Abstract
Caspase-3 (CASP3) is a major mediator of apoptosis activated during cellular exposure to cytotoxic drugs, radiotherapy, or immunotherapy. It is often used as a marker for efficacy of cancer therapy. However, recent reports indicate that caspase-3 also has non-apoptotic roles such as promotion of tumor relapse and tumor angiogenesis. Therefore, the roles of caspase-3 in tumor progression remains to be defined clearly. In this study, we established caspase-3 knockout (KO) colon cancer cell lines by use of the CRISPR technology. In vitro,caspase-3 knockout HCT116 cells were significantly less clonogenic in soft agar assays. They were also significantly less invasive and more sensitive to radiation and mitomycin C than control cells. In vivo, CASP3KO cells formed tumors at rates similar to control cells but were significantly more sensitive to radiotherapy. They were also less prone to pulmonary metastasis when inoculated either subcutaneously or intravenously. At the mechanistic level, caspase-3 gene knockout appeared to cause reduced EMT phenotypes when compared with parental HCT116 cells. Indeed, they showed significantly increased E-cadherin expression, reduced N-cadherin, Snail, Slug, and ZEB1 expression than control cells. Therefore, therapeutic targeting of caspase-3 may not only increase the sensitivity of cancer cell to chemotherapy and radiotherapy, but also inhibit cancer cell invasion and metastases.
Keywords: Caspase 3, invasion, metastasis, colon cancer, chemotherapy, radiotherapy
Caspases are a 15 member family of cysteine proteases playing essential roles in programmed cell death and inflammation.1 Among them, caspase 3 is a prototypical apoptotic executioner that, upon activation by initiator caspase 8 or caspase 9, cleaves many other functionally critical proteins within the cell, leading to apoptosis.2 Many anticancer therapies including cytotoxic drugs, radiotherapy, or immunotherapy are able to cause tumor cell death by activating caspase 3. As such, caspase 3 activation is used by numerous investigators as a surrogate marker for the efficacy of cancer treatment.
However, recent studies painted a more complicated picture of caspase 3 in cancer induction and treatment. One recent study shows that caspase 3, instead of acting as a tumor suppressor, promotes carcinogenesis after cellular exposure to chemicals and radiation.3 An earlier study showed that caspase 3 is involved in promoting tumor repopulation after radiotherapy through a paracrine signaling pathway.4 Another study showed that caspase 3 promotes tumor growth by providing a pro angiogenic microenvironment.5 Furthermore, dying bladder cancer cells promote tumor cell repopulation after chemotherapy.6 In human patients, Flanagan et al. reported that colon cancer patients with low levels of activated caspase 3 had a longer disease free survival time.7
Colon cancer is one of the 3 most prevalent cancers among both males and females.8 In 2017, there will be an estimated 95,520 new cases diagnosed in United States.8 According to statistics, about 25% of colon cancer patients will present with metastatic disease.9 Metastasis is the main obstacle in cancer treatment. The metastatic colon cancer often occurs in liver, lung, bone, lymph nodes and brain.10, 11 Surgery, chemotherapy, and radiotherapy are generally used to treat metastatic colon cancer. Nevertheless, despite advances in treatment, the overall 5 year survival rate for metastatic colon cancer is only 30% to 40%.12
In this study, we established caspase 3 knockout colon cancer cell lines by use of the CRISPR technology and compared their behavior in vitro and in vivo with control cells. Our results showed that caspase 3 not only contributes to drug and radiation resistance but is also involved in regulating colon cancer cell migration, invasion and metastasis.
Material and Methods
Cell lines
HCT116, HT29, and MBA MD231 cells were obtained from the Cell Culture Facility of Duke University School of Medicine. Their identities were verified by microsatellite characterization carried out by personnel at the Cell Culture Facility.
Establishment of caspase-3 knockout or knockdown cell lines
Caspase 3 knockout cell lines were established by use of a lentivirus based CRISPR/Cas9 system.13 Target single guided RNA (sgRNA) sequences were identified with an online CRISPR design software at http://crispr.mit.edu. The sgRNA sequence chosen is 5’TAGTTAATAAAGGTATCCA 3’, which was prepended with a G nucleotide for efficient U6 transcription. Annealed double stranded sgRNA oligoes were ligated into the lentiCRISPR v2 vector (a gift from Feng Zhang, Addgene plasmid # 52961) at the BsmBl site, which co express Cas9 and sgRNA in the same vector. The constructed lentivirus based CRISPR vectors were prepared, packaged according an established protocol.13 Subsequently HCT116 cells or MDA MB 231 cells were infected with sgRNA encoding lentivirus and cultured in DMEM medium supplemented with 10% FBS. The infected cells were then cultured in DMEN containing 1μg/ml puromycin for 14 days selection. Surviving cells were plated into 96 well plates with 1 cell per well. Colonies that emerged from single cells were selected and expanded for western blot analysis. Those clones without caspase 3 protein expression were selected for further analysis. The primers used to amplify caspase 3 gene sequences surrounding the target gene site were 5’GCAAAGAAATCATTATCCCCAG 3’ (Forward) and 5’ TTTGCTTATTACACATCCCCAT 3’ (Reverse). PCR products were purified and then subjected Sanger sequencing to verify gene disruption.
Caspase 3 knockdown HT29 cell lines were established by use of shRNA encoding lentivirus vectors purchased from Open Biosystems (now Thermo Fisher): Clone 1: V2LHS_15044. Clone 2:V2LHS_15045. HT29 cells were infected with shRNA encoding lentivirus and then cultured in in DMEN containing 1μg/ml puromycin for 14 days selection.
Western blot
Cells were washed with PBS, and then lysed in RIPA buffer supplemented with protease inhibitors. Equal amounts of proteins were separated by SDS PAGE and transferred to a PVDF membrane. Proteins were probed with specific antibodies followed by secondary antibodies conjugated with HRP. The HRP signal was developed by using ECL.
Growth curve
Cells were seeded into 96 well plates at increasing densities from 200 cells/well to 6400 cells/well. Growth curve for cells measured using the MTT assay. Briefly, cells were stained daily by use of the MTT (3 (4,5 Dimethylthiazol 2 yl) 2,5 Diphenyltetrazolium Bromide) reagent (ATCC). Cellular densities were then measured at 570 nm by use of a Biotek Synergy H1 plate reader. Five wells were plated for each seeding density of each cell type.
ELISA assay for prostaglandin E2 (PGE2) concentration
HCT116 caspase 3 KO or vector control cells were treated or untreated with X ray radiation at 10 Gy and plated in 6 well plates (1×106 cells/well) in 2% fetal bovine serum culture medium. Supernatant from the cells was collected 96 hours after radiation and diluted 8 fold. Cells were counted by using the Bio Rad cell counter in order to normalize PGE2 concentration to cell number. The concentration of PGE2 in the supernatant was measured following a protocol from an ELISA kit purchased from Cayman Chemical Company (Ann Arbor, MI).
Clonogenic survival assay
To measure cellular sensitivity to cytotoxic therapy such radiation and chemical treatments, clonogenic survival assay was performed according to an established protocol (44). Briefly, the cells were treated with mitomycin C at different concentration for 72 hours or irradiated with different doses of X rays. They were then plated in triplicate 10 cm dishes at different numbers according to mitomycin C concentration or radiation doses so that there would be 50 200 colonies form eventually in each well. After colonies are clearly visible (about 11 to 14 days), cells were fixed and stained with 0.5% crystal violet. Colony numbers are then counted and the surviving fractions were calculated according to the number of initially plated cell numbers and the clonogenicity (calculated as % of colonies formed from all inoculated cells) of untreated control cells. In some cases, cells were treated with caspase 3 inhibitor Z DEVD FMK at 15 μM for 4 hours and subsequently exposed to different concentrations of mitomycin C or different doses of X rays, and then incubated in the culture medium containing 1μM caspase 3 inhibitor Z DEVD FMK or vehicle for 14 days.
Soft agar colony formation assay
To measure the ability of cells to growth in 3D, soft agar assay was performed according to an established protocol.14 Cells were seeded at a density of 500 cells per well in 6 well plates in triplicate. After growth in the plates for 21 days, the colonies were fixed and stained with 0.005% crystal violet. Images of each well were taken and the number of colonies per well were counted.
Radiation exposure
Cells or mice were irradiated by use of a Precision X RAD 320 (Precision X Ray, Inc) machine operating at 320 kVp and 12.5 mA. During irradiation, a lead shield protected the mice’s vital organs while the hind limb is the only part of the mouse that was exposed.
Transwell migration and invasion assay
For transwell migration and invasion assay, 5×104 and 1×105 cells were suspended, respectively in 200μl serum free medium for 30 min, and then added to the upper chambers (Falcon™ Cell Culture Inserts, Corning, Inc.). Medium containing 10% FBS was added to the lower chambers (24 well plates, Corning, Inc.). After incubation for 24 hours (HCT116) or 40 hours (HT29) at 37°C in a humidified atmosphere with 5% CO2, cells were then fixed with 4% paraformaldehyde in PBS and stained with 1% crystal violet. The upper surfaces of the filters were scraped five times with cotton swabs to remove non migrated cells. The experiments were repeated in triplicate wells, and the migrated cells were counted microscopically in five randomly selected fields per filter under the microscope.
Scratch assay
Scratch assay was performed with slight modifications according to previous protocol.15 In brief, cells were seeding into 6 well plates at 2× 106 cells/well. Incubate the plates at 37 °C for 6 hours to allowing cells to adhere and spread completely. Use a P200 pipet tip to create a straight line in the cell monolayer. Wash the plates 3 times with PBS and replace with serum free medium. Observe the cells under a microscope and take pictures for further analysis. For each image, distances between one side of scratch and the other were measured.
Cell adhesion assay
CytoSelect™ 48 well cell adhesion assay (ECM Array, Colorimetric Format, Cell Biolabs Inc., San Diego, CA, USA) was performed according to manufacturer’s instructions. In brief, 5×104 cells were suspended in serum free medium, added into the wells (pre coated with adhesive substrates) and then incubated at 37˚C for 90 min in a cell culture incubator. After 5 times wash with PBS, adherent cells were stained and quantified at OD560 by using a microtiter plate reader (BioTek).
In vivo tumor invasion and metastasis analysis
Luciferase labeled caspase 3 knockout cells and controls cells were generated by infecting the cells with a lentivirus carrying a firefly luciferase gene. The luciferase signal of cultured cells was imaged by adding PBS with D luciferin (Caliper Life Sciences, Hopkinton, MA) at a concentration of 0.15 mg/ml. Single cell colonies exhibited similar luciferase signal were selected and expanded for later use. About 5×106/mice were injected to the tail veins of 6 week old female nude mice (4 mice per group) forin vivo tumor invasion and metastasis analysis. Prior to imaging, mice \ were injected with 150 mg/kg of D luciferin intraperitoneally in 200 μl of PBS and then anesthetized with continuous flow of isoflurane. Imaging of the mice was carried out 10 minutes later by use of the IVIS200 instrument (Caliper Life Sciences, Hopkinton, MA). Mice were imaged on day 0, 12, 20 and 32. Images were taken and analyzed by use of manufacturer supplied software for quantitative data. The body weight change of each mouse was measured every other day. Once any one of the mice’s body weight decreased by 15%, all mice in the group were sacrificed and lung weights of each mouse were recorded.
In vivo tumor growth delay
Tumor cells (1×106 cells in 50 μl PBS) were injected into the hind limb of the nude mice subcutaneously. The mice were then monitored for tumor growth every other day. Tumor sizes were measured by use of a caliper. When tumors reach 5 7 mm in diameter, some of them were exposed to x irradiation (2×8 Gy one day apart).
Immunofluorescence analysis
Cells grown in glass bottom dishes were fixed in 4% paraformaldehyde solution for 20 minutes. After three washes with PBS, cells were permeabilized and blocked with a PBS solution containing 1% Triton X 100, 5% donkey serum and 1% BSA for 1 hour. Thereafter, cells were incubated with primary antibody overnight at 4°C, followed by incubation with Alexa Fluor 488 conjugated secondary antibody for 1 hour at RT. After three washes with PBS, the stained cells were mounted with mounting medium (Vector Laboratories, CA) containing DAPI. Images were captured by use of confocal microscopy.
Antibodies
The caspase 3 (full length, clone 8G10) antibody and EMT antibody sampler Kit #9782 were purchased from Cell Signaling Technology.
Statistical analysis
Data were presented as mean ± SEM. One way ANOVA was performed for multiple group comparisons and comparisons between two groups were conducted using the LSD method (plating efficiencies, soft agar assay, growth curve, clonogenic survival assay, transwell migration and invasion assay). Statistical significance was also determined by paired t tests (In vivo tumor invasion and metastasis analysis, scratch assay) and log rank tests (survival curve). A P value less than 0.05 was considered statistically significant.
Results
Reduced in vitro tumorigenic abilities of HCT116-CASP3KO colon cancer cells
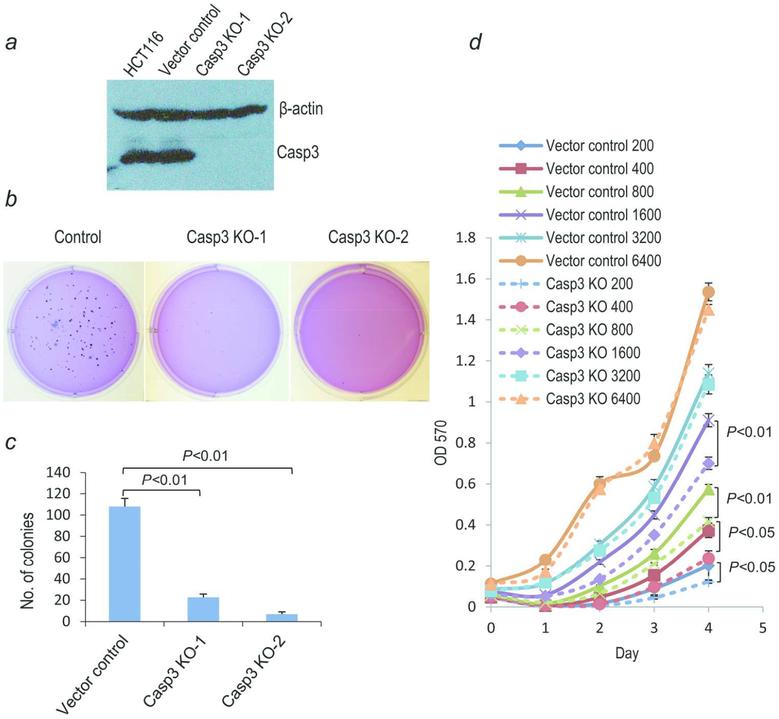
To investigate the biological roles of caspase 3 in colon cancer cells, we attempted to establish CASP3 knockout HCT116 cells by use of the lentivirus based CRISPR/Cas9 technology.13 HCT116 cells were infected with a lentivirus encoding Cas9 and sgRNAs targeting the caspase 3 gene. Individual clonal populations were then screened by Western blot analysis for caspase 3 expression. Two independent clones without caspase 3 expression were obtained (Fig. 1a). Sequencing results showed that the start codon ATG was deleted in both of the clones (Supporting Information Fig. S1). When we examined the effects of caspase 3 gene deficiencies on the tumorigenic abilities of the HCT116 cells in vitro by use of the soft agar assay, which measures the anchorage independent growth abilities of cells unique to cancer cells, our results indicated that CASP3 knockout cells had significantly reduced soft agar colony forming abilities (Fig. 1b, c) when compared with vector control cells. There were only less than 20 colonies per well on average in caspase 3 knockout cells (out of 500 cells plated) while more than 100 colonies/per well were observed in vector control cells. In order to determine if the growth properties were influenced by cellular density, we evaluated the cellular growth at different initiating cellular densities. When cells were seeded at high densities, the growth rates of CASP3 knockout cells and control cells were almost equal (Fig. 1d). However, CASP3 knockout cells grew more slowly than control cells when cells were plated at low densities (Fig. 1d). These results therefore showed that Caspase 3 plays important roles in promoting the tumorigenic abilities of colon cancer cells in vitro, especially at lower densities. It is interesting to note that soft agar colonies were also plated at very low cell densities. However, the difference observed in 3D soft agar colony assay (Fig. 1b,c) were much bigger than those observed for growth in 2D, suggesting that additional biological properties such as invasion and the ability to cut through extracellular matrix is also affected in CASP3KO cells. We also did experiments to determine if CASP3 deficiency caused any changes cellular abilities to adhere to matrix or Petri dish surface. Our resutls (Supporting Information Fig. S3) indicate no such differences exist. Furthermore, there was no difference in apoptosis rate between CASP3KO cells and vector control cells (Supporting Information Fig. S4).
Fig. 1. Casp-3 knockout and its effect on cell growth and soft agar colony forming abilities of HCT116 cells.
(a) Western blot analysis showing caspase 3 gene knockout in two clonal populations of HCT116. Vector control indicates HCT116 cells transduced with the lentiCRISPR v2 vector. (b) Photographic images of partial 6 well plates in which soft agar colony growth assays were carried out for control and CASP3KO HCT116 cells. (c) Quantitative estimates of the average number soft agar colonies per well for control and CASP3KO HCT116 cells. *P<0.01, Students’ t test, n=3. Error bars represent standard error of the mean. (d) Growth curves of control and Casp3 KO 1 HCT116 cells. Different number of cells were seeded into 96 well plates and then stained with MTT assay to measure cellular growth. N=3, Student’s t test. Error bars represent standard error of the mean.
Increased sensitivities to radiotherapy and chemotherapy in CASP3 knockout cells
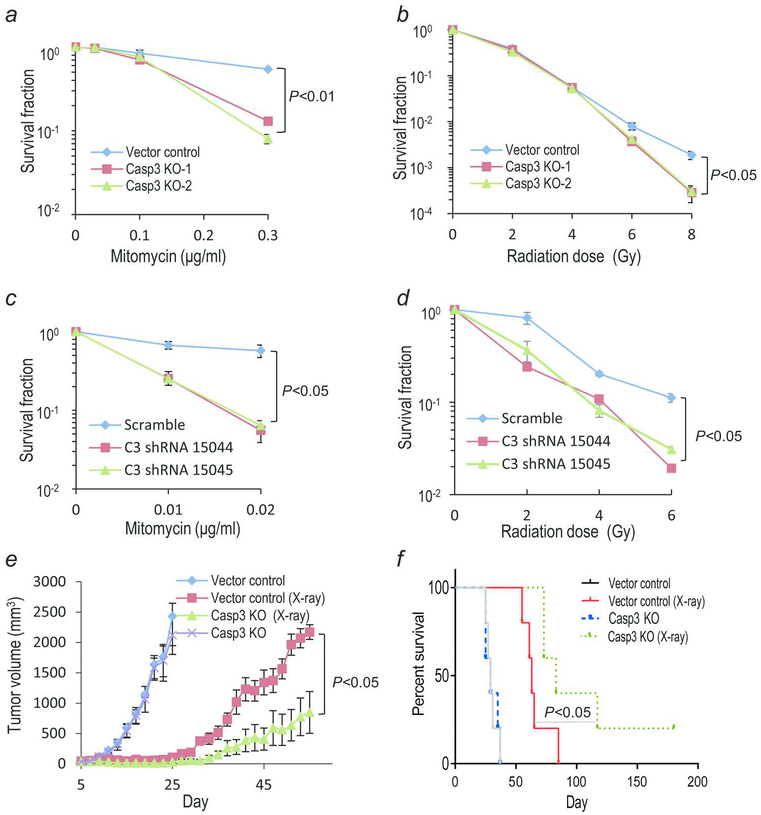
Chemotherapy and radiotherapy are commonly used to treat patients with colon cancer. To understand the function of caspase 3 during chemotherapy and radiotherapy, we first exposed CASP3KO HCT116 cells to anticancer drug mitomycin C at different concentrations. We then determined the clonogenic survival of the cells in Petri dish. Our results showed caspase 3 knockout cells were more sensitive to mitomycin C, especially when the concentration of mitomycin C was high (Fig. 2a). A similar phenotype was observed in colony forming assays conducted for cells exposed to X rays. Besides the lower clonogenicity (number of colonies formed from 100 seeded cells) of CASP3KO cells when compared with the vector control cells (Supporting Information Fig. S2), CASP3KO cells were significantly more sensitive to radiation than vector control cells only at high radiation doses (Fig. 2b). In order to verify these results in other cell lines, we established CASP3 knockdown HT29 cell line and a CASP3KO MDA MB 231 cell line (Supporting Information Fig. S5a,b). Consistent with results obtained in HCT116CASP3KO cells, enhanced radiation sensitivities could be observed in the CASP3KO MDA MB 231 cells (Supporting Information Fig. S5c). Furthermore, CASP3 knockdown HT29 cells were more sensitive to mitomycin C and X ray radiation (Fig. 2c, d)(Supporting Information Fig. S6). In a subsequent experiment, we treated the HCT116 cells with Caspase 3 inhibitor Z DEVD FMK and then exposed to different concentrations of mitomycin C or different doses of X rays. The results of clonogenic assay, as shown in Supporting Information Fig. S7, indicated that caspase 3 inhibitor could sensitize the cells to mitomycin C and X ray radiation. These results were very similar to those observed for CASP3KO HCT116 cells.
Fig. 2. Chemo- and radiation-sensitivities of CASP3KO HCT116 & shCASP3 HT29 colon cancer cells.
(a) Clonogenic assay results for vector control and CASP3KO HCT116 cells exposed to different concentrations mitomycin C. (b) Clonogenic assay results for vector control and CASP3KO HCT116 cells exposed to different doses of X rays. (c and d) Clonogenic assay results of control and HT29shCASP3 cells treated with different concentrations of mitomycin C (c) and different doses of X rays (d). (e)Tumor growth from control and CASP3KO HCT116 cells inoculated (at 1×10 cells/mouse) in nude mice with or without radiotherapy (2×8Gy of X rays). P<0.05, n=5 per group, Student’s t test. Error bars represent SEM. (f)Kaplan Meier survival curve of mice inoculated with control and CASP3KO HCT116 cells and treated with or without X rays, *P<0.001, log rank test, n=5. Lack of survival was defined as tumor volume ≥2,000 mm3 or when mice were sacrificed because they are moribund.
We next evaluated the growth rates of control and CASP3KO cells in vivo. Subcutaneously injected vector control and CASP3KO HCT116 cells formed tumor in nude mice at almost equal rates under non treated condition (Fig. 2e). However, radiotherapy caused significantly more growth delay in CASP3KO HCT116 tumors (Fig. 2e). These results indicate that caspase 3 gene plays a significant role in tumor response to radiotherapy. It is worth noting that CASP3KO caused significant attenuation in soft agar colony growth but makes no difference in in vivo tumor growth in the absence of radiotherapy (Fig. 1b, c vs Fig. 2e). The underlying reason is not entirely clear. We reason that it might be caused by the effects of cellular densities, similar to those observed for growth assay (Fig. 1d). Our previous data4, 16 showed that caspase 3 could mediate the production of prostaglandin E2 (PGE2), a growth regulator that increases tumor growth in many types of cancers, including colon cancer17–19. In order to determine if CASP3KO caused difference in PGE2 production, we measured PGE2 levels in supernatants from control and irradiated HCT116 cells. Our results indicate the significantly reduced level of PGE2 was found in HCT116 CASP3KO cells when compared with the control cells under both non irradiated and irradiated conditions (Supporting Information Fig. S8). Therefore, it is quite possible that the reduced level of PGE2 and other growth factors in CASP3KO cells is compensated at higher cellular densities in mice, where 1 million cells were injected per mouse. Another interesting observation from the experiments is that radiotherapy enabled mice bearing CASP3KO tumors to survive (Lack of survival was defined as death of host or tumor volume ≥2,000 mm3) significantly longer than those bearing control tumors (Fig 2f). Furthermore, two of the five mice bearing CASP3KO tumors treated with radiotherapy survive beyond 100 days while none of the mice bearing control tumors did (Fig. 2f).
Reduced invasiveness and metastatic potential of CASP3KO cells
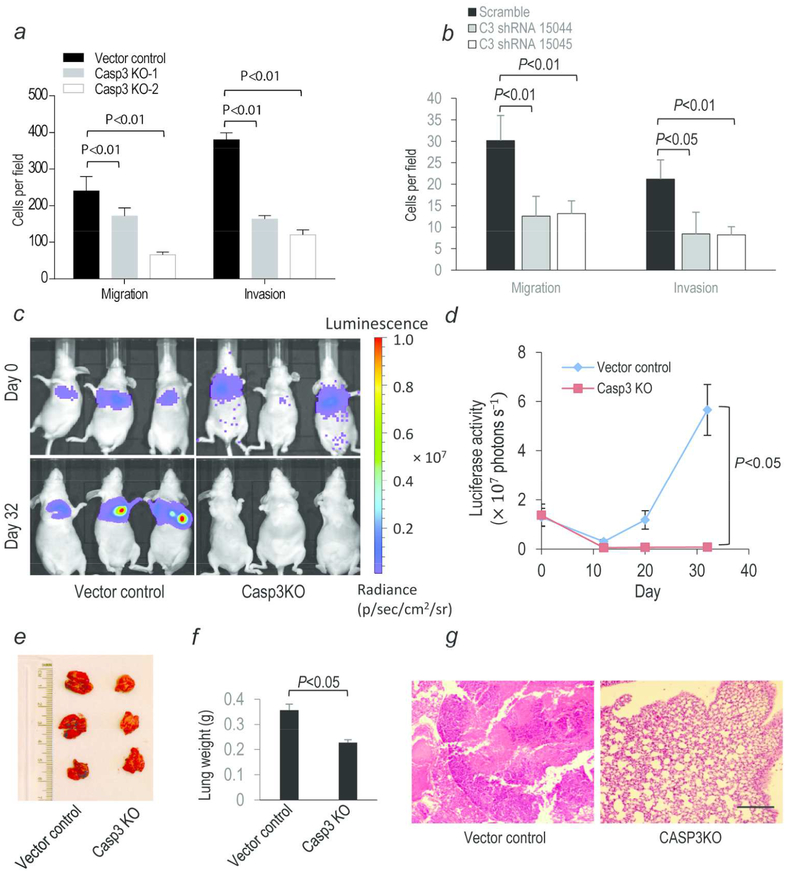
To investigate the role of caspase 3 in regulating cancer cell migration and invasion, we conducted migration and invasion assays by use of transwell plates. Our results showed that HCT116 CASP3KO or HT29 CASP3 knockdown cells possessed significantly less migration and invasion capacities (Fig. 3a, b and Supporting Information Fig. S9a, b). In addition, HCT116 CASP3KO they also showed significantly reduced migration ability in scratch assays (Supporting Information Fig. S10a, b). To validate our in vitro results, we established luciferase labeled CASP3KO and control cells. In vitro, the two cell lines have almost identical luciferase activities (Supporting Information Fig. S11). In addition, their luciferase activities showed a linear relationship with cell numbers (Supporting Information Fig. S11). The labeled cells were then intravenously injected into nude mice and observed for their abilities to establish metastasis by use of an in vivo optical imaging system. Our results showed that despite very similar luciferase activities right after tumor cell injection (day 0), the abilities of CASP3KO cells to establish lung metastases were significantly reduced compared with control cells (Fig. 3c, d). In fact, we couldn’t detect any luciferase signals in mice injected with CASP3KO cells 32 days after the tail vein injection while in mice injected with control cells, luciferase activities showed a steady increase from day 12 to day 32 (Fig 3c, d). Upon mouse sacrifice and examination at 32 days post inoculation, lungs of control group mice were full of metastatic nodules while those of CASP3KO mice had no nodules at all (Fig. 3e). Lung weights of control group mice were 50% higher than that of CASP3KO group mice (Fig. 3f).
Fig. 3. Invasiveness and metastasis of control and CASP3KO HCT116 cells in vitro and in vivo.
(a, b) Quantitative estimate of transwell migration and invasion assay. Each data point represents the average of five randomly selected fields. (c)In vivo metastases of control and CASP3KO HCT116 cells following trail vein inoculation of Luc labeled cells. (d)Quantitative estimates of the metastasis as measured by luciferase activities in control and CASP3KO HCT116 cells. N=3. (e)Presence of pulmonary metastatic nodules in nude mice 32 days after tail vein injection of the tumor cells. (f) Average weights of lungs from mice (n=3) injected with control or CASP3KO cells. Error bars represent standard error of the mean. In a, b, d, f, error bars represent standard error of the mean (SEM). (g) H&E staining of lung tissues taken from nude mice subcutaneously injected with either vector control (left panel) or CASP3KO (right panel) HCT116 cells. Scale bar represents 1 mm. Tumor tissue clearly visible in lung tissues from mice injected with control tumor cells.
Independently, reduced invasiveness and metastatic potential of CASP3KO cells were also observed in mice with subcutaneously established local tumors (mice from Fig. 2e, f). After radiotherapy, none (0 of 5) of the irradiated CASP3KO tumor bearing mice developed pulmonary metastasis while 60% (3 of 5) of irradiated control tumor bearing mice developed pulmonary metastasis when the lungs of the mice were visually examined upon their sacrifice (when they become morbid or when their tumors reach 2000 mm3 or bigger, whichever comes first). Furthermore, lung metastasis in the control tumor bearing mice was confirmed by H&E staining ((Fig. 3g). These results support a significant role for caspase 3 in mediating the metastatic abilities of colon cancer cells in vivo.
Roles of caspase-3 in mediating epithelial-to-mesenchymal transition (EMT) in colon cancer cells
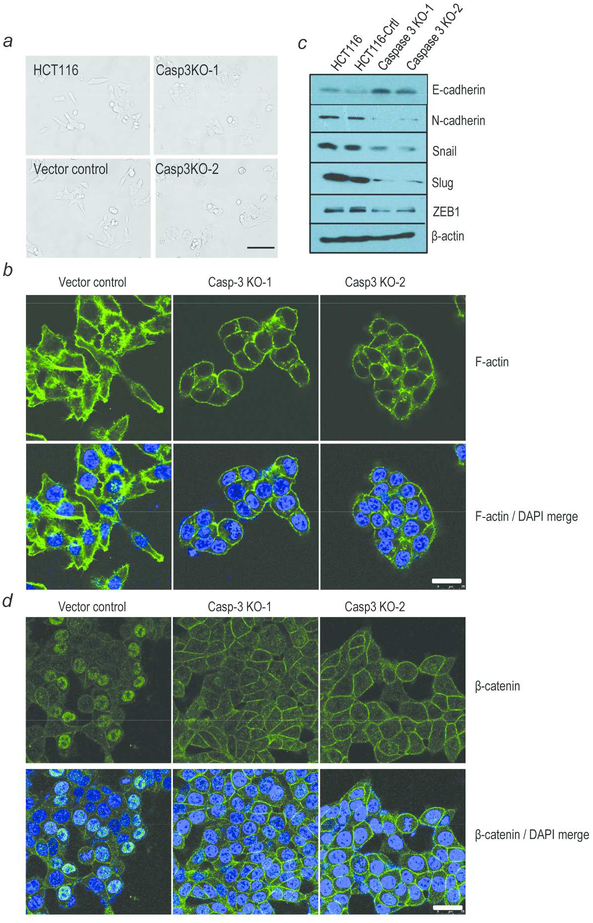
What are the mechanisms involved in CASP3 mediated cancer cell migration and invasion? Under the microscope, HCT116 CASP3KO cells showed altered cellular morphology with decreased lamellipodia and substantial reduction in cellular spreading when compared with the control cells (Fig 4a). The morphology observations were further confirmed with phalloidin staining of F actin (Fig. 4b).
Fig. 4. Characterization of EMT marker expression CASP3KO and control HCT116 cells.
(a) Morphology of parental, vector control, and CASP3KO HCT116 cells. Scale bar: 50 μm. (b) Vector control and CASP3KO HCT116 cells were stained with phalloidin to detect F actin and with DAPI to mark nuclei. Scale bar: 25 μm. (c) Western blot analysis of known proteins involved in EMT transition. (d) Immunofluorescence staining of β catenin in vector control and CASP3KO HCT116 cells. scale bar: 50 μm.
The observed morphology changes in HCT116 CASP3KO cells provided clues that Caspase 3 may contribute to epithelial to mesenchymal transition (EMT). To determine if this is true, we examined the EMT marker expression in control and CAPS3KO cells. Western blot results verified our speculation. Compared with control cells, epithelial marker E cadherin was expressed at higher levels in CASP3KO cells (Fig 4c). In comparison, mesenchymal marker N cadherin, snail, slug and ZEB1 were expressed at significantly reduced levels in CASP3KO cells (Fig 4c). Immunofluorescence assay further demonstrated CASP3 knockout cells with higher levels of E cadherin expression (Supporting Information Fig. S12). Furthermore, compared with the control cells, a reduction of beta catenin expression in the nuclei and an increase in the membrane protein location were detected by immunofluorescence (Fig. 4d). Activation and nuclear migration of β catenin has been reported to be another indication of EMT transition20, 21. Independently, upregulated epithelial marker (E cadherin) and downregulated mesenchymal markers (N cadherin, snail, slug and ZEB1) were also detected in CASP3 shRNA HT29 cells when compared with the scramble control (Supporting Information Fig. S13). Taking together, our results therefore suggest a role for caspase 3 in mediating an EMT phenotype in HCT116 and HT29 colon cancer cells
Discussion
Increased caspase 3 activities are generally considered as a sign of apoptosis and a positive indicator of efficacy in cancer treatment. Recently, there is increasing evidence that caspase 3 promotes stress induced cancer cell growth, cellular migration, invasiveness, and tumor angiogenesis.5, 22–24 In human cancer treatment, several studies reported that patients with higher levels procaspase 3 or active caspase 3 had worse prognosis than patients with lower levels of procaspase 3 or active caspase 3.7, 25–27 In this study, we explored the function of caspase 3 in colon cancer cells by comparing CASP3 knockout cells with vector control cells.
Our results from this study are consistent with the growing body of literature that supporting the non apoptotic roles of “apoptotic” caspases in mammalian biology. Indeed, in cancer biology, apoptosis may not be a dominant way of cell death even after cytotoxic therapy.28 This is in contrast to leukemia or lymphoma cells, where apoptosis plays a prominent role in tumor cell response to treatment. However, the lack of cell death through apoptosis in solid tumors does not mean that apoptotic caspases play no role in solid tumor biology. In contrast, caspases appear to play a significant number of non canonical roles in solid tumor biology.
Although CASP3 knockout cells are less tumorigenic in soft agar assays when compared with the control cells, their growth rates in vitro in 2D and in vivo are not affected when cells seeded or injected at high densities. Impaired growth of CASP3 knockout cells only found when cells seeded in low densities. Those observations are consistent with reduced PGE2 production in CASP3KO cells, which either alone or in combination with other growth factors may be responsible for the observed discrepancies between high and low density knockout cells. Furthermore, CASP3 knockout cells are more sensitive to mitomycin C and radiation than vector control cells, results that go against the conventional wisdom that activation of caspase 3 is good for killing cancer cells. Those in vitro results were further confirmed by in vivo experiments where CASP3KO cell derived tumors showed increased growth delay to radiotherapy. The most striking results were perhaps those demonstrating caspase 3 ablation completely abolished the abilities of HCT116 cells to metastasize to the lung, thereby demonstrating an important role for caspase 3 in colon cancer that has not been revealed before.
Our findings on the role of caspase 3 in metastasis of HCT116 colon cancer cells are important because metastasis accounts for the majority of failures in colon cancer treatment. At the mechanistic level, we showed that caspase 3 is involved in regulating EMT. EMT plays a major role in basal membrane degradation, cell migration and survival of invasion cells in a new environment.29 When the cells undergo EMT, they may change the morphology, lose their cell–cell junctions while retaining expression of migration promoting molecules.29 Our experiments clearly show that CASP3KO cells lost key EMT features that were prominent in parental HCT116 cells. These findings provide a rational explanation on why the loss of caspase 3 causes HCT116 colon cancer cells to lose their abilities to metastasize. Since caspase 3 promotes colon cancer resistance to radiotherapy and chemotherapy as well as colon cancer cells invasion and metastasis, combining chemotherapy or radiotherapy with caspase 3 targeted agents may thus be a promising approach for colon cancer treatment.
Supplementary Material
Novelty & Impact Statements:
Caspase 3 has long been established as an “executioner” caspase whose main function is to eliminate damaged cells. Thus it is generally perceived as an beneficial factor in cancer therapy. In this study, we found that, contrary to the established paradigm, Caspase 3 plays important roles in promoting colon cancer cell invasion and metastasis and can therefore serve as a potential target for colon cancer therapy.
Acknowledgements:
We thank Dr. Feng Zhang (MIT) for making their CRISPR/Cas9 plasmids available through Addgene (Cambridge, MA). This work was supported in part by National Natural Science Foundation of China grants (81472573 M. Zhou, 81120108017 and 81572951 to Q. Huang) and Shanghai Municipal Health Bureau project 20134043 (M.Z.). It was also supported in part by grants CA208852, CA216876, and ES024015 from the US National Institutes of Health (C. Li);
Abbreviations:
- CASP3
Caspase 3
- KO
Knockout
- PGE2
Prostaglandin E2
- EMT
Epithelial Mesenchymal Transition
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Eckhart L, Ballaun C, Uthman A, Kittel C, Stichenwirth M, Buchberger M, Fischer H, Sipos W, Tschachler E. Identification and characterization of a novel mammalian caspase with proapoptotic activity. J Biol Chem 2005;280: 35077–80. [DOI] [PubMed] [Google Scholar]
- 2.Galluzzi L, Lopez Soto A, Kumar S, Kroemer G. Caspases Connect Cell Death Signaling to Organismal Homeostasis. Immunity 2016;44: 221–31. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, He Y, Li F, Huang Q, Kato TA, Hall RP, Li CY. Caspase 3 promotes genetic instability and carcinogenesis. Mol Cell 2015;58: 284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Q, Li F, Liu X, Li W, Shi W, Liu FF, O’Sullivan B, He Z, Peng Y, Tan AC, Zhou L, Shen J, et al. Caspase 3 mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med 2011;17: 860–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng X, Yu Y, He S, Cheng J, Gong Y, Zhang Z, Yang X, Xu B, Liu X, Li CY, Tian L, Huang Q. Dying glioma cells establish a proangiogenic microenvironment through a caspase 3 dependent mechanism. Cancer Lett 2017;385: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtova AV, Xiao J, Mo Q, Pazhanisamy S, Krasnow R, Lerner SP, Chen F, Roh TT, Lay E, Ho PL, Chan KS. Blocking PGE2 induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 2015;517: 209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flanagan L, Meyer M, Fay J, Curry S, Bacon O, Duessmann H, John K, Boland KC, McNamara DA, Kay EW, Bantel H, Schulze Bergkamen H, et al. Low levels of Caspase 3 predict favourable response to 5FU based chemotherapy in advanced colorectal cancer: Caspase 3 inhibition as a therapeutic approach. Cell Death Dis 2016;7: e2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017. [DOI] [PubMed] [Google Scholar]
- 9.Manfredi S, Bouvier AM, Lepage C, Hatem C, Dancourt V, Faivre J. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg 2006;93: 1115–22. [DOI] [PubMed] [Google Scholar]
- 10.Qiu M, Hu J, Yang D, Cosgrove DP, Xu R. Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget 2015;6: 38658–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger AC, Sigurdson ER, LeVoyer T, Hanlon A, Mayer RJ, Macdonald JS, Catalano PJ, Haller DG. Colon cancer survival is associated with decreasing ratio of metastatic to examined lymph nodes. J Clin Oncol 2005;23: 8706–12. [DOI] [PubMed] [Google Scholar]
- 12.Bartlett DL, Chu E. Can metastatic colorectal cancer be cured? Oncology (Williston Park) 2012;26: 266–75. [PubMed] [Google Scholar]
- 13.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR Cas9 system. Nature Protocols 2013;8: 2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cifone MA, Fidler IJ. Correlation of patterns of anchorage independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc Natl Acad Sci U S A 1980;77: 1039–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2007;2: 329–33. [DOI] [PubMed] [Google Scholar]
- 16.Donato AL, Huang Q, Liu X, Li F, Zimmerman MA, Li CY. Caspase 3 promotes surviving melanoma tumor cell growth after cytotoxic therapy. J Invest Dermatol 2014;134: 1686–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rundhaug JE, Simper MS, Surh I, Fischer SM. The role of the EP receptors for prostaglandin E2 in skin and skin cancer. Cancer Metastasis Rev 2011;30: 465–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou PC, Li YH, Lin SC, Lin SC, Lee JC, Lin BW, Liou JP, Chang JY, Kuo CC, Liu YM, Sun HS, Tsai SJ. Hypoxia Induced Downregulation of DUSP 2 Phosphatase Drives Colon Cancer Stemness. Cancer Res 2017;77: 4305–16. [DOI] [PubMed] [Google Scholar]
- 19.Fang M, Li Y, Huang K, Qi S, Zhang J, Zgodzinski W, Majewski M, Wallner G, Gozdz S, Macek P, Kowalik A, Pasiarski M, et al. IL33 Promotes Colon Cancer Cell Stemness via JNK Activation and Macrophage Recruitment. Cancer Res 2017;77: 2735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz Schughart LA, Knuechel R, Kirchner T. Variable beta catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A 2001;98: 10356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He X Unwinding a path to nuclear beta catenin. Cell 2006;127: 40–2. [DOI] [PubMed] [Google Scholar]
- 22.Mukai M, Kusama T, Hamanaka Y, Koga T, Endo H, Tatsuta M, Inoue M. Cross talk between apoptosis and invasion signaling in cancer cells through caspase 3 activation. Cancer Res 2005;65: 9121–5. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Wang D, Zhao Z, Xiao Y, Sengupta S, Xiao Y, Zhang R, Lauber K, Wesselborg S, Feng L, Rose TM, Shen Y, et al. Caspase 3 dependent activation of calcium independent phospholipase A2 enhances cell migration in non apoptotic ovarian cancer cells. J Biol Chem 2006;281: 29357–68. [DOI] [PubMed] [Google Scholar]
- 24.Lauber K, Bohn E, Krober SM, Xiao YJ, Blumenthal SG, Lindemann RK, Marini P, Wiedig C, Zobywalski A, Baksh S, Xu Y, Autenrieth IB, et al. Apoptotic cells induce migration of phagocytes via caspase 3 mediated release of a lipid attraction signal. Cell 2003;113: 717–30. [DOI] [PubMed] [Google Scholar]
- 25.Estrov Z, Thall PF, Talpaz M, Estey EH, Kantarjian HM, Andreeff M, Harris D, Van Q, Walterscheid M, Kornblau SM. Caspase 2 and caspase 3 protein levels as predictors of survival in acute myelogenous leukemia. Blood 1998;92: 3090–7. [PubMed] [Google Scholar]
- 26.Faderl S, Thall PF, Kantarjian HM, Talpaz M, Harris D, Van Q, Beran M, Kornblau SM, Pierce S, Estrov Z. Caspase 2 and caspase 3 as predictors of complete remission and survival in adults with acute lymphoblastic leukemia. Clin Cancer Res 1999;5: 4041–7. [PubMed] [Google Scholar]
- 27.Jonges LE, Nagelkerke JF, Ensink NG, van der Velde EA, Tollenaar RA, Fleuren GJ, van de Velde CJ, Morreau H, Kuppen PJ. Caspase 3 activity as a prognostic factor in colorectal carcinoma. Lab Invest 2001;81: 681–8. [DOI] [PubMed] [Google Scholar]
- 28.Brown JM, Wouters BG. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res 1999;59: 1391–9. [PubMed] [Google Scholar]
- 29.Friedl P, Wolf K. Tumour cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003;3: 362–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.