Abstract
BACKGROUND & AIMS:
It has been a challenge to identify liver tumor suppressors or oncogenes due to the genetic heterogeneity of these tumors. We performed a genome-wide screen to identify suppressors of liver tumor formation in mice, using CRISPR-mediated genome editing.
METHODS:
We performed a genome-wide CRISPR/Cas9-based knockout screen of P53-null mouse embryonic liver progenitor cells that overexpressed MYC. We infected p53−/−;Myc;Cas9 hepatocytes with the mGeCKOa lentiviral library of 67,000 single-guide RNAs (sgRNAs), targeting 20,611 mouse genes, and transplanted the transduced cells subcutaneously into nude mice. Within 1 month, all the mice that received the sgRNA library developed subcutaneous tumors. We performed high-throughput sequencing of tumor DNA and identified sgRNAs increased at least 8-fold compared to the initial cell pool. To validate the top 10 candidate tumor suppressors from this screen, we collected data from patients with hepatocellular carcinoma (HCC) using the Cancer Genome Atlas and COSMIC databases. We used CRISPR to inactivate candidate tumor suppressor genes in p53−/−;Myc;Cas9 cells and transplanted them subcutaneously into nude mice; tumor formation was monitored and tumors were analyzed by histology and immunohistochemistry. Mice with liver-specific disruption of p53 were given hydrodynamic tail-vein injections of plasmids encoding Myc and sgRNA/Cas9 designed to disrupt candidate tumor suppressors; growth of tumors and metastases was monitored. We compared gene expression profiles of liver cells with vs without tumor suppressor gene disrupted by sgRNA/Cas9. Genes found to be upregulated following tumor suppressor loss were examined in liver cancer cell lines; their expression was knocked down using small hairpin RNAs, and tumor growth was examined in nude mice. Effects of the MEK inhibitors AZD6244, U0126, and trametinib, or the multi-kinase inhibitor sorafenib, were examined in human and mouse HCC cell lines.
RESULTS:
We identified 4 candidate liver tumor suppressor genes not previously associated with liver cancer (Nf1, Plxnb1, Flrt2, and B9d1). CRISPR-mediated knockout of Nf1, a negative regulator of RAS, accelerated liver tumor formation in mice. Loss of Nf1 or activation of RAS upregulated the liver progenitor cell markers HMGA2 and SOX9. RAS pathway inhibitors suppressed the activation of the Hmga2 and Sox9 genes that resulted from loss of Nf1 or oncogenic activation of RAS. Knockdown of HMGA2 delayed formation of xenograft tumors from cells that expressed oncogenic RAS. In human HCCs, low levels of NF1 mRNA or high levels of HMGA2 mRNA were associated with shorter patient survival time. Liver cancer cells with inactivation of Plxnb1, Flrt2, and B9d1 formed more tumors in mice and had increased levels of MAPK phosphorylation.
CONCLUSIONS:
Using a CRISPR-based strategy, we identified Nf1, Plxnb1, Flrt2, and B9d1 as suppressors of liver tumor formation. We validated the observation that RAS signaling, via MAPK, contributes to formation of liver tumors in mice. We associated decreased levels of NF1 and increased levels of its downstream protein HMGA2 with survival times of patients with HCC. Strategies to inhibit or reduce HMGA2 might be developed to treat patients with liver cancer.
Keywords: liver cancer, mouse model, CRISPR screen, tumor suppressor genes
INTRODUCTION
Liver cancer is the 2nd deadliest cancer type and one of the few that continues to increase in incidence and mortality 1-4. Ninety percent of patients die within 5 years of diagnosis, accounting for >27,000 deaths in the U.S. and >700,000 deaths worldwide each year. Despite the clinical need, only one drug—the multi-kinase inhibitor sorafenib—has been approved for patients with advanced liver cancer 5. Because sorafenib only extends patient survival by 3 months and the detailed anti-tumor mechanisms of this drug remain unclear 5, new biomarkers and drug targets for effective treatment are urgently needed for this disease.
The genetics of hepatocellular carcinoma (HCC)—which comprises 70% of liver cancer cases—remain poorly defined because the liver cancer genome is highly heterogeneous, with many potential cancer driver mutations 1, 6. Identification of bona fide liver tumor suppressors and oncogenes based on patient genomic data involves tedious functional analysis of individual candidates that could turn out to be passenger mutations. Thus unbiased genetic approaches will maximize the likelihood of identifying key liver cancer pathways.
Genetic screens have been widely used to identify genes that drive liver cancer. Transposon screens have been performed in a conditional dominant-negative p53 mouse model 9, MYC-induced liver cancer model 10, and hepatitis B mouse models 7. Lentiviral screen has also been applied in neonate mouse livers 11. Insertional mutagenesis can induce gain- or loss-of-function and allow the rapid identification of driver genes in liver cancer. However, these forward genetic screens have limitations such as insertional biases of transposon and lentivirus. RNA interference-based screens using short hairpin RNAs (shRNAs) have been applied to study tumorigenesis in liver progenitor cells 8 and in the liver via hydrodynamic injection 12, 13 using 631 shRNAs targeting 362 genes in genomic deletions of HCC. These focused shRNA libraries do not provide genome-scale coverage, and RNA interference typically results in incomplete knockdown of target genes. Moreover, screens using these approaches have identified different sets of candidate genes, which suggest that some cancer genes are context-dependent and that genetic screens have not saturated the identification of liver tumor suppressors.
CRISPR/Cas9 is a powerful reverse-genetic tool that can induce complete gene knockout 14. We have used CRISPR to study cancer drivers or disease genes in the mouse liver in vivo 15-18, and Weber et al. have also recently used the CRISPR system to target a series of known or reported tumor suppressor genes in adult mice that induce liver cancer 19. Moreover, CRISPR-based knockout screens have identified essential genes in cultured human cells 20, 21 and genes that mediate lung tumor metastasis in a xenograft mouse model 22. But an unbiased in vivo CRISPR screen in an HCC model has yet to be published.
Here we describe a genome-wide screen to identify liver tumor suppressors using CRISPR-mediated genome editing 14. This screen identified a number of candidate liver tumor suppressors, including some that have known tumor suppressor activity in other tissues and some that have not been described as tumor suppressors in any tissue. Mouse models and human HCC patient data support a role for NF1 (a tumor suppressor mutated in neurofibromatosis) as a tumor suppressor in liver. Mechanistically, loss of Nf1 or activation of Ras increases the expression of the liver progenitor-cell markers Hmga2 and Sox9. In human liver cancer patients, low NF1 or high HMGA2 mRNA levels predict poor survival. Treatment of human liver cancer cells with RAS pathway inhibitors including sorafenib suppresses HMGA2 and SOX9 expression, and knockdown of Hmga2 delays tumorigenesis driven by oncogenic RAS. Our data show that NF1 and the other MAPK regulators function as key liver tumor suppressors by negatively regulating Ras-dependent activation of Hmga2, and suggest that Nf1 and Hmga2 could be useful prognostic or therapeutic indicators.
RESULTS
Genome-wide CRISPR screen identifies NF1 and other candidate tumor suppressors
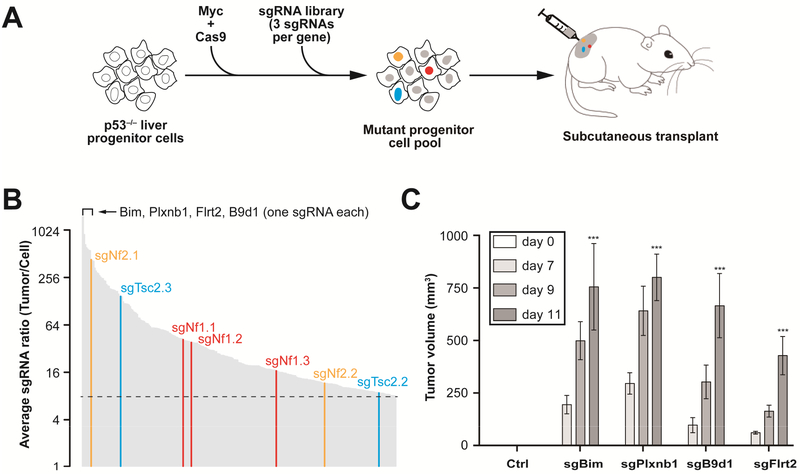
To identify functional liver tumor suppressors, we performed a genome-wide CRISPR/Cas9-based knockout screen in mouse embryonic liver progenitor cells lacking the tumor suppressor p53 and overproducing the Myc oncogene 8; ~30% of human HCC patients overexpress MYC, and p53 mutations or deletions are frequent in HCC 23. When transplanted under the skin of recipient mice, p53−/−; Myc cells form tumors slowly, but inactivation of additional tumor suppressors accelerates tumor formation 8. We therefore stably transduced p53−/−; Myc fetal hepatocytes with a lentivirus encoding Cas9 (Figure 1A). We infected the resulting p53−/−; Myc; Cas9 hepatocytes with the mGeCKOa lentiviral library of 67,000 single-guide RNA (sgRNA) targeting 20,611 mouse genes (~3 sgRNAs per gene; multiplicity of infection <1) 24, and transplanted 3 × 106 transduced cells (~45 cells per sgRNA) subcutaneously into immunocompromised Nu/Nu nude mice (Figure 1A). Within one month, 100% (n = 8) of mice that received the sgRNA library had developed subcutaneous tumors, whereas mice that received the control cells had not.
Figure 1. Genome-wide ex vivo CRISPR screen identifies new liver tumor suppressor genes.
(A) Outline of the screening strategy 8 sgRNAs targeting tumor suppressors accelerate formation of subcutaneous tumors and are enriched in the tumor.
(B) Average ratio of 267 individual sgRNAs enriched > 8-fold in tumors versus cell pool measured by high-throughput sequencing (n = 8). All three Nf1 sgRNAs (sgNf1.1, 2, 3) were enriched. Known liver tumor suppressors (Nf2, Tsc2) with two enriched sgRNAs, and new candidates (Bim, Plxnb1, etc) with one enriched sgRNA are highlighted.
(C) Validation of a subset of top-scoring sgRNAs in the subcutaneous tumor assay (n = 4 tumors). Average volume (n = 4) of tumors derived p53−/−; Myc; Cas9 cells infected with a control sgGFP (Ctrl) and a subset of top-scoring sgRNAs (sgBim, sgPlxnb1, sgB9d1, sgFlrt2). ***, P < .001. Error bars, mean ± s.e.m.
To identify candidate sgRNAs that drive tumor formation, we used high throughput sequencing to measure the representation of sgRNAs in all 8 tumors and the pre-transplantation cells, and calculated their average ratios in tumors to pre-transplantation cells (Supplementary Table S2). We identified 267 sgRNAs that were enriched at least 8-fold in tumors compared to the initial cell pool (Figure 1B; Supplementary Table S2). Our screen enriched for sgRNAs that inactivate known liver tumor suppressors (e.g., Nf2 and Tsc2) 1, tumor suppressors with demonstrated roles in other tissues—e.g., Nf1 25, Blc2l11 (i.e., Bim) 26, and Plxnb1 27, and genes not previously described as tumor suppressors (e.g., Flrt2 and B9d1). These results suggest that our approach is suited to identifying tumor suppressors, including those that function in liver.
Although the sgRNA library consists of ~3 sgRNAs per gene, Nf1 was the only target for which all three sgRNAs were enriched (41.5-, 38.3-, and 16.6-fold enrichment) (Figure 1B), and Nf2, Tsc2, Llgl1, and mir-6416 were the only targets for which two sgRNAs were enriched at least 8-fold (Supplementary Table S2). The remaining enriched sgRNAs each target unique genes. These results are consistent with a recent CRISPR screen in which only a subset of candidate genes were targeted by two or more independent sgRNAs using genome-wide library with high complexity 22. We don’t understand why only one sgRNA was enriched for so many targets. It remains possible that false-positive enrichment of an sgRNA could result from off-target editing of an unknown tumor suppressor. Alternatively, non-enriched sgRNAs might be inefficient or target exon sequences that are not essential for function 28. Two Bim sgRNAs (sgBim.1 and sgBim.2), for example, target sequences in the last exon of Bim and were not enriched in the screen (Supplementary Table S2), perhaps because premature termination codons in the last exons of mRNAs are often resistant to nonsense-mediated decay and result in weak loss of function or gain of function.
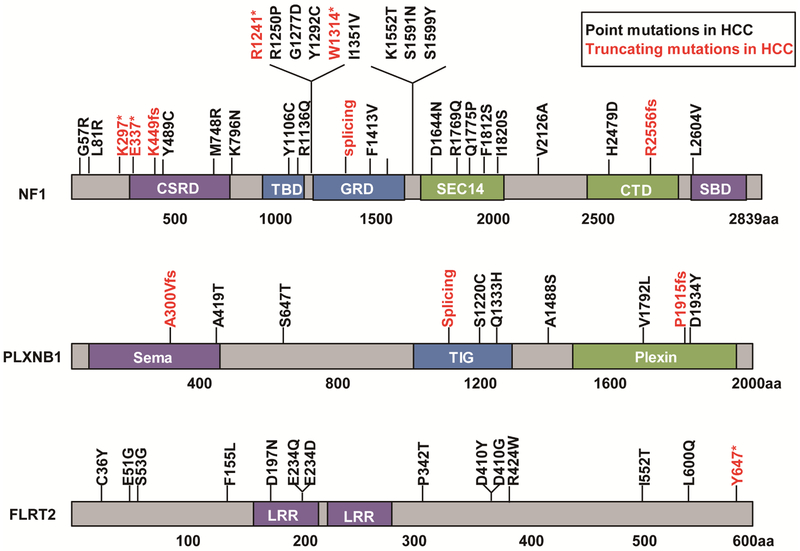
As an initial step toward validating the top 10 candidate tumor suppressors from our screen (Supplementary Table S2), we examined HCC patient data in the Cancer Genome Atlas (TCGA) 29 and COSMIC 30 databases and identified point mutations in NF2, NF1, PLXNB1, and FLRT2 (Figure 2, Supplementary Figure S1). Interestingly, mutations in NF1, PLXNB1, and FLRT2 are also found in human cholangiocarcinoma (CCA) (Supplementary Figure S2). When transduced into p53−/−; Myc; Cas9 cells, individual sgRNAs targeting Plxnb1, Bim, B9d1, or Flrt2 accelerated tumor growth in nude mice (Figure 1C), consistent with the idea that PLXNB1, BIM, B9D1, and FLRT2 are candidate tumor suppressors in liver cancer.
Figure 2. Point mutations in NF1, PLXNB1 and FLRT2 in human HCC.
* denotes nonsense mutation. Fs denotes frameshift mutation. CSRD, a cysteine-serine-rich domain; TBD, a tubulin-binding domain; GRD, a central GTPase-activating protein-related domain; SBD, a syndecan-binding domain; LRR, Leucine-rich repeat. Data are from TCGA and COSMIC.
Validation of Nf1 as a bona fide liver tumor suppressor in mouse models
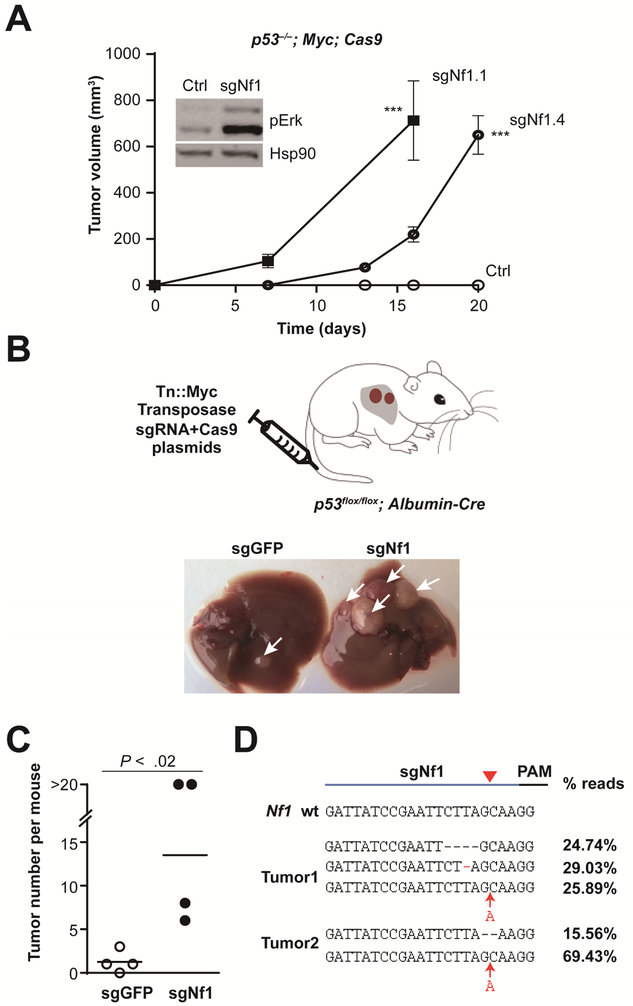
We focused our analyses on Nf1 because all three Nf1 sgRNAs were highly enriched in our screen (Figure 1B and Supplementary Table S2), and because NF1 encodes a GTPase-activating protein (GAP) that negatively regulates Ras 25. Mutations in NF1 underlie neurofibromatosis—i.e., neuronal tumors 25, but Nf1 has not been shown to function as a tumor suppressor in the liver. CRISPR-mediated inactivation of Nf1 in p53−/−; Myc; Cas9 cells using two individual Nf1 sgRNAs accelerated tumor formation in the subcutaneous transplant model (Figure 3A). Moreover, p53−/−; Myc; Cas9 cells infected with sgNf1 or the corresponding subcutaneous tumors show increased levels of phosphor-Erk (pErk), which is a downstream biomarker of the RAS-MAPK (Mitogen-activated protein kinase) pathway (Figure 3A and Supplementary Figure S3).
Figure 3. Nf1 is a bona fide liver tumor suppressor.
(A) Two individual Nf1 sgRNAs accelerated subcutaneous tumor growth. The tumors (n = 4) were derived from p53; Myc; Cas9 cells (Ctrl) infected with sgNf1.1 and sgNf1.4, independently. Error bars, mean ± s.e.m. The inset shows that sgNfl unregulated the phosphor-Erk (pErk) with Hsp90 as a loading control. ***, P < .001. Error bars, mean ± s.e.m.
(B) Schematic of hydrodynamic delivery of plasmids encoding Myc transposon (Tn) and Cas9/sgRNA (targeting Nf1, or GFP control) into liver-specific p53-knockout mouse. Representative images of livers from mice treated with sgGFP control (left) or sgNf1 (right) at 1 month after injection are shown. Arrows denote tumors.
(C) Quantification of tumor number per mouse (n = 4 mice) shown in (B).
(D) Sequences of Nf1 sgRNA target sites from representative liver tumors showing indel mutations and the fraction of reads for each.
Unlike lung cancer or pancreatic cancer, RAS gene mutations are rare in human liver cancer 31, but the inactivation or epigenetic silencing of RAS GAPs and other negative regulators of the RAS–MAPK pathway has been observed in liver cancer 32-35. However, RAS pathway has not been functionally validated in HCC using mouse models. We therefore sought to establish a genetic mouse model of Nf1 to study the mechanism of RAS signalling in liver cancer.
To test Nf1’s tumor suppressive role within the physiological environment of the mouse liver, we used hydrodynamic tail-vein injection to deliver plasmids encoding a transposon-derived Myc transgene and Nf1 sgRNA/Cas9 into liver-specific p53-knockout mice (Figure 3B) 15, 36, 37. As expected, sgNf1-injected mice developed significantly more liver tumors than mice that received the GFP sgRNA control (P = .02; Figure 3B-C). Using high-throughput sequencing, we verified the presence of bi-allelic Nf1 mutations proximal to the sgRNA target site in two different tumors (Figure 3D and Supplementary Table S3). The 1-nt, 2-nt or 4-nt indels are able to shift the reading frame of Nf1 and inactivate Nf1 by nonsense-mediated decay. To quantify potential off-target effect of sgNf1, we PCR amplified five of the top 10 predicted off-target sites from control and sgNf1 treated p53−/−; Myc; Cas9 cells. Deep sequencing revealed that the indels frequencies were < 1% for all five assayed sites (Supplementary Table S4).
Nf1 accelerates liver tumor formation in a multiplexed CRISPR mouse model
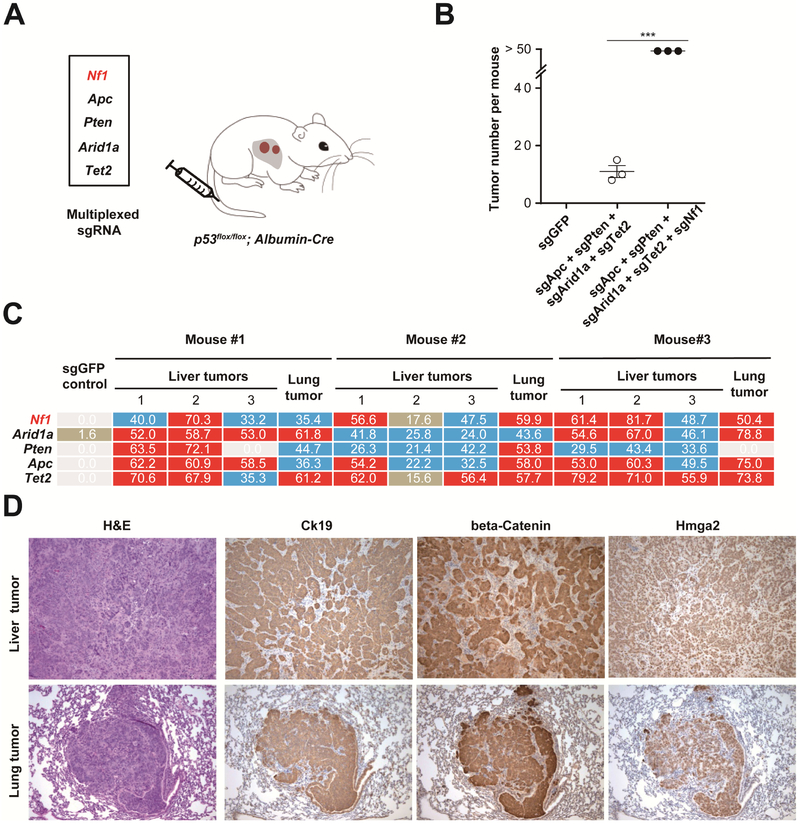
To assess NF1’s tumor suppressor activity in another genetic background, we combined Nf1 loss with four well-known tumor suppressors: Apc, Pten, Arid1a, and Tet2 19. Using hydrodynamic tail-vein injection in p53fox/flox; Albumin-Cre mice, we co-delivered Cas9/sgRNAs targeting Apc, Pten, Arid1a, and Tet2, with or without sgNf1 (Figure 4A). Within two months, the sgRNA multiplex with sgNf1 accelerated tumor formation (n = 3 per treatment; Figure 4B and Supplementary Figure S4A). At three months, all mice treated with the sgNf1-containing multiplex also developed lung tumors (n = 3) (Supplementary Figure S4B). Again high-throughput sequencing of sgRNA target genes revealed indels at each target site in most micro-dissected tumors (including tumor cells and stromal cells). Nf1 is consistently mutated in all tumors, whereas one liver tumor and one lung tumor lacked Pten indels (Figure 4C and Table S5). All liver and lung tumors were morphologically identical and positive for Hmga2 and beta-catenin (Figure 4D), consistent with Nf1 and Apc loss-of-function 38. Additionally, liver and lung tumors were also positive for the bile duct and cholangiocarcinoma marker Cytokeratin 19 (Ck19; Figure 4D) 19, suggesting that the lung tumors metastasize from primary cholangiocarcinomas. Together these data show that inactivation of Nf1 in the liver of an immunocompetent mouse promotes tumor formation. Thus NF1 is a bona fide tumor suppressor in the liver.
Figure 4. Multiplexed CRISPR accelerates tumor formation.
(A) sgRNA mixture was injected into p53flox/flox; Albumin-Cre mice by hydrodynamic injection.
(B) Quantification of tumor number per mouse (n = 3 mice) at 2 months shown in (A). ***, P < .001. Error bars, mean ± s.e.m.
(C) Deep sequencing for the representative liver and lung tumors at 3 months showing indel percentages at the assayed target sites (> 50 shown with red background, between 20 and 50 blue background, between 0 and 20 brown background, 0 white background). The varying indel rates may have been caused by different percentages of wild-type stromal cells within each tumor (see panel D). One liver tumor and one lung tumor had a low percentage of Pten indels.
(D) H&E and IHC. All lung tumors show Ck19 staining (10 × lens).
Nf1 negatively regulates Ras-dependent activation of liver progenitor cell markers Hmga2 and Sox9
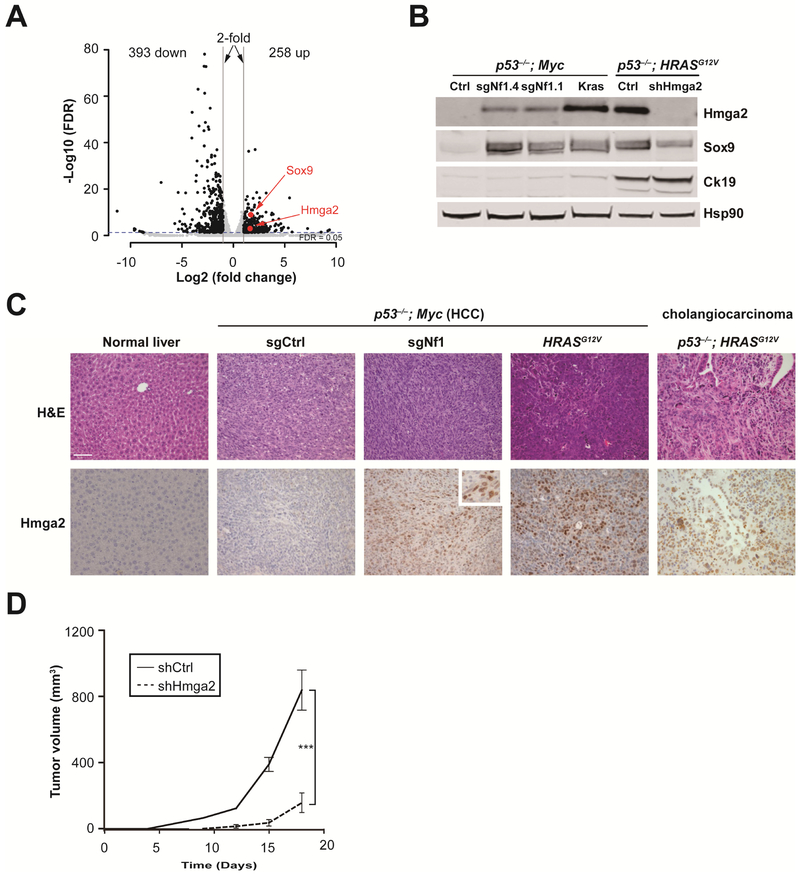
To identify genetic signatures regulated by Nf1 in mouse liver cells, we compared the mRNA profiles of control-treated and sgNf1-treated p53−/−; Myc; Cas9 liver cells. RNA-seq analysis identified 258 up-regulated and 393 down-regulated mRNAs (> 2-fold cutoff and FDR < 0.05) in sgNf1-treated cells compared to control-treated cells (Figure 5A and Supplementary Table S6). Consistent with the negative regulation of Ras by NF1, Gene Set Enrichment Analysis revealed a positive enrichment for the KRAS signalling pathway (Supplementary Figure S5A). Notably, Hmga2 and Sox9, two genes expressed in fetal liver and liver progenitor-like cells 3, 39, were significantly up-regulated in liver cells by Nf1 loss-of-function (Figure 5A-B) and by oncogenic Ras (KrasG12D or HRASG12V; Figure 5B). Hmga2 protein was also elevated in two Ras-dependent mouse models of cholangiocarcinoma (Figure 5C and Supplementary Figure S5B) 40, 41.
Figure 5. Nf1 negatively regulates Ras-dependent activation of liver progenitor cell markers Hmga2 and Sox9.
(A) Volcano plot of mRNA levels in Nf1 sgRNA-treated cells compared to the control sgRNA-treated cells (n = 3).
(B) Western blot shows increased Hmga2 and Sox9 levels upon sgRNA-inactivation of Nf1 or activation of Kras (KrasG12D cDNA) in p53−/−; Myc liver cells. Hmga2, Sox9 and Ck19 levels were high in p53−/−; HRASG12V cholangiocarcinoma cells.
(C) Immunohistochemistry of representative liver sections showing increased Hmga2 protein upon sgRNA-inactivation of Nf1 or activation of HRASG12V in p53−/−; Myc tumors, and in the p53−/−; HRASG12V cholangiocarcinoma (CCA) mouse model (n ≥ 3 mice per group). Normal liver serves as a control. Scale bar is 50 μm. Inset shows a high-magnification view.
(D) Graph of subcutaneous tumor volumes in nude mice receiving p53−/−; HRASG12V cells treated with Hmga2 shRNA (shHmga2) or control shRNA (shCtrl). ***, P < .001. Error bars are s.d. of mean (n = 4 tumors).
Hmga2 is predominantly expressed in embryonic and cancer stem cells 39, but its role in liver cancer remains uncharacterized. To determine whether suppression of Hmga2 can delay tumorigenesis, we infected p53−; HRASG12V mouse liver cancer cells expressing high levels of Hmga2 with lentivirus encoding a well-characterized Hmga2 shRNA (shHmga2) 40, 42 or a control shRNA (shCtrl) and transplanted in nude mice (Figure 5B). As shown in the Figure 5D, shHmga2 significantly suppressed tumor formation in nude mice (P < .001). Concordantly, shHmga2 suppressed proliferation of p53−/−; HRASG12V cells and of sgNf1-treated p53−/−; Myc; Cas9 cells (Supplementary Figure S5F). Notably, Nf1 knockout accelerated proliferation of p53−/−; Myc; Cas9 cells (Supplementary Figure S5F), consistent with the role of Nf1 as a tumor suppressor gene. These findings indicate that the RAS pathway drives liver tumor progression via Hmga2.
Sorafenib or MEK inhibitors can suppress HMGA2 and SOX9 expression in liver cancer cell lines
We found that HMGA2 and SOX9 mRNAs levels are high in Hep3B, HepG2, and Huh7 human HCC cell lines (Supplementary Figure S5C). HepG2 cells harbor the oncogenic NRASQ61L allele 43. In Hep3B and HepG2 cells, loss of the negative regulator of Ras, RASSF1A, stimulates the Ras–MAPK pathway 31. Consistent with Ras-dependent activation of HMGA2 and SOX9 (Figure 5), MEK inhibitors AZD6244 and U0126 31 or the multi-kinase inhibitor sorafenib—whose targets include RAF—suppressed both HMGA2 and SOX9 in all three human HCC cell lines (Figure 6A, 6C, and Supplementary Figure S5D, E). Sorafenib and AZD6244 also suppressed HMGA2 and SOX9 in mouse liver cancer cell lines derived from tumors that were isolated from p53−/−; Myc; Cas9; sgNf1 and p53−/−; HRASG12V mice (Figure 6B and6D). To further test whether MEK inhibitors can inhibit proliferation of liver cancer cells, we treated human and mouse cell lines with Trametinib, an FDA-approved MEK inhibitor for melanoma with BRAF mutations. All tested cell lines exhibited a delay in colony formation after incubating with 0.5 μM and 1 μM Trametinib (Figure 6E and 6F). Together these findings show that up-regulation of Hmga2 and Sox9 in liver cells by the Ras–MAPK pathway can be suppressed by Sorafenib or MEK inhibitors.
Figure 6. Sorafenib or MEK inhibitors can suppress HMGA2 and SOX9 expression in liver cancer cell lines.
(A) and (C) qPCR showing the levels of HMGA2 and SOX9 mRNA in Hep3B and Huh7 human HCC cells treated for 48 hours with 1 μM AZD6244, or 7.5 μM sorafenib relative to control-treated cells (Ctrl). (B) and (D) qPCR showing Hmga2 and Sox9 expression in p53−/−; HRASG12V and p53−/−; Myc; Cas9; sgNf1 mouse liver cells. Drug treatment was the same as in (A). Error bars are s.d. of mean (n = 3). ***, P < .001. (E) and (F) Trametinib, a MEK inhibitor, suppressed colony formation in both human and mouse liver cancer cell lines.
NF1 and HMGA2 correlate with patient survival in liver cancer
A previous study reported NF1 loss-of-heterozygosity in ~12.5% of HCC patients 32. Examining a published patient dataset 44, we found that HCC patients with low NF1 mRNA levels (bottom 40%) had a significantly shorter survival than those with high NF1 mRNA levels (top 40%, P = .005; Figure 7A). Conversely, high HMGA2 levels predict poor patient survival (P =.019; Figure 7B). Examining the TCGA liver cancer data, we found NF1 point mutations in 3.8% (14 out of 373) of HCC patients (Figure 7C and Supplementary Table S7). NF1 mRNA is not significantly associated with survival in the TCGA patient cohort (Supplementary Figure S6A), but high NRAS mRNA levels do predict poor survival (Supplementary Figure S6B). Moreover, high HMGA2 mRNA levels predict poor survival among liver cancer patients in the TCGA data and in another data set 45 (Supplementary Figure S6C-D). We also analyzed HMGA2 and NF1 gene expression in previously established molecular subtypes of HCC patients44, 46 Overall, high HMGA2 expression and low NF1 expression are associated with prognostic subtype A44, which correlates with poor patient survival. However, when comparing the expression of both genes between samples with hepatoblast subtype (HB) and hepatocyte subtype (HC)46 in prognostic subtype A, the gene expression patterns are similar (Supplementary Figure S7).
Figure 7. NF1 and HMGA2 correlate with patient survival in liver cancer.
(A-B) Survival curves of patients expressing high (Top 40%) or low (Bottom 40%) levels of NF1 mRNA (A) or HMGA2 mRNA (B). Based on data from a clinical study 44.
(C) Summary of NF1 and other selected RAS pathway mutations in TCGA liver cancer patients (n = 373). Each column (light grey bars) represents one patient. Dark grey denotes patients with mutations. RASA1 (p120 RAS GAP) and RP6SKA3 are inhibitors of RAS pathway. RP6SKA3 mutations have been reported in HCC 1.
(D) Schematic that loss of Nf1, Plxnb1, Flrt2 or B9d1 convergently activates the MAPK pathway and induces liver progenitor cell markers HMGA2/SOX9 in liver cancer. Green and red arrows denote decreased or increased activity.
Functional validation of additional candidate genes
To test whether the other candidate genes identified by our screen—B9d1, Plxnb1, and Flrt2—can function as tumor suppressors in vivo, we delivered plasmids encoding a transposon-derived Myc transgene and B9d1, Plxnb1, or Flrt2 sgRNA/Cas9 into liver-specific p53-knockout mice (p53flox/flox; Albumin-Cre; Supplementary Figure 8A) by hydrodynamic tail-vein injection. Within one month, the mice treated with sgRNAs targeting B9d1, Plxnb1, or Flrt2 developed more liver tumors than those with sgGFP controls (Supplementary Figure 8A and 8B). Because antibodies for mouse B9d1, Plxnb1 and Flrt2 are not available, we measured indel mutations at the sgRNA target genes in representative tumors by deep sequencing. We observed insertions and deletions (indels) clustered at the predicted Cas9 cleavage sites for all three genes (Supplementary Figure 8C), confirming the efficacy of these sgRNAs. Tumors also contain wild-type B9d1, Plxnb1 and Flrt2 sequence, which likely derives from normal stromal cells or endothelial cells that do not take up DNA from the hydrodynamic injection but do support tumor growth. Moreover, inactivation of Plxnb1, Flrt2 and B9d1 using two independent sgRNAs per gene also upregulated pErk (Supplementary Figure S9), indicate that these top candidate genes converge on the MAPK pathway.
Consistent with the activation of pErk by sgRNAs targeting Plxnb1, Flrt2 and B9d1, these sgRNAs also elevated Hmga2 in p53−/−; Myc; Cas9 cells using sgNf1 as a positive control (Supplementary Figure S9), indicating that these new candidate genes regulate Hmga2 through the MAPK pathway. Together, these data suggest that CRISPR-based screening can identify previously uncharacterized liver tumor suppressors.
DISCUSSION
Our results suggest that genetic screens to identify liver tumor suppressors are far from saturation. Nf1 and some candidate tumor suppressors uncovered by our genome-wide CRISPR screen have not been identified by previous transposon or RNAi-based screens 7, 8. Furthermore, although the genome-wide CRISPR library comprises 3 sgRNAs per gene, our screen and a recent screen in lung cancer cells only enriched one sgRNA for most of the candidates 22. An explanation for non-enriched sgRNAs in this library might be that they are inefficient or non-functional. Thus the CRISPR-based screen could benefit from additional optimization of screening parameters, including the use of later-generation sgRNA libraries. Our screen was based on liver progenitor cells with p53 loss-of-function and Myc overexpression. Some tumor suppressor genes may not score in this genetic context, so a different genetic background might help to identify context-dependent tumor suppressors. Moreover, CRISPR-based genetic screens performed directly in the mouse liver might help to identify tumor suppressor functions that depend on the tumor microenvironment compared to our ex vivo screen. Nevertheless, our screen provides a complementary approach to other genetic screens that seek to identify liver tumor suppressors.
Our study provides direct genetic evidence that Nf1 functions as a key tumor suppressor in liver. Regulation of liver cell lineage genes by RAS has not been reported in previous studies. Our data indicate that Nf1 and the other candidate genes negatively regulate RAS/MAPK-dependent activation of liver progenitor cell markers—HMGA2 and SOX9 (Figure 7D). HCC patients with high HMGA2 or SOX9 expression might therefore be good candidates for combined treatment with multiple RAS pathway inhibitors to inhibit cancer stem cell properties, or by combined treatment with a Ras pathway inhibitor and HMGA2 siRNA. Recent clinical success in RNAi-based therapeutics to treat liver disease makes the latter option quite attractive 47.
Our screen identified new candidate tumor suppressor genes that converge on the MAPK pathway (Figure 7D). Plxnb1 has been reported to be a GAP for R-Ras 48 Flrt2 interacts with fibroblast growth factor receptor (FGFR), which signals through MAPK pathway 49. B9d1 has not been implicated in MAPK signaling in the literature. Future studies are required to investigate how these new genes regulate the MAPK pathway. A recent transposon mutagenesis screen identified Ras genes as potential drivers in HCC50. This observation aligns with our study and highlights that Ras might be playing a broad role in HCC that is currently under-appreciated.
Thus CRISPR-based mouse models can be used to dissect tumor suppressor mechanisms in HCC. Because CRISPR induces somatic genetic mutations in a fraction of the time and cost of traditional mouse models 51, our method provides a flexible in vivo platform to rapidly identify tumor suppressor genes and to build precision mouse models for dissecting disease mechanisms.
Materials and Methods
CRISPR vectors
sgRNA sequences (Supplementary Table S1) were from the mGeCKOa library 24, or designed as described 52. sgNf1.1 was from the library and sgNf1.4 was designed to rule out off-target effects. sgRNA oligos (IDT) were annealed and cloned into the pX330 vector (addgene 42230) or lentiV2 (addgene 52961) 24 using standard Bbsl or BsmBI protocols.
Cell culture and infection
Cell culture conditions were as described 53 p53−/− fetal hepatocytes were harvested from ED = 18 p53−/− fetal liver 54 and infected with packaged retroviral Myc and lentiviral Cas9 twice and selected for with blasticidin. The cells were then infected with the lentiviral mGeCKOa sgRNA library 24. Virus was prepared by the UMass shRNA Viral Core. Puromycin-resistant cells were collected. 293fs cells were used to package lentivirus encoding individual sgRNA and Cas9. All data are representative of at least two independent infections. Sorafenib, selumetinib (AZD6244), Trametinib (GSK1120212) and U0126 were purchased from Selleck.
Animal experiments
All animal protocols were approved by the UMass Medical School Institutional Animal Care and Use Committee. Mice were humanely euthanized by CO2 asphyxiation. p53flox/flox mice were crossed with Albumin-Cre mice (Jackson Laboratories) 37. pX330.sgNf1 (20 μg), pT3-EF1α-Myc (5 μg), and CMV-SB10 transposase (1 μg) plasmids were delivered to ~8 week-old mice by hydrodynamic tail vein injection. Plasmid DNA was purified using an EndoFree Maxiprep DNA Kit (Qiagen). pX330.sgGFP (20 μg) was used as non-targeting control. For multiplexed CRISPR experiments, 10 μg of each pX330 sgRNA vector was injected. For mice livers with too many tumors to count, we defined that they have 20 tumors. For subcutaneous tumor growth, 3 × 106 (Figure 1) or 1 × 106 (Figure 3A) p53−/−; Myc; Cas9 cells were injected into flanks of 6-8-week old, female, NCI Nu/Nu mice (Taconic) and monitored as described 54. For p53−/−; HRASG12V cells, 50,000 (Figure 5D) cells/tumor were injected in Nu/Nu mice. KrasG12D;shp53 mouse cholangiocarcinoma was generated as in 41 using KrasLSL-G12D/+ liver progenitor cells which express endogenous level of KrasG12D.
Histology and Immunohistochemistry
Livers were fixed in 4% or 10% (v/v) formalin overnight, and embedded in paraffin. 4 μm liver sections were stained with hematoxylin and eosin (H&E) or with antibodies using standard immunohistochemistry protocols. The following antibodies and dilutions were used: 1:400 anti-Hmga2 (Biocheck), 1:100 anti-beta-Catenin (BD bioscience), 1:100 anti-Ck19 (Abcam). Histopathology of mouse liver tumors was evaluated by an experienced pathologist (A.F.).
sgRNA deep sequencing processing
High-Pure PCR Template Preparation Kit (Roche) was used for genomic DNA purification. sgRNA deep sequencing was performed as described 22. All sequencing datasets were evaluated using FastQC (version 0.11.2) to ensure high quality. We built a pipeline in Python (version 2.7.10) to process sgRNA raw sequence reads and derive a ranked list of genes with enriched sgRNAs, and the pipeline is summarized as follows. We first built an index using all sgRNAs in each sample and then used Bowtie (version 1.0.0) 55 to map the sgRNA sequences in the mGeCKOa library to the index in a strand-specific manner allowing up to one mismatch. We then counted mapped reads for each sample and normalized the abundance of each sgRNA to reads per million (RPM). The RPM of each tumor sample was divided by the control sample RPM to calculate a tumor:control ratio for each sgRNA.
CRISPR-induced insertion/deletion detection
Genomic DNA from tumors or cells was harvested. sgRNA target sites were PCR amplified and subjected to high throughput genomic DNA sequencing 53. We mapped the reads to the reference genomic sequence using BWA version 0.7.5 and SAMtools (version 0.1.19). We then used VarScan2 (version 2.3) to identify insertions and deletions with the ‘pileup2indel’ mode and parameters ‘--min-var-freq’, ‘--min-avg-qual', '--p-value’. The software can be found at: https://zlab.umassmed.edu/CIpipe/
RNA-sequencing and bioinformatics analysis
RNA-seq was performed as described 56. We first trimmed reads and removed PCR primers using Trimmomatic (v 0.30). We aligned RNA-seq reads to the mm10 genome using STAR (version 2.3.0e) with default parameters and selected only uniquely mapping reads. We removed redundant read pairs using SAMtools (version 0.0.19). For all genes annotated in GENCODE M7, we calculated the number of reads per gene using HTSeq. We then determined differential expression using DESeq (version 1.18.0) 57 and accounted for possible batch effects using a generalized linear model. In order to detect differentially expressed genes, we required the change in expression to be greater than two-fold and the false discovery rate (FDR) to be less than 0.05. Gene Set Enrichment Analysis (GSEA) was used to identify pathways enriched among differentially expressed genes.
Western blot analysis
Protein lysates from culture cells were harvested with RIPA buffer including proteinase and phosphatase inhibitors. Proteins were separated on 4-12% NuPage Bis-Tris gels (Life Technologies, NP0321), transferred to nitrocellulose membrane, and probed with antibodies. Blots were imaged using an Odyssey system (LICOR). The following antibodies and dilutions were used: 1:1000 anti-Hmga2 (Biocheck); 1:1000 anti-Sox9 (Millipore); 1:10,000 anti-Hsp90 (BD Biosciences); 1:1000 anti-Ck19 (Troma); 1:1000 anti-pErk (Cell Signaling).
Real-time PCR
Total RNA was purified using an RNeasy Mini Kit (Qiagen). First-strand cDNA was synthesized using Superscript (ABI) and used as template in TaqMan real-time PCR assays (Life technologies), according to manufacturer’s instructions. TaqMan probes are: HMGA2 Hs04397751_m1, SOX9 Hs01001343_g1, and ACTB (beta-ACTIN) Hs01060665_g1 (as a control).
Patient Survival Analysis
Expression array data of human hepatocellular carcinoma (HCC) and patient clinical information were obtained from the GEO record GSE1898 (Lee et al. 2004). The TCGA 29 human liver cancer dataset was downloaded via Firehose from the Broad Institute Genome Analysis Center (http://qdac.broadinstitute.org). The TCGA data comprises patient mRNA expression data (RSEM value, i.e., normalized RNA-seq by Expectation-Maximization) and clinical information. For each dataset, we separated patients into two groups, with high and low expression for each gene of interest. We then performed survival analysis and drew related graphs using the R (version 3.2.2) package ‘survival’. The results here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Statistics
Student’s t-test and Wilcoxon rank-sum test were used to determine P values.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Jacks, C. Mello, S. Lowe, P. Zamore, E. Sontheimer, V. Ambros, T. Flotte, P. Sharp, F. Sanchez-Rivera, M. Green, and M. Moore for their insightful comments and reagents. We thank the UMass Medical School shRNA Viral Core, Y. Liu in the Morphology Core, and E. Kittler in the Deep Sequencing Core for support. The pT3-Myc transposon vector was a kind gift of Dr. Xin Chen, UCSF.
Funding
H.Y. was supported by the Skoltech Center and the NCI in the MIT-Harvard Center of Cancer Nanotechnology Excellence (5-U54-CA151884-04). Yingxiang Li was supported by the China Scholarship Council (201506260151) and the Thousand Talent Plan funding to Z.W. from the Chinese government. This work was supported by grants from the NIH (R00CA169512, DP2HL137167 and P01HL131471), the Worcester Foundation, UMASS CCTS Pilot Project Program and the Lung Cancer Research Foundation to WX. Y.P. and X.W.W. were supported by grants (Z01-BC 010313 and Z01-BC 010876) from the intramural research program of the Center for Cancer Research, National Cancer institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest The authors disclose no conflicts.
Author names in bold designate shared co-first authorship.
References:
- 1.Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149:1226–1239. [DOI] [PubMed] [Google Scholar]
- 2.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674–87. [DOI] [PubMed] [Google Scholar]
- 3.Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer 2015;15:653–67. [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–90. [DOI] [PubMed] [Google Scholar]
- 6.Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–73. [DOI] [PubMed] [Google Scholar]
- 7.Bard-Chapeau EA, Nguyen AT, Rust AG, et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet 2014;46:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zender L, Xue W, Zuber J, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 2008;135:852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keng VW, Villanueva A, Chiang DY, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol 2009;27:264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell KA, Keng VW, York B, et al. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci U S A 2012;109:E1377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranzani M, Cesana D, Bartholomae CC, et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat Methods 2013;10:155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudalska R, Dauch D, Longerich T, et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med 2014;20:1138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wuestefeld T, Pesic M, Rudalska R, et al. A Direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell 2013;153:389–401. [DOI] [PubMed] [Google Scholar]
- 14.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- 15.Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 2014;514:380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014;32:551–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Mou H, Li S, et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin H, Song C-Q, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotech 2016;34:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber J, Ollinger R, Friedrich M, et al. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc Natl Acad Sci U S A 2015;112:13982–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shalem O, Sanjana NE, Hartenian E, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, Wei JJ, Sabatini DM, et al. Genetic Screens in Human Cells Using the CRISPR/Cas9 System. Science 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Sanjana NE, Zheng K, et al. Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell 2015;160:1246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet 2002;31:339–46. [DOI] [PubMed] [Google Scholar]
- 24.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 2015;15:290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czabotar PE, Lessene G, Strasser A, et al. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 2014;15:49–63. [DOI] [PubMed] [Google Scholar]
- 27.Gomez Roman JJ, Garay GO, Saenz P, et al. Plexin B1 is downregulated in renal cell carcinomas and modulates cell growth. Transl Res 2008;151:134–40. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, Wang E, Milazzo JP, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 2015;33:661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2014;43:D805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delire B, Starkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest 2015;45:609–23. [DOI] [PubMed] [Google Scholar]
- 32.Calvisi DF, Ladu S, Conner EA, et al. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol 2011;54:311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvisi DF, Ladu S, Gorden A, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 2006;130:1117–28. [DOI] [PubMed] [Google Scholar]
- 34.Ito Y, Sasaki Y, Horimoto M, et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology 1998;27:951–8. [DOI] [PubMed] [Google Scholar]
- 35.Li C, Wu X, Zhang W, et al. AEG-1 Promotes Metastasis Through Downstream AKR1C2 and NF1 in Liver Cancer. Oncol Res 2014;22:203–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CH, Lujambio A, Zuber J, et al. CDK9-mediated transcription elongation is required for mYc addiction in hepatocellular carcinoma. Genes Dev 2014;28:1800–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katz SF, Lechel A, Obenauf AC, et al. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology 2012;142:1229–1239. [DOI] [PubMed] [Google Scholar]
- 38.Moon RT, Kohn AD, De Ferrari GV, et al. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet 2004;5:691–701. [DOI] [PubMed] [Google Scholar]
- 39.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer 2007;7:899–910. [DOI] [PubMed] [Google Scholar]
- 40.Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007;445:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saborowski A, Saborowski M, Davare MA, et al. Mouse model of intrahepatic cholangiocarcinoma validates FIG-ROS as a potent fusion oncogene and therapeutic target. Proc Natl Acad Sci U S A 2013;110:19513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winslow MM, Dayton TL, Verhaak RG, et al. Suppression of lung adenocarcinoma progression by Nkx2–1. Nature 2011;473:101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breunig C, Mueller BJ, Umansky L, et al. BRaf and MEK inhibitors differentially regulate cell fate and microenvironment in human hepatocellular carcinoma. Clin Cancer Res 2014;20:2410–23. [DOI] [PubMed] [Google Scholar]
- 44.Lee JS, Chu IS, Mikaelyan A, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet 2004;36:1306–11. [DOI] [PubMed] [Google Scholar]
- 45.Roessler S, Long EL, Budhu A, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012;142:957–966 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006;12:410–6. [DOI] [PubMed] [Google Scholar]
- 47.Tabernero J, Shapiro GI, Lorusso PM, et al. First-in-Humans Trial of an RNA Interference Therapeutic Targeting VEGF and KSP in Cancer Patients with Liver Involvement. Cancer Discov 2013;3:406–417. [DOI] [PubMed] [Google Scholar]
- 48.Oinuma I, Ishikawa Y, Katoh H, et al. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science 2004;305:862–5. [DOI] [PubMed] [Google Scholar]
- 49.Haines BP, Wheldon LM, Summerbell D, et al. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev Biol 2006;297:14–25. [DOI] [PubMed] [Google Scholar]
- 50.Kodama T, Bard-Chapeau EA, Newberg JY, et al. Two-Step Forward Genetic Screen in Mice Identifies Ral GTPase-Activating Proteins as Suppressors of Hepatocellular Carcinoma. Gastroenterology 2016;151:324–337. e12. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer 2015;15:387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Park A, Mou H, et al. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biology 2015; 16:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zender L, Spector MS, Xue W, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 2006;125:1253–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shao DD, Xue W, Krall EB, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 2014;158:171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.