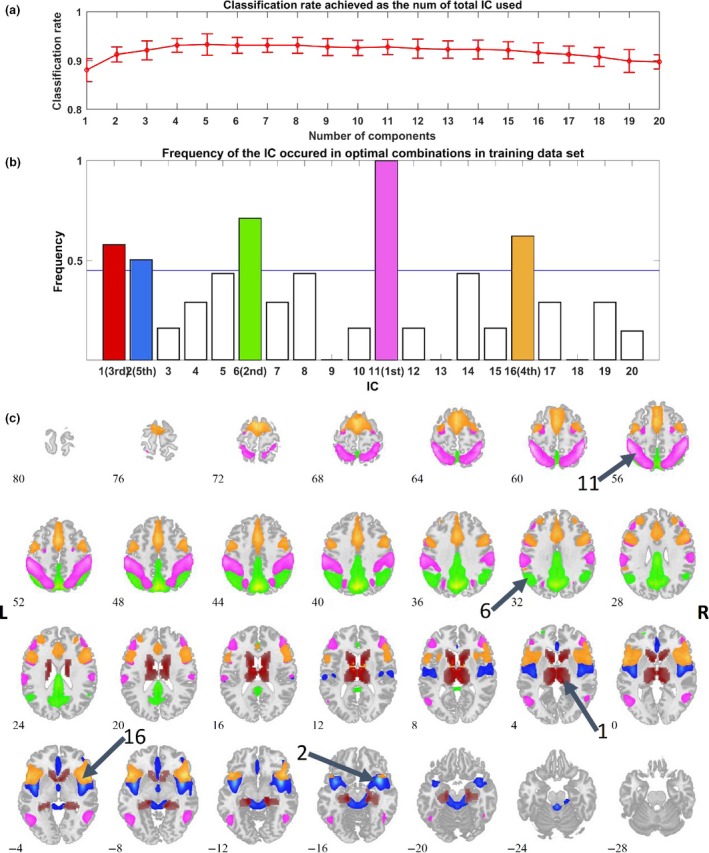
Classification Results. (a) The top line with error bars (1 standard deviation) shows classification rates achieved as a function of the total IC number used and illustrates that the optimal classification rate was achieved using five ICs. Yet using any number from 1 to 20 ICs for classification resulted in accuracy above 84%. (b) Frequency of the ICs as they occurred in 100 optimal combinations. We ran 10 times of 10‐fold nested cross‐validation. Each fold resulted in one optimal IC combination (selection and order of 20 ICs), yielding 100 combinations. This graph illustrates the ICs that were most contributory to the classification algorithm. (c) Spatial maps were overlaid with different colors, identical to those in (b). The sorting order of the five top selected ICs from high to low is 11, 6, 1, 16, and 2.
Discriminative components in known BD & MDD groups
There were 100 combinations of ICs yielded from ten times 10‐fold cross‐validations. The frequency of all 20 components in best combination is shown in Fig. 2(b). Five ICs were selected as the most discriminative functional networks for distinguishing BD from MDD (components 11,6,16,1,2; Fig. 2c). The five ICs cover several regions involved in classic brain networks including the dorsal attention network (DAN), default mode network (DMN), frontoparietal central executive network (CEN), and the salience network (SN),They also included an IC encompassing caudate, insula, and thalamus.
Classification of patients in the “unknown” group
Voting ratios for BD vs. MDD for each of the 12 UNK subjects, as well as the final classification prediction based on these ratios, clinical diagnoses and medication‐classes both during the scan and at sustained euthymia, and other variables are listed in Table 2. Each UNK subject was presumed as either a BD or MDD patient in our SVM diagnosis prediction with the proposed binary classification method.
Table 2.
DIGS Mood and Other Diagnoses, Clinical Mood and other Diagnoses & Medication‐Class and Vote of Classifier Algorithm. Shaded cases were scanned on the same medication‐class as that to achieve euthymia; thus, unshaded cases held higher test validity of the classification algorithm
| ID | Age | Sex | DIGS Mood Dx | Chart Mood Dx on Entry | Other Chart Dx's on Entry | Rx at Scan | Rx of Responsea | Days between scan and final chart review | BD (%) | MDD (%) | Voting Results |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1308 | 22 | M | BD‐I | MDD | MJ dependence; PTSD | AD | AP | 212 | 63.2 | 36.8 | BD |
| 1310 | 20 | M | BD‐I | MDD | MJ dependence; PTSD | AD | AD | 1631 | 16.4 | 83.6 | MDD |
| 1322 | 24 | M | MDD | MDD | Panic d/o | None | MS,AP | 153 | 64.9 | 35.1 | BD |
| 1325 | 18 | F | None | n.a. | n.a. | None | n.a. | n.a. | 5.6 | 94.4 | MDD |
| 1349b | 21 | M | BD‐I | BD‐I | Alcohol dependence | MS | MS,AP | 1348 | 32.9 | 67.1 | MDD |
| 1364 | 19 | F | MDD | MDD vs. BD | None | MS | MS | 1025 | 63.6 | 36.4 | BD |
| 1368 | 19 | M | MDD | MDD vs. BD | None | MS | MS | 288 | 61.2 | 38.8 | BD |
| 1372 | 19 | M | BD‐II | Nonec | Alcohol dependence; MJ dependence | None | None | 184 | 23.2 | 76.8 | MDD |
| 1378 | 26 | M | MDD | MDD vs. BD | None | AD | None | 275 | 45 | 55 | MDD |
| 1392 | 19 | F | BD‐II | MDD vs. BD | MJ dependence; PTSD | MS,AD | MS | 667 | 56.4 | 43.6 | BD |
| 1395 | 19 | F | MDD | MDD vs. BD | MJ dependence; PTSD | None | None | 532 | 28 | 72 | MDD |
| 1407 | 19 | M | BD‐II | MDD vs. BD | MJ dependence; PTSD | AD | AD | 377 | 17.4 | 82.6 | MDD |
Rx, medication; DIGS, Diagnostic Interview for Genetic Studies; Dx, diagnosis; d/o, disorder; PTSD, Posttraumatic Stress Disorder; MJ, Marijuana; AD, antidepressant; MS, mood stabilizer; AP, antipsychotic; n.a., not applicable.
The class of medication that was effective at the time the patient was stable, at the last time of evaluation as determined clinically and by chart review, to include only ADs, MSs, or APs.
This patient was ultimately diagnosed with schizoaffective disorder due to persistent paranoid ideation.
This patient was ultimately diagnosed with ADHD and GAD, but no mood disorder.
Details of several cases are noteworthy.
1308: treated as MDD by a psychiatrist in our clinic, but euthymia not sustained using any AD. Eventually, patient was stabilized on a single AP in another treatment setting. (The patient was never tried on a MS.) Algorithm voted BD.
1322: had over five trials of ADs from multiple classes, alone and in combination, in combinations with APs, MSs and light therapy as adjuncts; trial of electroconvulsive therapy (patient truncated after 8 sessions). Recovered only after all ADs were stopped to prepare for a monoamine oxidase inhibitor (MAOI); patient remained on a MS (carbamazepine) and an AP (quetiapine). Quickly recovered completely so the MAOI was never tried; returned to work and school. Patient spontaneously stopped medication many months later, when euthymic, and was scanned. Shortly after, returned to clinic with a relapse and again recovered completely with carbamazepine and quetiapine, for a second full remission. Total time in clinic was over 4.5 years. Algorithm vote BD, consistent with medication response. No clinical symptoms of BD ever by time of discharge.
1325: “healthy control” with first‐degree relative diagnosed with MDD after a suicide attempt. Follow‐up over 3 years after scan revealed a depressive episode.
1349: eventually diagnosed with schizoaffective disorder based on persistent paranoid ideation mentioned by mother after months of treatment at clinic. The one clear miss by the classification algorithm.
1368: like 1322, revealed no clinical evidence of BD. The patient demonstrated severe depression and agitated, angry outbursts. Several ADs trialed but failed. Lithium was tried as a last effort before deciding that the patient had challenges beyond a mood disorder, including possible Asperger's syndrome. Lithium led to full recovery and independence with gainful employment. Algorithm vote was again consistent with medication response.
Considering medication response as the “gold standard”, the algorithm was accurate for individual classification of 11 of 12 patients. It correctly categorized five patients as having brain function “more like BD” patients who were MS (plus one AP) responders; it correctly categorized as having brain function “more like MDD” two patients who were AD responders; and it conservatively categorized four individuals who had sustained euthymia on no medication as having brain function more like MDD. It miss‐categorized the one patient with schizoaffective disorder as more like MDD who actually required both an MS and an AP for response and had not responded to an AD.
Discussion
This classification algorithm demonstrated 92.4% accuracy between the known participant BD and MDD groups used to train it. In the group of 12 complex patients, representative of individuals for whom a medication response prediction algorithm is most needed, it correctly provided individual classification of the vast majority (91.7%), judging those with a sustained medication‐class of response to MS and/or AP medications as more like BD and those without such a response as more like MDD. While the use of brain scans and sophisticated classification algorithms is not practical in routine clinical care due to cost and time at present, advances in technology could change this. These findings also extend our understanding of the biological basis of medication‐class of responsiveness in mood disorders. Such knowledge can improve both diagnostic strategies administered in the clinic and treatment for patients with complex presentations of mood disorders who may otherwise go unsuccessfully treated for years.
Two of the most clinically interesting cases presented here are 1322 and 1368. There was no clear symptomatology of BD in either individual before, during or after treatment. Both patients had non‐response to ADs. Both would likely have been considered “treatment refractory” over time. In the case of 1322, it was only the removal of all ADs, with the retention of the MS and AP, which led to full remission. He was scanned medication free. For 1368 his depression was fully resolved with one medication, a MS (and he was scanned on that medication). It is unclear how many individuals categorized as treatment refractory in routine clinical care might exhibit the same brain pathophysiology and therefore the same algorithm classification and medication‐class of responsiveness as these patients.
There were six cases in which the psychiatrist was unclear of the specific mood diagnosis at the time of entry (1364, 1368, 1378, 1392, 1395, 1407). In four of those, the algorithm matched the medication‐class of sustained response, attesting to the accuracy of the algorithm for predicting medication responsiveness. In two of them the algorithm indicated the brain function more like MDD, which was matched to sustained recovery with no medication. Since the algorithm was binary for MDD or BD, this was the correct choice. MDD is more heterogeneous and more easily treated to remission without medication.
Taking a closer look at the individuals who were on no medication for a mood disorder at the time of sustained response (1325, 1372, 1378, 1395), participant 1325 was the “healthy control” with a first degree relative with MDD. This case had the lowest rating for BD‐like brain function by the algorithm and, interestingly, at post‐study follow‐up had had a depressive episode. Participant 1372 presented with complex symptoms and was diagnosed with generalized anxiety disorder and attention deficit hyperactivity disorder, but no primary mood disorder by the treating psychiatrist. Participants 1378 and 1395 were very similar to each other in having an unclear presentation per the psychiatrist, a DIGS diagnosis of MDD and an algorithm vote for brain function more like MDD. Neither required medication for sustained response; both had had a MS trial that was discontinued because in neither case was it particularly helpful. These four individual cases demonstrate the specificity of the algorithm for BD‐like brain function and the medication responsiveness compatible with that, exhibiting the algorithm's ability to correctly identify “not like BD” and “no MS required”.
There were five ICs most salient for classification. The first, IC11 in the algorithm, contained nodes of the DAN including the inferior parietal lobule bilaterally. The second most discriminatory component (IC6) involved largely posterior brain regions of the DMN. The SN node (anterior cingulate cortex) appeared in IC16, the third most distinguishing component, which also included frontal portions of the CEN. Several areas in IC16 have consistently been associated with BD in structural brain studies (28). The forth contributing IC (IC1) included caudate body, thalamus and parahippocampal gyrus. The thalamus is the major relay/association structure for all somatic signals and includes areas controlling arousal and alertness. The caudate is a dopaminergic region involved in goal‐directed action, emotion, motivation and memory and learning. Finally, the remainder of the SN, including the insula, appeared in IC2 (see Fig. 2c and supporting material).
This combination of ICs, providing a data‐driven grouping of regions, represents an interesting amalgamation of ‘classic’ large‐scale brain network nodes 30. Three of four ICs identified here had nodes of these networks that were split between different ICs. Only the DAN was preserved in one IC, though the frontal eye field regions were too small to reach threshold in this analysis (Fig. 2c, Table S1). Abnormalities in how these nodes sorted across ICs may pertain to the basic pathophysiology of mood disorders and may be particularly helpful for medication response prediction. Further research is needed to evaluate this. There were also major contributions from autonomic processing regions—bilateral thalamus and insula. These regions were identified in a recent meta‐analyses showing lack of specificity of some brain networks to the common mental illnesses 31. Additionally, our third most relevant IC showed strong connectivity within frontal regions, consistent with results by other researchers as regions of increase connectivity in BD patients anticipating a reward 32.
The advantage of our approach is the reduction of hypothesis‐bias so that the data can “draw its own conclusions” from the scans. The high accuracy of this algorithm for the known mood diagnoses and for the individual classification for medication responsiveness of the complex patients was achieved by not being restricted to established functional networks or theoretical biases.
There are limitations of this study. Although all patients were scanned in euthymia, most MDD/BD patients and some of the UNK patients were on either a MS or AD at the time of scanning. This raises the possibility that the algorithm was trained to detect the medication‐class that the patient was taking, rather than brain physiology related to either MDD or BD. Interestingly, however, three participants in the UNK group had a change in medication status from scan to sustained response (1308, 1322, 1378), and an additional four were on no medication when scanned. The one case incorrectly classified by the algorithm was on a MS, but the algorithm vote was for non‐BD‐like brain function. Sustained euthymia was defined clinically; future studies should define this more systematically. Additionally, patients were scanned in euthymia, but the highest need for response prediction is in the depressive state, when decisions about medication‐class are needed. Future studies should be done in medication‐free depressed patients to further test this approach.
In the group of complex patients presenting with unclear diagnoses, the algorithm described here provided correct individual classifications of medication response for 11 of 12 patients, with the remaining patient having a diagnosis that the algorithm was not trained to classify. The true test of a classification algorithm is in characterizing difficult patients at the individual level, where such a technique is most needed because the DSM is insufficient, and the question that any such algorithm most urgently needs to answer is, “What medication‐class is most likely to help my patient attain sustained recovery?” The algorithm presented here makes headway towards these goals. Further research can extend these findings to develop an accessible neuro‐functional biomarker for the diagnosis of mood and other psychiatric disorders.
Declaration of interest
No conflict of interest, actual or apparent, exists for this study for any of the authors.
Supporting information
Table S1 Talariach table of the spatial maps of the selected 5 ICs.
Funding
This project was supported by The National Institutes of Health grants P20GM103472, 1R01EB006841 and R01EB005846; the China National High‐Tech Development Plan (863 plan) No. 2015AA020513 and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB02060005); China National Natural Science Foundation (No. 81471367, 61773380, 61703253) and Natural Science Foundation of Shanxi Province (Grant No. 2016021077); the Lawson Health Research Institute, grant No. LHR D1374; Generous donations to FEMAP research made through the London Health Sciences Foundation; and a Pfizer Independent Investigator Award, grant No. WS2249136. In‐kind support came from the University of Western Ontario, Schulich School of Medicine and Dentistry, the London Health Sciences Centre, St. Joseph's Health Care, and the Lawson Health Research Institute. The funders played no role in study design, writing of this manuscript or the decision to submit for publication.
Author contributions
Elizabeth Osuch study design and oversight of subject recruitment and data collection, clinical care, data interpretation, analysis conceptualization, and manuscript writing/preparation. Shuang Gao conducted the data analysis and contributed to manuscript preparation. Michael Wammes participant recruitment, data collection and management/consolidation, manuscript preparation. Jean Théberge study design, imaging data collection design and oversight, manuscript preparation. Peter Williamson study design, interpretation, manuscript preparation. R.J. Neufeld study design, manuscript preparation. Yuhui Du interpretation, manuscript preparation. Jing Sui data analysis/oversight, data interpretation, manuscript writing/preparation. Vince Calhoun conceptualization of data analysis, manuscript writing/preparation.
Osuch E, Gao S, Wammes M, Théberge J, Williamson P, Neufeld RJ, Du Y, Sui J, Calhoun V. Complexity in mood disorder diagnosis: fMRI connectivity networks predicted medication‐class of response in complex patients.
Contributor Information
E. Osuch, Email: Elizabeth.osuch@lhsc.on.ca.
J. Sui, Email: jing.sui@nlpr.ia.ac.cn
References
- 1. Judd LL, Akiskal HS, Schettler PJ et al. The long‐term natural history of the weekly symptomatic status of bipolar I disorder. Arch Gen Psychiatry 2002;59:530–537. [DOI] [PubMed] [Google Scholar]
- 2. Keck PE, Kessler RC, Ross R. Clinical and economic effects of unrecognized or inadequately treated bipolar disorder. J. Psychiatric Pract 2008;14:31. [DOI] [PubMed] [Google Scholar]
- 3. Akiskal HS. The emergence of the bipolar spectrum: validation along clinical‐epidemiologic and familial‐genetic lines. Psychopharmacol Bull 2007;40:99–115. [PubMed] [Google Scholar]
- 4. El‐Mallakh RS, Vohringer PA, Ostacher MM et al. Antidepressants worsen rapid‐cycling course in bipolar depression: a STEP‐BD randomized clinical trial. J Affect Disord 2015;184:318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi L, Thiebaud P, McCombs JS. The impact of unrecognized bipolar disorders for patients treated for depression with antidepressants in the fee‐for‐services California Medicaid (Medi‐Cal) program. J Affect Disord 2004;82:373–383. [DOI] [PubMed] [Google Scholar]
- 6. Anand A, Li Y, Wang Y, Lowe MJ, Dzemidzic M. Resting state corticolimbic connectivity abnormalities in unmedicated bipolar disorder and unipolar depression. Psychiatry Res 2009;171:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goya‐Maldonado R, Brodmann K, Keil M, Trost S, Dechent P, Gruber O. Differentiating unipolar and bipolar depression by alterations in large‐scale brain networks. Hum Brain Mapp 2016;37:808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li M, Das T, Deng W et al. Clinical utility of a short resting‐state MRI scan in differentiating bipolar from unipolar depression. Acta Psychiatr Scand 2017;136:288–299. [DOI] [PubMed] [Google Scholar]
- 9. Ford KA, Theberge J, Neufeld RJ, Williamson PC, Osuch EA. Correlation of brain default mode network activation with bipolarity index in youth with mood disorders. J Affect Disord 2013;150:1174–1178. [DOI] [PubMed] [Google Scholar]
- 10. Jie NF, Zhu MH, Ma XY et al. Discriminating bipolar disorder from major depression based on SVM‐FoBa: efficient feature selection with multimodal brain imaging data. IEEE Trans Auton Ment Dev 2015;7:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He H, Yu Q, Du Y et al. Resting‐state functional network connectivity in prefrontal regions differs between unmedicated patients with bipolar and major depressive disorders. J Affect Disord 2016;190:483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He H, Sui J, Du Y et al. Co‐altered functional networks and brain structure in unmedicated patients with bipolar and major depressive disorders. Brain Struct Funct 2017;222:4051–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benazzi F, Koukopoulos A, Akiskal HS. Toward a validation of a new definition of agitated depression as a bipolar mixed state (mixed depression). Eur Psychiatry 2004;19:85–90. [DOI] [PubMed] [Google Scholar]
- 14. First MB, Spitzer RL, Gibbon M, Williams JBW. Structured clinical interview for DSM‐IV axis I disorders (SCID). New York: New York State Psychiatric Institute, Biometrics Research; 1997. [Google Scholar]
- 15. Nurnberger JI Jr, Blehar MC, Kaufmann CA et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry 1994;51:849–859; discussion 863‐844. [DOI] [PubMed] [Google Scholar]
- 16. Maxwell EM. Manual for the FIGS. Rockville, MD, Clinical Neurogenetics Branch, Intramural Research Program, National Institute of Mental Health; 1982.
- 17. Du Y, Pearlson GD, Liu J et al. A group ICA based framework for evaluating resting fMRI markers when disease categories are unclear: application to schizophrenia, bipolar, and schizoaffective disorders. NeuroImage 2015;122:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan Y, Liu Y, Wu H et al. Discriminant analysis of functional connectivity patterns on Grassmann manifold. NeuroImage 2011;56:2058–2067. [DOI] [PubMed] [Google Scholar]
- 19. Calhoun VD, Adali T. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev Biomed Eng 2012;5:60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp 2001;14:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du Y, Fan Y. Group information guided ICA for fMRI data analysis. NeuroImage 2013;69:157–197. [DOI] [PubMed] [Google Scholar]
- 22. Du Y, Allen EA, He H, Sui J, Wu L, Calhoun VD. Artifact removal in the context of group ICA: a comparison of single‐subject and group approaches. Hum Brain Mapp 2016;37:1005–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sui J, Adali T, Pearlson GD, Clark VP, Calhoun VD. A method for accurate group difference detection by constraining the mixing coefficients in an ICA framework. Hum Brain Mapp 2009;30:2953–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan Y, Liu Y, Wu H et al. Discriminant analysis of functional connectivity patterns on Grassmann manifold. NeuroImage 2011;56:2058–2067. [DOI] [PubMed] [Google Scholar]
- 25. Bjoerck A, Golub GH. Numerical methods for computing angles between linear subspaces. Math Comp 1973;27:579–594. [Google Scholar]
- 26. Hamm J, Lee DD. Grassmann discriminant analysis: a unifying view on subspace‐based learning. Proceedings of the 25th International Conference on Machine Learning Helsinki, Finland; 2008: 376–383.
- 27. Guyon I. An introduction to variable and feature selection. J Mach Learn Res 2003;3:1157–1182. [Google Scholar]
- 28. Chang C‐C, Lin C‐J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2011;2:1–27. [Google Scholar]
- 29. Bora E, Fornito A, Yucel M, Pantelis C. Voxelwise meta‐analysis of gray matter abnormalities in bipolar disorder. Biol Psychiatry 2010;67:1097–1105. [DOI] [PubMed] [Google Scholar]
- 30. Menon V. Large‐scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci 2011;15:483–506. [DOI] [PubMed] [Google Scholar]
- 31. Muller VI, Cieslik EC, Serbanescu I, Laird AR, Fox PT, Eickhoff SB. Altered brain activity in unipolar depression revisited: meta‐analyses of neuroimaging studies. JAMA Psychiatry 2017;74:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manelis A, Almeida JR, Stiffler R, Lockovich JC, Aslam HA, Phillips ML. Anticipation‐related brain connectivity in bipolar and unipolar depression: a graph theory approach. Brain 2016;139:2554–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Allen EA, Erhardt EB, Damaraju E et al. A baseline for the multivariate comparison of resting‐state networks. Front Syst Neurosci 2011;5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Talariach table of the spatial maps of the selected 5 ICs.


