Abstract
Background:
GCK gene variants have been reported to be associated with gestational diabetes mellitus (GDM) in the Caucasian population. There are no reports exploring this association in the Indian population.
Methods:
This cross-sectional study included subjects from Max Super Speciality Hospital, New Delhi, India, over a span of 6 months. Females diagnosed with GDM as per the International Association of the Diabetes and Pregnancy Study Groups (IADPSG) criteria were enrolled. Direct gene sequencing was performed to screen all 10 exons and promoter region of GCK gene.
Results:
Out of the total 1000 females screened, 154 subjects had any degree of hyperglycemia. GCK gene screening was done and we observed 11 variants in 80.4% (41/51) of the GDM subset and 89.6% (43/48) of the controls. Allele frequencies of observed variants were not different between the control subjects (12.5%) and those diagnosed with GDM (8.4%).
Conclusion:
To the best of our knowledge, this is the first report from north India exploring association of GCK variants with GDM and we do not observe any association of GCK variants with GDM in our study population.
CTRI Registration No: CTRI/2017/07/008964
Keywords: Gestational diabetes mellitus, glucokinase, GCK
Introduction
Gestational diabetes mellitus (GDM) is defined as any degree of glucose intolerance detected during pregnancy. As per the estimates of International Diabetes Federation (IDF), 21.3 million of the live births to women in 2017 had hyperglycemia in pregnancy and 86.4% of those were due to GDM.1 In India, its prevalence varies from 3.8% to 21%, which could be attributed to the regional variations and the diagnostic criteria used.2 Major risk factors for development of GDM include pre-pregnancy weight or obesity, older age, and family history of diabetes. Studies have suggested a possible association of GDM with the Glucokinase (GCK) gene.3–5 Glucokinase has an important role in the metabolism of glucose and catalyses the first step in the glycolytic pathway.6 Shaat et al7 has shown that common polymorphisms of the GCK gene increase the risk of GDM in Scandinavian women. The association of A allele with the increased risk of GDM has been reported in previous studies.7,8 A recent study conducted in Denmark reported a 5.8% prevalence of variants of GCK, HNF1A, HNF4A, HNF1B, or INS among women diagnosed with GDM.9 Heterozygous mutations in the GCK gene have also been reported to be a cause of a subtype of maturity-onset diabetes of the young (MODY), GCK-MODY or MODY 2.10 Data on the link between GDM and GCK gene is scarce in the Indian perspective. We, therefore, undertook this study to identify the GCK gene variants and assess their association with GDM in north Indian females.
Methods
Study population
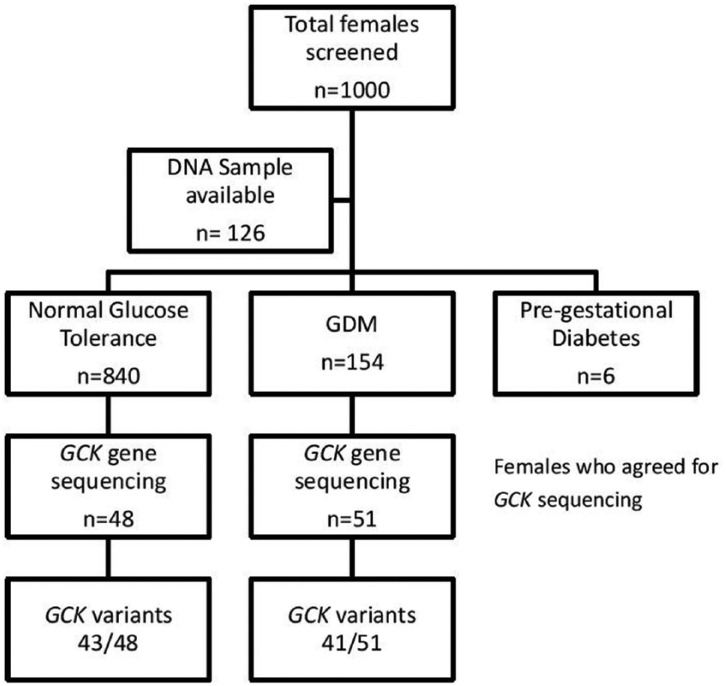
The present cross-sectional observational study was conducted at Max Super Speciality Hospital, Saket, New Delhi over a period of 6 months (January 2017 to June 2017). As per standard of care, all pregnant women attending our Gynecology and Obstetrics Unit are offered a 75 g oral glucose tolerance test (OGTT) at 24-28 weeks of gestation. Women at high risk (previous history of GDM or a family history of diabetes) are also offered a 75 g OGTT at 12-13 weeks. Those with test results meeting the International Association of the Diabetes and Pregnancy Study Groups (IADPSG) screening criteria for GDM, fasting blood sugar ≥92 mg/dL (5.1 mmol/L) or 1 hour blood sugar ≥180 mg/dL (10 mmol/L) or 2 hour blood sugar ≥153 mg/dL (5.3 mmol/L) are further referred for an endocrine evaluation.11 We screened 1000 pregnant women and those with any degree of glucose intolerance of age >18 years were recruited. We have observed in one of our previous studies, the average age of females diagnosed with GDM was 31.19 ± 3.66 years.12 Pregnant females (n = 840) with normal glucose tolerance and no family history of diabetes were recruited as controls (Figure 1). There was no consanguinity in the study groups and all members were of north Indian ethnicity.
Figure 1.
Algorithm of the study.
GDM, gestational diabetes mellitus.
Ethics approval
This study was approved by the Scientific and Institutional Ethics Committees of Max Super Speciality Hospital, Saket, New Delhi (Ref No.: TS/MSSH/DDF/ENDO/IEC/15-19). Written informed consent was taken from all the subjects and only those women who elected to participate were enrolled into the study.
Genetic analysis
Genomic DNA was extracted from ethylenediaminetetraacetic acid (EDTA)-anticoagulated peripheral blood by proteinase K digestion followed by the standard phenol-chloroform procedure.13 The concentration and purity of DNA were estimated on nanodrop1000 spectrophotometer. Exons 1a and 2-10 (β-cell GCK isoform), flanking intronic regions, and β-cell promoter of the GCK gene were bidirectionally sequenced in 99 subjects, using BigDye Terminator v3.1 chemistry (Applied Biosystems, Foster City, CA, USA). The data were analyzed both manually and electronically (using ABI SeqScape V2.7).
Bioinformatics analysis
Sequence information of genes was downloaded from the Public Databases like ENSEMBL and NCBI. Primer sequences were designed using Primer3 software. In silico predictions of the pathogenicity were carried out using the online tools SIFT,14 Polyphen-2,15 and Variation Taster.16
Statistical analysis
Gjesing et al9 reported a combined GCK, HNF1A, HNF4A, HNF1B, and INS variant prevalence of 5.9% in Danish women with GDM. We calculated the minimum required sample sizeas 15 using OpenEpi software version 3.01, with 95% confidence level, 5% precision and 80% power. Continuous variables were described using means and standard deviation (SD). All categorical variables were summarized by using frequencies and related percentages. The characteristic features of subjects with GDM and the controls were compared using Student t-test and Chi-square test. The data were analyzed using SPSS version 16.0.
Results
Population characteristics
A total of 1000 females were screened, of which 154 were diagnosed with GDM, 6 were diagnosed with diabetes prior to conception, and 840 were having normal glucose tolerance. GCK gene sequencing was performed in 51 females diagnosed with GDM and 48 healthy pregnant women with no family history of diabetes as controls. The baseline characteristics of study subjects are summarized in Table 1. Significant differences were noted in the subjects with GDM and those taken as controls in terms of age, prepregnancy body mass index (BMI), fetal bovine serum (FBS), and post prandial blood sugar (PPBS). We have identified a total of 11 variants in our study population. Table 2 represents the allelic distribution of these variants between GDM and control. Allele frequencies of these variants were not different between the control subjects (12.5%) and those with GDM (8.4%), P value <.000. GCK gene polymorphisms were observed in 80.4% (41/51) of the women with GDM and 89.6% (43/48) of the controls (Figure 1).
Table 1.
Characteristics of study population.
| S. No. | Parameters | GDM (n = 51) | Control (n = 48) | P value |
|---|---|---|---|---|
| 1. | Age (years) | 32.22 ± 4.13 | 29.23 ± 4.06 | <.001 |
| 2. | Height (cm) | 157.36 ± 5.83 | 157.99 ± 5.59 | .588 |
| 3. | Pre-pregnancy weight (kg) | 62.87 ± 9.85 | 59.24 ± 8.57 | .053 |
| 4. | BMI (kg/m2) | 25.38 ± 3.73 | 23.76 ± 3.49 | .028 |
| 5. | Week of gestation | 26.76 ± 5.99 | 24.71 ± 4.99 | .066 |
| 6. | BP (systolic) | 112.02 ± 10.93 | 109.7 ± 17.02 | .421 |
| 7. | BP (diastolic) | 71.71 ± 8.17 | 73.30 ± 7.09 | .305 |
| 8. | Primigravida | (28) 54.9% | (38) 79.17% | .027 |
| 9. | Multigravida (≥G2) | (23) 45.09% | (10) 20.83% | .027 |
| Clinical investigations | ||||
| 10. | Hb (g/dL) | 11.79 ± 1.35 | 11.53 ± 1.06 | .291 |
| 11. | FBS (mg/dL) | 102.73 ± 10.09 | 81.80 ± 6.53 | <.001 |
| 12. | PPBS (mg/dL) | 153.24 ± 26.39 | 106.44 ± 13.45 | <.001 |
| 13. | TSH (U/mL) | 2.36 ± 1.98 | 1.65 ± 1.29 | .036 |
| 14. | TLC | 10.27 ± 1.95 | 10.28 ± 1.78 | .966 |
| 15. | Platelet (109/L) | 234.84 ± 81.14 | 256.43 ± 56.28 | .135 |
| Delivery outcomes | ||||
| 16. | Week of delivery | 37.04 ± 3.53 | 38.38 ± 1.49 | .015 |
| 17. | Type of delivery LSCS Normal |
(39) 76.47% (11) 21.57% |
(30) 62.5% (18) 37.5% |
.072 |
| 18. | Baby weight (kg) | 2.98 ± 0.48 | 2.92 ± 0.43 | .542 |
| 19. | Baby gender Male Female |
(18) 35.29% (31) 60.78% |
(27) 56.25% (21) 43.75% |
.096 |
BMI, body mass index; BP, blood pressure; GDM, gestational diabetes mellitus; Hb, hemoglobin; FBS, fasting blood sugar; LSCS, lower segment Caesarean section; PPBS, post prandial blood sugar; TSH, thyroid stimulating hormone; TLC, total leukocyte count.
Boldface: p-value<0.05 is statistically significant.
Table 2.
List of observed GCK variants with their allele frequencies in the GDM and control group.
| S. No. | Region | Variation at cDNA level | Amino acid change | Zygosity | Allele frequency GDM (n = 51) |
Allele frequency Control (n = 48) |
P value |
|---|---|---|---|---|---|---|---|
| 1. | 5′-UTR | c.-84 C>G | - | Heterozygous | 5.9 | 9.4 | <.000 |
| 2. | Promoter | c.-516G>A | - | Homozygous | 13.7 | 18.8 | <.000 |
| 3. | Exon-3 | c.213C>T | p.Val71Val | Heterozygous | 0 | 1.0 | <.000 |
| 4. | Exon-3 | c.270G>A | p.Lys90Lys | Heterozygous | 0 | 2.1 | <.000 |
| 5. | Intron-3 | c.364-13G>A | - | Heterozygous | 0 | 1.0 | <.000 |
| 6. | Intron-4 | c.483+33G>A | - | Heterozygous | 0 | 1.0 | <.000 |
| 7. | Exon-6 | c.648 C>T | p.Tyr215Tyr | Heterozygous | 0.9 | 0 | <.000 |
| 8. | Intron-6 | c.676+38 T>C | - | Homozygous | 45.1 | 70.8 | <.000 |
| 9. | Intron-9 | c.1256+8C>T | - | Heterozygous | 22.5 | 27.1 | <.000 |
| 10. | Intron-9 | c.1256+49G>A | - | Heterozygous | 2.9 | 6.3 | <.000 |
| 11. | Exon-10 | c.1254-52to61delGTAATGAATG | - | Homozygous | 1.96 | 0 | <.000 |
cDNA, complementary DNA; GDM, gestational diabetes mellitus.
One of the variants, c.648 C>T (p.Tyr215Tyr), was observed only in a female diagnosed with GDM (MAF = 0.0091% ExAC, 0.0044% 1000 Genomes) at exon 6. The female carrying this variation was 28 years old with BMI 24.22 kg/m2 and was diagnosed with GDM at 25th week of gestation. Her fasting blood sugar (FBS) in an OGTT was 105 mg/dL (5.8 mmol/L) and PPBS was 117 mg/dL (6.5 mmol/L); she was prescribed insulin. She had a family history of diabetes and her pedigree chart is depicted in Figure 2A. She had a total weight gain of 11.1 kg during pregnancy and delivered a healthy baby girl at 40 weeks, weighing 3.0 kg. A deletion variant (c.1254-52to61delGTAATGAATG) was observed in another patient with GDM at exon 10 (MAF < 0.01) and not in any of the females in the control group. This variant is listed in the gnomAD browser, and it shows that it is found in 198/29,656 South Asian alleles. The female carrying this variant was 21 years old with BMI 21.22 kg/m2, FBS 97 mg/dL (5.4 mmol/L), and PPBS 126 mg/dL (7.0 mmol/L). She gained 10.7 kg during pregnancy and delivered a healthy baby girl weighing 3.1 kg at 38 weeks. Her family pedigree is depicted in Figure 2B.
Figure 2.
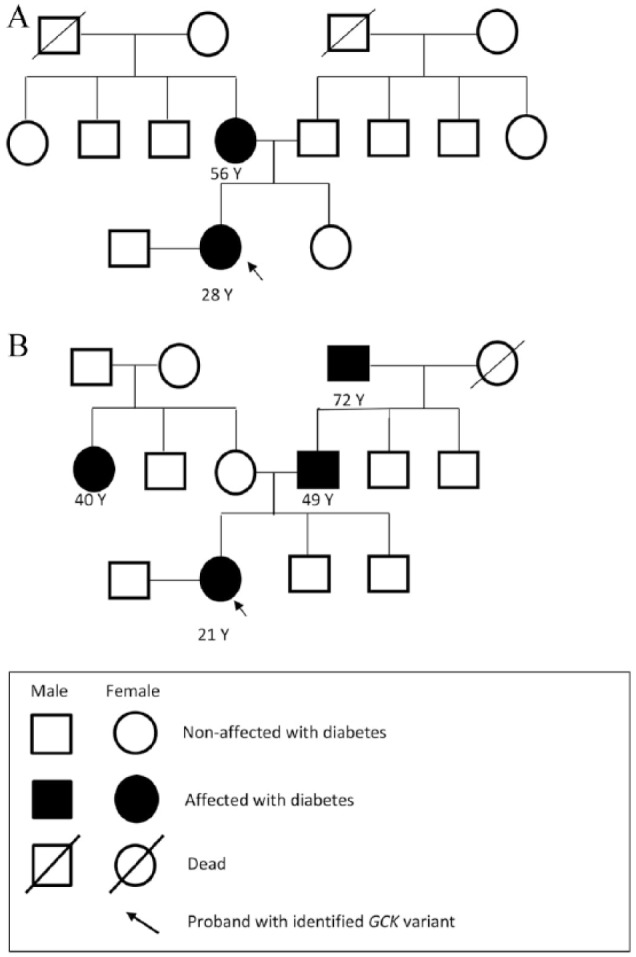
(A) Family pedigree of female carrying GCK variant c.648 C>T (p.Tyr215Tyr). (B) Family pedigree of female carrying GCK variant rs773255827 (c.1254-52to61delGTAATGAATG).
The polymorphisms, c.676+38 T>C (rs2268574) and c.-516G>A (rs1799884), have not been reported earlier in Indian population. We observed that their frequency was higher in the control subjects (70.8%, 18.8%) as compared with those having GDM (45.1%, 13.7%), respectively.
Discussion
There is evidence available for the genetic disposition of GDM similar to type 2 diabetes. A 2.3-fold increased risk of GDM has been reported by Williams et al,18 in females with a parental history of diabetes. A case-control study by Shaat et al7 has reported that common variants in the GCK and HNF1A genes increase the risk of GDM in Scandinavian women.
Findings of this study indicate that variations in GCK gene are not associated with GDM in north Indian females. Our study also indicates that there is a need for large-scale screening studies to be carried out in Indian females with GDM to further elucidate the link between gestational diabetes and GCK variants and their impact on the management of diabetes in pregnancy. There is less data available from northern India on this aspect.19 We observed the presence of GCK variants in 80.4% (41/51) of the women with GDM and 89.6% (43/48) of the controls. A total of 11 GCK variants were noted in our study population. These were common polymorphisms and were also present in the controls. c.648 C>T (p.Tyr215Tyr) is reported as a pathogenic variant and has been reported to be associated with MODY in literature.20–22 This variant was not present in the controls in our study, but found in one of the females with GDM. One of the polymorphisms (c.676+38 T>C) observed in our population subset has been previously reported by Frigeri et al17 to be associated with GDM, in Euro-Brazilian population. In contrast, we observed a higher frequency of this polymorphism in the subjects taken as controls as compared with the subjects with GDM. The association between GDM and rs1799884 (also known as -30G/A) have been studied extensively, and a meta-analysis of those studies has reported that the T allele of rs1799884 is associated with an increased risk of GDM.23 This variant has not been reported in Indian population, and in our study, we found it more in the controls as compared with the GDM subset. We did not find any definite association of GDM with GCK variants, which is in line with the study by Lukášová et al24 in Czech population. There is limited data on association of GCK variants and GDM from Indian population. A GCK variant c.1042T>A (Ile348Phe) has been recently reported by Doddabelavangala Mruthyunjaya et al19 in the Dravidian population with pre-gestational and overt GDM during comprehensive screening of MODY. However, we did not find this variation in our study group. As geographic variation and regional differences could be major factors in altering the spectrum of diabetes across India,25,26 it is natural to assume that the prevalence of variants will differ as well.
To the best of our knowledge, this is the first study from northern India exploring association of GCK variants with GDM and we do not observe any association of GCK variants with GDM in the population analyzed in the study. Also, we hereby report two common polymorphisms of the GCK gene in Indian population, reported in Caucasian population before.
Acknowledgments
We would like to thank the staff of MDRF Lab Chennai for sample analysis.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: SS involved in literature review, data collection, and result compilation and wrote the manuscript. SG contributed to genetic analysis and interpretation. SW, MP, and AB identified the subjects and reviewed the manuscript. VR, VM, and SD reviewed and edited the manuscript. SJ conceptualized the study and reviewed the manuscript.
Ethical Approval: This study was approved by the Scientific and Institutional Ethics Committees of Max Super Speciality Hospital, Saket, New Delhi (Ref No.: TS/MSSH/DDF/ENDO/IEC/15-19).
Informed Consent: Informed consent from each participant was taken.
References
- 1. International Diabetes Federation. IDF Diabetes Atlas. 8th ed. Brussels, Belgium: International Diabetes Federation; 2017. http://www.diabetesatlas.org [Google Scholar]
- 2. Seshiah V. Gestational diabetes mellitus—current guidelines for diagnosis & management. Med Updat. 2010;20:58–64. [Google Scholar]
- 3. Chiu KC, Go RC, Aoki M, et al. Glucokinase gene in gestational diabetes mellitus: population association study and molecular scanning. Diabetologia. 1994;37:104–110. [DOI] [PubMed] [Google Scholar]
- 4. Han X, Cui H, Chen X, Xie W, Chang Y. Association of the glucokinase gene promoter polymorphism -30G > A (rs1799884) with gestational diabetes mellitus susceptibility: a case–control study and meta-analysis. Arch Gynecol Obstet. 2015;292:291–298. [DOI] [PubMed] [Google Scholar]
- 5. Han H, Wang S, Ji L. Association of glucokinase gene with gestational diabetes mellitus in Chinese. Zhonghua Fu Chan Ke Za Zhi. 1999;34:23–26. [PubMed] [Google Scholar]
- 6. Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–980. [DOI] [PubMed] [Google Scholar]
- 7. Shaat N, Karlsson E, Lernmark Å, et al. Common variants in MODY genes increase the risk of gestational diabetes mellitus. Diabetologia. 2006;49:1545–1551. [DOI] [PubMed] [Google Scholar]
- 8. Weedon MN, Frayling TM, Shields B, et al. Genetic regulation of birth weight and fasting glucose by a common polymorphism in the islet cell promoter of the glucokinase gene. Diabetes. 2005;54:576–581. [DOI] [PubMed] [Google Scholar]
- 9. Gjesing AP, Rui G, Lauenborg J, et al. High prevalence of diabetes-predisposing variants in MODY genes among Danish women with gestational diabetes mellitus. J Endocr Soc. 2017;1:681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osbak KK, Colclough K, Saint-Martin C, et al. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum Mutat. 2009;30:1512–1526. [DOI] [PubMed] [Google Scholar]
- 11. International Association of Diabetes and Pregnancy Study Groups Consensus Panel. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vij P, Jha S, Gupta SK, et al. Comparison of DIPSI and IADPSG criteria for diagnosis of GDM: a study in a north Indian tertiary care center. Int J Diabetes Dev Countries. 2015;35:285–288. [Google Scholar]
- 13. Molecular Cloning. https://www.cshlpress.com/pdf/sample/2013/MC4/MC4FM.pdf
- 14. Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. [DOI] [PubMed] [Google Scholar]
- 17. Frigeri HR, Martins LT, Auwerter NC, et al. The polymorphism rs2268574 in Glucokinase gene is associated with gestational Diabetes mellitus. Clin Biochem. 2014;47:499–500. [DOI] [PubMed] [Google Scholar]
- 18. Williams MA, Qiu C, Dempsey JC, Luthy DA. Familial aggregation of type 2 diabetes and chronic hypertension in women with gestational diabetes mellitus. J Reprod Med. 2003; 48:955–962. [PubMed] [Google Scholar]
- 19. Doddabelavangala Mruthyunjaya M, Chapla A, Hesarghatta A, et al. Comprehensive Maturity Onset Diabetes of the Young (MODY) gene screening in pregnant women with diabetes in India. PLoS ONE 2017;12:e0168656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sagen JV, Odili S, Bjorkhaug L, et al. From clinicogenetic studies of maturity-onset diabetes of the young to unraveling complex mechanisms of glucokinase regulation. Diabetes. 2006;55:1713–1722. [DOI] [PubMed] [Google Scholar]
- 21. Ellard S, Beards F, Allen LIS, et al. A high prevalence of glucokinase mutations in gestational diabetic subjects selected by clinical criteria. Diabetologia. 2000;43:250–253. [DOI] [PubMed] [Google Scholar]
- 22. Sagen JV, Bjørkhaug L, Molnes J, et al. Diagnostic screening of MODY2/GCK mutations in the Norwegian MODY registry. Pediatr Diabetes. 2008;9:442–449. [DOI] [PubMed] [Google Scholar]
- 23. Zhang C, Bao W, Rong Y, et al. Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum Reprod Update. 2013;19:376–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lukášová P, Včelák J, Vaňková M, Vejražková D, Andělová K, Bendlová B. Screening of mutations and polymorphisms in the glucokinase gene in Czech diabetic and healthy control populations. Physiol Res. 2008;57:S99–S108. [DOI] [PubMed] [Google Scholar]
- 25. Unnikrishnan R, Anjana RM, Mohan V. Diabetes mellitus and its complications in India. Nat Rev Endocrinol. 2016;12:357–370. [DOI] [PubMed] [Google Scholar]
- 26. Unnikrishnan R, Pradeepa R, Joshi SR, Mohan V. Type 2 diabetes: demystifying the global epidemic. Diabetes. 2017;66:1432–1442. [DOI] [PubMed] [Google Scholar]