Abstract
Human adenovirus (Adv) infection is responsible for most community-acquired pneumonia in infants and children, which results in significant morbidity and mortality in children every year. MicroRNAs (miRNAs) are associated with viral replication and host immune response. Knowing the miRNA expression profile will help understand the role of miRNAs in modulating the host response to adenovirus infection and possibly improve the diagnosis of adenovirus-infected pneumonia. In our study, total RNA extracted from whole blood of adenovirus-infected pneumonia children and healthy controls were analyzed by small RNA deep sequencing. Expression profiles of whole blood microRNAs were altered and distinctly different in adenovirus-infected children. The top 3 upregulated miRNA (hsa-miR-127-3p, hsa-miR-493-5p, and hsa-miR-409-3p) were identified in adenovirus-infected children and provided a clear distinction between infected and healthy individuals. Potential host target genes were predicated and validated by qRT-PCR to study the impact of microRNAs on the host genes. Most of the target genes were involved in the MAPK signaling pathway and innate immune response. These highly upregulated microRNAs may have crucial roles in Adv pathogenesis and are potential biomarkers for adenovirus-infected pneumonia.
1. Introduction
Human adenovirus (Adv) infection is responsible for most community-acquired pneumonia in infants and children [1, 2]. Adv causes infections for 5–10% of upper and lower respiratory tract infections in children, which results in pneumonia and nearly 1.3 million deaths of children every year [3, 4]. The fatality rates for untreated severe pneumonia or disseminated disease caused by Adv may even exceed to 50% [5, 6]. There are no efficacious antiviral drugs for Adv treatment until now. Also, the traditional diagnosis of Adv infection is limited. Therefore, to discover the interaction between the virus and its host will help us to find novel treatment and diagnosis for Adv infection.
Human Advs are nonenveloped double-stranded DNA viruses and belong to the Adenoviridae family [7, 8]. Human Advs are divided into seven subgroups including 53 serotypes based on immunologic and biological characteristics. Adv replicates efficiently in human cells and triggers an innate immune response such as inflammatory response in the host cells. Also, viral infection has proved to have a great impact on cellular small RNA expression and gene expression [9–11]. Adv infects the host cells through binding to different cellular receptors such as coxsackievirus and adenovirus receptor (CAR) [12]. During Adv DNA replication, host cellular proteins such as nuclear factor I and POU2F1 are used by Adv [13, 14]. In turn, the host will trigger an innate immune response against Adv infection. However, the details of Adv and host interaction still remains poorly defined.
Small RNAs are important regulators that modulate development, proliferation, differentiation, and apoptosis of organisms [15, 16]. Small RNAs include microRNA (miRNA), siRNA, tRFs, piRNA, and rasiRNAs, which regulate gene expression in a wide range of processes such as viral replication and host immune response. miRNAs are the most well-studied small RNAs during the latest decades. miRNAs are very important regulators that modulate transcriptome changes [17]. miRNAs regulate gene expression in a wide range of physiological and pathological processes such as in immune response and viral replication [18]. Although miRNAs have been examined in Adv type 3-infected human laryngeal epithelial cells and Adv type 2-infected human lung fibroblast cells [19, 20], there is no research about small RNA profiling in whole blood of Adv-infected pneumonia children. In our study, we sought to present the different miRNA profiles between Adv-infected pneumonia children and healthy controls, identify candidate diagnostic biomarkers for pneumonia with Adv infection in children, and examine the role of miRNAs in host defense response in Adv-infected children.
2. Materials and Methods
2.1. Patients
The whole blood samples used in the study were obtained from Guangzhou Women and Children's Medical Center. Children diagnosed with human Adv pneumonia were included in the study. The diagnosis of human Adv pneumonia was considered certain when it was associated with the following criteria: (1) lower respiratory and/or systemic symptoms, (2) lung infiltration on chest radiography or computed tomography (CT) scan, and (3) positive results for human Adv IgM antibody in sera and/or human Adv DNA by PCR in throat swabs and/or bronchoalveolar lavage (BAL) fluid. A total of 33 samples from patients and 33 samples from healthy volunteers were used in the study. The ages of all patients (male or female) and healthy volunteers (male or female) range from one year to three years. The study was approved by the Ethics Committee at Guangzhou Women and Children's Medical Center (number 2014121815), and written informed consent was obtained from all guardians.
2.2. RNA Extraction and Small RNA Sequencing
The blood samples of the patients and volunteers were collected in anticoagulant tubes. The total RNA was isolated using RiboPure™ Blood RNA Isolation Kit (Ambion, USA) according to the manufacturer's protocol. The extracted small RNAs were treated with DNase before deep sequencing. RNA concentration was determined using a NanoDrop ND1000 system (Thermo Fisher Scientific, South San Francisco, CA), and their integrity was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Small RNA deep sequencing was performed as previously described [21]. A total of 3 samples from patients and 3 samples from healthy volunteers were used for small RNA deep sequencing in the study. The clinical characteristics of the 3 patients and 3 healthy volunteers are summarized in Table 1.
Table 1.
Clinical features of healthy volunteers and patients.
| Volunteer or patient | Age | Gender | Fever durationa (days) | Laboratory characteristicsb | Radiologye | ||||
|---|---|---|---|---|---|---|---|---|---|
| WBC (×109/L) (5–12) | HsCRP (mg/L)(<5) | Specific IgMc (IU/mL) | HAdV DNAd (throat swabs/BALF) | LDH (U/L) (159–322) | Consolidation | ||||
| V1 | 1y9m | male | − | 8.6 | − | − | − | − | − |
| V2 | 2y8m | female | − | 9.0 | − | − | − | − | − |
| V3 | 2y3m | male | − | 5.7 | − | − | − | − | − |
| A1 | 2y1m | male | 13 | 10.2 | 26.5 | + | +/+ | 919 | + |
| A2 | 1y9m | female | 16 | 8.3 | 51.14 | + | +/+ | 812 | + |
| A3 | 2y8m | female | 22 | 2.8 | 10.98 | + | +/+ | 948 | + |
aFever duration from onset to normothermia. bData extracted from the first test for the children on admission. cSpecific IgM antibodies against HAdV pneumoniae were detected in 2 mL of acute phase (on admission) patient serum using a commercial ELISA kit. dHAdV pneumonia DNA was detected by real-time PCR in throat swabs or bronchoalveolar lavage fluid (BALF). eJudged by chest radiograph or CT scan in whole course of the patients.
2.3. Cell and Virus Culture
Human primary lung fibroblasts (IMR-90) and human 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) (HyClone) supplemented with 10% fetal bovine serum (Invitrogen), streptomycin, and penicillin (Invitrogen). All virus infections were carried out in serum-free medium for 1 h, followed by addition of saved complete medium.
2.4. Transfection
miRNA inhibitors (GenePharma Co., China) were transfected into IMR-90 cells with Lipofectamine RNAiMAX (Invitrogen) at final concentrations of 100 nM according to the manufacturer's protocol.
2.5. Virus Growth Assay
IMR-90 cells transfected with miRNA inhibitors were infected with HAdV5 (isolated from patients) at an MOI of 10 in serum-free medium. Virus titers were determined 72 h after infection by plaque assays performed on 293T cells [22, 23].
2.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
A qRT-PCR experiment was performed using the Power SYBR Green PCR Master Mix. Each reaction was performed in a 10 μL volume system containing 0.5 μL of cDNA, 0.5 μL of each primer, 5 μL of Power SYBR Green PCR Master Mix, and 3.5 μL of ddH2O. The reactions were incubated in a 96-well plate at 95°C for 10 minutes, followed by 39 cycles of 95°C for 15 seconds and 62°C for 1 minute. For miRNA quantitation, 10 ng of total RNA was reverse-transcribed using specific stem-loop primers. U6 was used as an endogenous control.
2.7. Data Analysis
miRNA cluster and family information from miRBase (miRBase 20, http://www.mirbase.org/) was used to annotate the cluster of miRNAs. miRNA target genes were predicted based on two software: miRDB software (http://mirdb.org/) and TargetScan software (http://www.targetscan.org/).
2.8. Statistical Analysis
Data were analyzed using GraphPad Prism 6.0 software (La Jolla, CA, USA). The two-tailed Student's t-test was used to determine the significance of statistical data between two experimental groups. Data were considered significant at ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3. Results
3.1. Different Expressions of MicroRNAs in Adv-Infected Children vs. Healthy Children
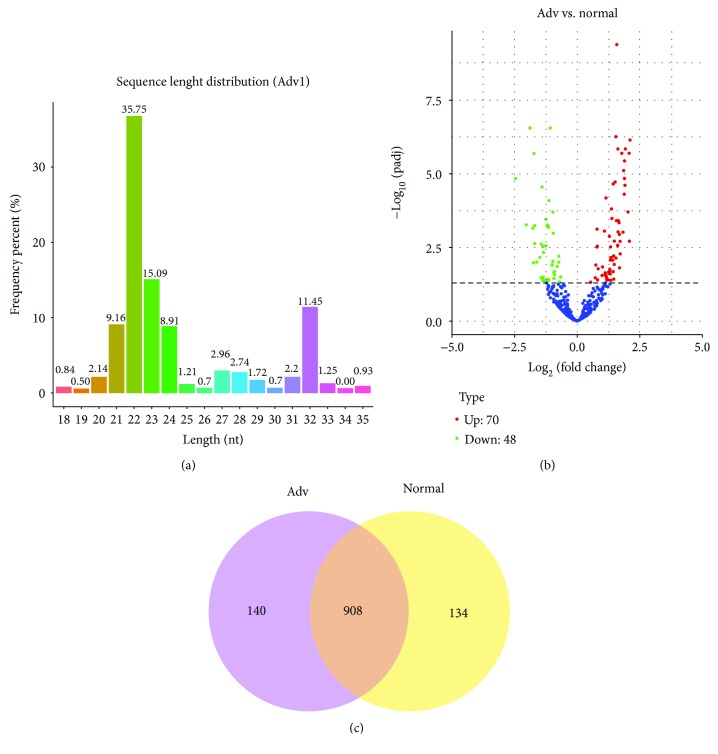
To study the impact of Adv infection on cellular small RNA expression in pneumonia children, deep sequencing of small RNAs was performed in our study. Accordingly, we found an apparent small RNA peak at 21–24 nt for miRNAs (Figure 1(a)). When we analyzed the differently expressed small RNAs, 118 miRNAs were found differently expressed in Adv-infected children vs. healthy children in a volcano plot (Figure 1(b)). We mapped the clean reads from each group to the known miRNA sequences and identified 908 miRNAs in Adv-infected children versus healthy controls (Figure 1(c)).
Figure 1.
miRNA expression profile of whole blood from adenovirus-infected pneumonia children. (a) Deep sequencing shows the distribution of small RNAs of whole blood from adenovirus-infected pneumonia children. The miRNA peaks appear around 22 nt. (b) Volcano plot of differently expressed miRNAs between Adv-infected children and healthy controls. (c) Venn diagram of differently expressed miRNAs between Adv-infected children and healthy controls.
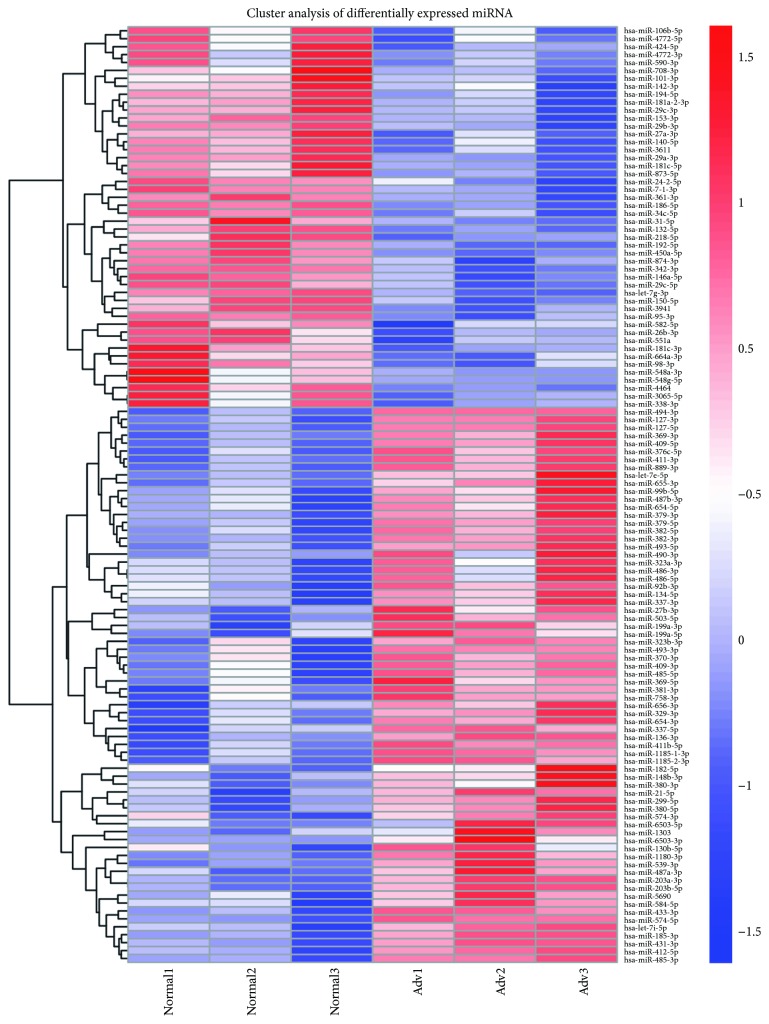
Furthermore, hierarchical cluster analysis of differentially expressed miRNA is shown in Figure 2. Specially, 77 differentially expressed miRNAs in the 6 samples passed our fold-change filter (log2 fold change > 1.0), among which 20 miRNAs that have high expression (reads up to 1000 in Adv samples) and showed significant different expressions were selected for further analysis (Table 2).
Figure 2.
Hierarchical cluster analysis of differentially expressed miRNAs.
Table 2.
Top 20 different expressed miRNAs.
| miRNA name | Reads in ADV | Reads in control | Log2 fold change | P value |
|---|---|---|---|---|
| has-miR-381-3p | 9358 | 3507 | 1.17 | 0.00511 |
| has-miR-486-5p | 5480 | 2240 | 1.12 | 0.00434 |
| has-miR-409-3p | 4916 | 1223 | 1.61 | 0.00023 |
| has-miR-486-3p | 4745 | 1922 | 1.12 | 0.00402 |
| has-miR-127-3p | 3950 | 753 | 2.10 | 6.35E − 09 |
| has-miR-182-5p | 2396 | 870 | 1.17 | 0.00762 |
| has-miR-99b-5p | 2241 | 650 | 1.46 | 0.00066 |
| has-miR-379-5p | 1687 | 378 | 1.88 | 3.31E − 07 |
| has-miR-370-3p | 1682 | 617 | 1.18 | 0.00621 |
| has-let-7e-5p | 1239 | 429 | 1.33 | 0.00030 |
| has-miR-493-5p | 1089 | 237 | 1.87 | 1.63E − 06 |
| has-miR-494-3p | 1070 | 342 | 1.54 | 3.94E − 09 |
| has-miR-101-3p | 57,893 | 150,004 | −1.12 | 0.00825 |
| has-miR-142-3p | 24,616 | 77,702 | −1.25 | 0.00841 |
| has-miR-150-5p | 15,854 | 41,850 | −1.25 | 0.00025 |
| has-miR-29a-3p | 6679 | 18,944 | −1.40 | 9.01E − 07 |
| has-miR-186-5p | 6093 | 13,066 | −1.07 | 1.49E − 09 |
| has-miR-27a-3p | 5558 | 11,953 | −1.00 | 0.00170 |
| has-miR-342-3p | 3373 | 13,980 | −1.87 | 1.37E − 09 |
| has-miR-29b-3p | 1339 | 3404 | −1.25 | 1.60E − 05 |
3.2. Validation of Differentially Expressed miRNAs
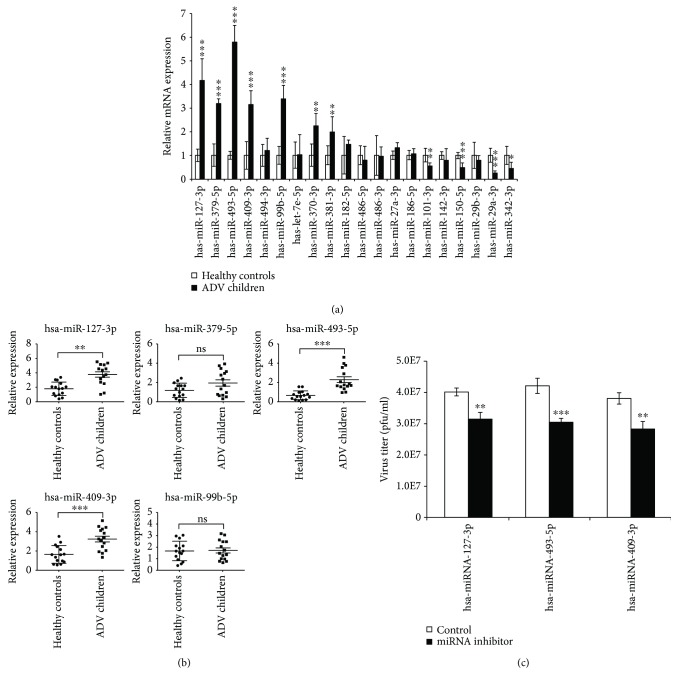
To confirm the differential expression of miRNAs in Adv patients vs. healthy controls, we performed qRT-PCR assays in our study. The result showed that hsa-miR-127-3p, hsa-miR-379-5p, hsa-miR-493-5p, hsa-miR-409-3p, hsa-miR-99b-5p, hsa-miR-370-3p, and hsa-miR-381-3p were upregulated in whole blood samples from 5 Adv-infected children vs. 5 healthy controls (Figure 3(a)), while hsa-miR-101-3p, hsa-miR-150-5p, hsa-miR-29a-3p, and hsa-miR-342-3p were downregulated in whole blood samples from 5 Adv-infected children vs. 5 healthy controls (Figure 3(a)), which was comparable with our sequencing data. To identify candidate diagnostic miRNA biomarkers, we focused on the upregulated miRNAs. More samples (15 Adv-infected children vs. 15 healthy controls) were collected to verify the expression of the upregulated miRNAs (Figure 3(b)). From the result, we found that hsa-miR-127-3p, hsa-miR-493-5p, and hsa-miR-409-3p were significantly increased. Also, after depleting these miRNAs with miRNA inhibitors, the viral replications were significantly decreased (Figure 3(c)). These findings imply that our selected miRNAs may reflect the infection of Adv, and such miRNAs can likely serve as biomarker candidates for Adv-infected patients.
Figure 3.
Quantification of miRNA expression levels by qRT-PCR. (a) Quantification of top 20 differently expressed miRNAs, including upregulated and downregulated miRNAs. Five samples from Adv-infected children and 5 from healthy controls were used in the experiment. (b) Quantification of top 5 upregulated miRNAs. 15 samples from Adv-infected children and 15 from healthy controls were used in the experiment. (c) The effect of miRNA inhibitors on the replication of Adv. After transfection with miRNA inhibitors, cells were infected with HAdV5 at an MOI of 10. Virus was harvested, and the titer was determined on 293T cells at the indicated time points. Data are shown as means ± SD from three independent experiments. ∗∗P < 0.01 and ∗∗∗P < 0.001 (Student t test).
3.3. Predict Target Genes of Differentially Expressed miRNAs
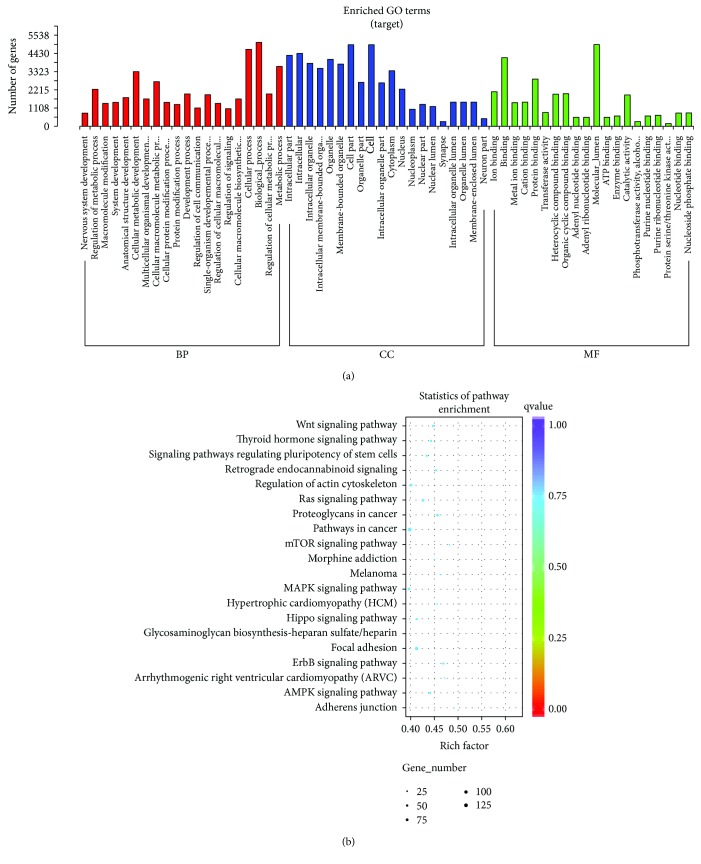
To study the biological significance of miRNAs, we then predicted the computational target genes of miRNAs. We focused on the targets of verified miRNAs with distinct expression profiles. Go enrichment of the predicted target genes of the miRNAs showed that the target genes were mainly involved in cellular process and molecular function (Figure 4(a)). In particular, most of the target genes participated in the MAPK signaling pathway and Ras signaling pathway (Figure 4(b)). The top five predicted target genes of hsa-miR-127-3p, hsa-miR-493-5p, or hsa-miR-409-3p are listed in Table 3 by the highest miRNA target score from two miRNA predictive software.
Figure 4.
Go enrichment of predicted target genes. (a) The GO classification enrichment of target genes. (b) The KEGG pathway scatterplot of target genes.
Table 3.
Top 5 predicted target genes.
| miRNAs | Target | Gene description |
|---|---|---|
| hsa-miR-127-3p | PSMB5 | Proteasome (prosome, macropain) subunit, beta type 5 |
| KIF3B | Kinesin family member 3B | |
| ITGA6 | Integrin, alpha 6 | |
| BCAS3 | Breast carcinoma-amplified sequence 3 | |
| MTSS1L | Metastasis suppressor 1-like | |
|
| ||
| hsa-miR-493-5p | SP3 | Sp3 transcription factor |
| MYCBP2 | MYC-binding protein 2, E3 ubiquitin protein ligase | |
| TCF7L2 | Transcription factor 7-like 2 (T-cell specific, HMG box) | |
| UBE2V2 | Ubiquitin-conjugating enzyme E2 variant 2 | |
| HIPK1 | Homeodomain-interacting protein kinase 1 | |
|
| ||
| hsa-miR-409-3p | RAB10 | RAB10, member RAS oncogene family |
| UBE2D2 | Ubiquitin-conjugating enzyme E2D 2 | |
| KANSL1 | KAT8 regulatory NSL complex subunit 1 | |
| MTF2 | Metal response element binding transcription factor 2 | |
| ELF2 | E74-like factor 2 (ETS domain transcription factor) | |
The target genes were selected for further validation with qRT-PCR, and we found that the mRNA expression of 8 genes (PSMB5, ITGA6, MYCBP2, TCF7L2, UBE2V2, HIPK1, UBE2D2, and KANSL1) were downregulated in Adv-infected patients compared to healthy controls (Figure 5). Most of them are transcription factors or factors involved in the ubiquitin pathway. The downregulated mRNAs may indicate the mechanism of Adv infection and Adv-induced pneumonia.
Figure 5.
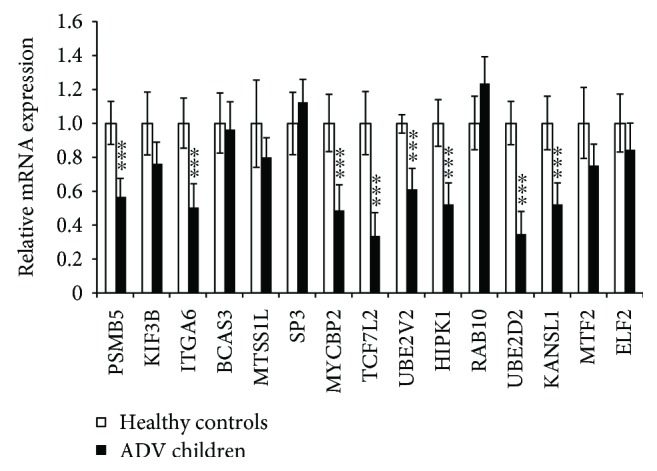
Validation of the expression of top 5 predicated target genes by qRT-PCR. Relative mRNA expression between 10 samples from Adv-infected children and 10 samples from healthy controls. Data are shown as means ± SD from three independent experiments. ∗∗∗P < 0.001 (Student t test).
4. Discussion
Human Advs are common causative pathogens of acute respiratory infections in children. The treatment of human Adv infections is limited because prospective, randomized therapeutic trials have not been done. Therefore, it is very important to discover the mechanism of Adv-infected pneumonia and search the biomarkers for Adv-infected pneumonia in children. The regulatory potential of miRNA is well defined, and the different profiles of miRNA expression are the result of diverse diseases including viral infections. In our study, we found that the miRNA profile in whole blood of Adv-infected children was different from that of healthy children. Blood samples of Adv-infected children reflect the associated pathology of Adv infection and thus provide a better understanding of the disease.
Profiling of miRNA expression from Adv-infected blood samples identified a cluster of 118 miRNAs significantly altered. The altered blood miRNA profile was similar to cells infected with Adv reported previously [19, 20], indicating that the different expressed miRNAs identified in our study could be taken as the candidate diagnostic biomarkers for pneumonia with Adv infection in children. Among these different expressed miRNAs, hsa-let-7e-5p was reported to involve in the replication of influenza infection [24]. hsa-miR-127-3p was reported to affect the Epstein-Barr virus-associated lymphoma through targeting the PTEN-AKT-mTOR pathway [25]. Altogether, these miRNAs play important roles in innate immune response or viral replication, thus affecting the outcome of the disease.
Through miRNA target gene GO analysis, we found that most target genes of different expressions of miRNAs were involved in the MAPK signaling pathway. The MAPK signaling pathway is activated by Toll-like receptors [26], which play important roles in innate immune response against viral infection. Also, the activation of the MAPK pathway will result in the activation of the NF-κB signal pathway, which will stimulate the production of inflammatory cytokines and MMPs [27] and finally lead to pneumonia. Thus, the altered miRNA expression profile of whole blood from Adv-infected children partly reflected the mechanism of Adv-infected pneumonia.
With a developmental framework or disease process, miRNAs exhibit dynamic expression patterns. In our study, we characterized the miRNA expression profile of Adv-infected children using deep sequencing analysis and identified that many miRNAs were differently expressed in Adv-infected children when compared with healthy children. Those with the greatest differences were chosen for further verification. In particular, the expressions of hsa-miR-127-3p, hsa-miR-493-5p, and hsa-miR-409-3p from 20 Adv-infected children were significantly higher than those from 20 healthy controls, indicating that these miRNAs could be taken as good diagnostic biomarkers for Adv-infected pneumonia.
To further explore the possible molecular mechanisms of the differently expressed miRNAs in Adv-infected children, we predicted the possible target genes of hsa-miR-127-3p, hsa-miR-493-5p, and hsa-miR-409-3p and found that most of them are transcription factors or factors involved in the ubiquitin pathway. Especially, after verifying the predicted target genes with qRT-PCR, 8 genes (PSMB5, ITGA6, MYCBP2, TCF7L2, UBE2V2, HIPK1, UBE2D2, and KANSL1) were found significantly downregulated in samples from Adv-infected children. In particular, MYCBP2 is an E3 ubiquitin protein ligase [28, 29], which regulated the cAMP and mTOR signaling pathway. The mTOR signaling pathway plays a critical role in effector T cell function, the downregulation of which will result in impaired cell cytolysis and incapability of virus elimination. UBE2V2 is an ubiquitin-conjugating enzyme and is involved in the differentiation of monocytes, which can produce proinflammatory cytokines like MIP-1β [30, 31]. Thus, the downregulated UBE2V2 will result in abnormal differentiation of monocytes and more proinflammatory will be produced, which in turn aggravates pneumonia. UBE2D2 is essential for the activation of MAVS and RIG-I in response to viral infection [32–34]. The downregulation of UBE2D2 will result in abnormal activation of MAVS and RIG-I signal and virus clearance. These findings imply that the miRNA expression profile changed in Adv-infected children resulting in different transcriptome profiles, which reflects the mechanism of Adv replication and the formation Adv-infected pneumonia.
5. Conclusions
In summary, we identified the 3 most markedly differently expressed miRNAs in whole blood from Adv-infected children, which can be taken as biomarkers for Adv-infected pneumonia. Simultaneously, based on target gene prediction and qRT-PCR analysis, we found that genes MYCBP2, UBE2V2, and UBE2D2 may play important roles in viral replication and Adv-induced pneumonia. However, additional studies are necessary to clarify their roles in these processes, which will provide a physiological basis for the treatment of Adv-infected pneumonia.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81701990, 81702327, and 81601759), the National Science and Technology Major Project (2018ZX10101004003001), and the General Projects of Guangzhou Scientific Research (201707010183).
Contributor Information
Xuefeng Li, Email: xuefengli@gzhmu.edu.cn.
Gen Lu, Email: lugen@gzhmu.edu.cn.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
Feng Huang and Junsong Zhang contributed equally to this work.
References
- 1.Nascimento-Carvalho C. M. Etiology of childhood community acquired pneumonia and its implications for vaccination. The Brazilian Journal of Infectious Diseases. 2001;5(2):87–97. doi: 10.1590/s1413-86702001000200007. [DOI] [PubMed] [Google Scholar]
- 2.Kim S. J., Kim K., Park S. B., Hong D. J., Jhun B. W. Outcomes of early administration of cidofovir in non-immunocompromised patients with severe adenovirus pneumonia. PLoS One. 2015;10(4, article e0122642) doi: 10.1371/journal.pone.0122642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhutta Z. A., Das J. K., Walker N., et al. Interventions to address deaths from childhood pneumonia and diarrhoea equitably: what works and at what cost? The Lancet. 2013;381(9875):1417–1429. doi: 10.1016/S0140-6736(13)60648-0. [DOI] [PubMed] [Google Scholar]
- 4.Agweyu A., Kibore M., Digolo L., et al. Prevalence and correlates of treatment failure among Kenyan children hospitalised with severe community-acquired pneumonia: a prospective study of the clinical effectiveness of WHO pneumonia case management guidelines. Tropical Medicine & International Health. 2014;19(11):1310–1320. doi: 10.1111/tmi.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch J. P., III, Kajon A. E. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Seminars in Respiratory and Critical Care Medicine. 2016;37(4):586–602. doi: 10.1055/s-0036-1584923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandkovsky U., Vargas L., Florescu D. F. Adenovirus: current epidemiology and emerging approaches to prevention and treatment. Current Infectious Disease Reports. 2014;16(8):p. 416. doi: 10.1007/s11908-014-0416-y. [DOI] [PubMed] [Google Scholar]
- 7.Ison M. G., Hayden R. T. Adenovirus. Microbiology Spectrum. 2016;4 doi: 10.1128/microbiolspec.DMIH2-0020-2015. [DOI] [PubMed] [Google Scholar]
- 8.Lynch J. P., Fishbein M., Echavarria M. Adenovirus. Seminars in Respiratory and Critical Care Medicine. 2011;32(4):494–511. doi: 10.1055/s-0031-1283287. [DOI] [PubMed] [Google Scholar]
- 9.Wu K. L., Zhang X., Zhang J., et al. Inhibition of hepatitis B virus gene expression by single and dual small interfering RNA treatment. Virus Research. 2005;112(1-2):100–107. doi: 10.1016/j.virusres.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan J., Cheung P. K. M., Zhang H. M., Chau D., Yang D. Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. Journal of Virology. 2005;79(4):2151–2159. doi: 10.1128/JVI.79.4.2151-2159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J., Wang F., Argyris E., et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nature Medicine. 2007;13(10):1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 12.Arnberg N. Adenovirus receptors: implications for targeting of viral vectors. Trends in Pharmacological Sciences. 2012;33(8):442–448. doi: 10.1016/j.tips.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Hartman Z. C., Black E. P., Amalfitano A. Adenoviral infection induces a multi-faceted innate cellular immune response that is mediated by the toll-like receptor pathway in A549 cells. Virology. 2007;358(2):357–372. doi: 10.1016/j.virol.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 14.Yoon J. S., Kim H. H., Lee Y., Lee J. S. Cytokine induction by respiratory syncytial virus and adenovirus in bronchial epithelial cells. Pediatric Pulmonology. 2007;42(3):277–282. doi: 10.1002/ppul.20574. [DOI] [PubMed] [Google Scholar]
- 15.Heintz-Buschart A., Yusuf D., Kaysen A., et al. Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biology. 2018;16(1):p. 52. doi: 10.1186/s12915-018-0522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed W., Razzaq M. RETRACTED: A small non-coding RNA AbcR2 regulate gntR transcription factor that modulate the intracellular survival of Brucella melitensis. Microbial Pathogenesis. 2018;118:118–125. doi: 10.1016/j.micpath.2018.03.025. [DOI] [PubMed] [Google Scholar]
- 17.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Piedade D., Azevedo-Pereira J. M. MicroRNAs as important players in host-adenovirus interactions. Frontiers in Microbiology. 2017;8, article 1324 doi: 10.3389/fmicb.2017.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi Y., Tu J., Cui L., et al. High-throughput sequencing of microRNAs in adenovirus type 3 infected human laryngeal epithelial cells. Journal of Biomedicine & Biotechnology. 2010;2010:8. doi: 10.1155/2010/915980.915980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H., Chen M., Tellgren-Roth C., Pettersson U. Fluctuating expression of microRNAs in adenovirus infected cells. Virology. 2015;478:99–111. doi: 10.1016/j.virol.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Zhong F., Zhou N., Wu K., et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Research. 2015;43(21):10474–10491. doi: 10.1093/nar/gkv954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frost J. R., Mendez M., Soriano A. M., et al. Adenovirus 5 E1A-mediated suppression of p 53 via FUBP1. Journal of Virology. 2018;92(14) doi: 10.1128/JVI.00439-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Huang F., Tan L., et al. Host protein Moloney leukemia virus 10 (MOV10) acts as a restriction factor of influenza a virus by inhibiting the nuclear import of the viral nucleoprotein. Journal of Virology. 2016;90(8):3966–3980. doi: 10.1128/JVI.03137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tambyah P. A., Sepramaniam S., Mohamed Ali J., et al. MicroRNAs in circulation are altered in response to influenza a virus infection in humans. PLoS One. 2013;8(10, article e76811) doi: 10.1371/journal.pone.0076811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onnis A., Navari M., Antonicelli G., et al. Epstein-Barr nuclear antigen 1 induces expression of the cellular microRNA hsa-miR-127 and impairing B-cell differentiation in EBV-infected memory B cells. New insights into the pathogenesis of Burkitt lymphoma. Blood Cancer Journal. 2012;2(8):p. e84. doi: 10.1038/bcj.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q.-W., Su Y., Sheng J.-T., et al. Anti-influenza A virus activity of rhein through regulating oxidative stress, TLR4, Akt, MAPK, and NF-κB signal pathways. PLoS One. 2018;13(1, article e0191793) doi: 10.1371/journal.pone.0191793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang W., Shang L., Wang T., Liu H., Ge S. Rho-kinase inhibitor Y-27632 downregulates LPS-induced IL-6 and IL-8 production via blocking p38 MAPK and NF-κB pathways in human gingival fibroblasts. Journal of Periodontology. 2018;89(7):883–893. doi: 10.1002/JPER.17-0571. [DOI] [PubMed] [Google Scholar]
- 28.James G., Key B., Beverdam A. The E3 ubiquitin ligase Mycbp 2 genetically interacts with Robo 2 to modulate axon guidance in the mouse olfactory system. Brain Structure & Function. 2014;219(3):861–874. doi: 10.1007/s00429-013-0540-8. [DOI] [PubMed] [Google Scholar]
- 29.Holland S., Scholich K. Regulation of neuronal functions by the E3-ubiquitinligase protein associated with MYC (MYCBP2) Communicative & Integrative Biology. 2014;4(5):513–515. doi: 10.4161/cib.15967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y., Long M. J. C., Wang Y., Zhang S., Aye Y. Ube2V2 is a Rosetta stone bridging redox and ubiquitin codes, coordinating DNA damage responses. ACS Central Science. 2018;4(2):246–259. doi: 10.1021/acscentsci.7b00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson L. J., Sale J. E. UBE2V2 (MMS2) is not required for effective immunoglobulin gene conversion or DNA damage tolerance in DT40. DNA Repair (Amst) 2005;4(4):503–510. doi: 10.1016/j.dnarep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Polge C., Koulmann N., Claustre A., et al. UBE2D2 is not involved in MuRF1-dependent muscle wasting during hindlimb suspension. The International Journal of Biochemistry & Cell Biology. 2016;79:488–493. doi: 10.1016/j.biocel.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 33.Geisler S., Vollmer S., Golombek S., Kahle P. J. The ubiquitin-conjugating enzymes UBE2N, UBE2L3 and UBE2D2/3 are essential for Parkin-dependent mitophagy. Journal of Cell Science. 2014;127(15):3280–3293. doi: 10.1242/jcs.146035. [DOI] [PubMed] [Google Scholar]
- 34.Chiang M.-H., Chen L.-F., Chen H. Ubiquitin-conjugating enzyme UBE2D2 is responsible for FBXW2 (F-box and WD repeat domain containing 2)-mediated human GCM1 (glial cell missing homolog 1) ubiquitination and degradation. Biology of Reproduction. 2008;79(5):914–920. doi: 10.1095/biolreprod.108.071407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.