Abstract
Background:
RNA is increasingly recognized as a powerful molecule that can be used to control gene expression. Sophisticated, well-engineered RNA-based regulators are being developed as oligotherapeutics.
Methods:
In particular, small activating RNAs (saRNAs) are promising therapeutic options for targeting human diseases. Numerous saRNAs targeting multiple cancers have been developed in preclinical models. One saRNA targeting C/EBPα is currently undergoing clinical trials in liver cancer.
Results and
Conclusion:
In this review, we describe the current working model of the intracellular mechanism of saRNA, discuss the recent progress of saRNA therapeutics in preclinical and clinical trials, and current advances in targeted delivery using aptamers in detail.
Keywords: saRNAs, therapeutics, human cancers, intracellular mechanism, targeted delivery, aptamers, clinic
1. INTRODUCTION
Small activating RNA (saRNA) is a type of double-stranded RNA that induces target gene expression. The concept of saRNA goes back to 2006, when saRNAs designed to target the gene promoters of p21, VEGF, and E-cadherin were shown to activate transcription [1]. Since the discovery of this novel concept, saRNAs have quickly emerged as powerful oligotherapeutics for targeting various human cancers. The mechanism of upregulation by small RNAs was not initially understood. However, many recent efforts have been made to understand the working mechanisms at the molecular and cellular level. Thus, in this review, we focus on the recent progress of saRNA gene upregulating mechanisms and preclinical models that may be candidates for future clinical trials.
2. DESIGN OF SARNAS
Current effective design of saRNAs on gene promoters is largely a hit-or-miss process. As transcriptional gene activation induced by saRNA is very sensitive to target gene promotor location [1], to design the effective saRNAs, it is noteworthy to follow the rules published by two groups. The
first group published the four rules to achieve a success rate of 10-20% [2]: 1) the sense DNA sequence of the promoter as the template; 2) the targets within a promoter region between -100 and -1000 bp upstream of the transcription start site (TSS); 3) 40-60% of GC content in 19-nt in size; 4) the lower thermodynamic stability at the 3´ end than the 5´ end of the saRNA duplexes.
The second group designed saRNAs following the four criteria based on the analysis of bioinformatics [3]: 1) overlapping the target’s promoter and the target mRNA´s 5´end; 2) overlapping the target mRNA; 3) targeting 200-100kb upstream of the target gene site; 4) targeting the 20-100kb downstream of the target gene’s polyadenylation site. Also, the single nucleotide mutation assay in the sense or antisense strand of saRNA showed that the seed region of the 5’-end of the antisense strand is particularly pivotal in strand selectivity for the upregulation of gene expression by saRNAs [4].
3. INTRACELLULAR WORKING MECHANISMS
Since the discovery of saRNAs, the working mechanism was not fully understood. But, the saRNAs are double-strand RNAs like small inhibiting RNAs (siRNAs), the role of Argonaute proteins (AGOs) has been implicated in the involvement of gene up-regulation. To investigate the involvement of four AGOs (AGO1, AGO2, AGO3, and AGO4) in the saRNA induced upregulation, the knockdown experiment of AGOs was performed in the treatment of E-cadherin and p21-saRNAs. It showed that the target expression was abolished by AGO2 depletion, whereas knockdown of the other AGOs did not significantly impair the saRNA activity [1, 5, 6]. For the direct assessment of AGOs with saRNA, the biotinylated CEBPA-saRNA was transfected into the cells. The CEBPA-saRNA protein complex was retrieved with streptavidin beads. The association of sense and antisense strand of CEBPA-saRNA with AGOs was confirmed with western blotting. It showed only AGO2 loaded strands of saRNAs, not AGO1, AGO3, and AGO4 [7], proving the importance of AGO2 for the saRNA activity.
To determine the AGO2 loading site of saRNA, biotin was linked to the 3’ end or 5’ end of termini within saRNAs. The modification of the 5’ end of antisense strand of p21-saRNA completely abolished its activating activity [8]. However, the modification of the 3’ end of both the sense and antisense strands [5] or either strand [8] had almost equivalent RNAs activity. These results confirmed that the modification of 5’end blocks the AGO2 loading. In the study of CEBPA-saRNA, the strand selectivity of AGO2 loading in CEBPA-saRNA has been determined using a 5’ inverted abasic modification on each strand [7]. The 5’ inverted abasic modification on the sense strand well tolerated the saRNA activity, but not 5’ inverted abasic modification on both the strands. It suggests that antisense is the guide strand loaded into AGO2.
As the saRNAs target specific promoter regions to stimulate gene expression at the transcriptional level, previous studies suggest the enrichment of RNA polymerase II (RNAP II) and histone methylation at lysine 4 (H3K4) at target promoters [9, 10]. To elucidate the transcriptional initiation and elongation by saRNAs, more detailed study has been performed on the biochemical, proteomic, and functional analysis [8]. The scanning ChIP analysis of RNAP II was performed on the target gene in the treatment of saRNAs [8]. The result of scanning ChIP showed the localization of the phosphorylation of RNAP II at serine 2 (Ser2P) accumulation surrounding the transcriptional starting site (TSS) in the treatment of saRNA [8]. Also, the massive increase in RNAP II surrounding the saRNA target site and the core promoter was observed, suggesting that saRNA-AGO2 complex targeting of promoters stabilizes or facilitates the transcription initiation complex by association with RNAP II [8]. To address the epigenetic changes, specially methylation, ChIP assay was performed using the antibodies of monoubiquitination of H2B (H2Bub1) [8], because the methylation of H3K4 is stimulated histone 2B (H2B) ubiquitination to lead active transcription [11]. The ChIP showed the enrichment of H2Bub1 in the treatment of saRNA, suggesting that the H2B ubiquitination is an early event of transcriptional/ epigenetic changes leading to transcriptional activation [8]. In the nucleosome positioning studies, the binding of saRNA to its target DNA induced the open chromatin structure, in which RNAP II can bind and initiate transcription by ChIP-PCR [12].
To discover other protein components involving saRNA effector complexes, a ChIP-based biotinylated RNA pull-down assay (ChIbRP) and mass spectrometry (MS) were used [8]. Of the 42 proteins, two proteins, RNA polymerase-associated protein CTR9 homolog (CTR9) and RHA (nuclear DNA helicase II) known transcriptional activators with DNA/RNA-unwinding/binding activity, were validated for the interactors with RNAP II. In the immunoprecipitation assays, two nuclear-localized proteins were co-immuno-precipitated with both AGO2 and RNAP II in the treatment of saRNAs, suggesting the strong association with saRNA-AGO2 complex, RHA, CTR9 and RNAP II [8].
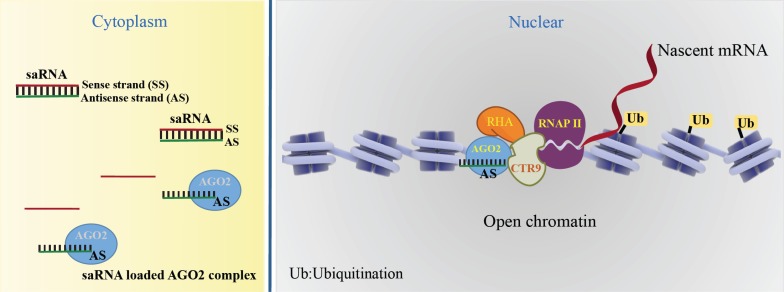
In summary, AGO2 loads the guide strand of saRNAs to form an active saRNA-AGO2 complex. The saRNA-loaded AGO2 binding to their promotor induces the open chromatin structure and recruitment of the complex of CTR9 and RHA. RNA-induced transcriptional activation (RITA), by AGO2 starts transcript initiation associated with phosphorylation of RNAP II on Ser2 and H2Bub1. Currently, the widely accepted working model of the molecular mechanisms of saRNA-induced gene activation is depicted in Fig. (1).
Fig. (1).
Canonical activating mechanism of saRNAs. A saRNA-loaded AGO2 complex binds at the target promoter and induces the open chromatin structure. The AGO2 recruits the complex of CTR9 and RHA, RNA-induced transcriptional activation (RITA), leading the transcript initiation associating with phosphorylation of RNAP II on Ser2 and H2Bub1.
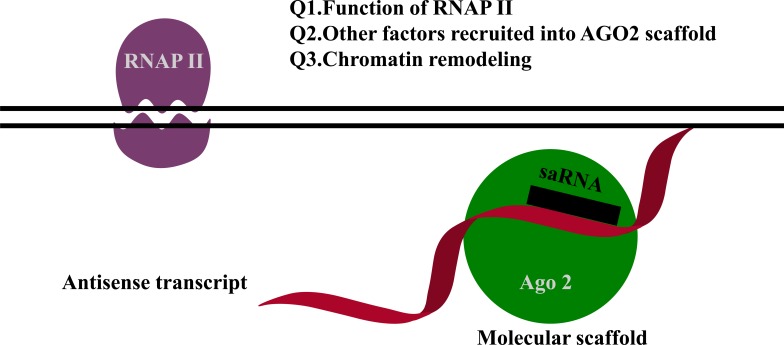
Another possible mechanism is that non-coding transcripts overlap with saRNA targets acting as binding motifs for saRNAs [13-15]. For example, for both the progesterone and the low-density lipoprotein receptor promoters, RNAs complementary to non-coding transcripts at the promoters activate gene expression in the recruitment of Ago2 [13-15]. The proposed mechanism is depicted in Fig. (2). However, several aspects of this proposed mechanism remain unresolved, such as the function of RNAP II, which other factors are recruited to the Ago2 scaffold complex, the role of chromatin remodeling, and the role of endogenous RNAs for gene activation. Currently, we do not have all of the answers to these questions. One possible explanation is that RNAP II interacts with Ago1 [16]. Furthermore, transcriptional gene activation by antisense non-coding RNAs occurs in cis and is not required for Ago2 cleavage activity [17].
Fig. (2).
Non-canonical activating mechanism of saRNAs. RNAs complementary to a non-coding transcript at the promoter activates gene expression in recruitment of Ago2. This model comes with unresolved questions that require further investigation, as indicated.
4. THERAPEUTIC POTENTIAL OF SaRNAs IN LIPOSOMAL NANOPARTICLES
Since the remarkable conceptual discovery of saRNAs, numerous therapeutic saRNAs have been developed to target mainly cancers. For the preclinical stage, a list of saRNAs currently being pursued in various human diseases is summarized in Table 1. Both in vitro and in vivo studies have shown convincing anti-tumor effects. These accumulated developments in the field of therapeutic saRNAs led to the development of the pharmaceutical company, MiNA Therapeutics Limited, which specializes in therapeutic saRNAs. In 2014, the MiNA therapeutics developed CEBPA-saRNA, which targets the transcription factor C/EBP-α, to improve liver function in liver cancer [18]. For in vivo delivery, CEBPA-saRNA complexed with dendrimer to test its effects in diethylnitrosamine (DEN)-induced cirrhotic rat liver tumor model. The CEBPA-saRNA-dendrimers have been injected via tail vein injection with 3 doses in one week. The in vivo results showed reduced tumor burden and improved liver function markers; albumin, bilirubin, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and ammonia [18].
Table 1.
Preclinical targets of therapeutic saRNAs in human cancers and renal disease.
| Gene Symbol | Full Name | Cancer | References |
|---|---|---|---|
| CDH1 | E-cadherin | Cervical cancer | [1] |
| CDKN1A (p21) | Cyclin-dependent kinase inhibitor 1A | Prostate cancer, Breast cancer, Cervical cancer, Bladder cancer Pancreatic cancer Non-small-cell lung carcinoma Liver cancer |
[1] [41] [42] [43] |
| VEGF | Vascular endothelial growth factor | Cervical cancer | [1] |
| PGR (PR) | Progesterone receptor | Breast cancer | [44] |
| MVP | Major vault protein | Breast cancer | [44] |
| KLF4 | Kruppel-like factor 4 | Prostate cancer Colorectal cancer |
[45] [46] |
| C/EBPα | CCAAT/enhancer-binding protein a | Liver cancer Pancreatic cancer |
[18, 47] [34] |
| PAWR | PRKC apoptosis WT1 regulator | Bladder cancer, prostate cancer | [48] |
| NKX3-1 | NK3 homeobox 1 | Prostate cancer | [49] |
| HIC-1 | HIC ZBTB transcriptional repressor | Breast cancer | [50] |
| NIS | Sodium/iodine symporter | Hepatocellular carcinoma | [51] |
| TP53 | Wild type p53 | Malignant pheochromocytoma | [52] |
| VHL | Von Hippel-Lindau | Renal cell carcinoma | [53] |
| TRPV5 | Transient receptor potential cation channel subfamily V member 5 | Nephrolithiasis | [39] |
In the following studies, CEBPA-saRNA was formulated with the liposomal nanoparticle, MTL-CEBPA, for the candidate of clinical trial [19]. In preclinical studies, MRL-CEBPA has been tested in multiple liver cancer models; DEN-induced cirrhotic rat liver cancer model, carbon-tetrachloride (CCl4)-induced rat liver cancer model, non-alcoholic methionine and cholic-deficient (MCD) diet-induced steatotic liver disease model. In DEN-induced cirrhotic rat liver tumor model, the MTL-CEBPA upregulated the expression of CEBPA and reduced the significant tumor burden, along with the improvement of the liver function markers [19]. In CCl4-induced rat liver cancer model, MTL-CEBPA improved the all five liver function markers. To assess the long-term benefits of MTL-CEBPA, the survival rate and liver function were determined in rats chronically exposed to CCl4 as advanced stages of liver model. The MTL-CEBPA showed the significant 78% increased survival benefit and reduction in liver damage markers. In MCD diet-induced animal model, the MTL-CEBPA improved the marker of liver damage, ALT and AST, to the normal level [19]. Currently, their lead therapeutic, MTL-CEBPA, is undergoing clinical trials for safety and tolerability in advanced liver cancer (ClinicalTrials.gov, NCT02716012). The MiNA’s achievement is a very big step toward opening a new chapter for saRNA in the field of therapeutics. We expect that the outcomes of MTL-CEBPA in patients will be published in the near future.
In other cancer models of saRNAs formulated with nanoparticles, p21-saRNAs have been formulated with nanoparticles to test the anti-tumor effects in xenografted prostate cancer model via intratumoral injection [20] and orthotopic bladder cancer model via intravesical injection [21]. The p21-saRNA-nanoparticles significantly inhibited the tumor growth in both the models. These preclinical results clearly indicate that saRNAs are very promising therapeutics in various cancers for the clinic.
5. THERAPEUTIC POTENTIAL OF SaRNAs IN APTAMER-MEDIATED TARGETED DELIVERY
To improve the therapeutic effects of saRNAs, targeted therapy is preferred. In recent decades, aptamers have become a popular tool for achieving targeted delivery. Aptamers are structured nucleic acid ligands that are isolated using the systematic evolution of ligands by exponential enrichment (“SELEX”) method [22]. Currently, there is no method
available for the rational design of aptamers by bioinformatics. It solely depends on the tedious lab-based selection. Theoretically, aptamers can be selected against any target, from small molecules to living cells [23-28]. As therapeutic molecules, they have excellent attributes, such as high specificity and affinity for their targets, structural stability, and limited toxicity and immunogenicity [29]. These features make them useful as targeted vehicles [30]. Chimeric constructs of aptamers and small interfering RNAs (siRNAs) are well-tolerated strategies for targeted downregulation of gene expression [31]. Given this, conjugation of promoter-targeted saRNA and functionalized aptamers is a promising option for targeted upregulation of gene expression.
Pancreatic cancer is one of the most aggressive types of cancers [32]. However, currently, there is no effective option for treating pancreatic cancer. Targeted delivery of saRNA using aptamers has been explored in a pancreatic cancer model. At the molecular and cellular level, loss of the histone demethylase KDB6B enhances the aggressiveness of pancreatic cancer via downregulation of C/EBPα [33], which suggests that C/EBPα is a potential therapeutic target in pancreatic cancer. In previous in vivo studies in liver cancer, saRNA targeting C/EBPα induced a potent antitumor effect through positive regulation of C/EBPα and its downstream targets, including cyclin-dependent kinase inhibitor 1 (p21) [18]. This compound is currently undergoing clinical trials as MTL-CEBPA. To apply targeted delivery of C/EBPα-saRNA to pancreatic cancer, pancreatic cancer-specific RNA aptamers have been isolated against naïve pancreatic cancer cells using cell-based SELEX. To ensure specificity for pancreatic cancer, liver cancer cells (Huh7) were used for counter SELEX. To develop constructs for targeted delivery of saRNAs, the pancreatic cancer-specific RNA aptamers were conjugated with C/EBPα-saRNA via sticky-bridge sequences [34]. The aptamer-conjugated C/EBPα-saRNA showed increased target gene and protein expression, and a strong anti-tumor effect in in vivo studies, without toxicity [34]. The successful results of targeted delivery of C/EBPα-saRNA in pancreatic cancer could act as a model for future studies in other cancers.
Other studies have been performed in a prostate cancer model. Prostate cancer has a high overall survival rate, but metastasis remains a major cause of mortality. The dihydropyrimidinase-related protein 3 (DPYSL3), also called collapsing response mediator protein 4 (CRMP4), is well known as a tumor metastasis suppressor in prostate cancer [35]. Thus, to inhibit prostate cancer cell invasion and metastasis, saRNA targeting the DPYSL3 promotor region has been developed and conjugated to prostate membrane antigen-specific aptamers for targeted delivery to upregulate target gene expression at the transcriptional level [36]. The aptamer-conjugated DPYSL3-saRNA induced the successful upregulation of target gene expression in vitro. It also completely suppressed distal metastasis of prostate cancer in orthotopic xenografted nude mice, with no side effects.
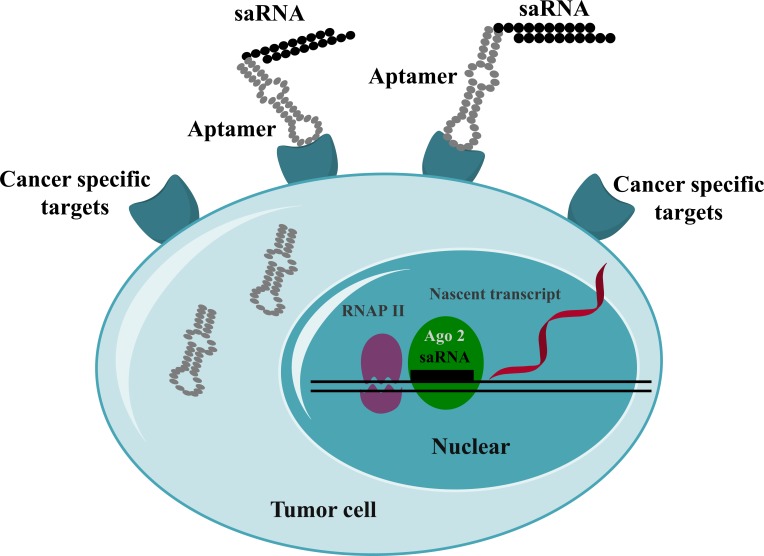
The main advantage of aptamers over nanoparticles for the delivery of saRNAs allows us for the targeted drug delivery into the target tissues specifically, leaving less unwanted effects in non-targeting tissues. The above mentioned two studies prove the successful delivery of saRNAs in preclinical cancer models. Hence, aptamer-mediated delivery of saRNAs might be another option to translate saRNAs into the clinic. The targeted delivery of saRNAs mediated by aptamers is depicted in Fig. (3).
Fig. (3).
Targeted delivery of saRNAs. Aptamer-conjugated saRNAs are internalized via cancer-specific receptors. The aptamer-saRNAs disassociate in the cytoplasm. The saRNAs are transported into the nucleus, where they induce the upregulation of target genes.
6. CHEMICAL MODIFICATION OF SARNAS
The chemical modification of siRNAs is a well-adopted strategy to improve endonuclease resistance and abrogate immune stimulation to translate to the clinic [37, 38]. To improve the therapeutic potential, the same strategy might be applied in saRNA therapeutic development. To determine the tolerance of saRNA molecules to 2´-O-methyl modification (2´-OMe), 2´-OMe modification in either the sense or antisense strand of p21-saRNA has been incorporated [5]. The 2´-OMe modification within the sense strand suppressed the activity about 50%, comparing naked saRNA and 2´-OMe modification in the antisense strand. In this study, it suggests that the modification of passenger stand in saRNA interferes in the saRNA activity [5].
In the study of CEBPA-saRNA, the variants of 2´-OMe modification were synthesized to prevent the immune stimulation. The heavy modification of 2´-OMe on sense strand was well tolerated the saRNA activity, but the 2´-OMe on antisense strand was not [7]. In summary, for optimizing the saRNA to develop the clinical candidates, the modification of AGO2 loading strand is very sensitive for saRNA activity.
CONCLUSION/REMARKS
saRNAs, which embody an opposite concept to siRNAs, are now considered mainstream oligotherapeutics in human diseases. The upregulation of target gene expression is AGO2 mediated transcriptional activation. Comparing siRNAs that show the effect within hours and disappearing when the exogenous siRNA is exhausted [5], saRNAs show the delayed effects by 48 hours [1, 5] or by 96 hours [7, 18]. The advantage of transcriptional up-regulation by saRNA is prolonged activation for nearly two weeks in some cells [1, 5], which is more beneficial for the candidates of clinical trials. Target diseases are mainly limited to cancers, but also include non-cancer conditions such as renal disease, in which saRNA against transient receptor potential cation channel of subfamily V member 5 (TRPV5) reduces the calcium oxalate (CaOx) crystals deposition in the kidney in vivo assay [39]. It shows great potential to expand the field of therapeutic saRNAs in other human diseases. Given that clinical trial of MTL-CEBPA has been initiated, we expect the continued active development of therapeutic saRNAs in additional human diseases. Along with this great achievement, cancer-specific aptamers are continuously under development [40]. To improve target specificity, we expect targeted delivery of saRNA using aptamers to be a focus in the near future, for improving the therapeutic index while minimizing off-target effects.
Consent for Publication
Not applicable.
Acknowledgements
This work was supported by NIH grant AI42552 to John J. Rossi. We thank Sarah T. Wilkinson, PhD, scientific writer at the City of Hope, for language editing.
Conflict of Interest
John J. Rossi is a co-founder of MiNA Therapeutics Limited.
REFERENCES
- 1.Li L.C., Okino S.T., Zhao H., Pookot D., Place R.F., Urakami S., Enokida H., Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA. 2006;103(46):17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang V., Qin Y., Wang J., Wang X., Place R.F., Lin G., Lue T.F., Li L.C. RNAa is conserved in mammalian cells. PLoS One. 2010;5(1):e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voutila J., Saetrom P., Mintz P., Sun G., Alluin J., Rossi J.J., Habib N.A., Kasahara N. Gene expression profile changes after short-activating RNA-mediated induction of endogenous pluripotency factors in human mesenchymal stem cells. Mol. Ther. Nucleic Acids. 2012;1:e35. doi: 10.1038/mtna.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng X., Jiang Q., Chang N., Wang X., Liu C., Xiong J., Cao H., Liang Z. Small activating RNA binds to the genomic target site in a seed-region-dependent manner. Nucleic Acids Res. 2016;44(5):2274–2282. doi: 10.1093/nar/gkw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Place R.F., Noonan E.J., Foldes-Papp Z., Li L.C. Defining features and exploring chemical modifications to manipulate RNAa activity. Curr. Pharm. Biotechnol. 2010;11(5):518–526. doi: 10.2174/138920110791591463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portnoy V., Huang V., Place R.F., Li L.C. Small RNA and transcriptional upregulation. Wiley Interdiscip. Rev. RNA. 2011;2(5):748–760. doi: 10.1002/wrna.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voutila J., Reebye V., Roberts T.C., Protopapa P., Andrikakou P., Blakey D.C., Habib R., Huber H., Saetrom P., Rossi J.J., Habib N.A. Development and mechanism of small activating RNA Targeting CEBPA, a novel therapeutic in clinical trials for liver cancer. Mol. Ther. 2017;25(12):2705–2714. doi: 10.1016/j.ymthe.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portnoy V., Lin S.H., Li K.H., Burlingame A., Hu Z.H., Li H., Li L.C. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016;26(3):320–335. doi: 10.1038/cr.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3(3):166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 10.Hu J., Chen Z., Xia D., Wu J., Xu H., Ye Z.Q. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochem. J. 2012;447(3):407–416. doi: 10.1042/BJ20120256. [DOI] [PubMed] [Google Scholar]
- 11.Weake V.M., Workman J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell. 2008;29(6):653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Wang B., Sun J., Shi J., Guo Q., Tong X., Zhang J., Hu N., Hu Y. Small-activating RNA can change nucleosome positioning in human fibroblasts. J. Biomol. Screen. 2016;21(6):634–642. doi: 10.1177/1087057116637562. [DOI] [PubMed] [Google Scholar]
- 13.Matsui M., Sakurai F., Elbashir S., Foster D.J., Manoharan M., Corey D.R. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem. Biol. 2010;17(12):1344–1355. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz J.C., Younger S.T., Nguyen N.B., Hardy D.B., Monia B.P., Corey D.R., Janowski B.A. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15(8):842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu Y., Yue X., Younger S.T., Janowski B.A., Corey D.R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38(21):7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang V., Zheng J., Qi Z., Wang J., Place R.F., Yu J., Li H., Li L.C. Ago1 Interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet. 2013;9(9):e1003821. doi: 10.1371/journal.pgen.1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X., Li H., Burnett J.C., Rossi J.J. The role of antisense long noncoding RNA in small RNA-triggered gene activation. RNA. 2014;20(12):1916–1928. doi: 10.1261/rna.043968.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reebye V., Saetrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D., Kostomitsopoulos N., Kasahara N., Nicholls J.P., Jiao L.R., Pai M., Spalding D.R., Mizandari M., Chikovani T., Emara M.M., Haoudi A., Tomalia D.A., Rossi J.J., Habib N.A. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59(1):216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reebye V., Huang K.W., Lin V., Jarvis S., Cutilas P., Dorman S., Ciriello S., Andrikakou P., Voutila J., Saetrom P., Mintz P.J., Reccia I., Rossi J.J., Huber H., Habib R., Kostomitsopoulos N., Blakey D.C., Habib N.A. Gene activation of CEBPA using saRNA: Preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene. 2018;37(24):3216–3228. doi: 10.1038/s41388-018-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Place R.F., Wang J., Noonan E.J., Meyers R., Manoharan M., Charisse K., Duncan R., Huang V., Wang X., Li L.C. Formulation of small activating RNA into lipidoid nanoparticles inhibits xenograft prostate tumor growth by inducing p21 expression. Mol. Ther. Nucleic Acids. 2012;1:e15. doi: 10.1038/mtna.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang M.R., Yang G., Place R.F., Charisse K., Epstein-Barash H., Manoharan M., Li L.C. Intravesical delivery of small activating RNA formulated into lipid nanoparticles inhibits orthotopic bladder tumor growth. Cancer Res. 2012;72(19):5069–5079. doi: 10.1158/0008-5472.CAN-12-1871. [DOI] [PubMed] [Google Scholar]
- 22.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 23.Ulrich H., Magdesian M.H., Alves M.J., Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J. Biol. Chem. 2002;277(23):20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Jiang H., Liu F. In vitro selection of novel RNA ligands that bind human cytomegalovirus and block viral infection. RNA. 2000;6(4):571–583. doi: 10.1017/s1355838200992215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blank M., Weinschenk T., Priemer M., Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001;276(19):16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 26.Daniels D.A., Chen H., Hicke B.J., Swiderek K.M., Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA. 2003;100(26):15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hicke B.J., Marion C., Chang Y.F., Gould T., Lynott C.K., Parma D., Schmidt P.G., Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001;276(52):48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 28.Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 29.Que-Gewirth N.S., Sullenger B.A. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14(4):283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J., Rossi J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017;16(3):181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J., Li H., Zhang J., Piotr S., Rossi J. Development of cell-type specific anti-HIV gp120 aptamers for siRNA delivery. J. Vis. Exp. 2011;(52):2954. doi: 10.3791/2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jemal A., Siegel R., Ward E., Murray T., Xu J., Smigal C., Thun M. J. Cancer statistics, 2006. CA Cancer J. Clin. 2006;56(2):106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto K., Tateishi K., Kudo Y., Sato T., Yamamoto S., Miyabayashi K., Matsusaka K., Asaoka Y., Ijichi H., Hirata Y., Otsuka M., Nakai Y., Isayama H., Ikenoue T., Kurokawa M., Fukayama M., Kokudo N., Omata M., Koike K. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPalpha. Carcinogenesis. 2014;35(11):2404–2414. doi: 10.1093/carcin/bgu136. [DOI] [PubMed] [Google Scholar]
- 34.Yoon S., Huang K.W., Reebye V., Mintz P., Tien Y.W., Lai H.S., Saetrom P., Reccia I., Swiderski P., Armstrong B., Jozwiak A., Spalding D., Jiao L., Habib N., Rossi J.J. Targeted delivery of C/EBPalpha -saRNA by pancreatic ductal adenocarcinoma-specific RNA aptamers inhibits tumor growth in vivo. Mol. The. 2016;24(6):1106–1116. doi: 10.1038/mt.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao X., Pang J., Li L.Y., Liu W.P., Di J.M., Sun Q.P., Fang Y.Q., Liu X.P., Pu X.Y., He D., Li M.T., Su Z.L., Li B.Y. Expression profiling identifies new function of collapsin response mediator protein 4 as a metastasis-suppressor in prostate cancer. Oncogene. 2010;29(32):4555–4566. doi: 10.1038/onc.2010.213. [DOI] [PubMed] [Google Scholar]
- 36.Li C., Jiang W., Hu Q., Li L.C., Dong L., Chen R., Zhang Y., Tang Y., Thrasher J.B., Liu C.B., Li B. Enhancing DPYSL3 gene expression via a promoter-targeted small activating RNA approach suppresses cancer cell motility and metastasis. Oncotarget. 2016;7(16):22893–22910. doi: 10.18632/oncotarget.8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czauderna F., Fechtner M., Dames S., Aygun H., Klippel A., Pronk G.J., Giese K., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31(11):2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sioud M., Furset G., Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 2007;361(1):122–126. doi: 10.1016/j.bbrc.2007.06.177. [DOI] [PubMed] [Google Scholar]
- 39.Zeng T., Duan X., Zhu W., Liu Y., Wu W., Zeng G. SaRNA-mediated activation of TRPV5 reduces renal calcium oxalate deposition in rat via decreasing urinary calcium excretion. Urolithiasis. 2017;46(3):271–278. doi: 10.1007/s00240-017-1004-z. [DOI] [PubMed] [Google Scholar]
- 40.Yoon S., Rossi J.J. Emerging cancer-specific therapeutic aptamers. Curr. Opin. Oncol. 2017;29(5):366–374. doi: 10.1097/CCO.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Wang Z., Liu X., Wang J., Li F., Li C., Shan B. Up-regulation of p21WAF1/CIP1 by small activating RNA inhibits the in vitro and in vivo growth of pancreatic cancer cells. Tumori. 2012;98(6):804–811. doi: 10.1177/030089161209800620. [DOI] [PubMed] [Google Scholar]
- 42.Wei J., Zhao J., Long M., Han Y., Wang X., Lin F., Ren J., He T., Zhang H. p21WAF1/CIP1 gene transcriptional activation exerts cell growth inhibition and enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC Cancer. 2010;10:632. doi: 10.1186/1471-2407-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosaka M., Kang M.R., Yang G., Li L.C. Targeted p21WAF1/ CIP1 activation by RNAa inhibits hepatocellular carcinoma cells. Nucleic Acid Ther. 2012;22(5):335–343. doi: 10.1089/nat.2012.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3(3):166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 45.Wang J., Place R.F., Huang V., Wang X., Noonan E.J., Magyar C.E., Huang J., Li L.C. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010;70(24):10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Q., Fan D., Huang K., Chen X., Chen Y., Mai Q. Activation of KLF4 expression by small activating RNA promotes migration and invasion in colorectal epithelial cells. Cell Biol. Int. 2017;42(4):495–503. doi: 10.1002/cbin.10926. [DOI] [PubMed] [Google Scholar]
- 47.Reebye V., Saetrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D., Kostomitsopoulos N., Kasahara N., Nicholls J.P., Jiao L.R., Pai M., Spalding D.R., Mizandari M., Chikovani T., Emara M.M., Haoudi A., Tomalia D.A., Rossi J.J., Habib N.A. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2013;59(1):216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang K., Shen J., Xie Y.Q., Lin Y.W., Qin J., Mao Q.Q., Zheng X.Y., Xie L.P. Promoter-targeted double-stranded small RNAs activate PAWR gene expression in human cancer cells. Int. J. Biochem. Cell Biol. 2013;45(7):1338–1346. doi: 10.1016/j.biocel.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 49.Ren S., Kang M.R., Wang J., Huang V., Place R.F., Sun Y., Li L.C. Targeted induction of endogenous NKX3-1 by small activating RNA inhibits prostate tumor growth. Prostate. 2013;73(14):1591–1601. doi: 10.1002/pros.22709. [DOI] [PubMed] [Google Scholar]
- 50.Zhao F., Pan S., Gu Y., Guo S., Dai Q., Yu Y., Zhang W. Small activating RNA restores the activity of the tumor suppressor HIC-1 on breast cancer. PLoS One. 2014;9(1):e86486. doi: 10.1371/journal.pone.0086486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia W., Li D., Wang G., Ni J., Zhuang J., Ha M., Wang J., Ye Y. Small activating RNA upregulates NIS expression: promising potential for hepatocellular carcinoma endoradiotherapy. Cancer Gene Ther. 2016;23(10):333–340. doi: 10.1038/cgt.2016.36. [DOI] [PubMed] [Google Scholar]
- 52.Lin D., Meng L., Xu F., Lian J., Xu Y., Xie X., Wang X., He H., Wang C., Zhu Y. Enhanced wild-type p53 expression by small activating RNA dsP53-285 induces cell cycle arrest and apoptosis in pheochromocytoma cell line PC12. Oncol. Rep. 2017;38(5):3160–3166. doi: 10.3892/or.2017.5993. [DOI] [PubMed] [Google Scholar]
- 53.Kang M.R., Park K.H., Lee C.W., Lee M.Y., Han S.B., Li L.C., Kang J.S. Small activating RNA induced expression of VHL gene in renal cell carcinoma. Int. J. Biochem. Cell Biol. 2018;97:36–42. doi: 10.1016/j.biocel.2018.02.002. [DOI] [PubMed] [Google Scholar]