Abstract
Objective
Hepatitis C virus (HCV) infection induces interferon-stimulated genes (ISGs) and downstream innate immune responses. This study investigated whether baseline and on-treatment differences in these responses predict response versus virological breakthrough during therapy with direct-acting antivirals (DAA).
Methods
Thirteen HCV genotype 1b-infected patients who had previously failed a course of peginterferon/ribavirin were re-treated with asunaprevir/daclatasvir for 24 weeks. After pre-treatment biopsy, patients were randomized to undergo a second biopsy at week 2 or 4 on-therapy. Microarray and NanoString analyses were performed on paired liver biopsies and analyzed using linear mixed models. As biomarkers for peripheral IFN responses, peripheral blood natural killer cells were assessed for pSTAT1 and TRAIL expression and degranulation.
Results
Nine (9/13, 69%) patients achieved sustained virological responses (SVR12) and four experienced virological breakthroughs between weeks 4–12. Patients who achieved SVR12 displayed higher ISG expression levels in baseline liver biopsies and a higher frequency of pSTAT1 and TRAIL-expressing, degranulating NK cells in baseline blood samples than those who experienced virological breakthrough. Comparing gene expression levels from baseline and on-therapy biopsies, 408 genes (±1.2 fold, p<0.01) were differentially expressed. Genes downregulated on treatment were predominantly ISGs. Downregulation of ISGs was rapid and correlated with HCV RNA suppression.
Conclusions
An enhanced interferon signature is observed at baseline in liver and blood of patients who achieve SVR12 as compared to those who experience a virological breakthrough. The findings suggest innate immunity may contribute to clearance of HCV during DAA therapy by preventing the emergence of resistance-associated substitutions that lead to viral breakthrough during DAA therapy.
Keywords: Hepatitis C Virus, Interferon-stimulated genes, Innate immunity, Liver biopsy, Natural killer cell
Introduction
Chronic hepatitis C virus (HCV) infection is characterized by a prominent type I and type III interferon (IFN) signature that is evident by increased expression of interferon-stimulated genes (ISGs) in the liver. Increased IFN-signaling is also detectable in innate immune cells, such as natural killer (NK) cells, which represent the most frequent innate immune cells in the liver and express the IFNα/β receptor. Because HCV persists at high titers in the presence of such immune responses, the contribution of innate immune activation to viral control is not clear.
Interestingly, high baseline ISG expression in the liver was shown to be associated with lower response to interferon-based therapy,1–3 which was the standard of care for patients with chronic HCV infection for more than 20 years. Elevated levels of ISG expression among persons with chronic HCV infection are partly genetically determined by single nucleotide polymorphisms rs12979860 and rs368234815 of the interferon lambda 3 and 4 genes, respectively, which are in strong linkage disequilibrium.4,5 Persons with the rs12979860[T] and rs368234815[ΔG] alleles have higher ISG expression but poorer response to interferon-based therapy.5, 6
The development of direct-acting antivirals (DAAs) has revolutionized the therapy of chronic HCV infection.7 Combinations of DAAs can lead to eradication of chronic HCV infection in 90–99% of individuals irrespective of host factors known to be negatively correlated with treatment response.8,9 However, some patients do fail DAA regimens. The presence of resistance-associated substitutions, cirrhosis, high BMI and non-compliance have been associated with higher failure rates during DAA therapy.10 Whether the endogenous intrahepatic interferon response plays a role in viral clearance during DAA therapy is not known. One prior study did report that ISGs were down-regulated with viral clearance during DAA therapy but did not assess the role of ISGs in the response to treatment.11 Furthermore, how rapidly ISGs are down-regulated in the liver and whether ISG levels at baseline or their decline during treatment are correlated with virological breakthrough or relapse remain unknown.
The aims of the current study were to investigate the early effects of viral suppression on ISG expression during therapy with an interferon-free, DAA-only regimen in subjects with chronic HCV infection who were prior partial or null responders to peginterferon (pegIFN)/ribavirin (RBV) therapy, and to determine whether baseline expression levels or early changes in ISG expression during therapy in the liver or response of interferon-sensitive innate immune cells in the blood were predictive of treatment outcome.
Materials and Methods
Study Population
Clinical data and research samples from patients participating in a clinical trial to evaluate asunaprevir (ASV) 100 mg twice daily and daclatasvir (DCV) 60 mg once daily for 24 weeks (Bristol-Myers Squibb, Raritan, NJ) who were prior partial or null responders to pegIFN/RBV with or without cirrhosis (ClinicalTrials.gov: NCT01888900) were used in this analysis. A partial response was defined as ≥2 log reduction by week 12 with detectable HCV RNA at week 24 during pegIFN/RBV therapy. A null response was defined as ≤2 log reduction by week 12 of IFN therapy. In the current study, a responder was defined as a patient who achieved undetectable HCV RNA 12 weeks post-therapy (Sustained Virological Response at week 12 off therapy [SVR12]). Virological breakthrough was defined as an increase in HCV RNA levels by ≥ 1 log from nadir while still on treatment.
Study Design
Patients underwent an initial liver biopsy within 12 weeks before starting therapy with ASV/DCV. Patients were randomized to undergo a second liver biopsy either at week 2 or week 4 following the start of therapy. The primary endpoint of the trial was SVR12. Post-therapy, all patients were monitored every 4 weeks until week 12 and then returned for a final visit at 24 weeks off treatment. All patients provided written informed consent and the protocol was approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases.
Serum HCV RNA Quantification
Serum HCV RNA level was quantitated using the Cobas TaqMan real-time polymerase chain reaction (Roche Molecular Diagnostics, Branchburg NJ) with a lower limit of detection of 10 IU/mL and a lower limit of quantification of 25 IU/mL.
Liver biopsy RNA Extraction and Microarray Analysis
Core liver biopsies were immediately placed in RNA later (Qiagen) and stored at −80°C until handling. Total RNA extraction from liver tissue was performed on paired liver biopsies using RNeasy kit (Qiagen). The quality of isolated total RNA was assessed using bioanalyzer, (Agilent Technologies, CA) and RIN (RNA Integrity Number) values were generally above 8.0 for all but three samples. Ten nanograms of total RNA from each sample was used for cDNA library preparation using Ovation Pico WTA system V2 kit (NuGEN Technologies Inc, CA). Ovation Pico WTA kit can successfully amplify samples with RIN numbers even as low as 2–5. For each array, 4 ug of cDNA was fragmented and biotin labeled using Encore Biotin module (NUGEN Technologies Inc, CA) and then added with hybridization control reagents (20× Eukaryotic controls and Control Oligo B2, Affymetrix Inc. CA). Samples were hybridized with Affymetrix Human Gene 2.0 ST arrays, incubated at 45°C for 18 hours rotating at 60 rpm in a GeneChip Hybridization oven (Affymetrix Inc, CA). Hybridized arrays were washed and stained with GeneChip HWS kit (Affymetrix Inc, CA) using Affymetrix 450 Fluidic Stations. Chips were scanned using Affymetrix GeneChip scanner 3000. To access the efficiency of cDNA synthesis, Poly A controls (dap, lys, phe, thr- Affymetrix Inc.) were spiked into the samples and hybridization controls (bioB, bioD, bioC and Cre, Affymetrix Inc.) were added to monitor labeling efficiency per the manufacturer’s instructions. Samples were processed as batches of 32 chips at a time. Raw files from Affymetrix Human Gene 2.0 array were preprocessed using RMA in R (Bioconductor oligo package, v.1.34.2), Supplementary Figure 1.12 Raw and processed data were deposited in GEO (GSE109278). Technical replicates were averaged for each sample and averages were used for further analysis by linear mixed models using the Bioconductor limma package (v.3.26.8).13 Pathway enrichment analyses were performed using the Fisher’s Exact Test on several databases (GO, Reactome, KEGG, WikiPathways). An adjusted p value of <0.05 was considered significant.
NanoString nCounter Gene Expression Assay
A customized probe set containing genes of interest from the 408 differentially expressed genes from the microarray data, in particular Cluster 1 (Supplementary Table 1), genes known to be associated with the innate immune response to HCV and the HCV life cycle based on a functional genomic screen14 and a comprehensive literature review,15 was designed and manufactured by NanoString and used for validation of microarray data. A codeset specific to a 100-base region of the target mRNA was designed using a (3′ biotin label) capture probe and a 5′ reporter probe tagged with a specific fluorescent barcode, creating two sequence-specific probes for each target transcript. Probes were hybridized to 100 ng of total RNA for 19 hours at 65 °C and applied to the nCounter™ Preparation Station for automated removal of excess probe and immobilization of probe-transcript complexes on a streptavidin-coated cartridge. Data were collected using the nCounter™ Digital Analyzer by counting the individual barcodes. NanoString data was normalized using the nSolver Analysis Software (NanoString Technologies Inc).
Genotyping
Genomic DNAs were used to identify the single nucleotide polymorphisms (SNP) associated with rs12979860 and rs368234815 (previously designated as ss469415590) as previously reported.4, 16
NK cell analysis
Peripheral blood mononuclear cells (PBMCs) were separated from heparin-anticoagulated blood on Ficoll Histopaque (Mediatech, Manassas, VA) density gradients, washed 3 times with phosphate-buffered saline (PBS, Mediatech) and cryopreserved in 70% fetal bovine serum (FBS, Serum Source International, Charlotte, NC), 20% RPMI1640 (Mediatech) and 10% DMSO (Sigma Aldrich, St. Louis, MO). Thawed PBMCs were processed and stained as described below prior to analysis on an LSRII flow cytometer using FacsDiva Version 6.1.3 (BD Biosciences, San Jose, CA) and FlowJo Version 8.8.2 software (Tree Star, Ashland, OR).
(i) TRAIL staining
Thawed PBMCs (1×106) were stained with ethidium monoazide (EMA) for 10 min on ice followed by staining with anti-CD3-AlexaFluor700, anti-CD19-PeCy5, anti-CD16-V500, anti-CD56-PeCy7 and anti-TRAIL-PE (all from BD Biosciences), and with anti-CD14-PECy5 (AbD, Raleigh, NC) for 25 min at 4°C.
(ii) NK cell degranulation
Thawed PBMCs were cultured overnight in RPMI 1640 with 10% FBS (Serum Source International), 1% penicillin/streptomycin, 2 mM L-glutamine and 10 mM HEPES (Mediatech). The next day, PBMCs were counted and cultured in the presence of anti-CD107-PE (BD Biosciences) with or without target K562 cells (ATCC, Manassas, VA) as described17 without addition of cytokines, then stained with EMA and lineage-specific antibodies as described above.
(iii) pSTAT1 staining
Thawed PBMCs (1.5×106) were rested in 0.5 ml RPMI 1640 with 10% FBS, 1% penicillin/streptomycin and 2 mM L-glutamine (Mediatech) for 2h. Thereafter, cells were fixed with BD Cytofix buffer for 10 min at 37°C and 5% CO2, centrifuged, permeabilized with BD Perm buffer III for 20 min on ice, washed twice and resuspended in BD Phosflow Buffer (all from BD Biosciences). Cells were subsequently stained with anti-CD56-PE (Beckman Coulter, Brea, CA), anti-CD3-APC, anti-CD20-PerCP/Cy5.5 and anti-pSTAT1-Alexa488 (all from BD Biosciences) for 20 min at room temperature.
Statistical Analysis
D’Agostino and Pearson omnibus normality tests and Mann–Whitney tests, were performed with GraphPad Prism 5.0a (GraphPad Software, La Jolla, CA). Two-sided P values less than .05 were considered significant.
Results
Study Cohort
Thirteen patients with HCV genotype 1b who were null responders (n=10) or partial responders (n=3) to a previous course of pegIFN/RBV therapy were enrolled into the study. The baseline clinical characteristics are shown in Table 1. There were six females, mean age 59 years; seven patients were Caucasian, four African-American and two Asian. Twelve of thirteen patients had the rs12979860-CT or -TT allele and twelve of thirteen patients had the rs368234815-ΔG genetic variant. Both variants are associated with an unfavorable response to interferon treatment. There was a 100% concordance between the two unfavorable SNPs. Mean serum alanine aminotransferase was 78 U/L and mean HCV RNA level was 6.7 log10 IU/mL. Five of thirteen (38%) patients had cirrhosis confirmed by liver biopsy (Ishak fibrosis score of 5–6), another four (31%) had advanced fibrosis (Ishak score of 3–4) and four (31%) had minimal or no liver fibrosis, (Ishak fibrosis score 0–2).
Table 1.
Baseline characteristics of the study population.
| Characteristics | Responders [n=9] |
Virological Breakthrough [n=4] |
|---|---|---|
| Mean Age (Range, Years) | 58 (39–75) | 63 (53–70) |
| Female (%) | 3 (33) | 3 (75) |
| Race (%) | ||
| White | 5 (56) | 2 (50) |
| African-American | 3 (33) | 1 (25) |
| Asian | 1 (11) | 1 (25) |
| IL28B rs12979860 Genotype | ||
| CC | 1 | 0 |
| CT or TT | 8 | 4 |
| IFNL4 rs368234815 Genotype | ||
| TT/TT | 1 | 0 |
| TT/ΔG | 7 | 3 |
| ΔG/ΔG | 1 | 1 |
| Mean ALT (Range, U/L) | 80 (37–172) | 75 (47–122) |
| Mean AST (Range, U/L) | 67 (21–190) | 55 (38–66) |
| HCV RNA log10(mean, IU/mL) | 6.6 | 7.0 |
| Ishak Fibrosis Score | ||
| 0–2 | 3 (33) | 1 (25) |
| 3–4 | 4 (45) | 0 (0) |
| 5–6 | 2 (22) | 3 (75) |
Virological Response
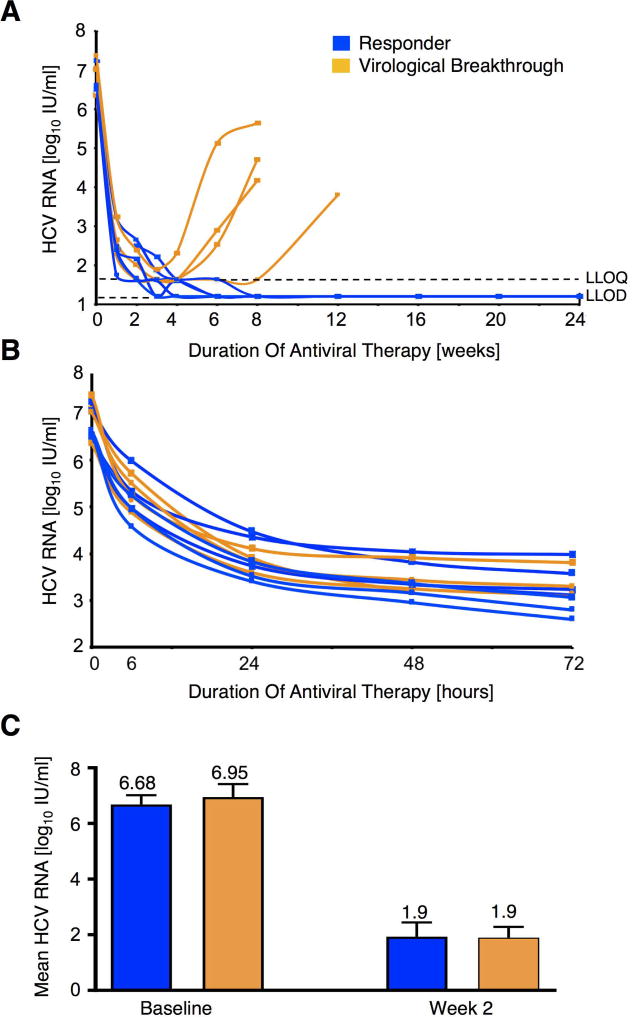
Among the 13 subjects treated with ASV/DCV, 9 achieved SVR12 and 4 experienced virological breakthrough, which occurred between weeks 4 and 12 of therapy (Figure 1A). All patients regardless of eventual response to DAA therapy experienced an initial rapid decline in HCV RNA level within the first 72 hours of therapy (Figure 1B). There was no difference in mean HCV RNA levels at baseline and at week 2 (earliest time point of second liver biopsy) between treatment responders and those with virological breakthrough (Figure 1C).
Figure 1. Viral kinetics during antiviral therapy.
(A) Eleven HCV genotype 1b partial or null-responders to interferon-based therapy were re-treated with 24 weeks of asunaprevir 100 mg (twice a day) and daclatasvir 60 mg (once daily). Seven patients responded to DAA therapy (blue) while four patients experienced virological breakthrough (orange) that occurred between weeks 4 and 12 of therapy. LLOQ, lower limit of quantitation; LLOD, lower limit of detection.
(B) HCV RNA levels (log10 IU/mL) declined rapidly within the first 72 hours of therapy in both DAA responders and those with subsequent virological breakthrough. The mean reduction in log10 HCV RNA IU/mL at 72 hours was 3.4 IU/mL.
(C) HCV RNA level (log10 IU/mL) did not differ at baseline and at week 2 of DAA therapy between patients that responded to DAA therapy and those subsequently experienced virological breakthrough. Mean and SEM are shown.
Changes in Global mRNA Expression During Therapy
In-vitro studies have reported that cellular factors play a pivotal role in the HCV life cycle.18–21 Therefore, we assessed changes in hepatic mRNA expression following DAA therapy by conducting a microarray-based transcriptome analysis using liver tissue from paired pre-treatment and on-treatment (week 2 or 4) biopsies. Eleven patients with paired liver biopsies were studied in the microarray analysis (two subjects with SVR12 who had not undergone the second biopsy at time of RNA extraction and microarray analysis were excluded). Thus, among the 11 patients included in the transcriptome analysis seven achieved SVR12 and 4 experienced virological breakthrough which occurred between treatment weeks 4 and 12. The expression data from the week 2 and 4 timepoints did not show any global segregation based on a similarity plot analysis (Supplementary Figure 2). Thus, for subsequent data analysis, data from both week 2 and 4 on-treatment biopsies were combined.
Comparing hepatic gene expression from biopsies obtained on-treatment to baseline, 408 genes (±1.2 fold, p<0.01) were differentially expressed (Supplementary Table 2). The majority of aberrantly expressed genes were downregulated upon therapy. Most of the significantly downregulated cellular pathways by GO, Reactome, KEGG, WikiPathways databases upon treatment were interferon signaling pathways, particularly those related to production of ISGs, and the top 25 downregulated genes were predominantly ISGs (Tables 2 and 3 and Supplementary Tables 2 and 3). Genes involved in mitochondrial and metabolic pathways (lipid and lipoprotein metabolism and oxidation) were enriched among those upregulated upon DAA therapy (Supplementary Table 4).
Table 2.
Selected pathways with greatest proportion of downregulated genes comparing baseline and on-treatment liver biopsies in gene ontology enrichment analysis of global mRNA expression.
| Gene Pathway | Proportion of downregulated genes [%] |
P-Value |
|---|---|---|
| Type I interferon signaling pathway | 37 | <10−9 |
| Interferon alpha/beta signaling | 31 | <10−9 |
| Response to type I interferon | 27 | <10−9 |
| Type II interferon signaling (IFNG) | 26 | <10−9 |
| Interferon-gamma-mediated signaling pathway | 25 | <10−9 |
| Negative regulation of viral genome replication | 22 | <10−9 |
| Interferon gamma signaling | 22 | <10−9 |
| Cellular response to type I interferon | 21 | <10−9 |
| Cellular response to interferon-gamma | 21 | <10−9 |
| Response to interferon-gamma | 21 | <10−9 |
Table 3.
Top 25 downregulated genes when comparing on-treatment to baseline biopsies.
| Gene | P-value | Fold change |
|---|---|---|
| ISG15 | 3.57E-09 | −4.084 |
| IFITM1 | 3.60E-08 | −1.789 |
| OASL | 6.29E-08 | −4.538 |
| ODF3B | 3.84E-07 | −1.499 |
| CMPK2 | 1.05E-06 | −2.917 |
| HERC6 | 1.47E-06 | −2.874 |
| CXCL11 | 1.83E-06 | −2.790 |
| MX1 | 2.62E-06 | −3.433 |
| NRIR | 2.92E-06 | −2.190 |
| EPSTI1 | 3.14E-06 | −2.600 |
| DDX60 | 3.42E-06 | −2.936 |
| IFI35 | 4.83E-06 | −1.635 |
| CXCL10 | 5.87E-06 | −4.782 |
| OAS3 | 9.71E-06 | −2.861 |
| SOCS1 | 1.28E-05 | −1.481 |
| STAT1 | 1.33E-05 | −2.207 |
| IFI44L | 1.40E-05 | −3.866 |
| RTP4 | 2.54E-05 | −2.259 |
| IFI6 | 3.80E-05 | −4.381 |
| PLSCR1 | 3.84E-05 | −1.588 |
| TAP2 | 4.62E-05 | −1.319 |
| HERC5 | 5.83E-05 | −2.600 |
| PPA1 | 5.84E-05 | −1.401 |
| MDK | 6.11E-05 | −1.657 |
| OAS2 | 7.98E-05 | −2.848 |
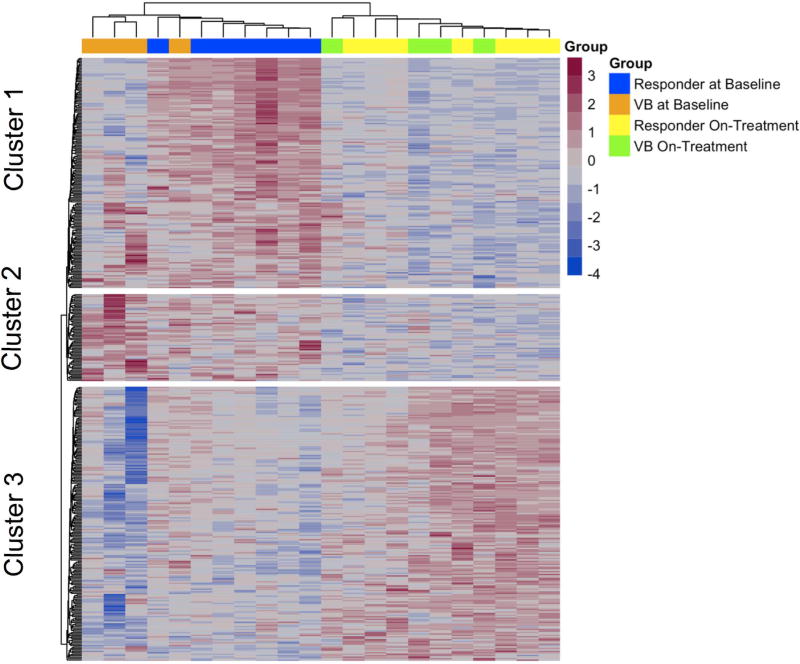
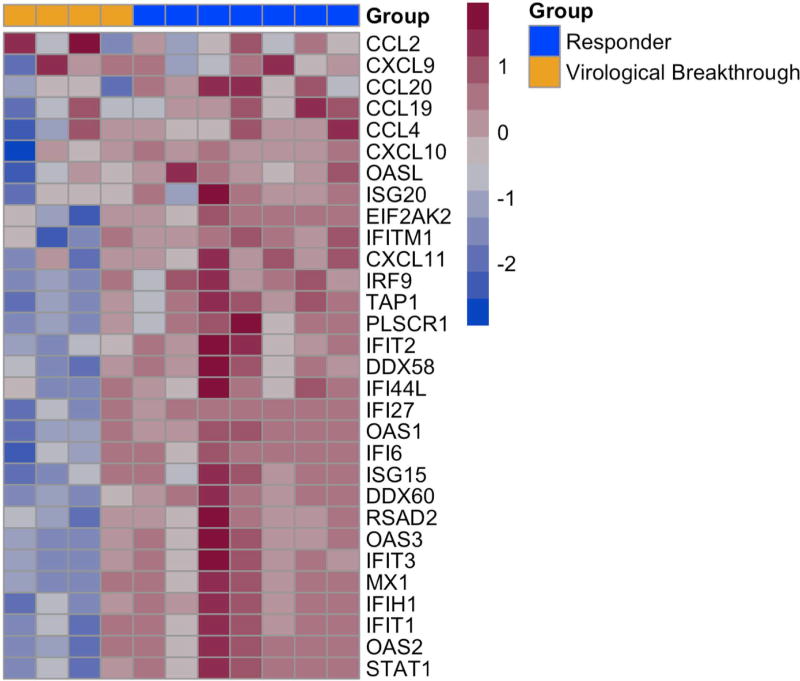
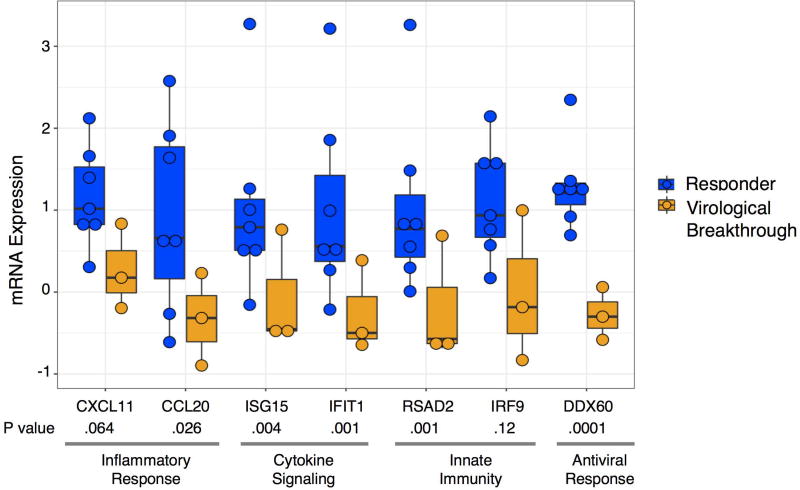
The heatmap of the transcriptomics data shows the expression levels of the 408 differentially expressed genes at the baseline and week 2/4 liver biopsies during DAA therapy (Figure 2). Among these 408 genes, there was a distinct cluster of genes on the baseline biopsy that separated patients who achieved SVR12 from those who experienced virological breakthrough, with the exception of a single patient (Figure 2, Cluster 1 and Supplementary Table 1). This led us to focus on the gene expression profiles at baseline between those who achieved SVR12 versus virological breakthrough. For this analysis, we selected genes of interest from Figure 2, Cluster 1 (listed in Supplementary Table 1), the most differentially expressed pathways including interferon signaling pathways, interferon signaling, defense response to virus, regulation of viral genome replication, metabolic pathways, from genes known to be associated with the innate immune response to HCV and the HCV life cycle based on a functional genomic screen14 and detailed literature review.15 Interestingly, patients who achieved SVR12 had higher pre-treatment expression levels of ISGs in comparison to patients with virological breakthrough, with the exception of a single patient (Figure 3). ISGs with greatest expression pre-treatment included ISGs with known antiviral function such as chemokine ligand 20 (CCL20), 2'-5'-oligoadenylate synthetase-like protein (OASL), C-X-C Motif Chemokine Ligand 10 (CXCL10), Myxovirus (Influenza) Resistance 1 (MX1), Interferon Induced proteins with Tetratricopeptide repeats 1 (IFIT1), and DEAD (Asp-Glu-Ala-Asp) Box Polypeptide 60 (DDX60). In addition, ISGs such as interferon-stimulated gene 15 (ISG15), Ubiquitin Specific Peptidase 18, (USP18) and Suppressor of Cytokine Signaling 1 (SOCS1), that inhibit type I interferon but not type III interferon signaling, were higher in the pre-treatment compared to the on-treatment biopsy (Supplementary Table 5). To validate these findings, we determined the expression level of 46 genes by n-Counter-based NanoString analysis. The nCounter NanoString Gene Expression Assay is based on direct digital detection of genes of interest by using target-specific capture probe and color coded reported probe pairs. The gene expression level is measured by tabulating the number of times the color-coded barcode for a gene is detected. The directionality and degree of fold changes obtained with NanoString were concordant with the microarray data (Supplementary Table 6). Seven ISGs representing the type I/III IFN signaling (CXCL11, CCL20), innate immunity (RSAD2), cytokine signaling (ISG15) and defense response to virus (DDX60 and IRF9) pathways are shown in Figure 4. Baseline expression levels of five of seven selected ISGs were significantly higher by NanoString (all were significant by microarray) in those with SVR12 compared to those with virological breakthrough (Figure 4 and Supplementary Table 7). In addition, there was greater decline in intrahepatic expression levels of these seven ISGs between baseline and on-treatment (weeks 2 or 4) time points in patients with SVR12 compared to patients with virological breakthrough (Supplementary Figure 3).
Figure 2. Expression data for the 408 differentially expressed genes from the pre-treatment and on-treatment week 2/4 liver biopsies during DAA therapy.
The heat map shows the 408 differentially expressed genes at baseline liver biopsy and on-treatment liver biopsies during DAA therapy. Blue bars above the heatmap indicate gene expression data at the baseline liver biopsy from patients who responded to DAA treatment. Orange bars indicate gene expression data at the baseline liver biopsy from patients who experienced virological breakthrough to DAA treatment. Yellow bars indicate gene expression data at the on-treatment liver biopsy from patients who responded to DAA treatment. Green bars indicate gene expression data at the on-treatment liver biopsy from patients who experienced virological breakthrough to DAA treatment. Expression of a distinct cluster of genes at baseline, Cluster 1, differed significantly between patients who achieved SVR12 (responders) compared to three of four patients that subsequently experienced a virological breakthrough during DAA therapy.
Figure 3. Gene expression profiles at baseline between those who achieved SVR12 versus virological breakthrough.
Genes included in this analysis were selected from Figure 2, Cluster 1 (listed in Supplementary Table 1), from the top differentially expressed pathways including interferon signaling pathways, interferon signaling, defense response to virus, regulation of viral genome replication, metabolic pathways and genes known to be associated with the innate immune response to HCV and the HCV life cycle based on a functional genomic screen and from a detailed literature review. Patients who achieved SVR12 had higher pre-treatment expression levels of ISGs in comparison to patients with virological breakthrough, with the exception of a single patient.
Figure 4. Gene expression by NanoString at baseline based on response to DAA treatment.
Shown are seven ISGs measured by NanoString representing different components of the innate immune response demonstrating higher expression among patients who achieved SVR12 compared to those with virological breakthrough. Each dot represents an individual patient. Note that the Nanostring data from one patient with virological breakthrough is missing from this analysis due to technical problems with the assay, which probably explains the lack of significance for CXCL11 and IRF9 with the NanoString data (expression levels were significant using microarray data). Patients who responded to DAA therapy are shown in blue and those who experienced virological breakthrough are shown in orange. The bar within the boxes represent the median gene expression and the upper and lower boundaries of the boxes represent the 75th and 25th percentiles of gene expression.
Baseline and Early On-treatment NK Cell Phenotype Differ Between Patients with SVR12 Versus Virological Breakthrough
Next, we assessed whether the noted ISG induction was observed not only at the mRNA but also at the protein level. We had previously reported that the phenotype and function of NK cells were influenced by chronic exposure to low levels of endogenous IFNs in patients with chronic HCV infection.22 This was evidenced by increased expression of pSTAT1, the phosphorylated state of a signaling molecule downstream of the IFNα/β receptor, and increased levels of NK cell cytotoxicity in HCV infected patients as compared to healthy controls.23 NK cells also expressed higher levels of TRAIL, an apoptosis-inducing effector molecule and itself an ISG.22, 23 This phenotype normalized after DAA-induced HCV clearance.24
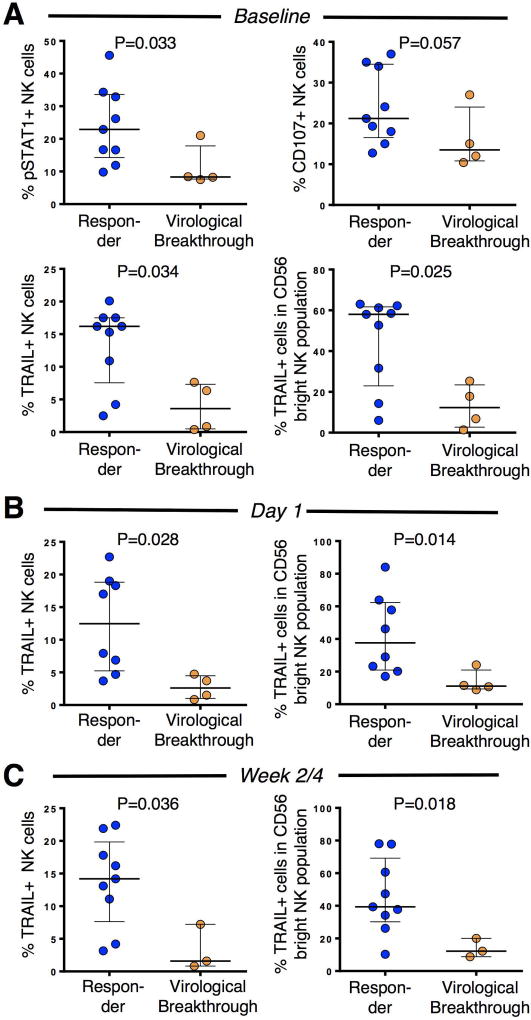
As such, we asked whether the baseline NK cell phenotype and the phenotypes at early on-treatment time points differed between patients who achieved SVR12 to DAA therapy and those who failed the treatment. The gating strategy for NK cell analysis by flow cytometry is shown in Supplementary figure 4. As shown in Figure 5A, NK cells of treatment responders displayed a higher frequency of pSTAT1+ NK cells than patients who failed the treatment (p= 0.03). This divergence was replicated by the observations of higher frequencies of TRAIL-expressing NK cells (p= 0.03) for treatment responders (Figure 5a) and a trend towards greater cytotoxicity as supported by increased cell surface expression of the degranulation marker CD107a+ NK cells (p=0.057). Because TRAIL (but not pSTAT1 and CD107a) is predominantly expressed in the CD56bright NK cell subpopulation, which constitute <10% of the peripheral blood NK cell population, we also confirmed the differential TRAIL expression by CD56 bright NK cells (Figure 5a, lower right panel). Differential TRAIL expression was maintained at day 1, and week 2/4 on-treatment (Fig. 5B, C), whereas differential expression of pSTAT1 was lost on treatment (not shown). Collectively, these results indicate an increased IFN-responsiveness at baseline in patients who subsequently mount an SVR12 as compared to those who experience a breakthrough.
Figure 5. The baseline and early on-treatment NK cell phenotype is consistent with an increased type I IFN signature in DAA responders and differs from NK cells of those with subsequent virological breakthrough.
(A) NK cells of DAA responders display higher baseline expression of the IFN-signaling molecule pSTAT1 and the ISG TRAIL, and have a higher capacity to degranulate compared to NK cells of patients who subsequently experience a virological breakthrough (orange).
(B, C) Differential TRAIL expression is maintained at day 1 (B) and at week 2/4 time of DAA therapy (C). At day 1, 8 out of 10 DAA responders were analyzed because blood samples of 2 responders were not collected. At week 2/4, only 3 out of 4 patients who failed treatment had still undetectable viremia. Median and IQR are shown. Statistical analysis: non-parametric unpaired Mann-Whitney test.
Discussion
HCV infection leads to activation of the innate immune response through sensing of extracellular and intracellular viral pathogen-associated molecular patterns (PAMPs) by multiple pathogen recognition receptors. This process results in production of type I and/or type III interferons and induction of various ISGs. This host defense response represents a strategy to contain the viral infection and restrict its spread to neighboring hepatocytes. However, in most cases, it is inadequate to eliminate acute HCV infection and HCV persists in the presence of increased ISG levels in a substantial proportion of chronically infected patients.1 This high ISG expression was shown to be associated with failure to respond to interferon-based treatment.1,25
In this study, we demonstrate that patients who responded to DAA therapy had higher baseline expression of ISGs compared to those who experienced a viral breakthrough with the exception of a single patient who failed therapy. This may have been due to the presence of many poor pre-treatment predictors of response to DAA therapy in this patient including presence of cirrhosis, African-American race, older age (70 years), high BMI (33) and the IFN lambda-3 genotype rs12979860 TT and IFN lambda-4 genotype rs368234815 ∆G/∆G genotype, associated with poor response to interferon therapy.6 This exception to the rule suggests that other host factors are important in predicting response to DAA therapy. Upregulation of antiviral ISGs was associated with increased IFN signaling in NK cells at baseline with downstream expression of the ISG and effector molecule TRAIL and cytotoxic effector functions. Thus, our results suggest that the innate immune response may contribute to viral clearance with an interferon-free regimen. This interpretation is supported by the comparison of NK cells from the responder and virological breakthrough groups. The observation that NK cells from patients who responded to DAA treatment displayed higher expression of STAT1 and TRAIL prior to treatment, at day 1 and weeks 2 and 4 on treatment suggests a greater type I interferon response in patients who responded compared to those who experienced virological breakthrough.
The results are notable because prior studies have demonstrated that an upregulated IFN response prior to treatment was associated with a high rate of treatment failure during interferon-based therapy.1,3,26 The mechanism for this paradox is not completely understood. Recently, it was demonstrated that type III interferon, produced by hepatocytes, may be the primary driver of ISG production in the chronically HCV-infected liver.27 The ISG profile of type III interferon is overlapping but not identical to that of type I interferon. In particular, strong induction of SOCS1 and USP18 that specifically inhibit the type I but not the type III interferon pathway has been reported.28 This is consistent with the finding that ISGs such as SOCS1 and USP18 were upregulated in baseline liver biopsies of our patient cohort. On the other hand, NK cells do not express the type III interferon receptor chain 1,29 suggesting that sufficient type I IFN is present to stimulate STAT1 phosphorylation and TRAIL expression in NK cells in our patient cohort.
Our study also demonstrates that ISG expression was rapidly downregulated following initiation of DAA therapy even in patients who previously failed an interferon-based regimen and typically express elevated ISG levels. This is consistent with a previous report that treated two HCV-infected chimpanzees with a miR122 antagonist and also found a rapid decline of hepatic ISG expression.30 Downregulation of ISG expression during DAA therapy was also reported in a patient study.11 However, that analysis utilized liver biopsies obtained prior to treatment and at the end of a 24-week course of treatment. Therefore, it was not possible to elucidate the effects of early viral suppression on ISG expression. In addition, the above-mentioned study included only patients who had never been treated previously and ribavirin was included in the treatment regimen, which has been shown to improve interferon signaling.25, 31 Collectively, our results extend the published data and suggest that viral replication is the inducer of high ISG expression. ISG expression data from the initial 72 hours, when there is the greatest decline in HCV RNA levels, would have provided stronger evidence. Unfortunately, we were unable to perform a third biopsy at this early treatment timepoint.
The study had several limitations. First, we only studied patients with HCV genotype 1b infection and it is not known whether the conclusions can be extrapolated to other HCV genotypes, However, the capacities in induction of ISGs by various HCV genotypes appear to be similar,32, 33 and as such, we would expect the results from genotype 1b-infected patients from this study to be broadly applicable to all other genotypes. Second, among any given population of HCV-infected patients there are some who are easy to treat and some who are difficult to treat. We selected patients who failed previous pegIFN-based therapy (rs12979860-TT/TC alleles, rs368234815-ΔG allele, high baseline viral load and advanced liver disease) as there is a pressing need to study this population. Whether the same findings apply to patients who are easy to treat is not yet clear.
In conclusion, we demonstrate that the baseline innate immune response contributes to a successful response to DAA therapy. We speculate that this innate immune response may prevent the emergence of resistance-associated substitutions that lead to viral breakthrough during DAA therapy.
Supplementary Material
Acknowledgments
Funding: This work was supported by the Intramural Research Program of NIDDK, NIH
We wish to acknowledge Ms. Kristin Valdez, LDB, NIDDK, NIH for her helpful discussions regarding the data analysis and careful review of the manuscript.
Abbreviations
- HCV
hepatitis C virus
- ISG
Interferon-stimulated gene
- DAA
direct-acting antiviral
- SVR
sustained virological response
- NK
natural killer
- TRAIL
TNF-related apoptosis-inducing ligand
Footnotes
Financial Disclosure: The authors are employees of the U.S. Government and have no financial conflicts of interest to disclose.
Authors’ contribution: Study concept & design: HA, MGG, QL; Acquisition of data: HA, CK, MGG, DS, FZ, ES, HP, NF, WH; analysis/interpretation: QL, ES, HP, BR; Drafting: HA, MGG, BR, ES, HP; Critical revision: TJL, QL, BR, MGG; Statistical analysis: MC, ES, EW.
References
- 1.Chen L, Borozan I, Feld J, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–44. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 2.Lanford RE, Guerra B, Bigger CB, et al. Lack of response to exogenous interferon-alpha in the liver of chimpanzees chronically infected with hepatitis C virus. Hepatology. 2007;46:999–1008. doi: 10.1002/hep.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feld JJ, Nanda S, Huang Y, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–63. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–71. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Brien TR, Pfeiffer RM, Paquin A, et al. Comparison of functional variants in IFNL4 and IFNL3 for association with HCV clearance. J Hepatol. 2015;63:1103–10. doi: 10.1016/j.jhep.2015.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heim MH, Bochud PY, George J. Host - hepatitis C viral interactions: The role of genetics. J Hepatol. 2016;65:S22–S32. doi: 10.1016/j.jhep.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 7.Ghany MG, Liang TJ. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;369:679–80. doi: 10.1056/NEJMc1307589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang TJ, Ghany MG. Therapy of hepatitis C--back to the future. N Engl J Med. 2014;370:2043–7. doi: 10.1056/NEJMe1403619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehermann B. HCV in 2015: Advances in hepatitis C research and treatment. Nat Rev Gastroenterol Hepatol. 2016;13:70–2. doi: 10.1038/nrgastro.2015.227. [DOI] [PubMed] [Google Scholar]
- 10.Pawlotsky JM. Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology. 2016;151:70–86. doi: 10.1053/j.gastro.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Meissner EG, Wu D, Osinusi A, et al. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. J Clin Invest. 2014;124:3352–63. doi: 10.1172/JCI75938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–7. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Zhang YY, Chiu S, et al. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog. 2014;10:e1004163. doi: 10.1371/journal.ppat.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horner SM, Gale M., Jr Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879–88. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noureddin M, Rotman Y, Zhang F, et al. Hepatic expression levels of interferons and interferon-stimulated genes in patients with chronic hepatitis C: A phenotype-genotype correlation study. Genes Immun. 2015;16:321–9. doi: 10.1038/gene.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahlenstiel G, Titerence RH, Koh C, et al. Natural killer cells are polarized toward cytotoxicity in chronic hepatitis C in an interferon-alfa-dependent manner. Gastroenterology. 2010;138:325–35. e1–2. doi: 10.1053/j.gastro.2009.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans MJ, von Hahn T, Tscherne DM, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–5. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 19.Ploss A, Evans MJ, Gaysinskaya VA, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–6. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawlotsky JM, Chevaliez S, McHutchison JG. The hepatitis C virus life cycle as a target for new antiviral therapies. Gastroenterology. 2007;132:1979–98. doi: 10.1053/j.gastro.2007.03.116. [DOI] [PubMed] [Google Scholar]
- 21.Foster TL, Gallay P, Stonehouse NJ, et al. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J Virol. 2011;85:7460–4. doi: 10.1128/JVI.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahlenstiel G, Edlich B, Hogdal LJ, et al. Early changes in natural killer cell function indicate virologic response to interferon therapy for hepatitis C. Gastroenterology. 2011;141:1231–9. 1239, e1–2. doi: 10.1053/j.gastro.2011.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edlich B, Ahlenstiel G, Zabaleta Azpiroz A, et al. Early changes in interferon signaling define natural killer cell response and refractoriness to interferon-based therapy of hepatitis C patients. Hepatology. 2012;55:39–48. doi: 10.1002/hep.24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serti E, Chepa-Lotrea X, Kim YJ, et al. Successful Interferon-Free Therapy of Chronic Hepatitis C Virus Infection Normalizes Natural Killer Cell Function. Gastroenterology. 2015;149:190–200. e2. doi: 10.1053/j.gastro.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feld JJ, Lutchman GA, Heller T, et al. Ribavirin improves early responses to peginterferon through improved interferon signaling. Gastroenterology. 2010;139:154–62. e4. doi: 10.1053/j.gastro.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Borozan I, Sun J, et al. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010;138:1123–33. e1–3. doi: 10.1053/j.gastro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 27.Thomas E, Gonzalez VD, Li Q, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978–88. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem. 1998;273:35056–62. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 29.Dickensheets H, Sheikh F, Park O, et al. Interferon-lambda (IFN-lambda) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. J Leukoc Biol. 2013;93:377–85. doi: 10.1189/jlb.0812395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas E, Feld JJ, Li Q, et al. Ribavirin potentiates interferon action by augmenting interferon-stimulated gene induction in hepatitis C virus cell culture models. Hepatology. 2011;53:32–41. doi: 10.1002/hep.23985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wieland S, Makowska Z, Campana B, et al. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology. 2014;59:2121–30. doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong MT, Chen SS. Emerging roles of interferon-stimulated genes in the innate immune response to hepatitis C virus infection. Cell Mol Immunol. 2016;13:11–35. doi: 10.1038/cmi.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.