Abstract
Background
Previous studies have shown an association with glutathione S-transferase (GST) gene polymorphisms in patients with non-small cell lung cancer (NSCLC) and treatment response. This study aimed to undertake a literature review and meta-analysis of GST gene polymorphisms, including GSTT1, GSTM1, and GSTP1 IIe105Val, and the treatment response to cisplatin-based chemotherapy in patients with NSCLC.
Material/Methods
A literature search was undertaken of the main medical publication databases for publications, up to March 2017, on the association between GSTT1, GSTM1, and GSTP1 IIe105Val polymorphisms and the clinical outcome in patients with NSCLC treated with cisplatin-based chemotherapy. A random fixed-effects model was used to calculate the pooled odds ratio (OR) and 95% confidence interval (CI) to evaluate the associations, considering multiple genetic models. A subgroup analysis according to ethnicity was performed.
Results
Twenty-three published studies were identified that showed that both the null GSTM1 and the GG genotype of GSTP1 IIe105Val were associated with improved treatment response to cisplatin-based chemotherapy (GSTT1 present/null: OR=1.328; 95% CI, 1.074–1.643) (GSTP1 GG + AG vs. AA: OR=0.596; 95% CI, 0.468–0.759). In subgroup analysis, the GSTP1 polymorphism was significantly associated with treatment response in East-Asian patients, but not in Caucasian patients.
Conclusions
Meta-analysis showed that the GG genotype of GSTP1 IIe105Val and the null GSTM1 genotype were associated with an improved treatment response to cisplatin-based chemotherapy in patients with NSCLC, especially in East-Asian patients.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; Chemotherapy, Adjuvant; Polymorphism, Genetic
Background
Worldwide, lung cancer has a high prevalence and is associated with a high mortality rate [1]. In the last century, the incidence of lung cancer has been rising rapidly, and it is estimated that there were about 8.8 million deaths of lung cancer in 2015 worldwide [2]. It has been reported that 85% of primary lung cancers are non-small cell lung cancer (NSCLC) and 80–85% of patients who are first diagnosed with NSCLC are at an advanced stage [3–5]. Except for a few patients who can undergo surgical treatment, most patients require comprehensive treatment based on systemic chemotherapy, with different individual responses to chemotherapy in terms of the tumor sensitivity to chemotherapy and the adverse reactions to chemotherapy. Recently published studies have provided evidence that hereditary factors play an important role in individual differences in the treatment response to chemotherapy, including the GSTM1, GSTP1, XRCC1, MRP2, and RRM1 genes [6–17]. Therefore, it is important to continue to undertake research to identify molecular markers for treatment response in patients with NSCLC.
Glutathione S-transferases (GSTs), and cellular glutathione (GSH), are members of a multigene family of phase II enzymes, which are associated with detoxifying chemotherapeutic agents. GSTs have been recognized as a significant factor related to the clinical outcome of cisplatin-based chemotherapy in patients with NSCLC. The GSTT1, GSTP1 and GSTP1 IIe105Val genes all belong to GST gene family [18]. In previously published studies, the role of GSTT1, GSTM1, and GSTP1 IIe105Val gene polymorphisms and the clinical outcome of patients with NSCLC treated with chemotherapy have demonstrated that the genetic variability of the GSTM1 and GSTT1 genes are linked to enzyme metabolizing activities [16]. The GSTP1 IIe105Val gene plays an important role in phase II metabolism of xenobiotics and is involved in the biological transformation of cisplatin in the body. With a substitution of adenine to guanine (A to G) at locus 313, the GSTP1 gene acquires isoleucine (lle) rather than valine (Val) at codon 105. The mutant alleles can increase intracellular cisplatin-GSH conjugates and suppress the metabolism of cisplatin, which will decrease the activity of GST and reduce the rate of the biological effect.
This study aimed to undertake a literature review and meta-analysis of GST gene polymorphisms, including GSTT1, GSTM1, and GSTP1 IIe105Val, and the treatment response to cisplatin-based chemotherapy in patients with NSCLC.
Material and Methods
Literature search and selection criteria
A systematic literature search was undertaken to include the main medical databases, including PubMed, Web of Science, EMBASE, the Cochrane database, and the China National Knowledge Internet (CNKI) database. The search covered all publications from September 2006 to March 2017. Keywords used included ‘GSTM1’ or ‘GSTT1’ or ‘GSTP1’ or ‘GSTs’ and ‘polymorphisms’ or ‘variation’ or ‘SNP’ and ‘lung cancer’ or ‘NSCLC’ and ‘chemotherapy’ or ‘platinum’ or ‘treatment.’ Publications from previous meta-analysis and retrieved articles were searched manually for additional studies.
Inclusion and exclusion criteria for publications that included patients with non-small cell lung cancer (NSCLC), glutathione S-transferase (GST) gene polymorphisms, and treatment response to platinum-based chemotherapy
The inclusion criteria for publications included studies that evaluated the association between GSTM1 or GSTT1 or GSTP1 gene polymorphisms and treatment response; patients with histologically-confirmed non-small cell lung cancer (NSCLC) treated with any combination of cisplatin-based chemotherapy, including cisplatin or gemcitabine-cisplatin or cisplatin (DDP); independent genotype data on GSTT1, GSTM1 and GSTP1 polymorphisms; the presence of treatment response and follow-up information that would allow for the calculation of the odds ratio (OR). The exclusion criteria for publications included studies that had insufficient data or lack of the necessary information; mixed data on cases of NSCLC and small cell lung cancer (SCLC), which would make extraction of data on patients with NSCLC difficult; review publications, meta-analysis, case reports, in vitro cell line studies, and in vivo animal studies.
Data extracted from each publication used in the meta-analysis included the details of the authors, the year of publication, the country where the study was undertaken, the race or ethnicity of the patient population, the study sample size, single nucleotide polymorphism (SNP) data, allele frequencies, and details on the platinum-based chemotherapy drug regime. Figure 1 shows the flowchart of the publication selection process
Figure 1.
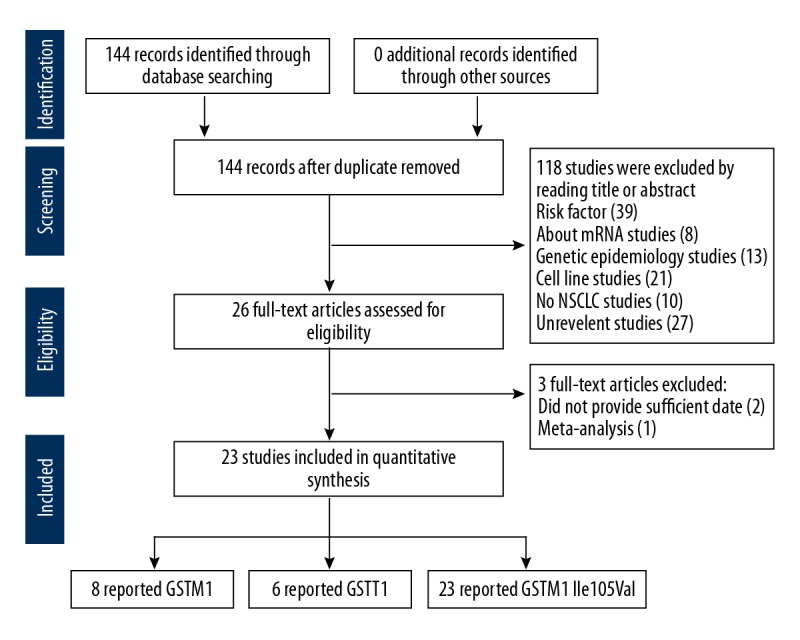
Flowchart of the publication selection process.
Definitions
The clinical data included patient treatment response and clinical outcome following chemotherapy, which was assessed at every two cycles of treatment, according to the World Health Organization (WHO) criteria and the response evaluation criteria in solid tumors (RECIST). On the basis of the response to chemotherapy, the patients were divided into two groups, the complete response (CR) group or the partial remission (PR) group. The objective response rate (ORR) was used to determine whether the patient has a ‘good response’ to chemotherapy (a chemo-sensitive group, CR + PR). Stable disease (SD) or progressive disease (PD) were considered ineffective or representing a poor response group (non-sensitive group, SD + PD).
Statistical analysis
Meta-analysis was performed to determine the association between GST gene polymorphisms and treatment outcome of cisplatin-based chemotherapy. Subgroup analysis was performed by patient ethnicity and included East-Asians and Caucasians. The potential heterogeneity between studies was analyzed. The association between GSTP1 IIe105Val and treatment response was performed using with five comparisons: the recessive genotype genetic model (GG vs. AG + AA); the dominant genotype genetic model (GG + AG vs. AA); a homozygous genotype genetic model (GG vs. AA); a heterozygous genotype genetic model (AG vs. AA); and allelic gene genetic model (G vs. A).
A formal Q statistical test and I2 statistical test were performed to assess the heterogeneity between studies, and P<0.05 was considered to be statistically significant. Depending on the heterogeneity results, the DerSimonian and Laird random-effects meta-analysis model or the Mantel-Haenszel fixed-effects model was used for the calculation of the pooled odds ratio (OR) with a 95% confidence interval (CI). A P-value <0.10 or I2 >50% was considered to support a fixed-effect model, with the alternative being a random-effects model. A sensitivity analysis was conducted, by sequential omission of each study, to validate the stability of all results. Both Egger’s regression test of asymmetry of the funnel plot and Begg’s adjusted rank correlation test were used to investigate publication bias. All statistical analysis was performed using STATA version 10.0 statistical software (STATA Corporation, College Station, TX, USA).
Results
Initially, a total of 144 publications were identified from the database search, after removing duplicate publications with all research based on univariate analysis. After carefully screening the titles and abstracts, 121 studies were excluded. Finally, 23 studies met the study selection criteria for meta-analysis [2,3,7–16,19–21,23–30] (Figure 1). Among the 23 studies, there were eight studies of non-small cell lung cancer (NSCLC) with 640 cases and 928 controls for the GSTM1 null genotype, six studies with 518 cases and 822 controls for the GSTT1 null genotype, 23 studies including 1733 cases and 1374 controls for the GSTP1 IIe105Val genotype. These studies contained two subgroups with 20 studies including patients from East-Asia and three studies that included Caucasian patients with NSCLC. The essential information of the included 23 publications are shown in Tables 1–5 [2,3,7–16,19–21,23–30]. Three individuals reviewed the studies to avoid potential bias in data interpretation.
Table 1.
Studies on GSTM1 polymorphisms and treatment response.
| Study | Year | Ethnicity/country | Chemotherapeutic drugs | Poor response present/null | Good response present/null | OR (95% CI) | Weight % | P-value | Study quality |
|---|---|---|---|---|---|---|---|---|---|
| Jia et al. [19] | 2016 | East-Asian/China | DDP | 118/61 | 33/32 | 1.876 (1.054, 3.337) | 11.21 | 0.03 | 7 |
| Xiao et al. [2] | 2016 | East-Asian/China | DDP | 83/36 | 80/63 | 1.816 (1.088, 3.029) | 14.94 | 0.02 | 7 |
| Chen et al. [20] | 2016 | East-Asian/China | DDP | 121/85 | 41/37 | 1.285 (0.761, 2.169) | 16.67 | 0.35 | 8 |
| Han Ba [11] | 2016 | East-Asian/China | DDP+ DOC/DDP+ NVB/Pt+ GEM | 48/34 | 36/38 | 1.490 (0.791, 2.807) | 10.66 | 0.16 | 9 |
| Wu et al. [21] | 2015 | East-Asian/China | DDP | 100/59 | 68/55 | 1.371 (0.849, 2.214) | 19.33 | 0.20 | 7 |
| Qiying et al. [30] | 2014 | East-Asian/China | TAX+ DDP/DDP+ NVB/DDP+ GEM/DDP+ VP-16 | 32/31 | 13/13 | 1.032 (0.414, 2.574) | 6.15 | 0.98 | 8 |
| Joerger et al. [23] | 2012 | Caucasian/ Netherlands | DDP+ GEM | 20/22 | 60/35 | 0.530 (0.254, 1.106) | 13.09 | 0.08 | 6 |
| Kalikaki et al. [25] | 2009 | Caucasian/ Greece | Pt/Pt-TXT-based | 49/29 | 23/13 | 0.955 (0.420, 2.170) | 7.95 | 0.34 | 8 |
| M-H pooled OR | 1.328 (1.074, 1.643) | 100 | 0.009 | ||||||
OR – odds ratio; 95% CI – 95%confidence interval; DDP – cisplatin; GEM – gemcitabine; NVB – vinorelbine; PEM – pemetrexed; TAX – taxol/paclitaxel; TXT – taxotere; DOC – docetaxel; VP-16 – etoposide; MMC – mitomycin; IFO – ifosfamide; VLB – vinblastine; Pt – platinum.
Table 2.
Subgroup analysis of the association between the GSTM1 polymorphisms with treatment response in patients with non-small cell lung cancer (NSCLC) receiving chemotherapy.
| Comparison | N | OR | 95% CI | I2 (%) | Ph | |
|---|---|---|---|---|---|---|
| All | 8 | 1.328 | 1.074 | 1.643 | 29.1 | 0.196 |
| Chinese | 6 | 1.498 | 1.188 | 1.889 | 0.0 | 0.817 |
| Caucasian | 2 | 0.691 | 0.400 | 1.194 | 8.7 | 0.295 |
N – number; OR – odds ratio; 95% CI – 95% confidence interval; I2 – I-squared; Ph – P-value of the heterogeneity test.
Table 3.
Characteristics of the studies evaluating the association between GSTT1 polymorphisms and treatment response.
| Study | Year | Ethnicity/country | Chemotherapeutic drugs | Poor response present/null | Good response present/null | OR (95% CI) | Weight % | P-value | Study quality |
|---|---|---|---|---|---|---|---|---|---|
| Jia et al. [19] | 2016 | East-Asian/China | DDP | 83/96 | 28/37 | 1.142 (0.645, 2.024) | 17.21 | 0.65 | 7 |
| Xiao et al. [2] | 2016 | East-Asian/China | DDP | 68/51 | 77/66 | 1.143 (0.700, 1.865) | 23.41 | 0.59 | 7 |
| Chen et al. [20] | 2016 | East-Asian/China | DDP | 118/88 | 37/41 | 1.486 (0.881, 2.507) | 17.91 | 0.14 | 8 |
| Han Ba [11] | 2016 | East-Asian/China | DDP+ DOC/DDP+ NVB/Pt+ GEM | 42/40 | 35/39 | 1.170 (0.624, 2.195) | 14.02 | 0.29 | 9 |
| Wu et al. [21] | 2015 | East-Asian/China | DDP | 92/67 | 69/54 | 1.075 (0.668, 1.729) | 25.61 | 0.77 | 7 |
| Kalikaki et al. [25] | 2009 | Caucasian/Greece | Pt/Pt-TXT-based | 73/4 | 33/2 | 1.106 (0.193, 6.342) | 1.84 | 0.72 | 8 |
| M-H pooled OR | 1.190 (0.941, 1.504) | 100 | 0.146 | ||||||
OR – odds ratio; 95% CI – 95%confidence interval; DDP – cisplatin; GEM – gemcitabine; NVB – vinorelbine; PEM – pemetrexed; TAX – taxol/paclitaxel; TXT – taxetere; DOC – docetaxel; VP-16 – etoposide; MMC – mitomycin; IFO – ifosfamide; VLB – vinblastine; Pt – platinum.
Table 4.
Characteristics of the studies evaluating the association between GSTP1 IIe105Val polymorphisms and treatment response.
| Study | Year | Ethnicity/country | Chemotherapeutic drugs | Poor response | Good response | OR (95% CI) | P-value* | Study quality | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Val/Val | Val/IIe | IIe/IIe | Val/Val | Val/IIe | IIe/IIe | |||||||
| Jia et al. [19] | 2016 | East-Asian/China | DDP | 22 | 77 | 80 | 16 | 28 | 21 | 0.591 (0.325, 1.074) | 0.085 | 7 |
| Xiao et al. [2] | 2016 | East-Asian/China | DDP | 33 | 30 | 56 | 45 | 36 | 62 | 0.861 (0.528, 1.404) | 0.549 | 7 |
| Bu et al. [13] | 2016 | East-Asian/China | DDP | 3 | 29 | 38 | 9 | 34 | 28 | 0.548 (0.281, 1.071) | 0.078 | 8 |
| Chen et al. [20] | 2016 | East-Asian/China | DDP | 23 | 84 | 99 | 25 | 37 | 16 | 0.279 (0.151, 0.515) | <0.001 | 8 |
| Yuan et al. [15] | 2016 | East-Asian/China | GEM+ DDP | 1 | 8 | 14 | 1 | 7 | 16 | 1.286 (0.390, 4.236) | 0.679 | 7 |
| Han Ba [11] | 2016 | East-Asian/China | DDP+ DOC/DDP+ NVB/Pt+ GEM | 3 | 33 | 46 | 12 | 36 | 26 | 0.424 (0.222, 0.809) | 0.009 | 9 |
| Xin et al. [29] | 2016 | East-Asian/China | GEM+ DDP | 6 | 7 | 20 | 4 | 11 | 36 | 1.560 (0.620, 0.923) | <0.001 | 7 |
| Zhao et al. [9] | 2015 | East-Asian/China | DDP | 5 | 40 | 55 | 16 | 54 | 36 | 0.421 (0.240, 0.739) | 0.003 | 8 |
| Wu et al. [21] | 2015 | East-Asian/China | DDP | 34 | 48 | 78 | 40 | 41 | 42 | 0.545 (0.336, 0.885) | 0.014 | 7 |
| Han et al. [16] | 2015 | East-Asian/China | DDP | 3 | 34 | 60 | 25 | 115 | 88 | 0.388 (0.238, 0.632) | <0.001 | 8 |
| Deng et al. [22] | 2015 | East-Asian/China | DDP+ GEM+ NVB+ TAX/DOC | 1 | 22 | 46 | 0 | 4 | 24 | 3.000 (0.930, 9.674) | 0.066 | 9 |
| Yuan et al. [8] | 2015 | East-Asian/China | GEM+ DDP | 1 | 8 | 14 | 1 | 7 | 16 | 1.286 (0.390, 4.236) | 0.679 | 8 |
| Zhigang et al. [7] | 2015 | East-Asian/China | TAX+ DDP/DDP/PEM + DDP | 8 | 20 | 25 | 7 | 16 | 9 | 0.438 (0.171, 1.123) | 0.086 | 7 |
| Liu et al. [12] | 2015 | East-Asian/China | GEM+ DDP | 1 | 8 | 15 | 1 | 10 | 17 | 0.927 (0.302, 2.847) | 0.895 | 8 |
| Qiying et al. [30] | 2014 | East-Asian/China | TAX+ DDP/DDP+ NVB/DDP+ GEM/DDP+ VP-16 | 0 | 19 | 44 | 0 | 8 | 18 | 0.972 (0.360, 2.619) | 0.955 | 8 |
| Joerger et al. [23] | 2012 | Caucasian/Netherlands | DDP+ GEM | 3 | 18 | 20 | 14 | 42 | 35 | 0.656 (0.312, 1.381) | 0.267 | 6 |
| Sun et al. [10] | 2010 | East-Asian/China | (DDP/CBP+ TAX/TXT/DOC)/(DDP/CBP+ GEM)/(DDP/CBP+ NVB) | 2 | 23 | 58 | 2 | 15 | 13 | 0.330 (0.139, 0.780) | 0.012 | 8 |
| Zheng et al. [24] | 2009 | East-Asian/China | DDP+ NVB | 0 | 6 | 22 | 2 | 11 | 15 | 0.315 (0.098, 1.013) | 0.053 | 7 |
| Kalikaki et al. [25] | 2009 | Caucasian/Greece | Pt/Pt-TXT-based | 0 | 30 | 48 | 0 | 12 | 25 | 1.302 (0.570, 2.973) | 0.531 | 8 |
| Booton et al. [26] | 2006 | Caucasian/UK | MMC+ IFO+ DDP/MMC+ VLB+ DDP | 12 | 23 | 25 | 4 | 9 | 13 | 1.400 (0.556, 3.528) | 0.475 | 8 |
| Jianhaoa et al. [31] | 2015 | East-Asian/China | PEM+ Platinum | 5 | 15 | 7 | 3 | 0.143 (0.026, 0.774) | 0.025 | 8 | ||
| Yapinga et al. [27] | 2012 | East-Asian/China | DDP+ NVB/DDP+ TAX/GEM+ DDPTAX/DDP | 11 | 35 | 9 | 7 | 0.244 (0.074, 0.810) | 0.021 | 6 | ||
| Zhoua et al. [14] | 2011 | East-Asian/China | Pt+ CBP+ DOC+ GEM+ NVB+ PEM | 26 | 50 | 22 | 13 | 0.307 (0.134, 0.707) | 0.005 | 7 | ||
| D+L pooled OR | 0.596 (0.468, 0.759) | <0.001 | ||||||||||
OR – odds ratio; 95% CI – 95%confidence interval; DDP – cisplatin; GEM – gemcitabine; NVB – vinorelbine; PEM – pemetrexed; TAX – taxol/paclitaxel; TXT – taxetere; DOC – docetaxel; VP-16 – etoposide; MMC – mitomycin; IFO – ifosfamide; VLB – vinblastine; Pt – platinum.
Integrate Val/Val with Val/IIe genotype.
Table 5.
Subgroup analysis of association between the GSTP1 IIe105Val polymorphisms with treatment response in patients with non-small cell lung cancer (NSCLC) receiving chemotherapy.
| Comparison | Studies | Genetic model | OR (95% CI) | P heterogeneity | I2 (%) | |
|---|---|---|---|---|---|---|
| All | 20 | Recessive model | GG vs. AG + AA | 0.510 (0.404–0.644)* | 0.174 | 23.7 |
| 23 | Dominant model | GG + AG vs. AA | 0.596 (0.468–0.759)# | 0.001 | 53.3 | |
| 20 | Homozygote model | GG vs. AA | 0.413 (0.273–0.623)# | 0.013 | 47.7 | |
| 20 | Heterozygote mode | AG vs. AA | 0.660 (0.557–0.782)* | 0.108 | 29.2 | |
| 20 | Allele | G vs. A | 0.683 (0.557–0.836)# | <0.001 | 59.4 | |
| East-Asian | 17 | Recessive model | GG vs. AG + AA | 0.492 (0.386–0.628)* | 0.182 | 24.0 |
| 20 | Dominant model | GG + AG vs. AA | 0.548 (0.426–0.704)# | 0.005 | 51.0 | |
| 17 | Homozygote model | GG vs. AA | 0.380 (0.246–0.587)# | 0.018 | 47.5 | |
| 17 | Heterozygote mode | AG vs. AA | 0.619 (0.516–0.742)* | 0.152 | 26.4 | |
| 17 | Allele | G vs. A | 0.639 (0.516–0.790)# | 0.001 | 58.3 | |
| Caucasian | 3 | Recessive model | GG vs. AG + AA | 0.770 (0.332–1.782)* | 0.209 | 36.7 |
| 3 | Dominant model | GG + AG vs. AA | 1.006 (0.628,1.611)* | 0.344 | 6.2 | |
| 3 | Homozygote model | GG vs. AA | 0.757 (0.314–1.827)* | 0.140 | 54.2 | |
| 3 | Heterozygote mode | AG vs. AA | 1.048 (0.641–1.714)* | 0.554 | 0.0 | |
| 3 | Allele | G vs. A | 0.954 (0.661–1.377)* | 0.227 | 32.6 | |
OR – odds ratio; 95% CI – 95% confidence interval; I2 – I-squared.
A fixed-effects model was constructed;
A random-effects model was constructed.
The Newcastle-Ottawa Scale (NOS) was used to evaluated the quality of these included studies, based on a scoring system with three components: the selection of the subjects (0–4 points); the comparability of the study groups (0–2 points); the determination of the outcome of interest (treatment response) (0–3 points). The highest NOS score for each publication was 9 points, and in this meta-analysis, two studies had a NOS score of 9 [11,31]; 11 studies had a NOS score of 8 [8–10,12,13,15,16,20,25,26,30]; eight studies had a NOS score of 7 [2,7,14,19,21,24,28,29]; and two studies had a NOS score of 6 [23,27].
The GSTM1 gene polymorphism
Eight studies described the relationship between the GSTM1 polymorphism and treatment outcome of chemotherapy in patients with NSCLC, which included 1,568 individuals (Table 1). A fixed-effects model was used so that no significant heterogeneity existed between the studies (I2=29.1%; P=0.196). The pooled meta-analysis showed that patients with the null GSTM1 genotype had a better clinical outcome following cisplatin-based combination chemotherapy, compared with the presence of GSTM1 (OR=1.328; 95% CI, 1.074–1.643) (Figure 2). A subgroup analysis was performed on the different ethnicities. The results showed that null GSTM1 was associated with an improved treatment response in Chinese patients with NSCLC (present vs. null: OR=1.498; 95% CI, 1.188–1.189), but not in Caucasian patients with NSCLC (present vs. null: OR=0.691; 95% CI, 0.400–1.194) (Table 2).
Figure 2.
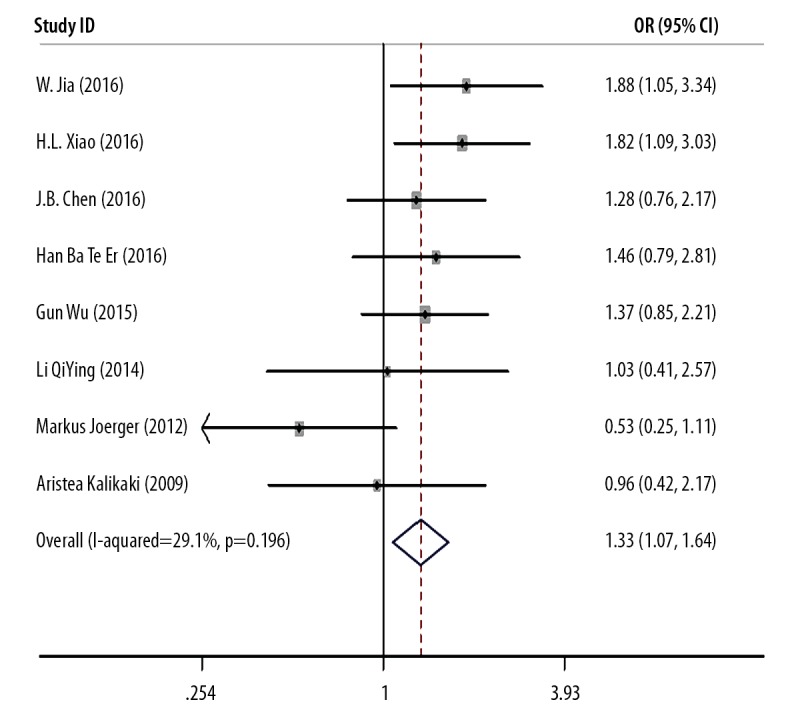
Meta-analysis of GSTM1 gene polymorphism and treatment response (TR).
The GSTT1 gene polymorphism
Six studies described the relationship between the GSTT1 polymorphism and the clinical outcome following chemotherapy in patients with NSCLC, which included 1,340 individuals (Table 3). Statistical heterogeneity was shown in the analysis (I2=0.0%; P=0.968). Therefore, a random-effects model was used. The pooled meta-analysis showed that there was no association between the GSTT1 polymorphism and the clinical outcome following chemotherapy in patients with NSCLC (OR=1.190; 95% CI, 0.941–1.504) (Figure 3).
Figure 3.
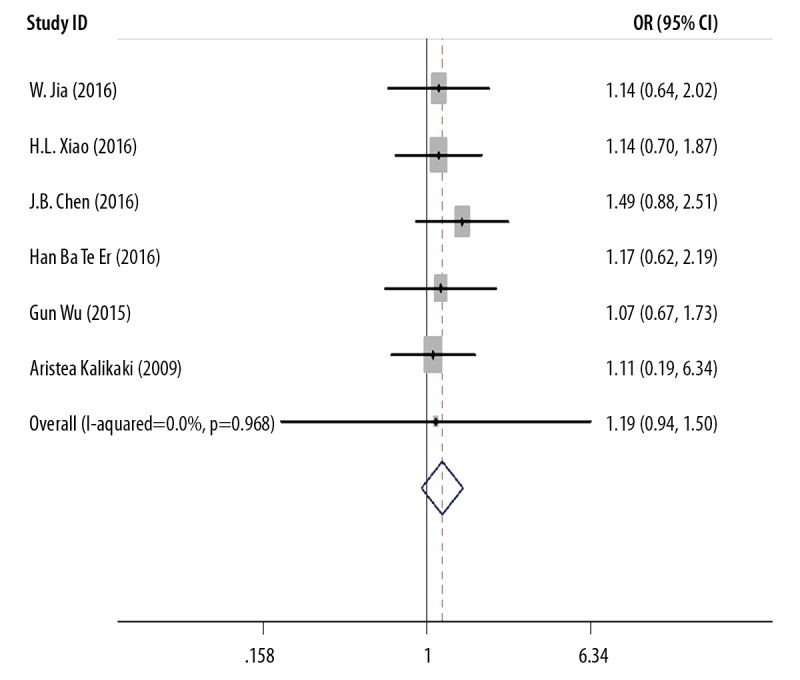
Meta-analysis of GSTT1 gene polymorphism and treatment response (TR).
The GSTP1 gene polymorphism
Twenty-three studies described the relationship between the GSTM1 IIe105Val polymorphism and treatment response to cisplatin-based chemotherapy in patients with NSCLC, which included 3,170 individuals. The main findings are summarized in Tables 4 and 5 and in Figure 4. The findings, according to the heterogeneity analysis with five comparison models included the recessive model: I2=23.7%, P=0.174; the dominant model: I2=53.3%, P=0.001; the heterozygous model: I2=29.2%, P=0.108; the homozygous model: I2=47.7%, P=0.013; the allele model: I2=59.4%, P=0.000, and the random-effects model was used.
Figure 4.
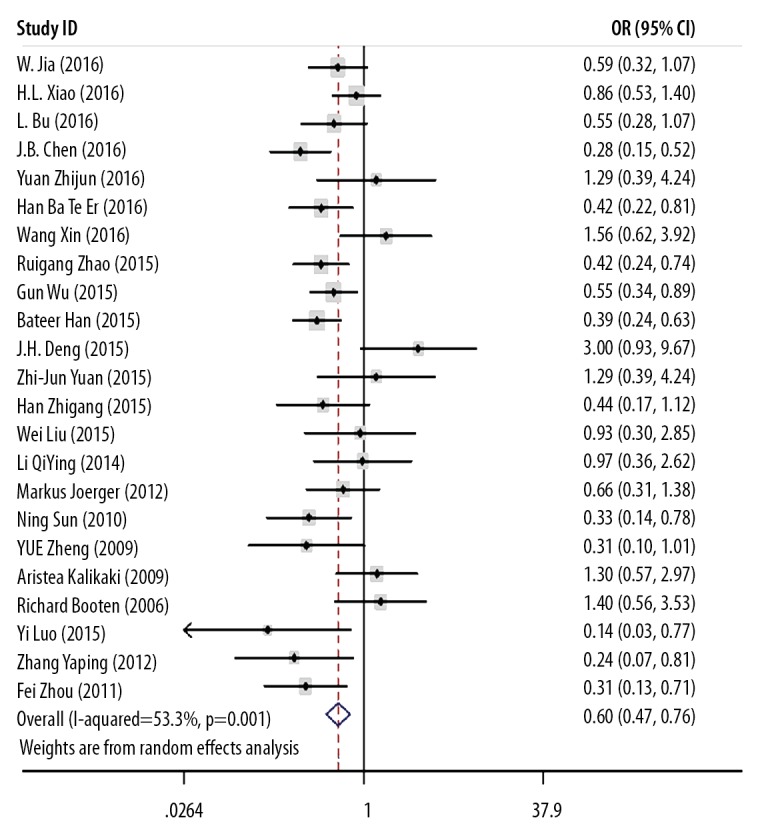
Meta-analysis of GSTP1 IIe105Val gene polymorphism and treatment response (TR).
The pooled meta-analysis showed that when compared to the AA genotype, the GG genotype of GSTP1 IIe105Val showed a better treatment response to chemotherapy in patients with NSCLC: the recessive model, GG vs. AG + AA: OR=0.510; 95% CI, 0.404–0.644; the dominant model, GG + AG vs. AA: OR=0.596; 95% CI, 0.468–0.759; the homozygous model, GG vs. AA: OR=0.413; 95% CI, 0.273–0.623; the heterozygous model, AG vs. AA: OR=0.660, 95% CI=0.557–0.782; the allele model, G vs. A: OR=0.683, 95% CI=0.557–0.836 (Figure 4).
A subgroup analysis was made on the different ethnicities. The variant G allele was associated with an improved response to chemotherapy in East-Asian patients: the recessive model, GG vs. AG + AA: OR=0.492; 95% CI, 0.386–0.628; the dominant model, GG + AG vs. AA: OR=0.548; 95% CI, 0.426–0.704; the homozygous model, GG vs. AA: OR=0.380; 95% CI, 0.246–0.587; the heterozygous model, AG vs. AA: OR=0.619; 95% CI, 0.516–0.742; the allele model, G vs. A: OR=0.639; 95% CI, 0.516–0.790. There was no association between the variant G allele and response to chemotherapy in Caucasian patients: the recessive model, GG vs. AG + AA: OR=0.770; 95% CI, 0.332–1.782; the dominant model, GG + AG vs. AA: OR=1.006; 95% CI, 0.628–1.611; the homozygous model, GG vs. AA: OR=0.757; 95% CI, 0.314–1.827; the heterozygous model, AG vs. AA: OR=1.048; 95% CI, 0.641–1.714; the allele model, G vs. A: OR=0.954; 95% CI, 0.661–1.377) (Table 5).
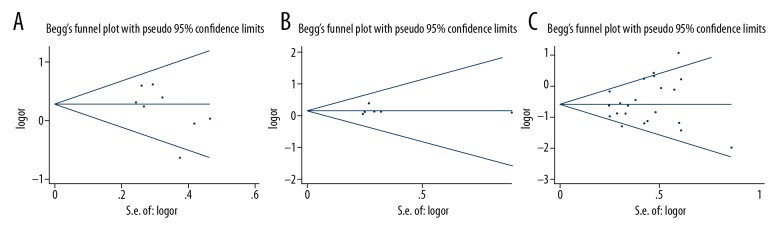
Publication bias
Figure 5A shows the shape of the funnel plot for the association between the GSTM1 polymorphism and the treatment response to chemotherapy. The P-values for Begg’s and Egger’s tests (0.174 and 0.117, respectively) indicated no publication bias.
Figure 5.
Funnel plot of glutathione S-transferase (GST) gene polymorphisms and treatment response (TR). (A) Funnel plot of GSTM1 gene polymorphism and treatment response (TR). (B) Funnel plot of GSTT1 gene polymorphism and treatment response. (C) Funnel plot of GSTP1 gene polymorphism and treatment response.
For GSTT, Egger’s test showed no evidence of publication bias (Figure 5). The P-value for Egger’s test was 0.947. Figure 5C shows the funnel plot for the association between the GSTP1 IIe105Val polymorphism and the treatment response to chemotherapy. There was no significant bias of the recessive model, dominant model, and homozygous model (the recessive model: PBegg=0.472, PEgger=0.893; the dominant model: PBegg=0.413, PEgger=0.317; the homozygous model: PBegg=0.344, PEgger=0.926). Two comparison models (the heterozygous model: PBegg=0.032, PEgger=0.020; the allele model: PBegg=0.023, PEgger=0.021) showed significant publication bias.
Sensitivity analysis
To investigate the causes of the study heterogeneity, a sensitivity analysis was performed by repeating the meta-analysis after sequentially removing each study. There was no significant effect in the odds ratios (ORs) by removing any individual study for GSTM1, GSTT1, and GSTP1 IIe105Val genotypes.
Discussion
Environmental and genetic factors are involved in the development and progression of non-small cell lung cancer (NSCLC), but the exact mechanisms of these factors remain unknown. There is growing evidence that indicates that the polymorphisms in genes involved in metabolism, signaling, DNA synthesis, and cellular response pathways may explain the individual variability of chemotherapy toxicity and treatment response. Therefore, it is important to assess the association between genetic polymorphisms and sensitivity or resistance to chemotherapy when selecting the most effective first-line treatment for patients with NSCLC.
The human glutathione S-transferases (GSTs) are a superfamily of dimeric phase II metabolic enzymes and currently are known to include eight dimeric isoenzymes: α-GST, β-GST, δ-GST, ɛ-GST, θ-GST, μ-GST, η-GST, and ω-GST. The GSTs are located on seven separate chromosomes with a highly homologous amino acid sequence trait. Each class has a specific substrate affinity and is encoded by one or several highly polymorphic genes. Several epidemiological studies have now reported that the GSTs may be associated with an improved treatment response to chemotherapeutic drugs, possibly due to their ability to detoxify the metabolites of chemotherapy drugs and carcinogens by catalyzing the reduction of these compounds through conjugation with glutathione. Although previously published studies have explored the relationship between GST polymorphisms and treatment response to chemotherapy in patients with NSCLC, the results have been inconclusive, possibly due to differences in response due to patient ethnicity and varied study design.
To quantify the strength of the association between GST polymorphisms and chemotherapy treatment response in patients with NSCLC, a meta-analysis of the published literature was performed. This meta-analysis examined the association between three polymorphisms of GSTs (T1, M1, and P1) and treatment response to chemotherapy in patients with NSCLC, including a total of 3,107 patients from 23 publications [2,3,7–16,19–21,23–30]. The results showed that the null GSTM1 and the GG genotype of GSTP1 (recessive model, dominant model, homozygous model, heterozygous model, and allele model) were both associated with an improved response to cisplatin-based chemotherapy.
The findings of this study on GST gene polymorphisms in patients with NSCLC are supported by the findings from other forms of malignancy, as GST gene polymorphisms have been shown to be associated with improved patient prognosis in colorectal cancer, bladder cancer, breast cancer, and osteosarcoma [32–36]. In 2013, Djukic et al. conducted a study in a Serbian population and showed that the presence of the GSTT1 genotype was associated with an increased risk of death in patients with muscle-invasive bladder cancer [32]. Oliveira et al. showed that the GSTT1, GSTM1, and GSTP1 IIe105Val gene polymorphisms were associated with a more favorable prognosis in patients with breast cancer in a Brazilian population [34]. Kap et al. studied 755 patients in Germany with colorectal cancer and found that the null GSTM1 genotype was associated with an improved treatment response to oxaliplatin [35]. In 2015, Goričar et al. reported a significant association between GSTP1 IIe105Val gene polymorphism and the survival and treatment outcomes of patients with osteosarcoma [33]. All of the above results indicate the GST genetic polymorphisms affect the treatment response and clinical outcome following chemotherapy in patients with malignant disease.
Concerning different ethnic groups, the results of the present study showed that the GG genotype of GSTP1 IIe105Val was associated with a better treatment response in East-Asians patients with NSCLC when compared with Caucasian patients with NSCLC. A further subgroup analysis was also conducted on the clinical outcomes associated with GSTM1, and the results showed that null GSTM1 was associated with an improved response in Chinese patients with NSCLC, but not in Caucasian patients with NSCLC. The correlation between GST polymorphisms and an improved therapeutic response was different in different ethnic groups, and the heterogeneity of these studies may be due to demographic background, the clinical condition of the patients, the tumor stage, or due to confounding factors. These factors can be regarded as limitations of the present study that require further investigation. However, in this study, there were insufficient numbers of samples, especially for Caucasian patients, as only three studies included Caucasian patients with a total of 333 patients with NSCLC in the analysis, which precluded further conclusions being made regarding genotype, ethnicity, and treatment response. Also, the lack of primary data limited the analysis of other parameters. However, the studies also showed heterogeneity and publication bias, for example, for gender, age, cigarette smoking, history histology type, tumor stage, and chemotherapy regime. The findings of this meta-analysis showed that the GG genotype of GSTP1 was associated with an improved treatment response to cisplatin-based chemotherapy in East-Asian patients with NSCLC. Also, the association with the null GSTM1 genotype and an improved treatment response and clinical outcome following chemotherapy was mainly observed in Asian patients, but not in Caucasian patients with NSCLC. However, these findings require support from further studies with larger patient sample sizes.
Conclusions
A meta-analysis of the published literature on glutathione S-transferase (GST) gene polymorphisms in patients with non-small cell lung cancer (NSCLC) showed that both the null GSTM1 and the GG genotype of GSTP1 IIe105Val were associated with improved treatment response to cisplatin-based chemotherapy. Subgroup analysis showed that the GSTP1 IIe105Val gene polymorphisms were more commonly found in East-Asian patients with NSCLC than in Caucasians, and an association between null GSTM1 polymorphisms and improved treatment response was found in Chinese patients with NSCLC. However, multi-center large-scale controlled clinical studies that include an ethnically diverse study population should be conducted to study the role of these GST polymorphisms further.
Footnotes
Conflict of interest
None.
Source of support: Funding for this study was provided by the Youth Fund Projects of the Education Department of Hebei Province (QN2015010) and the Scientific Research Project of the Hebei Education Department (Z2012079)
References
- 1.Kang X, Zhou L, Jian YM, et al. Effectiveness of antibody-drug conjugate (ADC): Results of in vitro and in vivo studies. Med Sci Monit. 2018;24:1408–16. doi: 10.12659/MSM.908971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao HL, Yang ZT, Han F, Wei HX. Association of glutathione S-transferase (GST) genetic polymorphisms with treatment outcome of cisplatin-based chemotherapy for advanced non-small cell lung cancer in a Chinese population. Genet Mol Res. 2016;15(2):1–7. doi: 10.4238/gmr.15027320. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Wang T, Wu Z, et al. Association between TERT rs2853669 polymorphism and cancer risk: A meta-analysis of 9,157 cases and 11,073 controls. PLoS One. 2018;13(3):e191560. doi: 10.1371/journal.pone.0191560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehua Z, Mingming C, Jisheng W. Meta-analysis of gemcitabine in brief versus prolonged low-dose infusion for advanced non-small cell lung cancer. PLoS One. 2018;13(3):e193814. doi: 10.1371/journal.pone.0193814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han ZG, Tao J, Yu TT, Shan L. Effect of GSTP1 and ABCC2 polymorphisms on treatment response in patients with advanced non-small cell lung cancer undergoing platinum-based chemotherapy: A study in a Chinese Uygur population. Med Sci Monit. 2017;23:1999–2006. doi: 10.12659/MSM.904156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Xian L. The association between the GSTP1 A313G and GSTM1 null/present polymorphisms and the treatment response of the platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients: A meta-analysis. Tumor Biol. 2014;35(7):6791–99. doi: 10.1007/s13277-014-1866-4. [DOI] [PubMed] [Google Scholar]
- 7.Zhigang H, Ling M, Jie T, Li S. [GSTP1 and ABCC2 polymorphisms with the sensitivity of platinum-based chemotherapy in advanced non-small cell lung cancer of Xinjiang Uygur]. Chinese Clinical Oncology. 2015;(01):23–29. [in Chinese] [Google Scholar]
- 8.Yuan S. [The correlative study between efficacy of pemetrexed combined platinum chemotherapy and polymorphism of TYMS GSTP1 gene in advanced non-small cell lung cancer]. Nan Hua University. 2015 [in Chinese] [Google Scholar]
- 9.Zhao R, Chen G. Role of GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms in the clinical outcome of advanced non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(11):14909–16. [PMC free article] [PubMed] [Google Scholar]
- 10.Sun N, Sun X, Chen B, et al. MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2010;65:437–46. doi: 10.1007/s00280-009-1046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Ba T. [Association of GSTP1 and XRCC1 gene polymorphisms with clinical outcome of advanced non-small cell lung cancer patients]. Southern Medical University. 2016 [in Chinese] [Google Scholar]
- 12.Liu W. [Relationship between in GSTP1, RRM1 genes and sensitivity to gemcitabine plus cisplatin chemotherapy (GP SCHEME) in patients with advanced non-small cell lung cancer]. Nan Hua University. 2015 [in Chinese] [Google Scholar]
- 13.Bu L, Zhang LB, Mao X, Wang P. GSTP1 Ile105Val and XRCC1 Arg399Gln gene polymorphisms contribute to the clinical outcome of patients with advanced non-small cell lung cancer. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15027611. [DOI] [PubMed] [Google Scholar]
- 14.Zhou F, Yu Z, Jiang T, et al. Genetic polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer (NSCLC) patients. Swiss Med Wkly. 2011;141:w13275. doi: 10.4414/smw.2011.13275. [DOI] [PubMed] [Google Scholar]
- 15.Yuan Z, Zhou W, Liu W, et al. Association of GSTP1 and RRM1 polymorphisms with the response and toxicity of gemcitabine-cisplatin combination chemotherapy in Chinese patients with non-small cell lung cancer. Asian Pac J Cancer Prev. 2015;16(10):4347–51. doi: 10.7314/apjcp.2015.16.10.4347. [DOI] [PubMed] [Google Scholar]
- 16.Han B, Guo Z, Ma Y, et al. Association of GSTP1 and XRCC1 gene polymorphisms with clinical outcome of advanced non-small cell lung cancer patients with cisplatin-based chemotherapy. Int J Clin Exp Pathol. 2015;8(4):4113–19. [PMC free article] [PubMed] [Google Scholar]
- 17.Gao ZJ, Yuan WD, Yuan JQ, et al. Downregulation of HIF-2alpha reverses the chemotherapy resistance of lung adenocarcinoma A549 cells to cisplatin. Med Sci Monit. 2018;24:1104–11. doi: 10.12659/MSM.906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikstacki A, Zakerska-Banaszak O, Skrzypczak-Zielinska M, et al. Glutathione S-transferase as a toxicity indicator in general anesthesia: Genetics and biochemical function. J Clin Anesth. 2015;27(1):73–79. doi: 10.1016/j.jclinane.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Jia W, Sun JY, Jia KY, Liu XC. Role of GSTM1, GSTT1, and GSTP1 IIe105Val gene polymorphisms in the response to chemotherapy and overall survival of advanced non-small cell lung cancer. Genet Mol Res. 2016;15(3) doi: 10.4238/gmr.15037668. [DOI] [PubMed] [Google Scholar]
- 20.Chen JB, Wang F, Wu JJ, Cai M. Glutathione S-transferase pi polymorphism contributes to the treatment outcomes of advanced non-small cell lung cancer patients in a Chinese population. Genet Mol Res. 2016;15(3) doi: 10.4238/gmr.15037498. [DOI] [PubMed] [Google Scholar]
- 21.Wu G, Jiang B, Liu X, et al. Association of GSTs gene polymorphisms with treatment outcome of advanced non-small cell lung cancer patients with cisplatin-based chemotherapy. Int J Clin Exp Pathol. 2015;8(10):13346–52. [PMC free article] [PubMed] [Google Scholar]
- 22.Deng JH, Deng J, Shi DH, et al. Clinical outcome of cisplatin-based chemotherapy is associated with the polymorphisms of GSTP1 and XRCC1 in advanced non-small cell lung cancer patients. Clin Transl Oncol. 2015;17(9):720–26. doi: 10.1007/s12094-015-1299-6. [DOI] [PubMed] [Google Scholar]
- 23.Joerger M, Burgers SA, Baas P, et al. Germline polymorphisms in patients with advanced non-small cell lung cancer receiving first-line platinum-gemcitabine chemotherapy. A Prospective Clinical Study. Cancer. 2012;118(9):2466–75. doi: 10.1002/cncr.26562. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Qian X, Ying X, et al. GSTP1 gene polymorphism and susceptibility as well as chemotherapy sensitivity to non-small cell lung cancer. Chin J Cancer Prev Treat. 2009;(19):1441–44. [Google Scholar]
- 25.Kalikaki A, Kanaki M, Vassalou H, et al. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10(2):118–23. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 26.Booton R, Ward T, Heighway J, et al. Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol. 2006;1(7):679–83. [PubMed] [Google Scholar]
- 27.Yaping Z, Guifeng S, Yongping L. [The relationship of GSTP1 and clinical response to platinum-based chemotherapy in advanced non-small lung cancer]. The Journal of Medical Theory and Practice. 2012;(24):3003–4. [in Chinese] [Google Scholar]
- 28.Zhijun Y, Wenwu Z, Wei L, et al. [Correlation of GSTP1/RRM1 gene polymorphisms with the clinical efficacy and toxicity of GP programs in the treatment of non-small cell lung cancer]. Anti-tumor Pharmacy. 2016;(02):136–41. [in Chinese] [Google Scholar]
- 29.Xin W, Ya-jun J, Du X. [Correlation of GSTP1/RRM1 gene polymorphisms with the clinical efficacy and prognosis of GP programs in the treatment of non-small cell lung cancer]. Oncology Progress. 2016;(11):1149–52. [in Chinese] [Google Scholar]
- 30.Qiying L, Yonghong T, Lumi H, et al. [Impact of GSTM1 and GSTP1 polymorphism on lung cancer patients treated with platinum-based chemotherapy]. Chong Qing Medicine. 2014;(20):2592–94. [in Chinese] [Google Scholar]
- 31.Jianhao D, Lianxi X, Jianhong L. [Meta-analysis on GSTP1 gene polymorphisms in a Chinese population with non-small cell lung cancer and platinum-based chemotherapy sensitivity]. China Pharmaceuticals. 2015;(08):53–56. [in Chinees] [Google Scholar]
- 32.Djukic TI, Savic-Radojevic AR, Pekmezovic TD, et al. Glutathione S-transferase T1, O1, and O2 polymorphisms are associated with survival in muscle-invasive bladder cancer patients. PLoS One. 2013;8(9):e74724. doi: 10.1371/journal.pone.0074724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goričar K, Kovač V, Jazbec J, et al. Genetic variability of DNA repair mechanisms and glutathione-S-transferase genes influences treatment outcome in osteosarcoma. Cancer Epidemiol. 2015;39(2):182–88. doi: 10.1016/j.canep.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira AL, Oliveira Rodrigues FF, Dos Santos RE, et al. GSTT1, GSTM1, and GSTP1 polymorphisms as a prognostic factor in women with breast cancer. Genet Mol Res. 2014;13(2):2521–30. doi: 10.4238/2014.January.22.9. [DOI] [PubMed] [Google Scholar]
- 35.Kap EJ, Richter S, Rudolph A, et al. Genetic variants in the glutathione S-transferase genes and survival in colorectal cancer patients after chemotherapy and differences according to treatment with oxaliplatin. Pharmacogenet Genomics. 2014;24(7):340–47. doi: 10.1097/FPC.0000000000000059. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Luo J, Wang Y, et al. Predictive potential role of glutathione S-transferases polymorphisms on prognosis of breast cancer. In J Clin Exp Pathol. 2014;7(12):8935–40. [PMC free article] [PubMed] [Google Scholar]