Abstract
Phase transitions that alter the physical state of ribonucleoprotein particles contribute to the spacial and temporal organization of the densely packed intracellular environment. This allows cells to organize biologically coupled processes as well as respond to environmental stimuli. RNA plays a key role in phase separation events that modulate various aspects of RNA metabolism. Here, we review the role that RNA plays in ribonucleoprotein phase separations.
Graphics Abstract
Organization of the densely packed intracellular environment requires compartmentalization. This is particularly important for gene expression as coordinated processes must occur in an ordered fashion. In eukaryotic cells, double stranded DNA (dsDNA) is sequestered in the nucleus and packaged in histones. Within the nucleus, DNA is organized into heterochromatin and euchromatin to control the relative access to the transcriptional machinery. Transcribed mRNA undergoes splicing, polyadenylation, and capping prior to export to the cytoplasm. Each of these processes is under spatiotemporal control that ensures correct processing and localization.
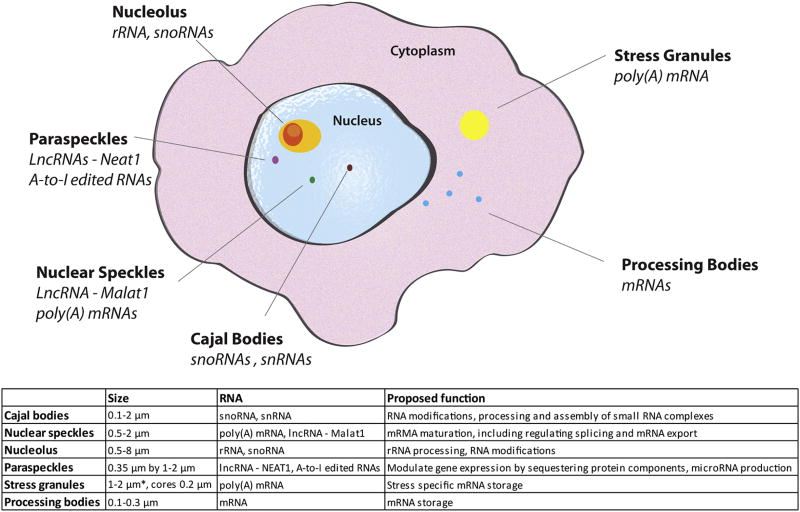
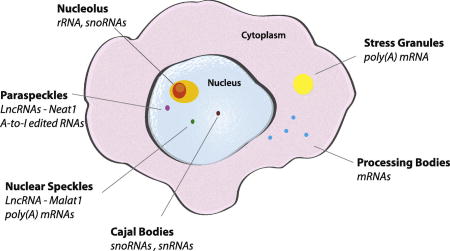
Just as membrane-enclosed organelles (e.g., nuclei, mitochondria, endoplasmic reticulum, golgi apparatus) serve to organize biological processes into discrete cellular domains, non-membrane enclosed domains play a similar role in the organization of biological activities throughout the cell. These are defined by the physical nature of their constituents and are referred to as membrane-less organelles (MLO) due to their ability to concentrate factors associated with a biological process, typically involving RNA metabolism (Figure 1). MLOs serve to concentrate their components in a way that facilitates processes such as transcription and ribosomal RNA (rRNA) processing in the nucleolus, or sequestration such as translation initiation complexes and signaling molecules in stress granules (SGs). MLOs can be visualized using phase contrast, immunofluorescence, or brightfield microscopy. The lack of a limiting membrane allows MLO constituents to rapidly exchange with the surrounding environment, allowing a dynamic response to changes in the cell state.
Figure 1. Membraneless organelles (MLOs).
Nuclear membraneless organelles include the nucleolus, paraspeckles, nuclear speckles and Cajal bodies. Cytoplasmic membraneless organelles including stress granules and processing bodies. * denotes that these values are reported for treatment with 0.5 mM sodium arsenite for 30 minutes, size of SGs can greatly change with time and stress used.
MLOs are assembled via a process of phase separation. During phase separation, a critical concentration of a given component (e.g., protein, RNA, ribonucleoprotein complex) allows a transition to a concentrated phase which is physically separated from a more dilute phase. This process is driven by multiple weak electrostatic, charge-charge and repetitive domain interactions that “demix” a homogenous state into two discrete phases. This process can produce a discrete liquid state within a liquid known as a liquid-liquid phase separation (LLPS), or a transition of a portion of the liquid into solid state causing a solid within a liquid. Molecules within a phase separated liquid domain which typically assumes a spherical shape, can continuously exchange with the surrounding solution while maintaining the phase separation. This is different than a transition to a solid state whose shape is internally defined by the patterning of its components which rarely exchange with the surrounding solution1; 2.
A major advance in our understanding of biological phase transitions came with the serendipitous discovery that biotinylated isoxazole (b-isox) forms a precipitate that selectively traps proteins associated with MLOs3; 4. These proteins possess low complexity domains (LCD) that are enriched in amino acids Ala, Arg, Gly, Gln, Ser, Pro, Glu, and Lys. A subset of LCDs are related to prion proteins that can adopt soluble and aggregation-prone conformations to modulate biological processes. In their native state, LCDs are intrinsically disordered, but they can assume a defined tertiary structure in response to post-transcriptional modifications or interactions with partner proteins or RNAs5. B-isox assumes a crystalline state comprised of a lattice-like structure that selectively templates LCD-containing proteins to assume a β-strand conformation4. B-isox condenses mRNAs in a 3’UTR length-dependent manner3; 4, probably reflecting the fact that longer mRNAs tend to have more binding sites for specific and non-specific RNA binding proteins. B-isox can condense mRNAs in a sequence selective manner: B-isox condenses RNAs bearing MS2 stem loops in the presence of a fusion protein composed of the MS2 stem loop-binding protein linked to a LCD that is known to phase separate in isolation3.
Purified LCDs can form in vitro droplets often using low salt concentrations or in the presence of molecular crowding agents such as Ficoll or polyethylene glycol. These droplets are formed via LLPS as they: (1) deform under shear force, (2) exchange with the surrounding environment, (3) have higher component concentrations than the surrounding environment, (4) fuse with other droplets, (5) are spherical, and (6) are temperature dependent2; 6. Importantly, not all LCDs have the same biophysical properties. Under conditions in which several LCDs promote LLPS, the TIA1-LCD (the prion-related domain of a ribosome recognition motif RNA-binding protein) does not undergo phase separation. In contrast, when the TIA1-LCD is expressed as a chimeric fusion protein with polypyrimidine tract-binding protein (PTB), PTB-TIA1-LCD undergoes a phase separation7. While RNA can coax PTB to phase separate in the absence of a LCD8, PTB-TIA1-LCD has a lower concentration threshold for phase separation7. Similarly, RNA can promote the phase separation of isolated RRM and LCD domains of hnRNPA1, but the concentration required is significantly higher than that of the full length hnRNPA1 protein9. Finally, RNA promotes FUS (a well-characterized, sequence non-specific, RNA binding protein associated with amyotrophic lateral sclerosis (ALS)) -mediated phase separation in a non-sequence specific manner10; 11. RNA can undergo a phase separation event in isolation12; 13. Disrupting RNA:RNA interactions inhibits this process13. Furthermore, there is a significant overlap between RNAs that undergo phase separation in vitro and RNAs that interact with SG components in vivo12. These findings suggest that RNA can promote phase separation events via RNA:protein and RNA:RNA interactions.
RNA lowers the concentration threshold for the phase separation of purified PGL-3 into P-granule-like droplets. mRNA is more effective at promoting phase separaton than in vitro transcribed rRNA. Yet, when heated this rRNA is effective at promoting PGL-3 condensation14, suggesting a role for complex RNA structures in granule formation. Neither the nature of these structures, nor the effect of RNA modifications (e.g., poly(A) tails, m7GTP caps, modified nucleotides) on phase separation events is known. Liquid droplets formed by the poly Q domain-containing RNA binding protein Whi3 have different biophysical properties depending on the associated mRNA15. Droplets formed in the presence of a formin transcript exhibit faster fusion and reduced viscosity than droplets formed in the presence of a cyclin transcript. Both mRNAs contain the same number of Whi3 binding sites yet the formin transcript is four times longer15. More work is needed to understand how specific mRNAs differ in their ability to stimulate phase separation events.
Aging of phase separated MLOs can reduce their dynamic properties and produce a solid-like state. Solid-like phase separations do not exchange with the surrounding solution and often (but not always) resemble amyloid-like structures. This is the case for FUS which undergoes an LLPS in vitro which ages into a fibrous, b-isox-like solid. This transition occurs more rapidly when recombinant FUS contains the ALS-associated mutations G156E or R244C16. Similarly, the ALS-associated mutant hnRNPA1-D262V expedites the transition to a fibrillar state9. The kinetics of liquid to solid transition for several LLPS chimeric proteins is increased by addition of RNA7, suggesting that RNA can drive the liquid to solid transition. In the case of TDP43, ALS-associated mutations enhance the assembly of amyloid-like β-sheet structures, a transition that is further enhanced in the presence of nucleic acids17. While it is clear that dynamic properties of droplets assembled in vitro are altered by age, mutations in component proteins, and specific RNAs, the relevance of these findings to MLOs in vivo remains to be determined.
Like RNA, DNA can modulate the propensity of LCDs to undergo phase separations. In the presence of dsDNA, a chimeric fusion protein between the FUS-LCD and the DNA-binding domain of the ETS transcription factor FLI assembles fibrillar structures that can be visualized using electron microscopy18. In contrast, in vitro droplets formed by the LCD of the RNA helicase DDX4 recruit ssDNA, but not dsDNA19, suggesting that individual unpaired bases may contribute to this phenomenon.
In vitro studies using proteins or protein domains in isolation have been used to make key observations about the biophysical properties of RNA granules. Yet, how this translates into the cellular context remains to be determined. Purified LCDs that phase separate in vitro, do not always drive granule assembly in cells20. Within the complex cytoplasmic milieu, factors such as protein and RNA interaction, subcellular localization, and post-translational modification can modulate phase separation events. Future studies are required to determine the significance of in vitro phase separation events and their link to specific biological functions.
Repeat expansion disorders and RNA repeat foci
While RNA repeats such as CAG, CUG, CCUG, and GGGGCC are found in the normal population, expansion of these repetitive nucleotides is associated with a subset of neurological diseases known as repeat expansion disorders. Nucleotide repeat expansions can be found in 5’ and 3’ untranslated regions, introns, and coding regions. In some cases, these repeats are translated into repetitive polypeptides (e.g., CAG repeats are translated into poly-glutamine in Huntingtin protein) (reviewed in21; 22; 23). Repeat expansion disorders are often associated with the appearance of RNA nuclear foci. RNA repeat expansions can recruit and sequester RNA binding proteins and directly contribute to disease pathogenesis.
In these conditions, a repeat threshold is typically associated with the assembly of RNA foci and disease pathogenesis. RNA repeats act as a template to recruit and sequester specific RNA-binding proteins. The resulting nucleoprotein complex can promote a phase separation event to produce pathological foci. CUG repeat expansions in the 3’UTR of the DM1 (also known as DMPK) mRNA24; 25; 26 and CCUG intronic repeat expansions in ZNF9 (also known as CNBP) cause myotonic dystrophy (DM) type 1 and 2, respectively27. Although these are different repeat expansions that affect different genes, both diseases display discrete nuclear RNA foci that disrupt RNA metabolism. This RNA gain-of-function mechanism causes the mislocalization of proteins including the splicing factor muscle-bind 1 (MBLN1)28. Sequestration of MBLN1 at RNA foci disrupts alternative RNA splicing in ways that contribute to DM1 and DM2 pathogenesis (reviewed in21; 23). MBLN1 knockout mice display a DM-like phenotype29 as does a mouse model that introduces 250 CUG repeats into a gene unrelated to DM130, implicating MBLN1 sequestration at CUG RNA foci in disease pathogenesis.
The intronic hexanucleotide GGGGCC repeat in the gene C9ORF72 is the most common cause of both inherited and sporadic ALS and frontotemporal dementia (FTD)31; 32. This RNA can form secondary structures including a G-quadruplex33; 34; 35; 36. A G-quadruplex is composed of stacked G-quartets which assemble when four guanosine residues hydrogen bond via Hoogsteen base pairing in a planar fashion. These planar structures are coordinated by specific monovalent cations and stack to form a G-quadruplex37. Uniquely, GGGGCC RNA can form RNA granules in vitro in the presence of cellular lysate in a manner similar to b-isox38. The G-quadruplex structure is required for GGGGCC RNA condensation from lysates38. This templated pattern drives phase separation in vitro and promotes the assembly of nuclear RNA foci and cytoplasmic SGs in cells38; 39. In C9-ALS/FTD patient derived cells, GGGGCC RNA nuclear foci are recognized by a G-quadruplex specific antibody39, suggesting that the G-quadruplex structure is preserved within RNA foci. Similarly, G-quadruplex structures encoded within 3’UTRs of selected mRNAs are enriched in neuronal granules which are targeted to, and translated in, neurites40. It has been estimated that a single C9-GGGGCC RNA transcript is sufficient to assemble a nuclear focus41, suggesting that intramolecular G-quadruplexes are involved in this process. Like G-quadruplexes with GGGGCC repeats, CUG/CCUG secondary structures are also thought to play a role in phase separation13; 23; 42.
Like LCD-containing proteins, GGGGCC, CAG and CUG repeat RNA can phase separate in vitro13. This occurs in a length dependent manner and is presumably due to base pair interactions and alternative structures including hairpins and G-quadruplex-like structures13. Repeat RNA-induced phase separations exhibit both liquid- and solid-like properties: they assume a spherical geometry, but rarely fuse with one another and do not exchange their constituent RNAs with the surrounding solution13. This is in contrast to RNA foci in cells: fluorescence recovery after photobleaching (FRAP) of MS2 tagged RNA reveals dynamic movement in and out of granules13.
Paraspeckles
While repeat expansion disorders provide examples of disease-associated RNAs driving phase separation events, the formation of paraspeckles is facilitated by the non-pathogenetic (normal) long non-coding RNA (lncRNA) NEAT1. Paraspeckles are non-essential but are proposed to have a role increasing microRNA production by allowing a platform for miRNA processing, and sequestering paraspeckle associated proteins (such as SFPQ) to modulate gene expression in different cellular contexts such as circadian cycling43.
Paraspeckles form in the interchromatin space that surrounds the lncRNA NEAT1 locus on chromosome 11. RNA polymerase (pol) II transcribes NEAT1.1 (3.7 kb) and NEAT1.2 (23 kb) from the same promoter. NEAT1.1 is poly-adenylated whereas NEAT1.2 encodes a tRNA-like structure at its 3’ end. This structure is cleaved by RNase P44 and the resulting 3’ end of NEAT1.2 forms a triplex structure that promotes nuclear retention45. Whereas paraspeckles are found in most cultured cell lines, they are only found in a subset of cells in mouse tissue46. Mice lacking NEAT1 are viable and fertile46 but show defects in mammary gland development and lactation47.
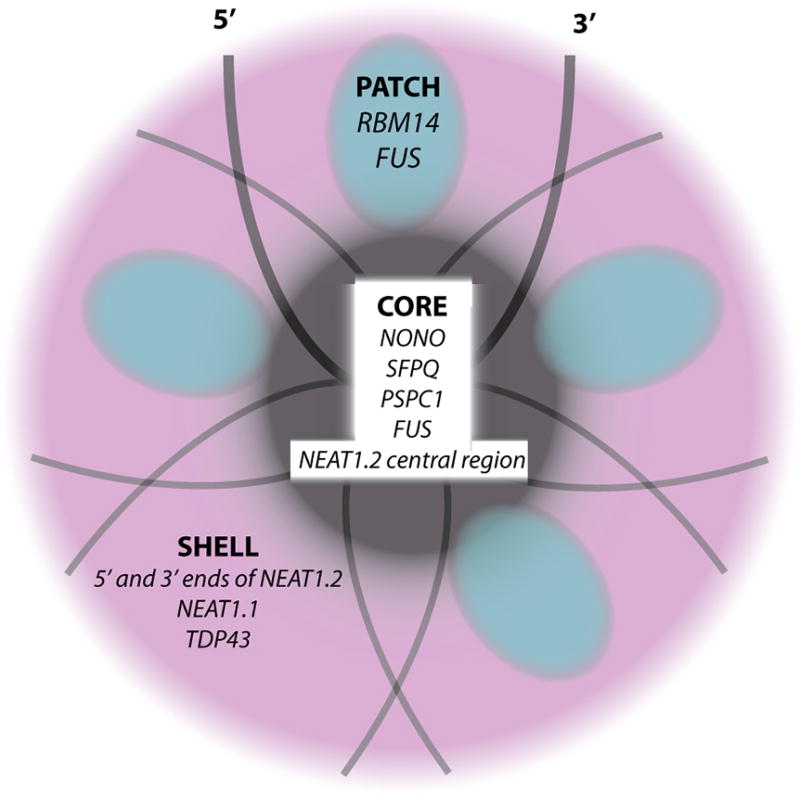
NEAT1.2 acts as a structural scaffold to promote paraspeckle formation44; 48; 49; 50. Whereas overexpression of NEAT1.1 enhances paraspeckle formation, it is unable to scaffold their formation on its own46. FRAP analysis reveals that MS2 tagged NEAT1 is relatively immobile compared with paraspeckle proteins that are in dynamic equilibrium with the nuclear matrix51. The 5’ and 3’ ends of NEAT1.2 and NEAT1.1 reside at the periphery of the paraspeckle surrounding a core composed of the central region of NEAT1.2 and the paraspeckle proteins NONO, FUS, and SFPQ (Figure 2)52; 53. The paraspeckle proteins NONO and SFPQ possess polymerization domains consisting of coiled-coil regions that come together to form long fibrils that are thought to coat NEAT1.254; 55. Surrounding the paraspeckle core is a shell comprised of NEAT1.1, RNAs (including mRNAs and introns) enriched in GA repeats, and TDP43, an ALS-associated protein known to have roles in other phase separations53. The core-shell structure is further held together by patches of prion-like domains (PLDs) found in the essential paraspeckle proteins FUS and RBM1456. The paraspeckle phase separation is so stable that its disruption requires Trizol extraction coupled with heat or extensive needle shearing57. The PLD of FUS is required for this extremely stable interaction as its deletion prevents paraspeckle assembly and eases extraction of NEAT1.257; 58. It is unknown whether specific sequences of NEAT1.2 or NEAT1.1 are required to promote phase separation during paraspeckle formation. Interestingly, TDP43 is recruited to paraspeckles, and binds specifically to (GU)x59. Several (GU)≥10 stretches are found in NEAT1.2, but their role in recruiting TDP43 is not known.
Figure 2. Paraspeckle Core-Shell Structure.
The lncRNA NEAT1.2 (grey lines) acts as structural scaffold, organizing the paraspeckle core (dark grey) around its central region and the shell around its 5’ and 3’ termini. Paraspeckles are further held together by patches (blue) of LCD containing proteins including RBM14 and FUS. NONO, SFPQ and PSPC1, proteins in the DBHS (Drosophila behavior/human splicing) family, as well as FUS make up the core while TDP43 is included in the shell.
Paraspeckle assembly is linked to the transcription of NEAT1.2 and pol II inhibitors disrupt paraspeckle formation51. Interestingly, the C-terminal domain (CTD) of Pol II contains 52 heptad repeats of the low complexity sequence YSPTSPS and this domain recruits other LCD containing proteins such as FUS18. It possible that the CTD of Pol II attracts the LCDs of paraspeckle proteins to seed a phase separation during NEAT1.2 transcription.
In addition to NEAT1 lncRNA, adenosine to inosine (A-to-I) edited RNAs are concentrated at paraspeckles48; 60. The main paraspeckle protein NONO (p54/nrb) has high binding affinity for A-to-I edited RNA61. A-to-I editing typically occurs on double stranded nuclear RNAs, most commonly associated with inverted Alu elements62; 63. Addition of Alu elements to the 3’UTR of a GFP reporter transcript causes NONO binding and nuclear retention62. Another target of A-to-I editing is a cationic amino acid transporter 2 (mCAT2) encoding transcript. The open reading frame encoding mCAT2 is included in two distinct transcripts that use different promoters and polyadenylation signals. The CAT2-transcribed nuclear RNA (CTN) uniquely undergoes A-to-I editing in its 3’UTR which results in nuclear retention. In response to stress, the 3’UTR of CTN transcripts is cleaved, allowing escape to the cytoplasm and translation of CAT2 protein60. The A-to-I edited CTN-RNA localizes to paraspeckle regions60 in a A-to-I independent manner64. In the absence of NEAT1 and paraspeckles, CTN-RNA interacts with paraspeckle proteins and accumulates in perinucleolar regions64. It is unclear whether paraspeckles contribute to nuclear retention of A-to-I edited RNAs or whether A-to-I edited RNAs modulate paraspeckle dynamics.
Interestingly, transfection of oligos with a phosphorothioate backbone can assemble paraspeckle-like structures that lack NEAT165. Phosphorothioate bonds include a non-bonding sulfur instead of an oxygen in the sugar linkages and are used to promote stability from nuclease degradation in synthetic oligos66. It is possible that increased stability allows for promiscuous binding to paraspeckle proteins. Transfection of these oligos also promotes cytoplasmic foci of unknown composition65.
Nuclear speckles
Interphase nuclei typically contain 20–50 dynamic phase dense foci known as nuclear speckles. Ultrastructural analysis indicates that nuclear speckles are composed of dense clusters that are connected by fibrils67. Nuclear speckles are proposed to have roles in promoting mRNA maturation and are localized with sites of active RNA pol II transcription. mRNA production and maturation, including transcription, splicing, polyadenylation, and mRNA export, are often coupled and the proteins involved in these processes localize to nuclear speckles68. The lncRNA Malat1 (also known as NEAT2) is concentrated at nuclear speckles but is not required for their assembly. The findings that splicing factors are concentrated at nuclear speckles and knockdown of the mRNA export complex TREX increases their size suggests possible roles in mRNA splicing and/or nuclear export68. Downregulation of Malat1 causes an increase in cytoplasmically localized poly(A) RNA69 suggesting that Malat1 may act at nuclear speckles to modulate splicing and/or nuclear retention of mRNA.
Amyloid (A) bodies
Recently described stress-induced nuclear foci known as amyloid (A) bodies assemble in response to the transcription of stress specific lncRNAs. These lncRNAs are derived from the ribosomal intergenic spacer (rIGS) region of the ribosomal DNA locus and are transcribed under specific stress conditions70. The acidic and hypoxic conditions found in the tumor microenvironment induce the assembly of A bodies. In a mouse model of tumorigenesis, knockdown of one of these lncRNAs prevents tumor growth and promotes a cellular state of dormancy70. A-bodies are named for their amyloid-like state. Amyloids are proteins that form tight interactions through β-sheets to generate fibrils that experimentally stain with amyloid specific stains such as CongoRed. Because of the fibril state and the lack of exchange with the surrounding environment, amyloids represent a solid-like state71. Whereas the assembly of most amyloids is irreversible, A-bodies disassemble in cells that recover from stress, suggesting they play a dynamic role in modulating the stress response program and cell survival.
Cajal bodies
In 1900, Ramon y Cajal first identified nuclear coiled bodies that were later renamed Cajal bodies after their discoverer72. Cajal bodies are sites of small nuclear ribonucleoprotein (snRNP) biogenesis, including the spliceosome and the telomerase RNP complex73. Cajal bodies are enriched in small nuclear RNAs (snRNAs), which includes small nucleolar RNAs (snoRNAs) and small Cajal body-specific RNAs (scaRNAs). snoRNAs and snaRNAs assemble into complexes with proteins to form snoRNPs and scaRNPs, respectively. These complexes mediate 2’-O-ribose methylation and pseudouridylation of nucleotides in rRNA and spliceosomal RNAs and are required for functional complexes. snoRNAs traffic through Cajal bodies enroute to the nucleolus and in some cell types, Cajal bodies are found in association with nucleoli. Like other nuclear MLOs, Cajal bodies are tied to transcriptional activity74. During the cell cycle, transcriptional arrest is accompanied by the disappearance of Cajal bodies75. Cajal bodies cluster around sites of snRNA transcription and intron-encoded snRNAs are then trafficked to Cajal bodies76.
Although Cajal bodies are not observed in all cells, both snoRNPs and snRNPs are required for assembly of ribosomes and spliceosomes, respectively. An essential step in the assembly of functional spliceosomal snRNPs requires the protein coilin. Coilin is also required for assembly of Cajal bodies, and loss of coilin results in defects in splicing, and snRNP assembly77. Coilin is not a component of snRNP complexes, but is thought to play a role in concentrating the proteins and RNAs required for their assembly, acting as an aggregating factor using its multimodular domains77. iCLIP, a method of using UV crosslinking and immunoprecipitation to identify direct protein-RNA interactions, shows that coilin interacts with hundreds of snRNAs, including those targeted to the nucleolus76, indicating coilin directly interacts with snRNA. Potentially, this direct interaction between coilin and snRNAs aids in the assembly of Cajal bodies. scaRNAs are sufficient to promote de novo Cajal body assembly, as an MS2-tagged scaRNA causes Cajal body formation78. This occurs in a manner similar to artificially tethering Cajal body proteins (coilin, SMN, etc) to DNA via a Lac operon to assess de novo Cajal body assembly78.
Unlike other MLOs, the primary sequence motifs that cause retention of scaRNAs in Cajal bodies have been identified and characterized. The protein WRD79 recognizes scaRNAs promoting their localization and retention in Cajal bodies. Box H/ACA scaRNAs require the CAB box (ugAG found in the loop at the 5’ or 3’ end of scaRNA) for Cajal body localization79. Addition of this motif to snoRNAs results in retention in Cajal bodies79 and mutation of this motif prevents Cajal body localization76; 79. Targeting of box C/D scaRNAs requires G:U wobble base pairing in a helical region of a hairpin yet is not dependent of the loop80. Potentially WRD79 recognizes this atypical helix caused by G:U wobble base pairs.
Nucleolus
Due to their easy detection by light microscopy, the size and number of nucleoli has been used as a cancer diagnostic for over 100 years. Only recently have we begun to appreciate the unique LLPS properties of nucleoli. Using Xenopus laevis oocyte germline vesicles which are similar in composition to somatic nucleoli, Brangwynne and colleagues were the first to demonstrate the dynamic and liquid-like properties of nucleoli81.
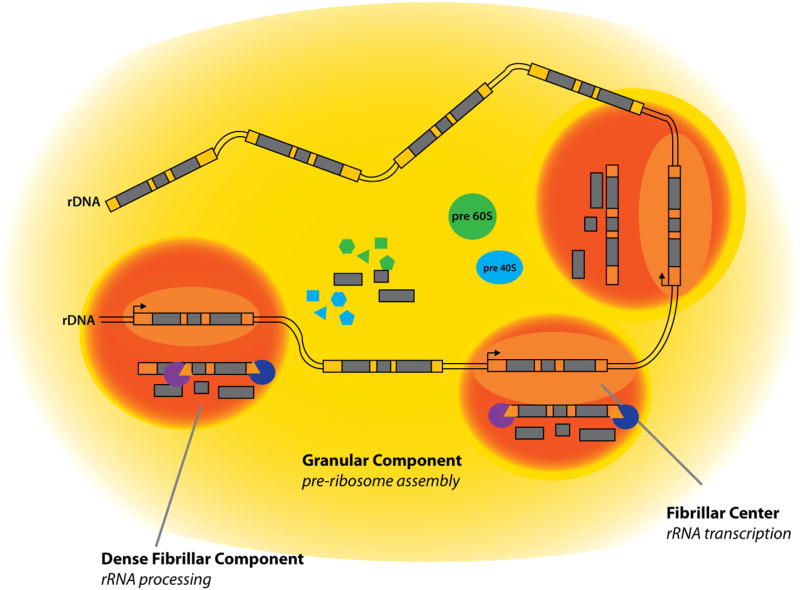
The nucleolus is located around clusters of ribosomal DNA (rDNA) repeats known as nucleolar organization regions. These genomic regions contain clusters of repeats that code for ribosomal RNA (rRNA). The nucleolus is further organized into multiple subregions that are defined by proteins involved in rRNA processing and ribosome assembly; rDNA is transcribed in the fibrillar center, rRNA processing (cleavage and modification) to produce 28S, 5.8S, and 18S rRNAs occurs in the dense fibrillar component and assembly into a pre-ribosome occurs in the granular component (Figure 3). Further processing takes place in the cytoplasm to generate fully competent 40S and 60S ribosomal subunits.
Figure 3. Nucleolar processes and structures.
The nucleolus is composed of three phase separations that separate the different processes in rRNA production and maturation and are structured around rDNA. Fibrillar center (light orange) is where rDNA is transcribed into rRNA, dense fibril component (orange) where rRNA is processed, granular component (yellow) where rRNA is further processed and assembled into pre-40S (blue oval) and pre-60S (green oval) ribosomal subunits with ribosomal proteins (blue and green shapes).
The multiple domains of the nucleolus couple processing events within distinct LLPS compartments that maintain boundaries for spatial and temporal processing of rRNA. As such, rRNA maturation and ribosome production take place within discrete phase separated domains. Just as concentration of the hammerhead ribozyme in an aqueous two phase system generated by the molecular crowding agents polyethylene glycol and dextran enhances activity by 70 fold82, the super concentration of enzymes and substrates within nucleoli is likely essential for efficient rRNA processing and ribosome subunit assembly. It would be interesting to use a similar system to assess rRNA processing, yet surprisingly, the detailed events of rRNA processing remain unclear in mammalian cells83.
The existence of nucleolar subregions raises the question of how multiple distinct phase separated domains can coexist. In vitro studies using purified nucleophosmin (NPM1) and fibrillarin (FIB1), proteins that localize to dense fibrillary and granular components, respectively, are instructive in this regard. In the presence of rRNA, purified NPM1 and FIB1 can independently assemble droplets in vitro. These droplets possess distinct biophysical properties: when mixed together, they do not fuse with one another but rather form two-layered droplets with the FIB1/rRNA phase inside an engulfing NPM1/rRNA phase, a restriction conferred by differential surface tensions associated with their respective RNA-binding domains84.
Over 4500 proteins have been identified as components of the nucleolus85. Scott et al analyzed the sequence and structural features of a large number of well curated nucleolar localization signals and, based on this, developed a bioinformatics predictor of nucleolar localization signals. Their analysis showed that these signals are enriched in basic residues that are located within solvent-accessible regions of proteins86. Mitrea and colleagues refined this view showing that the majority of nucleolar proteins exhibit multiple segments containing two or more closely spaced arginine residues within regions predicted to be intrinsically disordered87. They showed that proteins with multivalent arginine-motifs interact with the granular component, NPM1, which exhibits two highly acidic regions that interact with arginine-motifs in ribosomal and other nucleolar proteins. NPM1 also contains an RNA binding domain that is required for the nucleolar localization. Nucleolar components show specificity toward rRNA binding but how this is achieved remains unclear and is difficult to test in cells due to the necessity of rDNA/rRNA to the integrity of the cell and the large number of proteins associated with the nucleolus.
Stress granules (SGs)
Stress can trigger the assembly of several MLOs. Indeed, studies defining the principles of SG assembly are now accepted as those that define the major properties of MLOs: a discrete domain88 that is assembled by low affinity interactions between low complexity or prion related domains89 that are visible by light microscopy88, lack a limiting membrane89, show molecular constituents dynamically exchanged with the surrounding environment90. The subsequent realization that these properties are shared by several cellular entities revealed the importance of these properties for the organization of cellular processes.
SGs are cytoplasmic assemblies containing translation initiation factors, polyadenylated (poly(A)) mRNAs, 40S ribosomal subunits, RNA binding proteins, and selective signaling molecules (reviewed in91). SGs assemble in response to blocked translation initiation which results in a sudden increase in untranslated, non-polysomal mRNAs. This can result from stress-induced eIF2α phosphorylation or disruption of the eIF4F complex88. eIF2α phosphorylation depletes the initiator tRNA-methionine eIF2α-GTP complex, limiting translation initiation and causing downregulation of bulk translation92,93; 94; 95; 96. The eIF4F complex (composed of eIF4E, eIF4G and eIF4A) binds the m7GTP mRNA cap and under optimal growth conditions is the rate limiting step in translation initiation for most mRNAs (reviewed in92). Interestingly, not every stimulus that disrupts the eIF4F complex or causes eIF2α phosphorylation triggers SG formation: doxorubicin promotes robust eIF2α phosphorylation but not SG assembly97 and 4EGI-1 disrupts eIF4E:eIF4G interaction but does not induce robust SG assembly98, suggesting that additional changes are necessary to trigger the condensation step in SG assembly.
SGs are in dynamic equilibrium with active translation. SGs can be forcibly disassembled by adding emetine or cycloheximide, drugs that “freeze” ribosomes on polysomes. Conversely, SG assembly is enhanced by puromycin, a drug that disassembles polysomes90. It is important to note that puromycin requires a sub-SG inducing dose of stress to assemble SGs99. This implies that SGs are assembled when actively translating ribosomes “run off” the mRNA to release an excess of free mRNA100. Consistent with this, SGs are enriched in poly(A) mRNA and 40S, but not 60S, ribosomal subunits100; 101; 102. Thus, stalled 48S initiation complexes are core components of SGs. Interestingly, transfection of excess mRNA is sufficient to induce SGs but only in a subpopulation of cells102. Whether this requires the assembly of pre-initiation complexes on transfected mRNAs remains to be determined. Similarly, the G-quadruplex structure of the C9ORF72 ALS/FTD-associated GGGGCC repeat RNA or an intermolecular G-quadruplex formed from the 5’ fragments of cleaved tRNAsAla/Cys promote SG formation in a structure-dependent manner103; 104. These findings reveal a key role for RNA in SG assembly.
SGs are thought to sequester mRNAs during stress to preserve the transcriptome, allowing resumption of translation as cells repair stress-induced damage and recover. This should minimize energy expenditure in cells intermittently exposed to stress. This process also allows cells to partition mRNAs based on the current translational needs of the cell: mRNAs encoding proteins that repair stress-induced damage are excluded from SGs and translated, whereas “housekeeping” transcripts are translationally stalled and sequestered at SGs. Consistent with this notion, mRNAs encoding the heat shock chaperones Hsp70105 and Hsp 90106 are excluded from SGs, while abundant housekeeping mRNAs encoding c-myc and β-actin are concentrated at SGs (Table 1)106. Localization of specific mRNAs to SGs has been experimentally visualized using fluorescence in situ hybridization (FISH), tracking RNA via programmable RNA-targeting Cas9, and tagging RNA with the MS2 system105; 106; 107; 108; 109; 110; 111; 112.
Table 1.
| mRNAs localized to SGs | |||
|---|---|---|---|
| Symbol | mRNA name | Method of Detection | Ref. |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | FISH | 103 |
| H19 | Long noncoding RNA H19 | FISH | 103 |
| IGF-II | Insulin-like growth factor II | FISH | 103 |
| MYC | Myc proto-oncogene protein | FISH | 103 |
| ACTB | Beta actin | MS2, FISH and RCas9 | 103,105,106 |
| TFRC | Transferrin receptor protein 1 | FISH and RCas9 | 105 |
| CCNA2 | Cyclin-A2 | FISH and RCas9 | 105 |
| P21 | Cyclin-dependent kinase inhibitor 1 | FISH | 108 |
| AHNAK | Neuroblast differentiation-associated protein AHNAK | FISH | 104 |
| DYNC1H1 | Cytoplasmic dynein 1 heavy chain 1 | FISH | 104 |
| NORAD | Noncoding RNA activated by DNA damage | FISH | 104 |
| COX2 | Cyclooxygenase-2 | FISH | 109 |
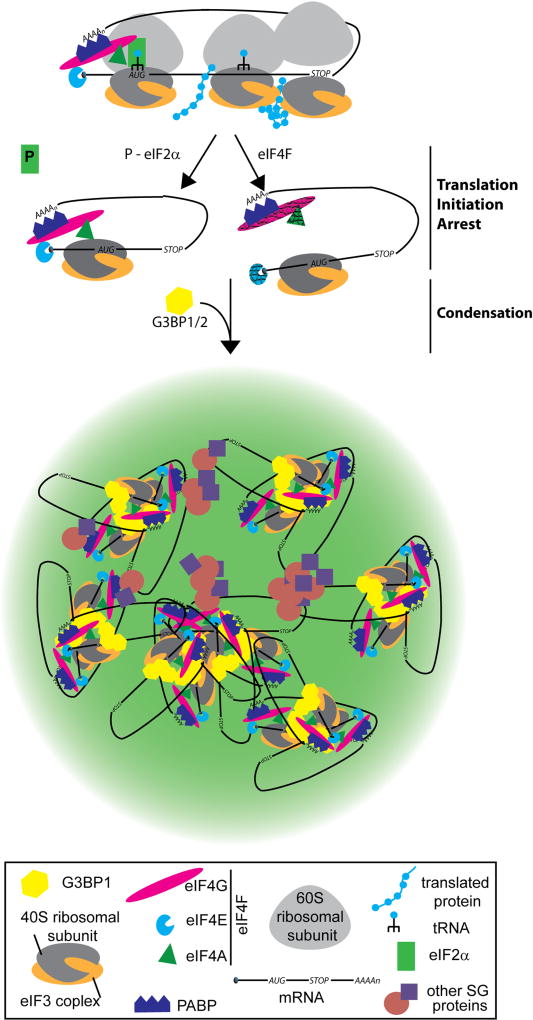
While translation initiation arrest and polysome disassembly are required for SG formation, the aggregation of untranslating mRNAs at SGs requires the related proteins G3BP1 and 2. Knocking out one of these proteins decreases SG assembly, but elimination of both G3BP1/2 completely blocks SG assembly in response to most stresses113; 114. Caprin1 and USP10 compete for G3BP1 binding to seed SG condensation - G3BP1:USP10 prevents while G3BP1:Caprin1 promotes this phase separation event114. Specifically, an FGDF motif in USP10 binds G3BP1 and this same motif is found in viral proteins that inhibit SG assembly to evade the cellular stress response114; 115. Furthermore, G3BP1/2 bind the 40S but not the 60S ribosomal subunit and this requires the G3BP1-RGG domain in an RNA dependent manner. This domain is also required for SG assembly114; 116, suggesting that RNA further contributes to SG condensation yet it is unclear how. This order of events is consistent with a two-step model requiring: (1) an excess of free cytoplasmic mRNA and (2) condensation by G3BP1/2 (Figure 4). G3BP1 has been implicated in the rearrangement of ALS-associated mRNP aggregates composed of mRNA and FUS or TDP43 as judged by atomic force microscopy102; 117, implying an ability to rearrange or disaggregate mRNPs. It remains to be determined whether this in vitro observation relates to SG formation in cells and furthermore whether the ability to rearrange mRNPs promotes or prevents SG formation.
Figure 4. Stress granule assembly.
Stress granules assembly requires two steps: (1) A block in translation initiation that is caused by eIF2α phosphorylation (denoted P) or modulation of the eIF4F complex (denoted with grey lines). This leads to ribosome run-off and an increase in mRNAs with translation initiation stalled 48S complexes. (2) Condensation of these translation initiation stalled mRNAs is then mediated by G3BP1. Other RNA binding proteins and LCD containing proteins are further recruited including signaling molecules.
G3BP1 has been reported to be post-translationally modified by methylation118, phosphorylation119, poly(ADP) ribosylation120, acetylation121, and ubiquitination122. PRMT1 and PRMT5 methylate multiple arginine residues in the RGG domain of G3BP1 and mutation of key Arg residues (429, 435, 443, 447, 460) to Lys prevents methylation and promotes SG formation. In contrast, Arg to Phe mutants serve as methylation mimics that prevent SG formation123. Consistently, inhibition of asymmetric arginine methylation, a modification most likely added by PRMT1, promotes SG formation. PRMT1, but not PRMT5, is recruited to SGs123. Potentially differential methylation/demethylation modulates SG assembly and shuttling in and out of SGs. As such, modulating arginine methylation may be involved in recruiting proteins to SGs124. Arginine methylation has been shown to alter subcellular localization as well as interactions with RNA and/or proteins125; 126. Gly-Arg-Gly is the consensus arginine methylation site that is enriched in RNA binding domains and LCD125; 126, possibly linking arginine methylation to the process of phase separation.
Studies using high resolution microscopy show that SGs are structurally heterologous with central regions of higher density101; 127. Biochemical purification of SG cores using differential centrifugation and G3BP1 immunoaffinity purification identifies an mRNP interactome that is likely related to SGs128. BioID/APEX labeling, methods that allow the identification of proteins in close proximity, have uncovered an interactome that is largely pre-assembled in the absence of stress129; 130. In the presence of stress, the recruitment of several key proteins and RNAs may be essential for phase separation into SGs. Importantly, interactions between G3BP1 and the eIF3 complex were shown to be stress specific129. Knockdown of the eIF3 complex was previously shown to block SG assembly131, further validating the importance of this interaction. Approximately 20% of G3BP1-associated proteins were found to be specific to the stress or cell type used, indicating a previously unappreciated level of heterogeneity in SGs129.
Yeast SGs and mammalian SGs have some distinctive differences including their composition and biophysical properties: whereas yeast SGs adopt a more solid-like state, human SGs adopt a more liquid-like state132. Pab1, yeast PABP, acts as a stress sensing signal that alters its binding interaction with RNA in response to heat shock and low pH133. Interestingly, RNA prevents Pab1 phase separation in vitro. Mutations that decrease Pab1 phase separation cause deleterious effects on the ability of yeast to grow under stress conditions, consistent with Pab1 acting as a stress sensor that modulates its biophysical properties133. It is unknown whether this mechanism of stress sensing occurs in other proteins or whether this mechanism is conserved in mammals.
Different stresses trigger unique stress responses and not all stress-induced cytoplasmic foci are SGs99. For example, hyperosmolar conditions (as experimentally modeled by increases in NaCl) induce cytoplasmic foci that contain classical SG proteins (G3BP1, eIF3b, mRNAs) but are not in dynamic equilibrium with active translation as judged by the effects of cycloheximide and puromycin. This complicates the prospect of therapeutically targeting SGs (or more generally phase separations in disease states such as cancer or neurodegeneration) as careful analysis of the compositional and functional effects of candidate drugs will be required.
Processing bodies (P-bodies)
While P-bodies house many mRNA decay factors including the decapping enzyme DCP1 and the exonulease Xrn1134; 135, their assembly is not required for mRNA decay. Translation repression and decay are still carried out in yeast lacking key P-body components. Similarly, in cells lacking P-bodies, bulk mRNA decay, non-sense mediated mRNA decay, and RNA-mediated gene silencing still occur136; 137.
Recent evidence suggests that P-bodies are primarily sites of mRNA storage. Using a method of particle sorting to purifying P-bodies, Hubstenberger et al show that approximately one third of all mRNAs are recruited to P-bodies138. These mRNAs are translationally repressed with a large fraction comprising mRNA regulons linked by a common biological process such as chromatin remodeling138. This is consistent with earlier reports that linked P-bodies to sites of translational repression139. P-body size and number increases with stress and decreases with removal of stress, correlating P-bodies with translational repression. mRNAs from P-bodies can then return to active translation140. Like SGs, P-bodies decrease in response to cycloheximide, which “freezes” polysomes resulting in a dynamic disassembly of P-bodies. This freezing of polysomes depletes the pool of non-ribosome bound mRNAs in the cytoplasm decreasing the mRNAs available to promote SGs and P-bodies. Recent evidence questions the role of P-bodies as sites of mRNA storage. Using a modified MS2 system (MBSV6) to label RNAs, Tutucci and colleagues found that during stress two mRNAs do not localized to P-bodies for storage141. Potentially, this indicates that the pool of mRNAs that localize to P-bodies does not change after stress, yet more experiments are needed to confirm this hypothesis.
RNA is required for the assembly and structural integrity of P-bodies. RNase treatment of purified P-bodies leads to their disruption142. In addition, DDX6 is required for P-body formation143 suggesting that rearrangement of RNA is required for P-body assembly. FRAP analysis using MS2-YFP tagged mRNAs suggests that a population of mRNA is immobile or slowly exchanges with the surrounding cytoplasm and that this increases with stress. mRNAs recovered from purified P-bodies do in fact have poly(A) tails of various lengths138. This is contrary to observations made using FISH where poly(A) RNA is not detected in P-bodies144. Moreover, immunofluorescence does not detect PABP at P-bodies. These discrepancies could be due to differential accessibility of antibodies and/or oligonucleotides used as P-body markers. Alternatively, post-translational modification of PABP, post-transcriptional RNA modification of poly(A) tails or extensive poly(A) tail base pairing with other RNAs could account for these experimental variations. Interestingly, when sequestered into phase separated domains in vitro miRISC more efficiently deadentylates a target RNA145, yet whether this occurs in a cellular compartment such as P-bodies remains to be determined.
P-bodies and SGs can dock and the extent of docking is largely dependent on the stress conditions146. Docking can be stabilized by overexpression of TTP or BRF1147. TTP localization to SGs is stress specific and regulated by its interaction with 14-3-3. TTP binds to 14-3-3 when phosphorylated by MK2146; 148. Interestingly, in the absence of stress overexpression of cytoplasmic polyadenylation element-binding protein 1 (CPEB1) causes localization of P-body components to SGs149 and similarly overexpression of p54/Rck drives fusion of P-bodies and SGs150, suggesting these granules have similar properties that when modulated can be tipped to promote fusion. Furthermore some protein components are shared between SGs and P-bodies, including YB-1 and TIA1/TIAR151; 152. It is unclear whether mRNAs are shared or transferred between these compartments and whether RNA plays an active role in docking.
PARP
The amino acids lysine, arginine and glutamic acid can be post-translationally modified with ADP-ribose: mono(ADP)-ribosylation typically targets arginine whereas poly(ADP)-ribosylation (PAR) typically targets lysine and glutamic acid. PAR modifications can cause 2–200 (ADP)-ribose units to be added in a branching pattern. These modifications are added by poly(ADP)-ribose polymerases (PARP) and are removed by poly(ADP)-ribose glycohydrolase (PARG)153. Several PARP and PARG enzymes are recruited to SGs and PAR-modified proteins are enriched in SGs120. Similarly, a comparison of PAR modified proteins and proteins condensed by b-isox reveals a significant overlap153; 154, indicating an enrichment in PAR modification within RNA granules.
PAR is negatively charged like RNA and can act like RNA to decrease the phase boundary and promote in vitro phase separation16; 154. PAR causes a high local density of negative charges that recruits positively charged arginine residues in RGG domains154. In this manner, PAR modifications can act as a de novo seed for phase separation. Such enrichment occurs at sites of DNA damage where PARP1 is rapidly recruited16; 154. This model can explain the earliest stages of the DNA damage response in which an initial recruitment of PARP1 causes rapid PAR modifications that recruit LCD proteins such as FUS, EWS, and TAF15. In cells, FUS and other LCD-containing proteins are recruited to sites of DNA damage in a PAR-dependent manner16, suggesting PAR modifications nucleate a phase separated state at the site of DNA damage. Additional post-translational modifications, including phosphorylation and ubiquitination, play important roles in the DNA damage response. It has been suggested, but not tested, that phosphorylation by DNA damage response kinases further regulates the PAR-induced DNA damage phase separation. It has been proposed that negative charges introduced by phosphorylation reverse the phase separation, allowing the PAR-mediated DNA damage phase separation to be transient in nature154. Phosphorylation has been shown to control other phase separations8; 18. Furthermore, many granule related proteins bind to PAR, including the SG regulating proteins G3BP1/2, and the paraspeckle proteins NONO and SFPQ. G3BP1 binds the PAR modification via its glycine-arginine rich domain155.
RNA contribution
Recent work has greatly expanded our understanding of MLOs. With the general concepts of phase separation and the necessity of multivalent weak interactions for MLO formation in place, the field needs to address the specificity of phase separation events. If the only factors involved were weak multivalent interactions and surface tension then presumably all phase separated domains within a cell would fuse together. Yet MLOs, such as docked SGs and P bodies, appear to maintain their separate biophysical properties. Understanding what is driving these differences is a key question moving forward.
MLOs are composed of different RNA species and these RNA species contribute to the differences in cellular phase separations. A more thorough understanding of the cis elements or RNA secondary structures that direct MLO targeting or assembly is needed, such as the identified motifs in scaRNAs that allow for Cajal body targeting and G-quadruplexes in forming SGs. Similarly, an understanding of how LCD and RNA binding domains contribute to the assembly of distinct phase separated domains is required. While we know that LCDs are enriched in sites that can be post-translationally modified, a more thorough understanding of when post-translational modifications occur and how these post-translational modifications impact phase separation will be important moving forward. Recent connections between MLO and diseases including neurodegeneration, cancer and viral infection, have promoted further interest that will aid in expanding our knowledge.
Acknowledgments
We thank Anderson and Pavel Ivanov lab members for discussion. This work was supported by the NIH (GM111700 and CA168872 to PA, and AI007306 to MMF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 3.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–79. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013;1834:932–51. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–19. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–40. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–25. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Schwartz JC, Cech TR. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015;43:7535–43. doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A. 2018;115:2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, Hein MY, Osborne-Nishimura E, Mahamid J, Jahnel M, Jawerth L, Pozniakovski A, Eckmann CR, Julicher F, Hyman AA. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell. 2016;166:1572–1584. e16. doi: 10.1016/j.cell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–30. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–77. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 17.Lim L, Wei Y, Lu Y, Song J. ALS-Causing Mutations Significantly Perturb the Self-Assembly and Interaction with Nucleic Acid of the Intrinsically Disordered Prion-Like Domain of TDP-43. PLoS Biol. 2016;14:e1002338. doi: 10.1371/journal.pbio.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–47. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R. Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep. 2018;22:1401–1412. doi: 10.1016/j.celrep.2018.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–58. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22:R45–51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–77. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 24.Aslanidis C, Jansen G, Amemiya C, Shutler G, Mahadevan M, Tsilfidis C, Chen C, Alleman J, Wormskamp NG, Vooijs M, et al. Cloning of the essential myotonic dystrophy region and mapping of the putative defect. Nature. 1992;355:548–51. doi: 10.1038/355548a0. [DOI] [PubMed] [Google Scholar]
- 25.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 26.Harley HG, Brook JD, Rundle SA, Crow S, Reardon W, Buckler AJ, Harper PS, Housman DE, Shaw DJ. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992;355:545–6. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- 27.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–7. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 28.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–70. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 29.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–80. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 30.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–73. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 31.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Consortium I, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–6. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, Johnston A, Overstreet K, Oh SY, Todd PK, Berry JD, Cudkowicz ME, Boeve BF, Dickson D, Floeter MK, Traynor BJ, Morelli C, Ratti A, Silani V, Rademakers R, Brown RH, Rothstein JD, Boylan KB, Petrucelli L, Disney MD. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–50. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidle S. Therapeutic applications of quadruplex nucleic acids. 1. Elsevier/Academic Press; London ; Waltham, MA: 2012. [Google Scholar]
- 38.Fay MM, Anderson PJ, Ivanov P. ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Rep. 2017;21:3573–3584. doi: 10.1016/j.celrep.2017.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, Manley JL. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016;5 doi: 10.7554/eLife.17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Hu J, Ludlow AT, Pham JT, Shay JW, Rothstein JD, Corey DR. c9orf72 Disease-Related Foci Are Each Composed of One Mutant Expanded Repeat RNA. Cell Chem Biol. 2017;24:141–148. doi: 10.1016/j.chembiol.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciesiolka A, Jazurek M, Drazkowska K, Krzyzosiak WJ. Structural Characteristics of Simple RNA Repeats Associated with Disease and their Deleterious Protein Interactions. Front Cell Neurosci. 2017;11:97. doi: 10.3389/fncel.2017.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem Sci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–9. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20:1844–9. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–78. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–7. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, Yanaka K, Kingston RE, Hirose T, Bond C, Fox A, Nakagawa S. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214:817–30. doi: 10.1083/jcb.201601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobson L, Nyitray L, Gaspari Z. A conserved charged single alpha-helix with a putative steric role in paraspeckle formation. RNA. 2015;21:2023–9. doi: 10.1261/rna.053058.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee M, Sadowska A, Bekere I, Ho D, Gully BS, Lu Y, Iyer KS, Trewhella J, Fox AH, Bond CS. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 2015;43:3826–40. doi: 10.1093/nar/gkv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, Hosoki K, Goshima N, Kawaguchi T, Hatters D, Trinkle-Mulcahy L, Hirose T, Bond CS, Fox AH. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol. 2015;210:529–39. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chujo T, Yamazaki T, Kawaguchi T, Kurosaka S, Takumi T, Nakagawa S, Hirose T. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 2017;36:1447–1462. doi: 10.15252/embj.201695848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shelkovnikova TA, Robinson HK, Troakes C, Ninkina N, Buchman VL. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum Mol Genet. 2014;23:2298–312. doi: 10.1093/hmg/ddt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, Ratti A. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem. 2012;287:15635–47. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–63. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–75. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 62.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest AR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 64.Anantharaman A, Jadaliha M, Tripathi V, Nakagawa S, Hirose T, Jantsch MF, Prasanth SG, Prasanth KV. Paraspeckles modulate the intranuclear distribution of paraspeckle-associated Ctn RNA. Sci Rep. 2016;6:34043. doi: 10.1038/srep34043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen W, Liang XH, Crooke ST. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014;42:8648–62. doi: 10.1093/nar/gku579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shaw JP, Kent K, Bird J, Fishback J, Froehler B. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 1991;19:747–50. doi: 10.1093/nar/19.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fei J, Jadaliha M, Harmon TS, Li ITS, Hua B, Hao Q, Holehouse AS, Reyer M, Sun Q, Freier SM, Pappu RV, Prasanth KV, Ha T. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci. 2017;130:4180–4192. doi: 10.1242/jcs.206854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galganski L, Urbanek MO, Krzyzosiak WJ. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017;45:10350–10368. doi: 10.1093/nar/gkx759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Audas TE, Audas DE, Jacob MD, Ho JJ, Khacho M, Wang M, Perera JK, Gardiner C, Bennett CA, Head T, Kryvenko ON, Jorda M, Daunert S, Malhotra A, Trinkle-Mulcahy L, Gonzalgo ML, Lee S. Adaptation to Stressors by Systemic Protein Amyloidogenesis. Dev Cell. 2016;39:155–168. doi: 10.1016/j.devcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 73.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA. 2013;4:17–34. doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- 74.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis--evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;120:841–52. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell. 2014;56:389–99. doi: 10.1016/j.molcel.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 77.Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17:403–9. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- 78.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–7. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 79.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–93. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marnef A, Richard P, Pinzon N, Kiss T. Targeting vertebrate intron-encoded box C/D 2'-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res. 2014;42:6616–29. doi: 10.1093/nar/gku287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–9. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nat Chem. 2012;4:941–6. doi: 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]
- 83.Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6:225–42. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. NOPdb: Nucleolar Proteome Database-2008 update. Nucleic Acids Res. 2009;37:D181–4. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 2010;38:7388–99. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife. 2016;5 doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–98. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–68. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Srivastava SP, Kumar KU, Kaufman RJ. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–23. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 94.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 96.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280:16925–33. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 97.Szaflarski W, Fay MM, Kedersha N, Zabel M, Anderson P, Ivanov P. Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget. 2016;7:30307–22. doi: 10.18632/oncotarget.8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mokas S, Mills JR, Garreau C, Fournier MJ, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell. 2009;20:2673–83. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci. 2017;130:927–937. doi: 10.1242/jcs.199240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci. 2009;122:3619–26. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- 102.Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Pietrement O, Pastre D. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 2014;42:8678–91. doi: 10.1093/nar/gku582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–23. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017;8:1127. doi: 10.1038/s41467-017-01278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–9. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 106.Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol. 2006;175:527–34. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell. 2017;68:808–820. e5. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nelles DA, Fang MY, O'Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–96. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zurla C, Lifland AW, Santangelo PJ. Characterizing mRNA interactions with RNA granules during translation initiation inhibition. PLoS One. 2011;6:e19727. doi: 10.1371/journal.pone.0019727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Unsworth H, Raguz S, Edwards HJ, Higgins CF, Yague E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24:3370–80. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- 111.Gareau C, Fournier MJ, Filion C, Coudert L, Martel D, Labelle Y, Mazroui R. p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS One. 2011;6:e20254. doi: 10.1371/journal.pone.0020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ansari MY, Haqqi TM. Interleukin-1beta induced Stress Granules Sequester COX-2 mRNA and Regulates its Stability and Translation in Human OA Chondrocytes. Sci Rep. 2016;6:27611. doi: 10.1038/srep27611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 2013;18:135–46. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 114.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–60. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Panas MD, Schulte T, Thaa B, Sandalova T, Kedersha N, Achour A, McInerney GM. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog. 2015;11:e1004659. doi: 10.1371/journal.ppat.1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol. 2016;215:313–323. doi: 10.1083/jcb.201609081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Abrakhi S, Kretov DA, Desforges B, Dobra I, Bouhss A, Pastre D, Hamon L. Nanoscale Analysis Reveals the Maturation of Neurodegeneration-Associated Protein Aggregates: Grown in mRNA Granules then Released by Stress Granule Proteins. ACS Nano. 2017;11:7189–7200. doi: 10.1021/acsnano.7b03071. [DOI] [PubMed] [Google Scholar]
- 118.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–26. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 119.Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony JP, Tocque B, Tazi J. A novel phosphorylation-dependent RNase activity of GAPSH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol. 1998;18:3956–65. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–99. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 122.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.013284. M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, Lloyd RE. Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J Biol Chem. 2016;291:22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xie W, Denman RB. Protein methylation and stress granules: posttranslational remodeler or innocent bystander? Mol Biol Int. 2011;2011:137459. doi: 10.4061/2011/137459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 127.Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. Elife. 2016;5 doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016 doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao FB, Bennett EJ, Lecuyer E, Yeo GW. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell. 2018;172:590–604. e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, Angers S, Morris Q, Fabian M, Cote JF, Gingras AC. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell. 2018;69:517–532. e11. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 131.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–31. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell. 2017;168:1028–1040. e19. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–24. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–501. [PMC free article] [PubMed] [Google Scholar]
- 136.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–49. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–81. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, Daunesse M, Bertrand E, Pierron G, Mozziconacci J, Kress M, Weil D. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell. 2017;68:144–157. e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 139.Lui J, Castelli LM, Pizzinga M, Simpson CE, Hoyle NP, Bailey KL, Campbell SG, Ashe MP. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 2014;9:944–54. doi: 10.1016/j.celrep.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–9. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tutucci E, Vera M, Biswas J, Garcia J, Parker R, Singer RH. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods. 2018;15:81–89. doi: 10.1038/nmeth.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–82. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ayache J, Benard M, Ernoult-Lange M, Minshall N, Standart N, Kress M, Weil D. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol Biol Cell. 2015;26:2579–95. doi: 10.1091/mbc.E15-03-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Aizer A, Kalo A, Kafri P, Shraga A, Ben-Yishay R, Jacob A, Kinor N, Shav-Tal Y. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sci. 2014;127:4443–56. doi: 10.1242/jcs.152975. [DOI] [PubMed] [Google Scholar]
- 145.Sheu-Gruttadauria J, MacRae IJ. Phase Transitions in the Assembly and Function of Human miRISC. Cell. 2018 doi: 10.1016/j.cell.2018.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stoecklin G, Kedersha N. Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol. 2013;768:197–211. doi: 10.1007/978-1-4614-5107-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–24. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci. 2005;118:981–92. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 150.Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell. 2008;19:4469–79. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 153.Leung AK. Poly(ADP-ribose): an organizer of cellular architecture. J Cell Biol. 2014;205:613–9. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MB, Streicher W, Jungmichel S, Nielsen ML, Lukas J. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Isabelle M, Gagne JP, Gallouzi IE, Poirier GG. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J Cell Sci. 2012;125:4555–66. doi: 10.1242/jcs.106963. [DOI] [PubMed] [Google Scholar]