Abstract
Eukaryotic cells are known to contain a wide variety of RNA-protein assemblies, collectively referred to as RNP granules. RNP granules form from a combination of RNA-RNA, protein-RNA and protein-protein interactions. In addition, RNP granules are enriched in proteins with intrinsically disordered regions (IDRs), which are frequently appended to a well folded domain of the same protein. This structural organization of RNP granule components allows for a diverse set of protein-protein interactions including traditional structured interactions between well folded domains, interactions of short linear motifs (SLiMs) in IDRs with the surface of well folded domains, interactions of short motifs within IDRs that weakly interact with related motifs, and weak interactions involving at most transient ordering of IDRs and folded domains with other components. In addition, both well-folded domains, and IDRs, in granule components frequently interact with RNA, and thereby can contribute to RNP granule assembly. We discuss the contribution of these interactions to liquid-liquid phase separation and the possible role of phase separation in the assembly of RNP granules. We expect that these principles also apply to other non-membrane bound organelles and large assemblies in the cell.
Introduction
RNP granules are a ubiquitous feature of eukaryotic cells. Nuclear RNP granules include the nucleolus, paraspeckles, and Cajal bodies [1]. In the cytoplasm, common RNP granules are P-bodies, which contain untranslating mRNAs and components of the translation repression and mRNA degradation machinery, and stress granules, which form from untranslating mRNAs and contain some RNA binding proteins and translation factors [2,3]. Related granules also exist in oocytes and embryos, where they play a role in sequestering maternal mRNAs [4], and in neurons [5], where such neuronal RNP granules can affect at least some forms of synaptic plasticity [6].
RNP granule assembly occurs by a combination of protein-protein, protein-RNA and RNA-RNA interactions. Several lines of evidence argue that intermolecular RNA-RNA interactions promote RNP granules assembly [7]. For example, the stress granule transcriptome can be largely recapitulated through RNA self-assembly in vitro [8], specific basepairing between the OSCAR or BICOID mRNAs can target those mRNAs into maternal RNP granules in Drosophila [9,10], diverse RNAs are effective at self-assembly in vitro [8,11], and aspects of RNA structure can affect the specificity of mRNA targeting to RNP granules in Ashbya gossypii. [12]
RNA-RNA interactions appear to act in summation with protein-protein interactions to promote RNP granule assembly [7,8]. Evidence for protein interactions promoting granule formation has come from genetic experiments. For example, the Edc3 and Lsm4 proteins are important in the assembly of yeast P-bodies [13], the G3BP protein promotes the assembly of mammalian stress granules [14,15], and the Meg proteins are involved in the formation of P-granules in C. elegans [16]. Protein-RNA interactions also play an important role in RNP granule formation by coupling the assembly of RNA binding proteins to the self-assembly of RNA. In this review, we focus on the manners by which protein-protein interactions affect RNP granule assembly.
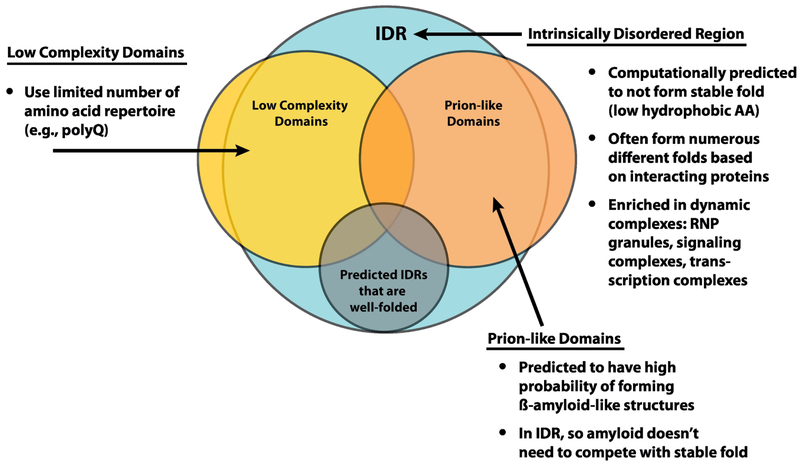
An interesting feature of RNP granules is that many of their components contain intrinsically disordered regions (IDRs), which are regions of polypeptide chains that do not adopt a unique folded structure but interconvert between many non-globular conformations. Explicit experimental data shows the disordered character for some of these protein regions in vitro [17–21], and many more are computationally predicted to be disordered. Some IDRs are also referred to as low-complexity domains since IDRs typically are reduced in hydrophobic residues and sometimes show a strong bias in amino acids composition. IDRs also frequently contain amino acid sequences predicted to have a propensity to adopt cross-beta structures analogous to the protein folds seen in prions, and as such many RNP granule proteins also contain prionlike domains. Thus, IDRs, low-complexity domains, and prion-like domains form a set of interrelated protein domains (Figure 1). It remains possible that some polypeptides predicted to be disordered by computational methods may be reasonably well-folded. The ability of IDRs to intercovert between many relatively weakly folded structures allows for the surfaces of binding partners to provide interactions that mediate binding dependent folding of the IDR [22]. However, it should be noted that order-to-disorder transitions mediated by binding have also been described [23]. Eventually, we will need context-dependent disorder predictors. Throughout this review, we will generally refer to intrinsically disordered regions of proteins as IDRs.
FIGURE 1:
Schematic to show overlap of intrinsically disordered proteins, defined by computational predictions, low complexity domains, defined by a skewed representation of amino acid composition, and prion-like domains, which are computationally predicted to have high probability of forming cross β-sheet structures based on similarities to known prion domains. The size of circles and overlapping regions do not indicate exact proportions.
The IDRs of RNP granule components are of interest for three broad reasons. First, genetics has shown that IDRs of RNP granule components can contribute to RNP granule assembly. For example, while the wild-type TIA1 will trigger stress granule assembly as assessed by multiple markers when over-expressed, a variant lacking the prion-like IDR of TIA1 fails to do so [24]. Similarly, in strains partially defective in P-body assembly, deleting the C-terminal IDRs of the yeast Lsm4 or Dhh1 proteins decreases P-body assembly [13,25]. Moreover, in all three cases these IDRs can be replaced with some other IDRs suggesting the ability to promote RNP granule assembly is a property conferred by multiple IDRs [13,24,26]. Thus, IDRs can play a role in RNP granule assembly.
IDRs have drawn a lot of attention with regards to granule assembly since many IDRs have been seen to undergo various forms of self-assembly in vitro. For example, IDRs from many RNA binding proteins in RNP granules are able to undergo liquid-liquid phase separation in vitro and thereby form a second separate phase in solution that might be similar to liquid-like RNP granules [17,27–30]. Similarly, IDRs, either with prolonged incubation, or at high local concentrations within a liquid dense phase, can transition to an amyloid-like or fibrillar assembly, or dense phases can gel [27,28,30–32]. Although how these particular self-assembly reactions relate to their roles in RNP granule assembly is debated [19,29,33–35], the observation that the IDRs of RNP granule components can undergo self-assembly is an intriguing correlation with their in vivo formation of RNP granules and may explain at least part of the mechanisms by which IDRs can contribute to RNP granule assembly.
The IDRs of RNP granule components are also of interest since multiple mutations in these regions contribute to degenerative diseases. For example, mutations in the IDRs of TDP-43, TIA1, hnRNPA1, or hnRNPA2B1 can be causative in various muscular or neurodegenerative diseases [36]. Interestingly, in these cases the mutations typically cause the proteins to hyper-aggregate or lead to increased rates of fibrilization in vitro [27,28,30,37,38], suggesting a model where increasing the ability of these IDRs to form a prion-like structure can lead to degenerative disease [39,40].
Herein, we discuss how IDRs, folded domains and RNA synergize in the assembly of dynamic RNP granules, which protein architectures are particularly suitable for mediating these types of interactions, and how the interactions may be altered in pathogenic conditions.
Protein-Protein interactions in RNP granule assembly
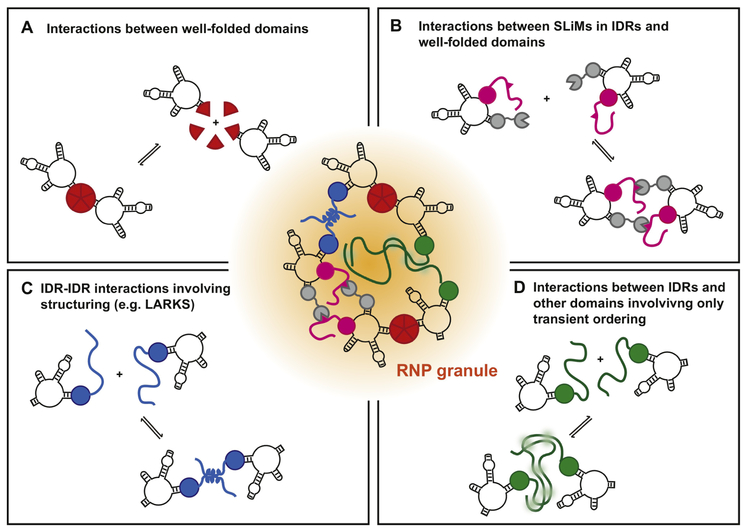
Protein-protein interactions contribute to RNP granule formation in several modes (Figure 2). One manner by which protein-protein interactions contribute to RNP granule assembly is through structured interactions between well-folded protein domains (Figure 2a). For example, formation of G3BP dimers is important for stress granule assembly in mammals [14]. In another example, point mutations in the dimerization interface of the yeast Edc3 protein prevent its ability to promote P-body assembly [41]. Similarly, the N-terminus of coilin forms an oligomerization structure that promotes Cajal body formation [42].
FIGURE 2:
The figure illustrates the general classes of protein-protein interactions that can contribute to RNP granule assembly including interactions between well-folded domains, SLiMs interacting with the surface of well-folded domains, specific interactions between local structural modules of IDRs such as LARKS, and disordered interactions between IDRs and well-folded domains based on interactions that involve at most transient ordering such as charge-charge, π-π or cation-π interactions. RNA molecules are depicted as black curves, protein molecules as colored shapes and lines. RNA-RNA interactions also play roles in RNP granule assembly and are not depicted explicitly here.
Structured interactions between well-folded protein domains of RNP granule components and short linear motifs (SLiMs), which typically fit specifically into a surface groove of a well-folded protein domain (Davey et al., 2010), can also contribute to RNP granule assembly (Figure 2b). For example, in the assembly of P-bodies in S. pombe a series of SLiMs in the Dcp2 C-terminal domain interact with a surface groove on the Edc3 Lsm domain and contribute to P-body assembly [43]. Such SLiMs are common in facilitating P-body assembly and highlight this mechanism by which IDRs can promote RNP granule assembly [44].
IDRs can also facilitate RNP granule assembly through short stretches of amino acids that form local structures that can self-interact. For example, the C-terminal IDR in the TDP-43 protein has a region that forms a local alpha-helix, which can self-associate and thereby could affect RNP granule assembly [20]. Similarly, many RNP granule components are enriched in short motifs that include aromatic residues. Specific types of such motifs, so-called low-complexity aromatic-rich kinked segments (LARKS) [34], can interact and form connections either within a given protein, or between different proteins with similar LARKS. LARKS can form amyloid-like stacked β-sheet structures, potentially with multiple LARKS in tandem forming a more extensive structure as has been seen with the N-terminal domain of FUS, at least in vitro [45]. Whether pairs of LARKS can interact as discrete dimers and whether multiple LARKS in an IDR can mediate a network of weak interactions that drive the assembly of liquid droplets or RNP granules, is an open question. In conclusion, some regions of IDRs can form local structural elements that can interact with related elements in other RNP granule components (Figure 2c).
Finally, IDRs can interact with themselves or other proteins without resulting in observable structuring [19,21,33] (Figure 2d). These IDRs seem to remain largely disordered even in macroscopic liquid assemblies, at least under in vitro conditions where the structure can be directly assessed. One mechanism by which such interactions could occur would be via π-π or cation-π interactions. Many RNP granule components are rich in amino acids capable of such interactions, which may also explain their sequence bias. These interactions may occur between many types of IDRs, and in some cases even with the surface of well folded domains [46]. This ability of IDRs to mediate weak interactions involving at most transient structural ordering provides a possible explanation for why different IDRs can often functionally substitute for a natural IDR in granule assembly [13,24,26]. This may also explain why IDR phase separation in vitro can be disrupted in the presence of competitor proteins, and other proteins can be recruited into IDR driven dense phases or hydrogels [26,27,31]. How specific or non-specific these merely transiently ordering interactions are is currently unclear; it may be possible to encode a spectrum of specificities in such sequences. Several such interactions may generate specificity collectively, e.g. if they can individually interact with various other sequences but have overlap in their preference for only few sequences. Transiently ordering interactions of IDRs can synergize with structured interactions by being linked to those well-folded domains and providing additional avidity for assembly of the granule (Figure 2d) [26].
The utilization of IDRs has unique advantages for modulating the assembly of large complexes [26]. Because IDR-mediated interactions are not limited to individual components or spatial arrangements, and IDRs can instead have many SLiMs in a short sequence and bridge different spatial arrangements due to their flexibility, they can increase avidity between multiple different subcomplexes within larger assemblies. Similarly, IDRs are preferred targets for post-translational modifications thus allowing their functional modulation by signaling pathways [47].
IDRs on RNA binding proteins and RNP granule components are frequently additional sites of RNA binding. This is demonstrated by the fact that numerous IDRs cross –link directly to RNA in cells [48], and by the biochemical demonstration that numerous purified IDRs can bind RNA in vitro [27,28]. It is not yet clear if IDR binding to RNA involves structuring of the IDR or can occur while maintaining disorder, although basic IDRs of the Mss116 and CYT-19 proteins can bind nucleic acids in multiple conformation suggesting IDRs can be plastic in their interactions with RNA [49]. In any case, the additional binding of IDRs to RNAs provides another source of interactions that can stabilize and/or promote RNP granule assembly.
An important question is, how the individual interactions mentioned above can cooperate to form micrometer-sized RNP granules. To form such large assemblies through typical interactions with defined stoichiometry, these interactions would have to occur at very high affinity and would render the assemblies solid. Large solid assemblies can also be generated through polymerization and kinetic trapping. However, while some RNP granules may have solid cores [50,51], at least their outer shell has liquid character, allowing them to fuse and coalesce. Moreover, RNP granules are dynamic assemblies of proteins, and individual molecules can enter and leave them within seconds, reviewed in [52]. Thus, the formation of RNP granules must be mediated by weak but highly cooperative interactions that give rise to collective assembly properties or be subject to continual remodeling by energy-driven cellular machines.
RNP Granule formation modeled as liquid-liquid phase separation.
A popular, and potentially illuminating, model is to consider RNP granules as arising through liquid-liquid phase separation (LLPS). LLPS is a process in which two liquids demix from each other. In biologically relevant phase separation processes, macromolecules in solution condense into dense liquid droplets. The droplets contain high concentrations of the macromolecule and co-exist with a light phase, which has a low concentration of the macromolecule. LLPS occurs when the interactions of components for each other is sufficiently more energetically favorable than interactions of building blocks with the solvent such that the total free energy of the system is favorable for the demixing [53]. This process depends on the volume fractions of the components and can also depend on environmental conditions such as temperature, pH, salt concentration and the presence of other solutes. LLPS is well known to crystallographers; their attempts to crystallize proteins often result in condensed liquid droplets rather than crystals [54,55], but knowledge of how supersaturation drives nucleation and crystal growth can also be exploited to generate optimal crystallization conditions [56]. In liquid droplets, the protein molecules show no order over longer length scales, whereas in a crystal lattice, the orientation of all molecules to each other is related via defined symmetry operations. Protein molecules, but also nucleic acids such as RNA and DNA, and other biomacromolecules, can self-assemble into solid assemblies (such as crystals, amyloids or amorphous aggregates) and dense liquid assemblies. Notably, these liquid assemblies produced in vitro share some properties with RNP granules in cells. Further, LLPS results from weak multivalent interactions that form highly cooperatively and therefore have the necessary properties to form large, dynamic assemblies as discussed above.
The hypothesis that RNP granules arise by LLPS originally arose from the observation that P granules in the gonads of worms [57] and nucleoli [58] behave like liquid droplets. This liquid-like behavior is also observed for many non-membrane-bound organelles, including stress granules, nuclear speckles, PML bodies and others [28,30,59,60]. The co-existence of two liquid phases, i.e. the co-existence of liquid organelles with the cytoplasm or nucleoplasm, depending on where they are located, is suggestive of LLPS as a mechanism contributing to their formation. Indeed, the growth of certain liquid droplets in cells (e.g. those formed by an IDR of Ddx4 in the nuclei of HeLa cells [17]) and the formation of nucleoli after cell division [61] follows the behavior expected for phase separated systems. Moreover, the concentration dependence of the formation of nucleoli reveals a coexistence concentration as expected for LLPS [62]. However, other work shows that some nucleolar proteins do not follow behavior expected for phase separation, but rather behave as if controlled by an active process [63]. This may indicate that nucleolar assembly is regulated overall, or that the incorporation of some proteins into the nucleous is independent of the formation of the overall structure. Additional support for considering RNP granules as arising by LLPS comes from the observation that many proteins localizing to non-membrane bound organelles in the cell can phase separate under close to physiological conditions in vitro [18,19,27,28,30,43,51,60,64–67]. These observations support the model that RNP granules arise via LLPS.
Some properties of RNP granules in cells are inconsistent with being classical liquids. For example, many RNP granules, including stress granules, P-bodies, and P-granules are quite stable once liberated from the cell [50,68]. Since LLPS should be sensitive to dilution, this suggests that if these RNP granules form by LLPS, they rapidly transition to a more stable and permanent assembly. Moreover, where it has been examined, the liquid-like behavior of stress granules and nucleoli requires ATP [50,58]. These observations are potentially consistent with two possible interpretations. (1) These assemblies are held together by a summation of interactions that together form a stable assembly and ATP is required to remodel them continuously to yield liquid properties in cells. (2) Alternatively, the liquid assemblies make a transition over time to a more energetically stable state, perhaps involving amyloid-like fibrillization. There is evidence for transitions of condensed liquid droplets into more stable states in vitro through protein fibrillization or gelation [27,28,30,69,70] and in cells experimentally induced LLPS assemblies can be observed to become less dynamic over time, although the specific interactions driving that transition remain to be determined [71–73]. In synthetic so-called “optoDroplets” in cells in which light-induced oligomerization is used to drive LLPS, less dynamic assemblies can be produced below a certain quench depth and their rigidity may thus result from gelation [73]. The presence of ATP-consuming chaperones and helicases in RNP granules [50], and the ability of ATP to destabilize dense liquid phases even without enzymes [74] potentially supports the first interpretation.
Similarly, while the theory of LLPS predicts that no RNP granules should exist until a saturation concentration is reached, where this issue has been examined in cells, smaller assemblies form even at lower concentrations of constituents [25]. One possibility is that RNPs can form higher-order assemblies through a variety of interactions, and once those assemblies become quite large, they can transition to the very large RNP granules sometimes seen in cells (Figure 3). This observation is thus in agreement with the model that smaller assemblies form through multivalent interactions and non-covalent crosslinking and once those assemblies are large enough, the weak interactions between them, which can be of a different molecular nature than the interactions driving the smaller scale assembly, are of sufficient energy in summation to allow phase separation [65,75]. Nevertheless, whether RNP granules are classical phase separated assemblies in cells or whether additional processes contribute to their formation or maturation, the model of self-assembly in vitro through LLPS has been useful to learn about the self-interactions of RNP granule components and how those might inform us on RNP granule assembly in cells. Below, we will discuss how different protein architectures give rise to or enhance LLPS.
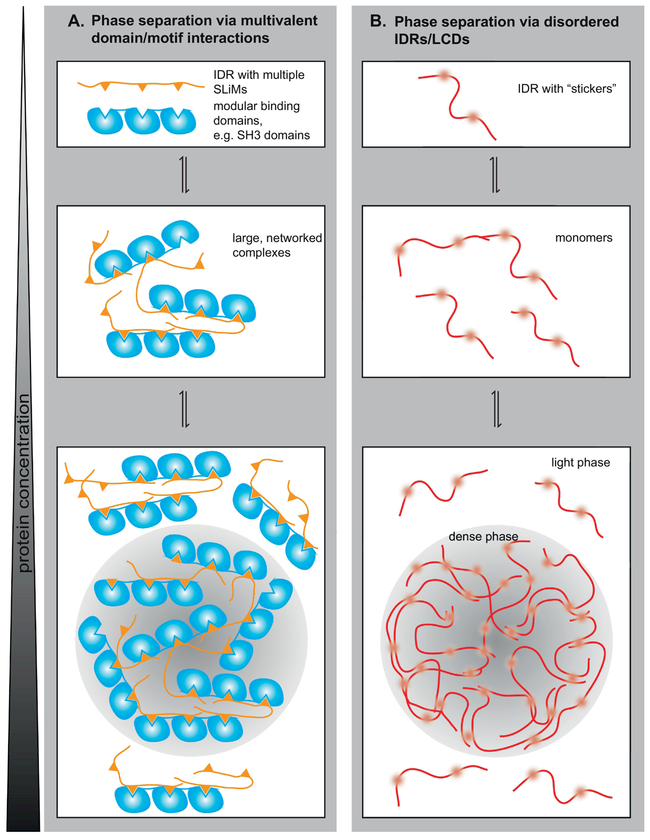
FIGURE 3: Two archetypes of interactions mediate LLPS.
A. The presence of multiple modular binding domains in one protein, and of multiple SLiMs for these modular binding domains in the binding partner, encodes multivalent domain/motif interactions. The resulting multivalent interactions mediate the formation of large, networked complexes, which phase separate from the solution into a dense phase [53], B. IDRs with “stickers”, i.e. with interacting motifs, can undergo phase separation [16, 68]. They likely remain monomeric or form only small complexes before undergoing a highly cooperative phase transition. Polyelectrolyte IDRs can interact with oppositely charged IDRs (or nucleic acids) and undergo complex coacervation, a type of LLPS.
LLPS is mediated by weak multivalent interactions
LLPS is mediated by weak multivalent interactions that do not have to involve extensive structuring (Figure 3). Such types of multivalent interactions can lead to the formation of large networks of protein molecules instead of complexes with defined stoichiometry. As discussed below, multivalency can occur by a number of different manners including oligomerization, proteins with multiple sites of interactions with other partners, or through IDRs that have multiple sites of interactions with other components of the assembly. Only if the multivalency results in the formation of large networks in three dimensions, is it able to mediate LLPS. Interactions that are individually weak allow for the repetitive formation and release of interactions and therefore permit the mobility of molecules within the dense phase and between dense and light phases. This allows dense phases to be dynamic. Note that in RNP granules in cells, stronger interactions could be used and their dynamics then modulated by ATP driven machines that promote transitions in protein-protein, RNA-protein, or RNA-RNA interactions.
Mike Rosen and coworkers characterized a classic system that exhibits such weak, multivalent interactions; the protein Nck contains several SH3 domains, which can interact with several proline-rich motifs (PRMs) in N-WASP. This architecture promotes the formation of large networked complexes which phase separate from solution into a dense phase [65] (Figure 3a). Notably, the ability to form a dense phase strongly increases with the valency of the binding partners; at higher valency, lower protein concentrations are sufficient to form a dense phase and, equivalently, a lower protein concentration remains in the light phase.
Such multivalent domain/motif interactions are not the only interactions that are able to mediate phase separation under close-to-physiological conditions. Certain IDRs can also mediate phase separation efficiently at least in vitro (Figure 3b). The IDRs of Ddx4 and Laf-1, proteins in nuage organelles and P granules, are necessary and sufficient for phase separation of these proteins [17,18]. Similar findings were also reported for the disordered regions of a number of other proteins, including FUS, hnRNPA1, hnRNPA2B1, Pub1, Whi3 and eIF4G [19,21,27,30]. The ability of these IDRs to mediate LLPS in vitro demonstrates that they are capable of mediating weak multivalent interactions with themselves and/or other proteins. The interacting motifs are often called “stickers”, the intervening sequences “linkers” [75].
Multivalent domain/motif interactions and IDRs represent two archetypes of protein architectures that mediate phase separation of proteins under close-to-physiological conditions. We will describe below which variants of these archetypes exist and how they cooperate towards the functional compartmentalization of the cell.
Interactions in IDRs
IDRs are a common feature of phase separating proteins, but the motifs driving their condensation into dense droplets remain poorly understood. The current knowledge has recently been reviewed [76,77], and we thus only briefly describe the sequence features known to contribute to phase separation.
IDRs often contain short motifs mediating multiple, weak interactions that drive phase separation, so-called stickers. The compositional bias and specific patterning of IDRs likely contains information on the relevant stickers. IDRs can be rich in small polar residues including glycine (which is counted as polar residue because of its polar backbone and the lack of modifying sidechain), serine, asparagine and glutamine. Deviations from these residues include aromatic, hydrophobic and charged residues, often arginines. These residues are typically sprinkled in the sequence with regular spacing, suggesting that they may be part of the interacting motifs. Indeed, aromatic residues are essential for the phase separation and hydrogel formation of several IDRs [17,31,33,78,79]. Electrostatic interactions can contribute to phase separation if charged residues are distributed in blocks of opposite charges [17,80]. Aromatic/arginine interactions could contribute to cation-π or π-π interactions. The latter have been proposed to be ubiquitous in phase separating proteins, and may also explain the high frequency of glutamine and asparagine residues with conjugated π systems in their sidechains [46]. What types of structural features IDRs adopt in the condensed phase remains unclear so far. While several studies report the lack of structuring of IDRs [19,29,33–35], others report that motifs in IDRs can adopt kinked cross-β-type structures [34,45], which may contribute to the internal structure of condensed droplets [29].
Protein architectures encoding multivalent domain/motif interactions
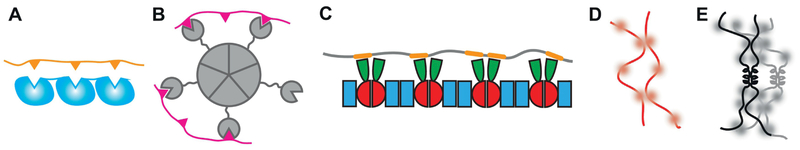
Repeats of folded interaction domains in one binding partner (e.g. SH3 domains in Nck) and repeats of SLiMs in the other binding partner (e.g. PRMs in N-WASP) give rise to archetypal multivalent domain/motif interactions (Figure 4a). These also appear in other systems, including in RNA-binding proteins that contain repeats of folded RNA recognition motifs (RRMs) and interact with multiple motifs in RNA [27,28,65]. The multivalency is linearly encoded in a pair of monomeric proteins in this case, but other multivalent architectures occur in natural proteins as well.
FIGURE 4: Protein architectures encoding multivalency.
Multivalent domain/motif interactions can be encoded via (A) repeats of modular binding domains in a single protein chain and repeats of SLiMs in the binding partner, as in the Nck/WASP system [53], via (B) discrete oligomerization of a protein with a modular binding domain, as in Npm1 [55]m, or via (C) dimerization through two interfaces, as in SPOP [48] and TDP-43. Multivalency can also be manifest as (D) multiple “stickers” (blurry dots) on an IDR [16,68]. This multivalency can be enhanced via (E) oligomerization of the IDR, e.g. through coiled coil formation, as in TDP-43 [19].
Some proteins gain multivalency through oligomerization, which can then promote higher order assembly through additional interactions. If a protein has only one modular binding domain, i.e. a valency of one, it can increase its valency through oligomerization. Npm1, a protein mediating phase separation in the granular component of the nucleolus, binds RNA through a C-terminal nucleic acid binding domain and interacts with arginine-rich motifs in nucleolar proteins. Npm1 monomers, which are the stable form when phosphorylated, thus encode a valency of 2 for interaction partners, which is typically not sufficient for phase separation. But Npm1 monomers oligomerize in the nonphosphorylated form through their N-termini to form a pentameric ring structure [81], resulting in a defined multivalency of 10 for RNA-protein binding, and mediating phase separation [66] (Figure 4b).
Some phase-separating proteins use linear self-association to increase their valency. In the simplest type of linear self-association, each addition of a monomer occurs with the same dissociation constant. The resulting linear oligomers (Figure 4c) span a range of different oligomeric sizes, and therefore valencies, and a higher protein concentration increases their size and valency. TDP-43, an RNA-binding protein implicated in the pathology of Amyotrophic Lateral Sclerosis, Frontotemporal Dementia and Multisystem Proteinopathy, self-associates weakly through its N-terminal domain in this linear manner [64]. TDP-43 phase separates through its C-terminal IDR [20]; the N-terminal linear oligomerization effectively generates an oligomer with several IDRs, increasing the valency of interacting motifs within these IDRs. Disrupting the self-association ability of the N-terminal domain via mutations reduces the propensity of TDP-43 to phase separate and results in a splicing defect [64,82]. Similarly, the Speckle-type POZ protein (SPOP), a substrate recruiting subunit of a ubiquitin ligase, self-associates linearly via two dimerization domains [59] (Figure 4c). The resulting linear polymers are multivalent for substrate binding because each monomer recognizes a SLiM in substrates [83]. Disrupting the self-association ability of SPOP prevents it from phase separating with its substrates.
Folded proteins and their domains can contribute to phase separation through weak electrostatic or hydrophobic interactions on their surfaces, without a need to oligomerize through defined interfaces. This is the nature of the interactions driving phase separation of folded proteins sometimes observed in crystallization trials. These types of interactions may also mediate physiological phase separation, as seems to be the case in the yeast PolyA-binding protein Pab1. While Pab1 has a IDR that modifies the temperature-dependence of phase separation, the folded RRM domains are required for Pab1 phase separation, and do so even in the absence of RNA as a function of elevated temperature or decreased pH [70]. The interacting surfaces mediating this phase separation remain to be discovered.
Discrete oligomerization that contributes to phase separation can also be encoded within IDRs. The C-terminal IDR of TDP-43 is largely disordered and mediates phase separation through stickers (Figure 4d). However, a 30-residue segment attains helical structure at increasing protein concentrations and forms helical bundles via homo-oligomerization (Figure 4e). This oligomerization enhances phase separation by increasing the valency of the TDP-43 IDR. Disrupting the helical bundle via mutations reduces phase separation of TDP-43 as expected [20].
All of these protein architectures have in common that they increase the valency of interacting domains or motifs, and thereby enhance phase separation. The different architectures that encode multivalency offer different modes of regulation; e.g. oligomerization of a protein can be enhanced or decreased through post-translational modifications [64]. The different architectures result in an increase of the valency with different concentration dependences, as expected from biophysical principles of oligomerization. These differences may turn out to be exploited for the regulation of RNP granule assembly.
Weak multivalent interactions that mediate the formation of non-covalently cross-linked complexes do not necessarily have to mediate LLPS. They can instead lead to gelation, i.e. the formation of a system-spanning network of macromolecules [84]. In order for phase separation to occur, a driving force for condensation into droplets must be encoded in the proteins in addition to multivalent interactions. This tendency for condensation, i.e. surface tension, requires the release of solvent and can be conveniently encoded in IDRs of phase separating proteins, although folded proteins are able to condense into droplets as well. The propensity of IDRs to interact with the solvent is encoded in their primary sequence. For some IDRs, water is a good solvent, and they adopt extended conformations. For other IDRs, water is a poor solvent and they adopt more collapsed conformations because they trade protein/solvent with protein/protein interactions [85]. It seems likely that IDRs with such sequence/conformation properties can mediate the condensation of cross-linked complexes into condensed droplets with a surface tension [64]. E.g., the linker between the first two SH3 domains in Nck enhances phase separation of Nck with N-WASP [86]. Coarse-grained simulations show that collapsed disordered linkers drive phase separation, whereas expanded linkers can mediate gelation [75].
It is noteworthy that not all IDRs are expected to undergo phase separation by themselves. Slightly collapsed IDRs are likely the flavor that can undergo phase separation. More expanded IDRs may gel instead, which refers to the formation of a system-spanning gel instead of condensed droplets [75]. IDRs with high net charge and therefore likely large global dimensions can form condensed droplets with oppositely charged polymers via a process called complex coacervation. As an example, polyamines can undergo complex coacervation with RNA. Compartmentalization through complex coacervation has even been implicated in the origin of life [87]. Hence, different flavors of IDRs can mediate condensation, which may explain the observation that IDRs in RNPs can be replaced by some others. While this observation may suggest that the effect of IDRs on RNP granule assembly are not due to a specific sequence in the IDR, the known relationships between sequence, conformation and function in IDRs points towards the importance of specific sequence features that require further exploration.
The role of domain/motif interactions vs IDRs in the cell
Multivalent domain/motif interactions and motifs in IDRs can mediate phase separation, and proteins encoding both types of interactions are abundant in the cell. Do these different types of multivalent interactions mediate different functions, or are they the result of convergent evolution, i.e. they both serve similar purposes? This question is difficult to answer given the sparse experimental data; little data exist on the relative contribution of both types of interactions for the formation of non-membrane-bound organelles. Recent data suggested that the presence of high concentrations of other proteins, e.g. cell lysates, can interfere with the formation of condensed IDR droplets, while it enhances phase separation of multivalent domain/motif-containing proteins [26]. This may indicate that multivalent domain/motif interactions are important for determining the identity of phase separated structures, while IDR-containing proteins are recruited to preexisting structures.
The structural basis of domain/motif interactions are vastly better understood than the merely transiently ordered interactions in IDRs. Crystal structures of motifs bound to domains are available for many relevant proteins, thus making it tempting to speculate that multivalent domain/motif interactions are more specific than interactions in potentially disordered IDRs. However, domain/motif interactions that drive phase separation often have affinities as weak as hundreds of micromolar or millimolar [65] and may thus be in a similar range as weak interactions driven by short motifs in disordered IDRs. Specificity is a measure of the affinity of the native interaction relative to all non-native interactions; as long as non-native interactions are weaker, the native interaction is specific. Whether specificity can be achieved through disordered interactions, as formed by phase separating IDRs in the absence of large-scale or persistent structuring, remains largely untested. IDRs seem to have a preference to colocalize with certain IDRs and exclude others, but these tendencies seem relatively weak, i.e. they result in small differences in partitioning coefficients [27]. Whether they are sufficient to provide specificity of localization to different non-membrane-bound organelles in the cell remains to be explored.
The possible role of phase separation for hyperaggregation of disease mutants
As mentioned above, the IDRs of RNP granule components are interesting also from a disease point of view. Mutations in the IDRs of TDP-43, TIA1, hnRNPA1, hnRNPA2B1 or FUS can lead to muscular or neurodegenerative diseases, apparently due to their hyperaggregation. Recently, it has been reported for all of these proteins that they can condense into liquid droplets via LLPS [27,28,30,64,72]. These droplets maturate into more solid assemblies over time, or the proteins even form amyloid-like fibrils within the droplets. These processes are enhanced for disease mutants of the proteins [27,28,30,72], possibly because they introduce new LARKS or amyloid-forming steric zippers. These observations may mean that interactions leading to fibrillar assemblies do not represent the physiological state and are only accessed when stabilized via mutations or through persistent stress granules. However, more work is needed to be answer this question with confidence.
Future Questions
Given the results described above, there is now a wealth of data on the specific interactions that promote RNP granule assembly. However, several key issues remain to be resolved. First, is the process of LLPS a good model for RNP granule formation, and/or at what scale does LLPS contribute to RNP granule assembly? This is an issue since smaller RNP assemblies appear to form under conditions where no large granules exist, and might serve as precursor for a LLPS transition that leads to the larger assembly. Second, what is the function of large RNP granules, and at what size scale is that function satisfied? This is an issue since experiments to disrupt RNP granules have often failed to identify clear functions in the control of RNA metabolism [13,88]. Finally, a key issue will be to discern the fundamental principles of the formation of large biological condensates and what predictive properties those allow.
Box
Liquid-liquid phase separation:
LLPS is a process in which two liquids demix from each other. In biologically relevant phase separation processes, macromolecules in solution condense into dense liquid droplets. The droplets contain high concentrations of the macromolecule and coexist with a light phase, which contains a low concentration of the macromolecule. LLPS occurs when the interactions of components for each other are more energetically favorable than interactions of building blocks with the solvent such that demixing leads to a decrease in the total free energy of the system. LLPS depends on the volume fractions of the components and can also depend on environmental conditions such as temperature, pH, salt concentration and the presence of other solutes. In a scenario in which the macromolecular building blocks were progressively concentrated in solution, the system would demix into a dense and a light phase upon reaching the saturation concentration. A further increase of the building block concentration would result in an increase of the volume fraction of the dense phase, at the expense of the light phase, while the concentrations in the dense and light phase would remain constant.
Oligomerization:
Oligomerization refers to the formation of molecular complexes with defined stoichiometry and typically a few monomer units.
Amyloid fibrils are polymers that form via cross-β interactions and are implicated in the pathogenesis of neurodegenerative (and other) diseases. Some amyloid-like assemblies can also have normal functions as in melanin synthesis (Fowler PLoS 2006), and translational control (Berchowitz Cell 2015, Carpenter Dev Cell 2018).
Liquid-like properties:
Many RNP granules have now been shown to fuse and coalesce, or even flow and drip, which are classic liquid properties, but they have not been characterized in enough detail to exclude the possibility that they have viscoelastic instead of purely viscous properties. We hence describe them as liquid-like.
Gelation describes the formation of system-spanning networks and can be coupled to LLPS or happen independently. Gelled dense phases loose their purely liquid properties.
Intrinsically disordered protein region (IDR):
IDRs are regions of proteins that do not adopt a single structured state but rather interconvert between many different conformations devoid of much secondary or tertiary structure. Many IDRs undergo disorder-to-order transitions upon interacting with binding partners, often referred to as folding-upon-binding. Other IDRs stay largely disordered even in complexes. These behaviors are encoded in the IDR sequences and in that of their binding partners. The global dimensions and propensity for local secondary or non-local tertiary contacts varies widely between IDRs.
Low-complexity domain (LCD):
LCDs are characterized by a biased amino acid composition with overrepresentation of certain amino acids and underrepresentation of others, resulting in a low sequence complexity. Low sequence complexity can generate intrinsic disorder if the hydrophobic residues essential for a folded protein core are underrepresented. However, some proteins with low sequence complexity, e.g. leucine-rich repeats with a bias towards leucines, are well-folded proteins. The IDRs of many RNA-binding proteins are typically referred to as LCDs; they are rich in glycines, small polar residues, aromatics and arginines, and undergo phase separation under physiologically relevant conditions. Other types of LCDs, e.g. elastin-like sequences can also undergo LLPS, but not all proteins with low sequence complexity do so.
Prion-like domain (PrLD):
PrLDs are LCDs with a similar amino acid composition as yeast prion domains, i.e. domains of proteins such as Sup35 and Rnq1 that are essential for forming non-Mendelian inheritance elements.
Highlights.
Well-folded and disordered domains contribute to RNP granule assembly.
Macroscopically, liquid-liquid phase separation results in dense liquid assemblies that share liquid properties, viscosity and surface tension with RNP granules.
Microscopically, archetypal multivalent domain/motif interactions and interactions of motifs within intrinsically disordered protein regions can mediate liquid-liquid phase separation.
RNP granules may arise through liquid-liquid phase separation but show some differences from classical phase separation.
Acknowledgements
This work was funded by R01GM112846, the American Lebanese Syrian Associated Charities and St. Jude Children’s Research Hospital (to T.M.) and the Howard Hughes Medical Institute (to R.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mao YS, Zhang B, Spector DL, Biogenesis and function of nuclear bodies, Trends Genet 27 (2011) 295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Protter DS, Parker R, Principles and Properties of Stress Granules, Trends Cell Biol 26 (2016) 668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Parker R, Sheth U, P bodies and the control of mRNA translation and degradation, Mol Cell. 25 (2007) 635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- [4].Schisa JA, New insights into the regulation of RNP granule assembly in oocytes, Int Rev Cell Mol Biol 295 (2012) 233–289. doi: 10.1016/B978-0-12-394306-4.00013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Buchan JR, mRNP granules. Assembly, function, and connections with disease, RNA Biol 11 (2014) 1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M, The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation, Proc Natl Acad Sci U S A. 108 (2011) E655–62. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Van Treeck RPB, Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies, Cell (in Press. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R, RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome, Proc Natl Acad Sci U S A. 115 (2018) 2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jambor H, Brunei C, Ephrussi A, Dimerization of oskar 3’ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte, RNA. 17 (2011) 2049–2057. doi: 10.1261/rna.2686411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C, RNA-RNA interaction is required for the formation of specific bicoid mRNA 3’ UTR-STAUFEN ribonucleoprotein particles, EMBO J 16 (1997) 1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Aumiller WM, Pir Cakmak F, Davis BW, Keating CD, RNA-Based Coacervates as a Model for Membraneless Organelles: Formation, Properties, and Interfacial Liposome Assembly, Langmuir 32 (2016) 10042–10053. doi: 10.1021/acs.langmuir.6b02499. [DOI] [PubMed] [Google Scholar]
- [12].Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, Myong S, Gladfelter AS, mRNA structure determines specificity of a polyQ-driven phase separation, Science (80-. ). 360 (2018) 922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Decker CJ, Teixeira D, Parker R, Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae, J Cell Biol 179 (2007) 437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J, The RasGAP-associated endoribonuclease G3BP assembles stress granules, J Cell Biol 160 (2003) 823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [15].Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, Mclnerney GM, Ivanov P, Anderson P, G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits, J Cell Biol 212 (2016) 845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G, Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans, Elife. 3 (2014) e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ, Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles, Mol Cell. 57 (2015) 936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP, The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics, Proc Natl Acad Sci U S A. 112 (2015) 7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Burke KA, Janke AM, Rhine CL, Fawzi NL, Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II, Mol Cell. 60 (2015) 231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Conicella AE, Zerze GH, Mittal J, Fawzi NL, ALS Mutations Disrupt Phase Separation Mediated by alpha-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain, Structure. 24 (2016) 1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ryan VH, Dignon GL, Zerze GH, V Chabata C, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, Fawzi NL, Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation, Mol Cell. 69 (2018) 465–479e7. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wright PE, Dyson HJ, Linking folding and binding, Curr. Opin. Struct. Biol 19 (2009) 31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD, Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch, Nature. 519 (2015) 106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- [24].Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P, Stress granule assembly is mediated by prion-like aggregation of TIA-1, Mol Biol Cell. 15 (2004) 5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rao BS, Parker R, Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae, Proc Natl Acad Sci U S A. 114 (2017) E9569–E9578. doi: 10.1073/pnas.1712396114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R, Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly, Cell Rep 22 (2018) 1401–1412. doi: 10.1016/j.celrep.2018.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lin Y, Protter DS, Rosen MK, Parker R, Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins, Mol Cell. 60 (2015) 208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP, Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization, Cell. 163 (2015) 123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL, The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei, Cell. 163 (2015) 829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S, A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation, Cell. 162 (2015) 1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- [31].Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, V Grishin N, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL, Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels, Cell. 149 (2012) 753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, Hosoki K, Goshima N, Kawaguchi T, Hatters D, Trinkle-Mulcahy L, Hirose T, Bond CS, Fox AH, Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles, J Cell Biol 210 (2015) 529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, Kay LE, Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation, Proc Natl Acad Sci U S A. 114(2017) E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, Eisenberg DS, Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks, Science (80-. ). 359 (2018) 698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Luo Y, Na Z, Slavoff SA, P-Bodies: Composition, Properties, and Functions, Biochemistry. (2018). doi: 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Taylor JP, Brown RH Jr., D.W. Cleveland, Decoding ALS: from genes to mechanism, Nature. 539 (2016) 197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP, Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS, Nature. 495 (2013) 467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guo W, Chen Y, Zhou X, Kar A, Ray P, Chen X, Rao EJ, Yang M, Ye H, Zhu L, Liu J, Xu M, Yang Y, Wang C, Zhang D, Bigio EH, Mesulam M, Shen Y, Xu Q, Fushimi K, Wu JY, An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity, Nat Struct Mol Biol 18 (2011) 822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ramaswami M, Taylor JP, Parker R, Altered ribostasis: RNA-protein granules in degenerative disorders, Cell. 154 (2013) 727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li YR, King OD, Shorter J, Gitler AD, Stress granules as crucibles of ALS pathogenesis, J Cell Biol 201 (2013) 361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ling SH, Decker CJ, Walsh MA, She M, Parker R, Song H, Crystal structure of human Edc3 and its functional implications, Mol Cell Biol 28 (2008) 5965–5976. doi: 10.1128/MCB.00761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hebert MD, Matera AG, Self-association of coilin reveals a common theme in nuclear body localization, Mol Biol Cell. 11 (2000) 4159–4171. http://www.ncbi.nlm.nih.gov/pubmed/11102515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fromm SA, Kamenz J, Noldeke ER, Neu A, Zocher G, Sprangers R, In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery, Angew Chem Int Ed Engl 53 (2014) 7354–7359. doi: 10.1002/anie.201402885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jonas S, Izaurralde E, The role of disordered protein regions in the assembly of decapping complexes and RNP granules, Genes Dev 27 (2013) 2628–2641. doi: 10.1101/gad.227843.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R, Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains, Cell. 171 (2017) 615–627 e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, Forman-Kay JD, Pi-Pi contacts are an overlooked protein feature relevant to phase separation, Elife. 7 (2018). doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Csizmok V, Forman-Kay JD, Complex regulatory mechanisms mediated by the interplay of multiple post-translational modifications, Curr. Opin. Struct. Biol 48 (2018) 58–67. doi: 10.1016/j.sbi.2017.10.013. [DOI] [PubMed] [Google Scholar]
- [48].Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW, Comprehensive Identification of RNA-Binding Domains in Human Cells, Mol Cell. 63 (2016) 696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM, Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail, Proc. Natl. Acad. Sci 108 (2011) 12254–12259. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R, ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure, Cell. 164 (2016) 487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, V Pappu R, Brangwynne CP, Coexisting Liquid Phases Underlie Nucleolar Subcompartments, Cell. 165 (2016) 1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Buchan JR, Parker R, Eukaryotic Stress Granules: The Ins and Outs of Translation, Mol. Cell 36 (2009) 932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry, Nat. Rev. Mol. Cell Biol 18 (2017) 285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Muschol M, Rosenberger F, Liquid-liquid phase separation in supersaturated lysozyme solutions and associated precipitate formation/crystallization, J. Chem. Phys 107 (1997) 1953–1962. doi: 10.1063/1.474547. [DOI] [Google Scholar]
- [55].Raut AS, Kalonia DS, Pharmaceutical Perspective on Opalescence and Liquid–Liquid Phase Separation in Protein Solutions, Mol. Pharm 13 (2016) 1431–1444. doi: 10.1021/acs.molpharmaceut.5b00937. [DOI] [PubMed] [Google Scholar]
- [56].Heymann M, Opthalage A, Wierman JL, Akella S, Szebenyi DME, Gruner SM, Fraden S, Room-temperature serial crystallography using a kinetically optimized microfluidic device for protein crystallization and on-chip X-ray diffraction, IUCrJ. 1 (2014) 349–360. doi: 10.1107/S2052252514016960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA, Germline P granules are liquid droplets that localize by controlled dissolution/condensation, Science (80-. ). 324 (2009) 1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- [58].Brangwynne CP, Mitchison TJ, Hyman AA, Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes, Proc Natl Acad Sci U S A. 108 (2011) 4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, Ben-Nissan G, Kolaitis RM, Peters JL, Pounds S, Errington WJ, Prive GG, Taylor JP, Sharon M, Schuck P, Ogden SK, Mittag T, Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles, EMBO J 35 (2016) 1254–1275. doi: 10.15252/embj.201593169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK, Compositional Control of Phase-Separated Cellular Bodies, Cell. 166 (2016) 651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP, RNA transcription modulates phase transition-driven nuclear body assembly, Proc Natl Acad Sci U S A. 112 (2015) E5237–45. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Weber SC, Brangwynne CP, Inverse size scaling of the nucleolus by a concentration-dependent phase transition, Curr Biol 25 (2015) 641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Falahati H, Wieschaus E, Independent active and thermodynamic processes govern the nucleolus assembly in vivo, Proc. Natl. Acad. Sci 114 (2017) 1335–1340. doi: 10.1073/pnas.1615395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, Shewmaker F, Naik MT, Mittag T, Ayala YM, Fawzi NL, A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing, EMBO J 37 (2018). doi: 10.15252/embj.201797452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, V Hollingsworth J, King DS, Banani SF, Russo PS, Jiang GX, Nixon BT, Rosen MK, Phase transitions in the assembly of multivalent signalling proteins, Nature. 483 (2012) 336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW, Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA, Elife. 5 (2016). doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nuske E, Richter D, Baumeister W, Grill SW, V Pappu R, Hyman AA, Alberti S, Phase separation of a yeast prion protein promotes cellular fitness, Science (80-. ). 359 (2018). doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- [68].TEIXEIRA D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R, Processing bodies require RNA for assembly and contain nontranslating mRNAs, RNA. 11 (2005) 371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, Kronenberg-Versteeg D, Li Y, Yang SP, Wakutani Y, Meadows W, Ferry RR, Dong L, Tartaglia GG, Favrin G, Lin WL, Dickson DW, Zhen M, Ron D, Schmitt-Ulms G, Fraser PE, Shneider NA, Holt C, Vendruscolo M, Kaminski CF, St George-Hyslop P, ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function, Neuron. 88 (2015) 678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Riback JA, Katanski CD, Kear-Scott JL, V Pilipenko E, Rojek AE, Sosnick TR, Drummond DA, Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response, Cell. 168 (2017) 1028–1040 e19. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS, RNA Controls PolyQ Protein Phase Transitions, Mol Cell. 60 (2015) 220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, Matchett BJ, Mittag T, Temirov J, Hsiung GR, Krieger C, Murray ME, Kato M, Fryer JD, Petrucelli L, Zinman L, Weintraub S, Mesulam M, Keith J, Zivkovic SA, Hirsch-Reinshagen V, Roos RP, Zuchner S, Graff-Radford NR, Petersen RC, Caselli RJ, Wszolek ZK, Finger E, Lippa C, Lacomis D, Stewart H, Dickson DW, Kim HJ, Rogaeva E, Bigio E, Boylan KB, Taylor JP, Rademakers R, TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics, Neuron. 95 (2017) 808–816 e9. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP, Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets, Cell. 168 (2017) 159–171 e14. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA, ATP as a biological hydrotrope, Science (80-. ). 356 (2017) 753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- [75].Harmon TS, Holehouse AS, Rosen MK, V Pappu R, Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins, Elife. 6 (2017). doi: 10.7554/eLife.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Martin EW, Mittag T, Relationship of Sequence and Phase Separation in Protein Low-Complexity Regions, Biochemistry. (2018). doi: 10.1021/acs.biochem.8b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lin YH, Forman-Kay JD, Chan HS, Theories for Sequence-Dependent Phase Behaviors of Biomolecular Condensates, Biochemistry. (2018). doi: 10.1021/acs.biochem.8b00058. [DOI] [PubMed] [Google Scholar]
- [78].Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, AN R, Yunus AA, Liu DR, V Pappu R, Rosen MK, Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein, Mol Cell. 63 (2016) 72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lin Y, Currie SL, Rosen MK, Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs, J Biol Chem 292 (2017) 19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lin YH, Forman-Kay JD, Chan HS, Sequence-Specific Polyampholyte Phase Separation in Membraneless Organelles, Phys Rev Lett 117 (2016) 178101. doi: 10.1103/PhysRevLett.117.178101. [DOI] [PubMed] [Google Scholar]
- [81].Mitrea DM, Grace CR, Buljan M, Yun M-K, Pytel NJ, Satumba J, Nourse A, Park C-G, Madan Babu M, White SW, Kriwacki RW, Structural polymorphism in the N-terminal oligomerization domain of NPM1, Proc. Natl. Acad. Sci 111 (2014) 4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Afroz T, Hock EM, Ernst P, Foglieni C, Jambeau M, Gilhespy LAB, Laferriere F, Maniecka Z, Pluckthun A, Mittl P, Paganetti P, Allain FHT, Polymenidou M, Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation, Nat Commun 8 (2017) 45. doi: 10.1038/s41467-017-00062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Pierce WK, Grace CR, Lee J, Nourse A, Marzahn MR, Watson ER, High AA, Peng J, Schulman BA, Mittag T, Multiple Weak Linear Motifs Enhance Recruitment and Processivity in SPOP-Mediated Substrate Ubiquitination, J Mol Biol 428 (2016) 1256–1271. doi: 10.1016/j.jmb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wong Po Foo CT, Lee JS, Mulyasasmita W, Parisi-Amon A, Heilshorn SC, Two-component protein-engineered physical hydrogels for cell encapsulation, Proc Natl Acad Sci U S A. 106 (2009) 22067–22072. doi: 10.1073/pnas.0904851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hofmann H, Soranno A, Borgia A, Gast K, Nettels D, Schuler B, Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy, Proc. Natl. Acad. Sci 109 (2012) 16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Banjade S, Wu Q, Mittal A, Peeples WB, V Pappu R, Rosen MK, Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck, Proc Natl Acad Sci U S A. 112 (2015) E6426–35. doi: 10.1073/pnas.1508778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Poudyal RR, PirCakmak F, Keating CD, Bevilacqua PC, Physical Principles and Extant Biology Reveal Roles for RNA-Containing Membraneless Compartments in Origins of Life Chemistry, Biochemistry. (2018). doi: 10.1021/acs.biochem.8b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E, P-Body Formation Is a Consequence, Not the Cause, of RNA-Mediated Gene Silencing, Mol. Cell. Biol 27 (2007) 3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]