Abstract
Background:
Oligonucleotide drug development has revolutionised the drug discovery field. Within this field, ‘small’ or ‘short’ activating RNAs (saRNA) are a more recently discovered category of short double-stranded RNA with clinical potential. saRNAs promote transcription from target loci, a phenomenon widely observed in mammals known as RNA activation (RNAa).
Objective:
The ability to target a particular gene is dependent on the sequence of the saRNA. Hence, the potential clinical application of saRNAs is to increase target gene expression in a sequence-specific man-ner. saRNA-based therapeutics present opportunities for expanding the “druggable genome” with partic-ular areas of interest including transcription factor activation and cases of haploinsufficiency.
Results and
Conclusion:
In this mini-review, we describe the pre-clinical development of the first saRNA drug to enter the clinic. This saRNA, referred to as MTL-CEBPA, induces increased expression of the transcription factor CCAAT/enhancer-binding protein alpha (CEBPα), a tumour suppressor and critical regulator of hepatocyte function. MTL-CEBPA is presently in Phase I clinical trials for hepatocel-lular carcinoma (HCC). The clinical development of MTL-CEBPA will demonstrate “proof of concept” that saRNAs can provide the basis for drugs which enhance target gene expression and consequently improve treatment outcome in patients
Keywords: MiNA therapeutics, MTL-CEBPA, CEBPα, saRNA, hepatocellular carcinoma, liver, RNA therapeutics
1. INTRODUCTION
With the recent FDA approvals of several oligonucleotide drugs, the RNA therapeutics field has finally made its mark [1]. After decades of setbacks associated with stability, off-target effects, and delivery the fields of chemistry, RNA biology, and genome sequencing have revolutionised RNA therapeutics in terms of possible drug compositions, mechanism of actions, and target indications. Furthermore, the ability of scientists and manufacturers of oligonucleotide drugs to utilize the same production platform and developmental profiles (e.g. toxicology and pharmacokinetic) should help further reduce the bench-to-bedside timeline [2]. The recent expansion of targeting approaches for selective delivery of oligonucleotide drugs, [3] has also dramatically improved the potential of this technology. Together, these advancements will likely lead to a rapid and robust pipeline of RNA therapeutics.
Approved antisense oligonucleotide (ASO) drugs function via a variety of mechanisms ranging from exon-skipping to RNase-H induced mRNA cleavage [4, 5]. In addition, late-stage clinical trials have also included ASOs which bind to and inhibit microRNAs (miRNA) [5].
Most ASO and double-stranded RNA (dsRNA) drugs directly inhibit gene expression. The development of oligonucleotide drugs which have the ability to directly enhance gene expression also holds great promise. These potential drugs will be able to piggyback on the strategies (chemistry, delivery, targeting) that have been developed for other oligonucleotide therapeutics. Several companies are currently developing gene activation technologies including CRISPR/Cas9 and small activating RNAs (saRNAs) [6, 7], which will complement mRNA therapeutics [8, 9]. In 2016, MTL-CEBPA was the first RNA activating oligonucleotide drug to enter clinical development. MTL-CEBPA is being developed by MiNA Therapeutics (London) to treat liver disease among other indications by directly activating the transcription of CCAAT/Enhancer-Binding Protein Alpha (CEBPα). This mini-review will briefly introduce the field of saRNAs, CEBPα as a transcription factor, and conclude with the research and pre-clinical development of MTL-CEBPA for treatment of liver cancer.
1.1. SARNAs
‘Small’ or ‘short’ activating RNAs (saRNA) induce long-lasting and sequence-specific expression of their target gene (Table 1). This remarkable finding was made in the mid 2000’s by Long-Chen Li and colleagues when they observed short double-stranded RNAs (dsRNA) targeting gene promoter sequences activated, rather than suppressed, transcription of p21WAF1/CIP1 (p21), VEGFA, and E-cadherin [10]. This newly discovered phenomenon was named RNA activation (RNAa) and the dsRNAs responsible were subsequently termed ‘saRNAs’ to differentiate them from short interfering RNAs (siRNA). Shortly after, Bethany Janowski et al. showed that progesterone receptor (PR) expression could also be induced by dsRNA targeting PR promoter [11]. The works of Li et al. and Janowski et al. were further supported by the finding that endogenous miRNA miR-373 targets the promoter regions of e-cadherin and CSDC2 which results in increased transcription from both genes [12]. Together, these studies laid the groundwork for understanding how RNAa influences gene regulation and pointed to the potential for saRNA in the clinic [13, 14]. saRNAs are structurally identical to siRNA. Both are double-stranded, ~21-mer, RNA oligonucleotides. The critical design difference between the two is the intended target. saRNAs act in the nucleus and are typically designed to contain sequences complementary to regions near or within gene promoters, [10, 11] while siRNAs are complementary to mRNA (Fig. 1). Short dsRNAs that are introduced into a cell, or endogenously generated in the case of miRNA, are recognized in the cytoplasm by dsRNA loading factors and subsequently loaded into one of the four Argonaute (AGO) proteins (Fig. 1) [15]. The guide strand (complementary to the target RNA of interest) is retained upon loading while the passenger strand (matching the target RNA sequence) is discarded. Critically, loading of AGO2 is required for RNAa [10, 11]. RNA-AGO complexes have post-transcriptional gene silencing (PTGS) potential in agreement with their canonical role in the RNA induced silencing complex (RISC) [15, 16]. In addition to PTGS in both the cytoplasm and nucleus, [17, 18] AGO2 can induce gene-specific transcriptional activation [10, 11] or suppression [19] when present in the nucleus [20] (Fig. 1). Nuclear AGO2 is also involved in DNA repair [21] and alternative-splicing [22]. AGO proteins do not contain a nuclear localization sequence (NLS). Nuclear-cytoplasmic shuttling of AGO2 appears to be, in part, dependent on importin-8 [23] as knockdown of this importin reduces the nuclear pool of AGO2 [24]. However, importin-8 knockout does not completely exclude nuclear translocation of AGO2. This indicates other import factors may also shuttle AGO2 [25, 26]. Alternatively, passive accumulation in the nucleus may occur during mitosis if AGO2 binds to chromatin or chromatin-bound RNA prior to the nuclear membrane reforming.
Table 1.
Multiple genes have been targeted by saRNAs.
| Target Gene | Protein | Reported |
|---|---|---|
| CDKN1A | p21WAF1/CIP1 | 2006 [10] |
| VEGFA | Vascular endothelial growth factor A (VEGFA) | 2006 [10] |
| CDH1 | E-cadherin | 2006 [10] |
| PGR | Progesterone Receptor (PR) | 2007 [11] |
| NANOG | Nanog | 2012 [38] |
| KLF4 | Kruppel-like factor 4 (KLF4) | 2012 [39] |
| MYC | MYC | 2012 [39] |
| MafA | MafA | 2013 [40] |
| NOS2 | Nitric Oxide Synthase, Inducible (iNOS) | 2013 [41] |
| NKX3–1 | Homeobox protein Nkx-3.1 | 2013 [42] |
| CEBPA | CCAAT/enhancer-binding protein alpha (CEBPA) | 2014 [7] |
| OCT4 | Octamer-binding transcription factor 4 (OCT4) | 2015 [43] |
| GJA1 | Gap junction alpha-1 protein (GJA1) | 2015 [44] |
| DPYSL3 | Dihydropyrimidinase Like 3 | 2016 [45] |
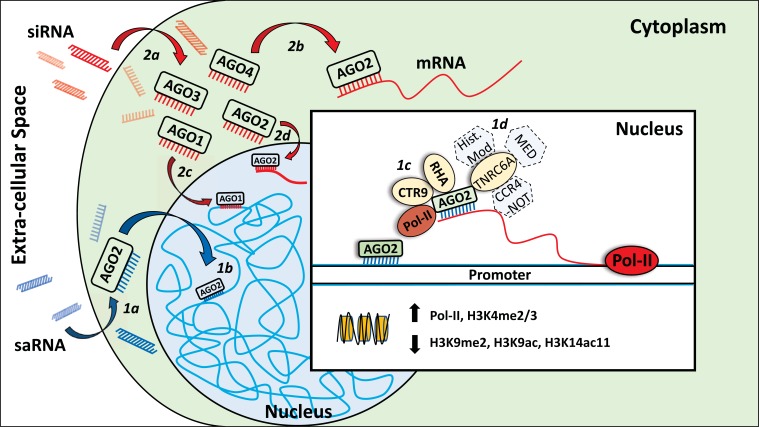
Fig. (1).
Known and proposed mechanisms for saRNA- and siRNA-mediated gene regulation. (1a) Upon entering the cytoplasm, saRNA is loaded into AGO2. (1b) AGO2 is translocated into the nucleus with the help of binding partners (not depicted) and import factors such as Importin-8 (not depicted). In the nucleus, AGO2-bound saRNA binds either directly to DNA or to chromatin-bound RNA such as promoter-associated transcripts or long non-coding RNA. Following saRNA treatment, target promoters are more transcriptionally active and show: decreased H3K9me2, reduced acetylation of H3K9ac and H3K14ac, increased H3K4me2/3 and increased RNA pol-II occupancy. (1c) AGO2 recruits RHA and CTR9, both of which have known roles in transcription. (1d) Emerging evidence also suggests TNCR6A as a critical RNA activation factor. TNCR6 proteins bind strongly to nuclear AGO2. Nuclear TNCR6 proteins have also been shown to bind to proteins involved in histone modification, mediator complex, and the CCR4-NOTcomplex. (2a) In contrast to saRNAs, siRNAs are loaded into the RISC complex (not depicted) via any of the four AGO proteins in the cytoplasm. (2b) The siRNA acts as a guide to target the RISC to complementary RNA. Once bound by RISC the target RNA is degraded. (2c) Alternatively, siRNA-bound AGO1 can be transported into the nucleus and induce transcriptional gene silencing. (2d) siRNA-bound AGO2 can be transported into the nucleus and induce nuclear post-transcriptional gene silencing via a nuclear RISC (not depicted) which might differ in composition to the cytoplasmic RISC.
Once inside the nucleus, AGO2 can bind promoter-associated transcripts [27, 28] that contain a complementary sequence or RNAs that are transcribed through a promoter region with a complementary sequence (Fig. 1) [10, 29]. SaRNAs are also capable of inducing RNAa of genes when designed to target RNA transcribed from outside a gene promoter such as long non-coding RNAs (lncRNA) [7, 30]. Here, the targeted nascent RNA likely acts as a ‘tether’ for the saRNA-AGO2 complex keeping the complex in close proximity to the target gene promoter and allowing physical contact between the two (Fig. 1). Supporting an RNA targeting model, knockdown of these RNAs by antisense oligonucleotides (ASOs) abolishes the effect of saRNA [29]. When RISC is loaded with a guide siRNA that is perfectly complimentary to a target cytoplasmic or nuclear RNA, AGO2 can induce cleavage leading to rapid degradation of the target RNA [20, 31, 32]. AGO2 is unique among the mammalian AGO proteins due to this ‘slicer activity’. However, this activity appears to be dispensable in RNAa [13, 33]. In addition to saRNA targeting RNA, chromatin immunoprecipitation (ChIP) studies suggest that biotinylated saRNA might also bind directly to DNA [13, 34].
A nuclear saRNA-AGO2 complex is believed to recruit proteins that decrease H3K9me2 [10], reduce acetylation of H3K9ac and H3K14ac [11], increase H3K4me2/3, [10, 11] and increase RNA pol-II occupancy [34-36] when localized to a target gene (Fig. 1). This alternative cast of nuclear localized proteins interacting with AGO2 suggests an alternative protein complex which is distinct from cytoplasmic and nuclear RISC. However, the mechanism by which a saRNA-AGO2 complex is potentially involved in these changes remains incomplete. While increased Pol-II occupancy, decreased H3K9me2 (repressive modification) [37], and increased H3K4me2/3 (marker of active transcription) [37] likely contribute to gene activation, decreased H3K9ac and H3K14ac (markers of active transcription) [37] might be envisioned to repress transcription. Nevertheless, the net effect of saRNA is activation of target genes and the loss of both repressive and active histone 3 modifications might indicate a blockage of proteins involved in H3 modification. Timing further complicates mechanistic studies and poses a major challenge should recruitment or denial of any of these modifying factors be a transient process. Observable effects of RNAa on gene expression takes anywhere between 24 to 48 hours, depending on cell line, and can last up to 2 weeks [10, 11, 36]. This is in contrast to cytoplasmic and nuclear RNAi which is observed in under 24 hours [18]. The lag time of RNAa compared to RNAi might hint at a greater time requirement for RNAa-based epigenetic changes to be established at gene promoters.
The question remains as to whether RNAa-mediated protein recruitment proceeds through direct or indirect binding to AGO2. Through mass spectrometry and immunoblotting, a recent study identified the TNCR6A/B/C proteins and AGO3 as direct binding partners of nuclear AGO2 [46]. A follow-up study performed in a similar manner provided further evidence that nuclear TNCR6A is tightly-associated
with AGO2 and is also associated with proteins involved in: histone modification, mediator complex, and CCR4-NOT complex (Fig. 1) [47]. This finding is tantalizing given the role of histone modifications, mediator complex, and CCR4-NOT in general transcription. Further, mass spectrometry and immunoblotting of nuclei-isolated biotinylated guide saRNAs identified AGO2, RNA Helicase A (RHA), and CTR9 as proteins associated with saRNAs (Fig. 1) [34]. AGO2 was shown to directly associate with RHA and CTR9 in the presence of saRNA [34]. This proposed complex was termed the RNA-induced Transcriptional Activation (RITA) complex [34]. Efficient knockdown of RITA complex components RHA or CTR9 by siRNA inhibited RNAa of p21 [34] and CEBPA [13]. Conspicuously, RHA and Pol-II are binding partners [48]. Increased Pol-II occupancy following saRNA treatment might then be partly due to saRNA-AGO2 recruiting RHA-Pol-II to the target promoter.
Additionally, CTR9 is part of the polymerase-associated factor 1 (PAF1) complex which has a general role in transcription [49] including regulation of H3K4 methylation [50]. However, due to CTR9 being part of a complex involved in transcription, the exact role of CTR9 in RNAa has yet to be fully defined. Increased occupancy of gene promoters with transcription-associated machinery might be a result of their recruitment in a saRNA-mediated fashion or a downstream consequence of a promoter that has been made transcriptionally active by RNAa. While these findings are exciting advances in our understanding of RNAa, further studies will be needed to confirm the possible roles for TNCR6A-associated proteins and other factors beyond RHA and CTR9 in the RNAa process.
2. saRNAS AS THERAPEUTICS
The potential clinical application and limitations of saRNAs has been suggested since their initial discovery. Gene-specific activation of a target might prove useful in treating diseases where inhibition of transcription has been identified as an underlying cause for disease or in cases where over-expression might be potentially beneficial. To this end, multiple saRNAs have been screened at the preclinical level and include but are not limited to: VEGFA [51], p21 [52], and more recently CEBPα [7, 13, 53]. However, limitations to saRNA therapeutics should be considered before attempting clinical development. The most important limitation being saRNA intervention is less useful or even futile for diseases in which the gene product to be upregulated has become defective due to mutation. Additionally, given their structural similarity and a requirement for AGO proteins, saRNAs and siRNAs share many of the same limitations for use in a therapeutic context. saRNA therapeutics have thus far required a higher concentration of molecules (nM) compared to siRNA (pM - nM) to produce therapeutic effects. Dosing requirements have recently been commented on [54]. Targeting of oligonucleotides, in general, to specific tissues also represents a major challenge and is complicated by the dosing requirement [55]. For these reasons, targeting liver and using lipid nanostructure-oligonucleotide formulations has been considered the ‘low-hanging fruit’ in the field due to a higher accumulation of lipid nanostructures in the liver than in most other tissues. Accumulation of lipid nanostructure-oligonucleotide in the liver ultimately leads to higher concentrations of delivered therapeutic. However, a higher concentration of oligonucleotide drug in a cell also increases the potential for off-target effects and should be taken into account during screening. While these limitations are not insurmountable, significant effort is required to bring saRNAs and other oligonucleotide-based therapeutics to the clinic.
3. CCAAT/Enhancer- Binding Protein Alpha (CEBPA)
CCAAT enhancer-binding protein family members (CEBPs) are a family of six transcription factors (α - ζ) that regulate genes involved in: cellular differentiation, proliferation, metabolism, and immunity [56-59]. CEBPs are a sub-group of the basic region leucine zipper transcription factors (bZIPs) family. CEBPs are characterized as having a N-terminal transactivation domain(s) (TAD) [60-62], a DNA binding domain which recognizes the sequence (G/A)TTGCG(T/C)AA(T/C) or, broadly, the promoter CCAAT box sequence [63], and a C-terminal bZIP domain containing a conserved leucine-rich dimerization domain [64-66]. The formation of homo- or hetero-dimers between CEBP family members and bZIP extended family members is a requirement for DNA binding [67].
CEBPα is an intronless gene located on chromosome 19q13.1. It encodes the protein CEBPα and is transcribed from the reverse strand [68]. The resulting transcript contains a primary and alternative translation initiation codon and is translated into one of two isoforms, 42 kDa (p42) or 30 kDa (p30) [69-71]. In some instances, the two isoforms exhibit similar biological effects. For example, both can act as transactivators of the CEBPα and adipocyte protein 2 (aP2) promoters [70]. Additionally, both isoforms have inhibitory potential through a negative regulatory domain found N-terminal to TAD3 [69, 70, 72]. However, functional differences between the two isoforms do exist. CEBPα p42 is generally more abundant [71], is more often associated with transactivation of target genes owing to its three TADs (TAD1-3) [69-71], and possesses anti-mitotic activity due in part to N-terminal-binding of retinoblastoma (Rb) and p21 (Cip1) [73-75]. In contrast, CEBPα p30 does not possess anti-mitotic activity and lacks TAD1-2 [69, 70]. Consequently, CEBPα p30 cannot bind with TATA box-binding protein (TBP), [62] TFIIB, [62] or the histone acetyltransferase CREB-binding Protein (CBP) [76] resulting in severely reduced transactivation potential compared to CEBPα p42 [69, 70]. In certain contexts, the isoforms have opposite effects; in the case of G-CSF receptor expression CEBPα p30 is inhibitory [77] while CEBPα p42 promotes expression [78]. Consequently, if mutation of one allele results in the production of a defective CEBPα p42 protein, CEBPα p30 will act in a dominant-negative fashion by competing for CEBPα p42 binding partners [79, 80].
CEBPα is expressed across a wide range of human solid tissues and blood cells albeit at differing levels. The solid tissues with highest expression based on mRNA-seq are liver, skin, adipose, and breast followed by small intestine and lung [81]. Multiple regulatory mechanisms have evolved to maintain tight control over CEBPα expression in tissues at the transcriptional, post-transcriptional, and post-translational levels. At the transcriptional level, CEBPα is regulated by two promoters, a core promoter (-437 to +4bp from the transcriptional start site (TSS)) and distal promoter (-1400 to -600bp from the TSS), both of which lie within a CpG island [82, 83]. Regardless of high or low expression, the core CEBPα promoter is generally free of DNA methylation [84], which potentially provides a platform for temporary transcription factor binding depending on stimuli. In contrast, distal promoter methylation is inversely-correlated with CEBPα expression such that expression is repressed in cells that do not continuously express CEBPα [84]. Enhancers have profound impact on development and cell-type specific regulation of proteins [85]. Enhancer-promoter associations have been described in both general and cell-type specific CEBPα regulation [86, 87]. LncRNAs transcribed from the CEBPα locus have also been shown to have regulatory effects on CEBPα [88]. The antisense CEBPα transcript ‘adipogenic differentiation-induced noncoding RNA’ (ADINR) was recently shown to stimulate CEBPα expression in cis during adipogenesis in human mesenchymal stem cells by directly binding PA1 which in turn recruits MLL3/4 histone methyl-transferase complexes [89]. Another lncRNA, extra-coding CEBPα (ecCEBPα), is transcribed upstream and through the gene body of CEBPα and has been proposed to block DNA methyltransferase 1 (DNMT1) from performing maintenance methylation on the CEBPα promoter [90].
CEBPα regulates genes involved in cellular differentiation, proliferation, metabolism, and immunity. Regulation can be general or cell type specific. (+) CEBPα activates target gene expression. (-) CEBPα represses target gene expression. (*) Differential expressions upon CEBPA saRNA treatment based on a cancer-specific 84 gene microarray assay or direct RT-qPCR analysis.
Post-transcriptionally and post-translationally, the turnover of CEBPα mRNA and protein in cultured cells is rapid [91, 92]. For mRNA, a 70-80% reduction was observed after 2 hours of actinomycin D treatment in hepatocytes [91]. CEBPα protein half-life was found to be 100 minutes, as measured by pulse chase in HL-60 cells [92]. miRNA mediated degradation could partially be responsible, miR-182 targets the 3’ UTR of CEBPα mRNA resulting in lower CEBPα expression [93]. Additional modes of CEBPα regulation involve RNA-binding proteins (e.g. calreticulin (CRT)) which bind stem-loop structures within the CEBPα transcript inhibiting translation [94] and post-translational modifications which effect CEBPα stability (e.g. phosphorylation by c-Jun N-terminal kinase (JNK1)) [92, 95, 96]. Unsurprisingly, misregulation of CEBPα by defects in these regulatory mechanisms correlates with disease states in afflicted tissues [80, 81, 83, 97-99].
4. CEBPA IN LIVER FUNCTION
The high abundance of CEBPα in the liver suggests a critical role in liver function. Indeed, CEBPα -/- murine neonates fail to store hepatic glycogen and die rapidly (within 8 hours) after birth due to hypoglycemia [100]. Livers from these CEBPα-/- neonates displayed reduced albumin, glycogen synthase, PEPCK, GLUT2, and G6Pase mRNA levels compared to WT or heterozygous mice. In addition, CEBPα -/- mice develop hyperammonemia resulting from impaired expression of ornithine cycle enzymes [101]. These findings along with the observations of anti-mitotic effects of CEBPα [73-75], confirm a major role for CEBPα in liver physiology.
5. CEBPA IN LIVER Disease AND LIVER CANCER
Common liver diseases include: non-alcoholic steatohepatitis (NASH) or Fatty Liver Disease, alcohol-associated steatohepatitis (ASH), and hepatitis B/C; all of which increase the risk of impaired liver function [102-105]. If left untreated, these diseases can also contribute to the development of liver cirrhosis and liver cancer [102-107]. Liver cancer is the second-most common cause of cancer-associated death worldwide in men and the sixth in women where HCC accounts for 70-80% of all cases [108, 109]. Surgical resection of tumors is the preferred method of treatment. While resection has the potential to cure patients and improve long-term survival compared to other methods, this option is only possible in about 5-15% of cases [110]. Poor liver function or existence of tumors that have invaded into the surrounding vasculature can prohibit surgery which would further compromise the liver [111]. Consequently, barring a liver transplant, prognosis for patients is generally poor with an overall 5-year survival rate ranging from 31% in early disease to 3% in metastatic disease [112]. Currently, sorafenib [113], a multikinase inhibitor of Raf-1, B-Raf, VEGFR, and PDGFR, represents the standard-of-care treatment for advanced HCC. However, treatment only prolongs the median life expectancy by approximately 3 months [114, 115]. Therefore, there is still significant unmet medical need in the treatment of HCC.
In the field of oncology, CEBPα is known as a tumour-suppressor gene. Established hepatocyte cultures derived from CEBPα -/- mice exhibit rapid growth, accumulation of chromosomal abnormalities, and form nodules when placed into abdominal subcutaneous tissue of nude mice [121]. Mutations in either the TADs or bZIP domains (common in leukemias) which diminish or abolish CEBPα function, along with general repression of CEBPα expression, can promote tumorigenesis and tumor progression [80, 81, 97-99]. Increases in oncogenic miRNAs such as miR-182, which silences CEBPα mRNA, [93] and down-regulation of CEBPα due to distal promoter hyper-methylation are common in many cancers [122-124] including hepatocellular carcinoma [83]. In general, solid tumors exhibit deficient CEBPα expression rather than loss-of-function mutations [81]. Thus, targeted restoration of CEBPα in HCC holds clinical promise.
Beyond HCC, functional inhibition of CEBPα in hepatocytes might be partially responsible for the lipid accumulation observed in NASH. PPARγ and CEBPα promote lipolysis and decrease triglyceride content in fully-differentiated adipocytes [118] and might function similarly in hepatocytes. Finally, given the role for CEBPα in gluconeogensis [100] in the liver and in adipogenesis, [125] it will be of interest to determine what effects, if any, CEBPα has on resting glucose levels and insulin resistance with regard to type II diabetes. Taken together, restoration of CEBPα function in liver disease has powerful therapeutic potential.
6. DEVELOPMENT OF CEBPα saRNA FOR Liver disease
6.1. saRNA design
CEBPα saRNAs were designed against the CEBPα promoter or coding regions as described by Voutila et al. [39] and Reebye et al. [40]. Four key parameters were considered: gene annotations of target from UCSC RefSeq database, annotated sequences of antisense RNA, promoter or coding region selections overlapping with antisense sequences, and identification of candidate short activating RNAs. CEBPα saRNA selection also considers factors such as the removal of polymeric motifs, Guanine-Cytosine (GC) content, off-target effects, target composition, and predicted saRNA activity.
6.2. Proof of Concept Studies
CEBPα saRNA transfection into HepG2 increased CEBPα mRNA in a dose-dependent manner [7]. saRNA transfection also induced albumin mRNA expression and protein secretion, consistent with the role of CEBPα in liver function. A reduction in methylation at CpG islands in both CEBPα and albumin promoter regions was observed, where CEBPα binding motifs were also present. The expression of other downstream markers (e.g. ornithine transcarbamylase (OTC) and Alpha-fetoprotein (AFP)) were also altered as expected due to their role in hepatocyte maintenance and liver differentiation. Microarray analysis of 84 liver specific genes identified additional factors altered upon CEBPα saRNA treatment. Down-regulated genes were strongly enriched in functions related to inhibition of apoptosis or driving of cell cycle, whereas up-regulated genes were enriched in functions related to regulation of cell differentiation. For oncogenes c-Myc and STAT3, an increase in the methylation state of their promoters was suggested to be responsible for their reduced expression; as would be predicted, CEBPα binding motifs were located in their promoter regions. Phenotypically, CEBPα saRNA reduced proliferation of HepG2 cells by 50% in a dose-dependent manner. This study demonstrated that a saRNA targeting the CEBPα locus can increase CEBPα levels and consequently improve surrogate markers of liver function in a HCC cell system.
Intravenous delivery of the CEBPα saRNA was first tested using dendrimer delivery [7]. saRNA was complexed with polyamidoamine (PAMAM) dendrimer, a nanoparticle which preferentially accumulates in PBMCs and the liver [126]. The complex was injected three times over one week into the tail vein of diethylnitrosamine (DEN)-induced HCC model rats [7]. Up regulation of CEBPα and albumin mRNA, as well as altered expression of critical hepatocyte markers including HNF1α and HNF4α in liver tissue was shown when compared to the scrambled saRNA control group. Furthermore, increases in serum albumin and decreases in serum bilirubin, aspartate aminotransferase (AST), and alanine aminotransferase (ALT) were also observed. Histological examination and immunohistology studies of the liver showed a CEBPα saRNA-induced reduction in tumour burden by 80% and preneoplastic marker GST-P levels by 40%. This exciting study confirmed that CEBPα saRNA is functional in vivo and provided the basis to further develop CEBPα saRNA for clinical use.
6.3. SaRNA Optimisation
Prior to pre-clinical studies, the CEBPα saRNA compound was optimised with regards to its composition and target CEBPα sequence. Bioinformatic analysis of the CEBPα locus identified two hot spots for saRNA activity [13]. Both hot spots were located in the coding region of CEBPα, within GENEBANK ID: AW665812, where at least a portion of an antisense ncRNA is also transcribed. Nucleotide walk across these two hotspots produced several candidates whose activities were then assessed in HepG2 cells. The candidate with the strongest potency was selected (CEBPα-51; 2.5-fold) and a dose response observed with an EC50 of 5.36 nM. This saRNA target site was located within the same hot spot of the original saRNA previously described (CEBPA-AVI [7]). Corresponding increases in CEBPα protein were observed as well as an increase in the downstream CEBPα marker albumin [13]. As shown for CEBPA-AVI, phenotypically, CEBPA-51 reduced proliferation of HepG2 and Hep3B cells. Potential for off-target effects were assessed in a variety of species. The target site for CEBPA-51 was highly-conserved between human, non-human primate, and rodent. Furthermore, CEBPα saRNA activity was also conserved across these species. Although one or two mismatched targets were identified, no significant effects on six relevant liver or cancer targets were observed upon CEBPα saRNA treatment [13].
To further develop CEBPA-51 as a clinical candidate, the effect of chemical modifications on activity and immune stimulation in primary human PMBCs was assessed. 2`-O-methyl modifications on the sense strand were well tolerated: upon treatment, one compound limited TNF-α and IFN-α secretion was observed. This suggested that particular 2`-O-methyl modification patterns within saRNAs display limited immune activation. To improve guide-strand loading bias, addition of a 5`inverted abasic modification [127] on the sense strand further enhanced activity through directing AGO2 loading towards the functional antisense strand [13]. Based on these promising observations, the chemically-improved CEBPA-51 saRNA was taken forward into pre-clinical trials for diseases of the liver.
6.4. Mechanistic Investigations
Investigations into the mechanism of action of CEBPα saRNAs, predominantly in HepG2 and Hep3B cell lines, largely agree with the saRNA mechanistic studies discussed previously [13]. CEBPα saRNA was capable of driving CEBPα expression at both the mRNA (RT-qPCR) and protein levels (western blot and luciferase reporter assay). A nuclear run-on assay showed increased levels of nascent CEBPα mRNA transcription following saRNA treatment. The RNAa active guide strand was determined by biasing strand-loading through a 5’ inverted abasic modification in either the sense (SS) or antisense (AS) strands of the saRNA. This modification lowers the ability of the modified strand to be loaded into AGO proteins. Here, the SS strand is complementary to CEBPα mRNA while the AS is complementary to the antisense ncRNA AW665812. Biasing the loading towards AS improved the potency of CEBPA saRNA, while biasing towards SS resulted in no activation. Further, mutation of the seed sequence of the AS diminished RNAa activity. Thus, the active guide strand was shown to be the antisense strand. Although still not understood, the level of the antisense RNA was also upregulated. The increase of antisense RNA along with CEBPα mRNA might indicate that the CEBPα locus becomes more transcriptionally permissive as a whole. It is noteworthy that AGO2-mediated cleavage of the antisense RNA was not observed which suggests that CEBPα transcriptional activation is not dependent on the depletion of the antisense transcript. Like previous studies, AGO2 was shown to be critical for saRNA activity. AGO2 knockout mouse embryonic fibroblasts (MEFs) showed no RNAa activity when transfected with CEBPα saRNA compared to a 2.3-fold increase in CEBPα mRNA in WT MEFs. This result was further validated by a co-IP experiment using biotinylated saRNA. Biotinylated saRNA only showed an association with AGO2 when western blots were performed to determine potential associations with AGO1-4. While CEBPA-51 likely targets the antisense transcript, AW665812, it is also possible that it binds to chromatin as well. ChIP experiments found enrichment of biotinylated saRNA within the general CEBPα locus [13, 53]. In agreement with its role in RNAa [34], CTR9 was found to be required for CEBPα saRNA activity. Cumulatively, these experiments show that CEBPA-51 acts as a bonafide saRNA.
6.5. Pre-clinical and Clinical
For efficient delivery, CEBPA-51 is encapsulated in Marina Biotech’s liposomal carrier molecules (SMARTICLES) and the resulting formulation is referred to as MTL-CEBPA. SMARTICLES are amphoteric liposomes with anionic and cationic groups. They provide a pH-triggered endosomal escape for intracellular delivery of the double-stranded saRNA payload. SMARTICLES also enhance the serum stability of saRNAs.
Delivery of MTL-CEBPA into a diethylnitrosamine (DEN)-induced cirrhotic HCC rat model [53] resulted in increased CEBPα mRNA expression, an 80% reduction in tumour size, and a decrease in relevant liver parameters (i.e. AST, ALT) one week after treatment, compared to the negative control oligonucleotide. Additional studies of MTL-CEBPA treatment in CCl4-induced liver failure rat models showed significant improvements in clinically-relevant parameters including an increase in serum albumin and other markers for liver function (e.g. AST and ALT). Overall survival was also significantly improved following MTL-CEBPA treatment [53]. In vivo studies involving methionine choline-deficient diet-induced non-alcoholic steatohepatitis mice and orthotopic HCC xenograft nude mice [53, 128] demonstrated that MTL-CEBPA treatment improves a variety of different liver parameters in a diverse range of models. Furthermore, liver, intrahepatic, and distant lung tumour formation were lower in MTL-CEBPA-treated mice [128]. Combined, these studies strongly supported clinical exploration of CEBPA saRNA in treatment of HCC, liver fibrosis, and other liver diseases. MTL-CEBPA is presently being assessed in a Phase I clinical trial for patients with advanced liver cancer. This clinical study represents the first-in-human trial of a small activating RNA therapeutic.
CONCLUSION
The mono-therapeutic one drug-one target nature of oligonucleotide drugs has created an obstacle for their use in multigenic diseases, such as cancer, where the pathogenic state is made up of multiple dysregulated and, often, redundant pathways. Except in rare circumstances, (e.g. BCR-ABL positive CML) inhibition of a single gene product typically has negligible effects in cancer treatment. Targeting transcription factors, which have multiple connections linking redundant pathways acting in union to drive disease state, allows such drugs to function as widespread pathway regulators – despite targeting a single gene. saRNAs are part of a restricted repertoire of drugs which can selectively enhance transcription factor expression, modulating their extensive yet directed network. CEBPα is an attractive transcription factor target for saRNA development in advanced HCC, which at present has limited treatment options due to poor liver function or large unresectable tumours which makes patients ineligible for surgical intervention. CEBPα has proven anticancer properties, strong associations with disease severity in patients, and is critical for hepatocyte function. The findings that a saRNA targeting CEBPα reduced tumour burden and improved liver function in animal models provided the basis for its advancement into the clinic. We hope that by targeting tumours and improving general liver function, saRNA mediated up-regulation of CEBPα will improve the outcomes for patients who are ineligible for surgical intervention, as well as positively impact liver function in a range of additional liver-related indications.
Consent for Publication
Not applicable.
Table 2.
A non-exhaustive list of genes regulated by CEBPα.
| Regulation | Gene | Function |
|---|---|---|
| + | ALB [100] * | Plasma colloid osmotic pressure, carrier protein |
| - | c-MYC [116] * | Cell cycle, apoptosis, transformation |
| + | CEBPA [117]* | Cell cycle, adipogenesis, liver function, cell type specific differentiation, apoptosis |
| + | ATGL [118] | Lipolysis |
| + | CSF3R [119] | Receptor for granulocyte colony stimulating factor |
| + | CDKN1A [74] * | Cell cycle |
| + | GYS1 [100] | Glycogen synthesis |
| - | INFG [120] | Cytokine / immune response |
Acknowledgements
We would like to thank Elizabeth W. Epps for proofreading this submission.
Conflict of Interest
John J. Rossi and Nagy A. Habib are shareholders of MiNA (Holdings) Limited.
REFERENCES
- 1.Stein C.A., Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017;25(5):1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44(14):6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 5.Lightfoot H.L., Hall J. Target mRNA inhibition by oligonucleotide drugs in man. Nucleic Acids Res. 2012;40(21):10585–10595. doi: 10.1093/nar/gks861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539(7630):479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 7.Reebye V., Saetrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D., Kostomitsopoulos N., Kasahara N., Nicholls J.P., Jiao L.R., Pai M., Spalding D.R., Mizandari M., Chikovani T., Emara M.M., Haoudi A., Tomalia D.A., Rossi J.J., Habib N.A. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59(1):216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itaka K. Development of mRNA-based therapeutics. Nippon Yakurigaku Zasshi. 2016;148(4):190–196. doi: 10.1254/fpj.148.190. [DOI] [PubMed] [Google Scholar]
- 9.Sergeeva O.V., Koteliansky V.E., Zatsepin T.S. mRNA-Based therapeutics - advances and perspectives. Biochemistry (Mosc.) 2016;81(7):709–722. doi: 10.1134/S0006297916070075. [DOI] [PubMed] [Google Scholar]
- 10.Li L.C., Okino S.T., Zhao H., Pookot D., Place R.F., Urakami S., Enokida H., Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA. 2006;103(46):17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3(3):166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 12.Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voutila J., Reebye V., Roberts T.C., Protopapa P., Andrikakou P., Blakey D.C., Habib R., Huber H., Saetrom P., Rossi J.J., Habib N.A. Development and mechanism of small activating RNA targeting CEBPA, a novel therapeutic in clinical trials for liver cancer. Mol. Ther. 2017;25(12):2705–2714. doi: 10.1016/j.ymthe.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarker D., Plummer E.R., Basu B., Meyer T., Huang K.-W., Evans T.R.J., Spalding D., Ma Y.T., Palmer D.H., Chee C.E., Toh H.C., Habib N.A. 2017.
- 15.Nakanishi K. Anatomy of RISC: How do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip. Rev. RNA. 2016;7(5):637–660. doi: 10.1002/wrna.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannon G.J. RNA interference. Nature. 2002;418(6894):244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 17.Robb G.B., Brown K.M., Khurana J., Rana T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12(2):133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 18.Lennox K.A., Behlke M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44(2):863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Younger S.T., Corey D.R. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39(13):5682–5691. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R. RNAi factors are present and active in human cell nuclei. Cell Reports. 2014;6(1):211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao M., Wei W., Li M.M., Wu Y.S., Ba Z., Jin K.X., Li M.M., Liao Y.Q., Adhikari S., Chong Z., Zhang T., Guo C.X., Tang T.S., Zhu B.T., Xu X.Z., Mailand N., Yang Y.G., Qi Y., Rendtlew Danielsen J.M. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014;24(5):532–541. doi: 10.1038/cr.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.C., Mathieu J., Hamiche A., Ait-Si-Ali S., Muchardt C., Batsche E., Harel-Bellan A. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012;19(10):998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 23.Weinmann L., Hock J., Ivacevic T., Ohrt T., Mutze J., Schwille P., Kremmer E., Benes V., Urlaub H., Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136(3):496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 24.Wei Y., Li L., Wang D., Zhang C.Y., Zen K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biologic. Chem. 2014;289(15):10270–10275. doi: 10.1074/jbc.C113.541417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishi K., Nishi A., Nagasawa T., Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013;19(1):17–35. doi: 10.1261/rna.034769.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schraivogel D., Schindler S.G., Danner J., Kremmer E., Pfaff J., Hannus S., Depping R., Meister G. Importin-beta facilitates nuclear import of human GW proteins and balances cytoplasmic gene silencing protein levels. Nucleic Acids Res. 2015;43(15):7447–7461. doi: 10.1093/nar/gkv705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A., Sharp P.A. Divergent transcription from active promoters. Science. 2008;322(5909):1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz J.C., Younger S.T., Nguyen N.B., Hardy D.B., Monia B.P., Corey D.R., Janowski B.A. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15(8):842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yue X., Schwartz J.C., Chu Y., Younger S.T., Gagnon K.T., Elbashir S., Janowski B.A., Corey D.R. Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat. Chem. Biol. 2010;6(8):621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 32.Haaland R.E., Herrmann C.H., Rice A.P. siRNA depletion of 7SK snRNA induces apoptosis but does not affect expression of the HIV-1 LTR or P-TEFb-dependent cellular genes. J. Cell. Physiol. 2005;205(3):463–470. doi: 10.1002/jcp.20528. [DOI] [PubMed] [Google Scholar]
- 33.Matsui M., Chu Y., Zhang H., Gagnon K.T., Shaikh S., Kuchimanchi S., Manoharan M., Corey D.R., Janowski B.A. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41(22):10086–10109. doi: 10.1093/nar/gkt777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portnoy V., Lin S.H., Li K.H., Burlingame A., Hu Z.H., Li H., Li L.C. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016;26(3):320–335. doi: 10.1038/cr.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu Y., Yue X., Younger S.T., Janowski B.A., Corey D.R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38(21):7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J., Chen Z., Xia D., Wu J., Xu H., Ye Z.Q. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochem. J. 2012;447(3):407–416. doi: 10.1042/BJ20120256. [DOI] [PubMed] [Google Scholar]
- 37.Lawrence M., Daujat S., Schneider R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet.: TIG. 2016;32(1):42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Wang X., Wang J., Huang V., Place R.F., Li L.C. Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem. J. 2012;443(3):821–828. doi: 10.1042/BJ20111491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voutila J., Saetrom P., Mintz P., Sun G., Alluin J., Rossi J.J., Habib N.A., Kasahara N. Gene expression profile changes after short-activating RNA-mediated induction of endogenous pluripotency factors in human mesenchymal stem cells. Mol. Ther. Nucleic Acids. 2012;1:e35. doi: 10.1038/mtna.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reebye V., Saetrom P., Mintz P.J., Rossi J.J., Kasahara N., Nteliopoulos G., Nicholls J., Haoudi A., Gordon M., Habib N.A. A Short-activating RNA oligonucleotide targeting the islet beta-cell transcriptional factor MafA in CD34(+) Cells. Mol. Ther. Nucleic Acids. 2013;2:e97. doi: 10.1038/mtna.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T., Li M., Yuan H., Zhan Y., Xu H., Wang S., Yang W., Liu J., Ye Z., Li L.C. saRNA guided iNOS up-regulation improves erectile function of diabetic rats. J. Urol. 2013;190(2):790–798. doi: 10.1016/j.juro.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 42.Ren S., Kang M.R., Wang J., Huang V., Place R.F., Sun Y., Li L.C. Targeted induction of endogenous NKX3-1 by small activating RNA inhibits prostate tumor growth. Prostate. 2013;73(14):1591–1601. doi: 10.1002/pros.22709. [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Huang V., Ye L., Barcena A., Lin G., Lue T.F., Li L.C. Identification of small activating RNAs that enhance endogenous OCT4 expression in human mesenchymal stem cells. Stem Cells Dev. 2015;24(3):345–353. doi: 10.1089/scd.2014.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esseltine J.L., Shao Q., Huang T., Kelly J.J., Sampson J., Laird D.W. Manipulating Cx43 expression triggers gene reprogramming events in dermal fibroblasts from oculodentodigital dysplasia patients. Biochem. J. 2015;472(1):55–69. doi: 10.1042/BJ20150652. [DOI] [PubMed] [Google Scholar]
- 45.Li C., Jiang W., Hu Q., Li L.C., Dong L., Chen R., Zhang Y., Tang Y., Thrasher J.B., Liu C.B., Li B. Enhancing DPYSL3 gene expression via a promoter-targeted small activating RNA approach suppresses cancer cell motility and metastasis. Oncotarget. 2016;7(16):22893–22910. doi: 10.18632/oncotarget.8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalantari R., Hicks J.A., Li L., Gagnon K.T., Sridhara V., Lemoff A., Mirzaei H., Corey D.R. Stable association of RNAi machinery is conserved between the cytoplasm and nucleus of human cells. RNA. 2016;22(7):1085–1098. doi: 10.1261/rna.056499.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hicks J.A., Li L., Matsui M., Chu Y., Volkov O., Johnson K.C., Corey D.R. Human GW182 paralogs are the central organizers for RNA-mediated control of transcription. Cell Reports. 2017;20(7):1543–1552. doi: 10.1016/j.celrep.2017.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakajima T., Uchida C., Anderson S.F., Lee C.G., Hurwitz J., Parvin J.D., Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90(6):1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 49.Jaehning J.A. The Paf1 complex: Platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta. 2010;1799(5-6):379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogan N.J., Dover J., Wood A., Schneider J., Heidt J., Boateng M.A., Dean K., Ryan O.W., Golshani A., Johnston M., Greenblatt J.F., Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 51.Turunen M.P., Lehtola T., Heinonen S.E., Assefa G.S., Korpisalo P., Girnary R., Glass C.K., Vaisanen S., Yla-Herttuala S. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circulat. Res. 2009;105(6):604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- 52.Place R.F., Wang J., Noonan E.J., Meyers R., Manoharan M., Charisse K., Duncan R., Huang V., Wang X., Li L.C. Formulation of small activating RNA into lipidoid nanoparticles inhibits xenograft prostate tumor growth by inducing p21 expression. Mol. Ther. Nucleic Acids. 2012;1:e15. doi: 10.1038/mtna.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reebye V.H.K., Lin V., Jarvis S., Cutilas P., Dorman S., Ciriello S., Andrikakou P., Voutila J., Saetrom P., Mintz P.J., Reccia I., Rossi J.J., Huber H., Habib R., Kostomitsopoulos N., Blakey D., Habib N. Gene activation of CEBPA using saRNA: Preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene. doi: 10.1038/s41388-018-0126-2. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L-C. Advances in Experimental Medicine and Biology, RNA activation. 2017. [Google Scholar]
- 55.Tatiparti K., Sau S., Kashaw S.K., Iyer A.K. siRNA Delivery Strategies: A comprehensive review of recent developments. Nanomaterials (Basel) 2017;7(4):E77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lekstrom-Himes J., Xanthopoulos K.G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biologic. Chem. 1998;273(44):28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 57.Ramji D.P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365(Pt 3):561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukada J., Yoshida Y., Kominato Y., Auron P.E. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54(1):6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 59.Nerlov C. The C/EBP family of transcription factors: A paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17(7):318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Friedman A.D., McKnight S.L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4(8):1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 61.Nerlov C., Ziff E.B. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes Dev. 1994;8(3):350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 62.Nerlov C., Ziff E.B. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14(17):4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osada S., Yamamoto H., Nishihara T., Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J. Biologic. Chem. 1996;271(7):3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 64.Landschulz W.H., Johnson P.F., McKnight S.L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 65.Agre P., Johnson P.F., McKnight S.L. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science. 1989;246(4932):922–926. doi: 10.1126/science.2530632. [DOI] [PubMed] [Google Scholar]
- 66.Vinson C.R., Hai T., Boyd S.M. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: Prediction and rational design. Genes Dev. 1993;7(6):1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 67.Newman J.R., Keating A.E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300(5628):2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 68.Hendricks-Taylor L.R., Bachinski L.L., Siciliano M.J., Fertitta A., Trask B., de Jong P.J., Ledbetter D.H., Darlington G.J. The CCAAT/enhancer binding protein (C/EBP alpha) gene (CEBPA) maps to human chromosome 19q13.1 and the related nuclear factor NF-IL6 (C/EBP beta) gene (CEBPB) maps to human chromosome 20q13.1. Genomics. 1992;14(1):12–17. doi: 10.1016/s0888-7543(05)80276-9. [DOI] [PubMed] [Google Scholar]
- 69.Ossipow V., Descombes P., Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl. Acad. Sci. USA. 1993;90(17):8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin F.T., MacDougald O.A., Diehl A.M., Lane M.D.A. 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: Transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA. 1993;90(20):9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calkhoven C.F., Muller C., Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14(15):1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 72.Chattopadhyay S., Gong E.Y., Hwang M., Park E., Lee H.J., Hong C.Y., Choi H.S., Cheong J.H., Kwon H.B., Lee K. The CCAAT enhancer-binding protein-alpha negatively regulates the transactivation of androgen receptor in prostate cancer cells. Mol. Endocrinol. 2006;20(5):984–995. doi: 10.1210/me.2005-0240. [DOI] [PubMed] [Google Scholar]
- 73.Timchenko N.A., Wilde M., Darlington G.J. C/EBPalpha regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol. Cell. Biol. 1999;19(4):2936–245. doi: 10.1128/mcb.19.4.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Timchenko N.A., Wilde M., Nakanishi M., Smith J.R., Darlington G.J. CCAAT/enhancer-binding protein alpha(C/EBP alpha) inhibits cell proliferation through the p21(WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10(7):804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 75.Timchenko N., Wilde M. Iakova, P.; Albrecht, J.H.; Darlington, GJ. E2F/p107 and E2F/p130 complexes are regulated by C/EBPalpha in 3T3-L1 adipocytes. Nucleic Acids Res. 1999;27(17):3621–3630. doi: 10.1093/nar/27.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kovacs K.A., Steinmann M., Magistretti P.J., Halfon O., Cardinaux J.R. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J. Biologic. Chem. 2003;278(38):36959–36965. doi: 10.1074/jbc.M303147200. [DOI] [PubMed] [Google Scholar]
- 77.Cleaves R., Wang Q.F., Friedman A.D. C/EBPalphap30, a myeloid leukemia oncoprotein, limits G-CSF receptor expression but not terminal granulopoiesis via site-selective inhibition of C/EBP DNA binding. Oncogene. 2004;23(3):716–725. doi: 10.1038/sj.onc.1207172. [DOI] [PubMed] [Google Scholar]
- 78.Wang W., Wang X., Ward A.C., Touw I.P., Friedman A.D. C/EBPalpha and G-CSF receptor signals cooperate to induce the myeloperoxidase and neutrophil elastase genes. Leukemia. 2001;15(5):779–786. doi: 10.1038/sj.leu.2402094. [DOI] [PubMed] [Google Scholar]
- 79.Pabst T., Mueller B.U., Zhang P., Radomska H.S., Narravula S., Schnittger S., Behre G., Hiddemann W., Tenen D.G. Dominant-negative mutations of CEBPA, encoding CCAAT/ enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat. Genet. 2001;27(3):263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 80.Gombart A.F., Hofmann W.K., Kawano S., Takeuchi S., Krug U., Kwok S.H., Larsen R.J., Asou H., Miller C.W., Hoelzer D., Koeffler H.P. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99(4):1332–1340. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- 81.Lourenco A.R., Coffer P.J. A tumor suppressor role for C/EBPalpha in solid tumors: More than fat and blood. Oncogene. 2017;36(37):5221–5230. doi: 10.1038/onc.2017.151. [DOI] [PubMed] [Google Scholar]
- 82.Tada Y., Brena R.M., Hackanson B., Morrison C., Otterson G.A., Plass C. Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein alpha activity in lung cancer. J. Natl. Cancer Inst. 2006;98(6):396–406. doi: 10.1093/jnci/djj093. [DOI] [PubMed] [Google Scholar]
- 83.Lu G.D., Leung C.H., Yan B., Tan C.M., Low S.Y., Aung M.O., Salto-Tellez M., Lim S.G., Hooi S.C. 2010. [DOI] [PubMed]
- 84.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., Gnirke A., Jaenisch R., Lander E.S. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ong C.T., Corces V.G. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Avellino R., Havermans M., Erpelinck C., Sanders M.A., Hoogenboezem R., van de Werken H.J., Rombouts E., van Lom K., van Strien P.M., Gebhard C., Rehli M., Pimanda J., Beck D., Erkeland S., Kuiken T., de Looper H., Groschel S., Touw I., Bindels E., Delwel R. An autonomous CEBPA enhancer specific for myeloid-lineage priming and neutrophilic differentiation. Blood. 2016;127(24):2991–3003. doi: 10.1182/blood-2016-01-695759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo H., Ma O., Friedman A.D. The Cebpa +37-kb enhancer directs transgene expression to myeloid progenitors and to long-term hematopoietic stem cells. J. Leukoc. Biol. 2014;96(3):419–426. doi: 10.1189/jlb.2AB0314-145R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao T., Liu L., Li H., Sun Y., Luo H., Li T., Wang S., Dalton S., Zhao R.C., Chen R. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha. Stem Cell Reports. 2015;5(5):856–865. doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Di Ruscio A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P., Wu M., D’Alo F., Melnick A., Leone G., Ebralidze K.K., Pradhan S., Rinn J.L., Tenen D.G. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mischoulon D., Rana B., Bucher N.L., Farmer S.R. Growth-dependent inhibition of CCAAT enhancer-binding protein (C/EBP alpha) gene expression during hepatocyte proliferation in the regenerating liver and in culture. Molecul. Cellul. Biol. 1992;12(6):2553–2560. doi: 10.1128/mcb.12.6.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trivedi A.K., Bararia D., Christopeit M., Peerzada A.A., Singh S.M., Kieser A., Hiddemann W., Behre H.M., Behre G. Proteomic identification of C/EBP-DBD multiprotein complex: JNK1 activates stem cell regulator C/EBPalpha by inhibiting its ubiquitination. Oncogene. 2007;26(12):1789–1801. doi: 10.1038/sj.onc.1209964. [DOI] [PubMed] [Google Scholar]
- 93.Wang C., Ren R., Hu H., Tan C., Han M., Wang X., Zheng Y. MiR-182 is up-regulated and targeting Cebpa in hepatocellular carcinoma. Chin. J. Cancer Res. 2014;26(1):17–29. doi: 10.3978/j.issn.1000-9604.2014.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Timchenko L.T., Iakova P., Welm A.L., Cai Z.J., Timchenko N.A. Calreticulin interacts with C/EBPalpha and C/EBPbeta mRNAs and represses translation of C/EBP proteins. Mol. Cell. Biol. 2002;22(20):7242–7257. doi: 10.1128/MCB.22.20.7242-7257.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hattori T., Ohoka N., Inoue Y., Hayashi H., Onozaki K. C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene. 2003;22(9):1273–1280. doi: 10.1038/sj.onc.1206204. [DOI] [PubMed] [Google Scholar]
- 96.Subramanian L., Benson M.D., Iniguez-Lluhi J.A. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biologic. Chem. 2003;278(11):9134–9141. doi: 10.1074/jbc.M210440200. [DOI] [PubMed] [Google Scholar]
- 97.Miller M., Shuman J.D., Sebastian T., Dauter Z., Johnson P.F. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. J. Biologic. Chem. 2003;278(17):15178–15184. doi: 10.1074/jbc.M300417200. [DOI] [PubMed] [Google Scholar]
- 98.Kato N., Kitaura J., Doki N., Komeno Y., Watanabe-Okochi N., Togami K., Nakahara F., Oki T., Enomoto Y., Fukuchi Y., Nakajima H., Harada Y., Harada H., Kitamura T. Two types of C/EBPalpha mutations play distinct but collaborative roles in leukemogenesis: Lessons from clinical data and BMT models. Blood. 2011;117(1):221–233. doi: 10.1182/blood-2010-02-270181. [DOI] [PubMed] [Google Scholar]
- 99.Leroy H., Roumier C., Huyghe P., Biggio V., Fenaux P., Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19(3):329–334. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 100.Wang N.D., Finegold M.J., Bradley A., Ou C.N., Abdelsayed S.V., Wilde M.D., Taylor L.R., Wilson D.R., Darlington G.J. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269(5227):1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 101.Kimura T., Christoffels V.M., Chowdhury S., Iwase K., Matsuzaki H., Mori M., Lamers W.H., Darlington G.J., Takiguchi M. Hypoglycemia-associated hyperammonemia caused by impaired expression of ornithine cycle enzyme genes in C/EBPalpha knockout mice. J. Biologic. Chem. 1998;273(42):27505–27510. doi: 10.1074/jbc.273.42.27505. [DOI] [PubMed] [Google Scholar]
- 102.Benedict M., Zhang X. Non-alcoholic fatty liver disease: An expanded review. World J. Hepatol. 2017;9(16):715–732. doi: 10.4254/wjh.v9.i16.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rajbhandari R., Chung R.T. Treatment of hepatitis B: A concise review. Clin. Transl. Gastroenterol. 2016;7(9):e190. doi: 10.1038/ctg.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Manns M.P., Buti M., Gane E., Pawlotsky J.M., Razavi H., Terrault N., Younossi Z. Hepatitis C virus infection. Nat. Rev. Dis. Primers. 2017;3:17006. doi: 10.1038/nrdp.2017.6. [DOI] [PubMed] [Google Scholar]
- 105.Rasineni K., Casey C.A. Molecular mechanism of alcoholic fatty liver. Int. J. Pharmacol. 2012;44(3):299–303. doi: 10.4103/0253-7613.96297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsochatzis E.A., Bosch J., Burroughs A.K. Liver cirrhosis. Lancet. 2014;383(9930):1749–1761. doi: 10.1016/S0140-6736(14)60121-5. [DOI] [PubMed] [Google Scholar]
- 107.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 108.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA (Edinb.) 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 109.London W.T.M.K. 2006. [Google Scholar]
- 110.El-Serag H.B., Marrero J.A., Rudolph L., Reddy K.R. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134(6):1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 111.Fan S.T. Hepatocellular carcinoma--resection or transplant? Nat. Rev. Gastroenterol. Hepatol. 2012;9(12):732–737. doi: 10.1038/nrgastro.2012.158. [DOI] [PubMed] [Google Scholar]
- 112.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2017. CA (Edinb.) 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 113.Adnane L., Trail P.A., Taylor I., Wilhelm S.M. Sorafenib(BAY 43-9006, Nexavar), a dual-action inhibitor that targets RAF/MEK/ERK pathway in tumor cells and tyrosine kinases VEGFR/PDGFR in tumor vasculature. Methods Enzymol. 2006;407:597–612. doi: 10.1016/S0076-6879(05)07047-3. [DOI] [PubMed] [Google Scholar]
- 114.Sanoff H.K., Chang Y., Lund J.L., O’Neil B.H., Dusetzina S.B. Sorafenib effectiveness in advanced hepatocellular carcinoma. Oncologist. 2016;21(9):1113–1120. doi: 10.1634/theoncologist.2015-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Parikh N.D., Marshall V.D., Singal A.G., Nathan H., Lok A.S., Balkrishnan R., Shahinian V. Survival and cost-effectiveness of sorafenib therapy in advanced hepatocellular carcinoma: An analysis of the SEER-Medicare database. Hepatology. 2017;65(1):122–133. doi: 10.1002/hep.28881. [DOI] [PubMed] [Google Scholar]
- 116.Johansen L.M., Iwama A., Lodie T.A., Sasaki K., Felsher D.W., Golub T.R., Tenen D.G. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol. Cell. Biol. 2001;21(11):3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Timchenko N., Wilson D.R., Taylor L.R., Abdelsayed S., Wilde M., Sawadogo M., Darlington G.J. Autoregulation of the human C/EBP alpha gene by stimulation of upstream stimulatory factor binding. Mol. Cell. Biol. 1995;15(3):1192–1202. doi: 10.1128/mcb.15.3.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yogosawa S., Mizutani S., Ogawa Y., Izumi T. Activin receptor-like kinase 7 suppresses lipolysis to accumulate fat in obesity through downregulation of peroxisome proliferator-activated receptor gamma and C/EBPalpha. Diabetes. 2013;62(1):115–123. doi: 10.2337/db12-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith L.T., Hohaus S., Gonzalez D.A., Dziennis S.E., Tenen D.G.P.U. 1(Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88(4):1234–1247. [PubMed] [Google Scholar]
- 120.Tanaka S., Tanaka K., Magnusson F., Chung Y., Martinez G. J., Wang Y.-H., Nurieva R.I., Kurosaki T., Dong C. 2014.
- 121.Soriano H.E., Kang D.C., Finegold M.J., Hicks M.J., Wang N.D., Harrison W., Darlington G.J. Lack of C/EBP alpha gene expression results in increased DNA synthesis and an increased frequency of immortalization of freshly isolated mice [correction of rat] hepatocytes. Hepatology. 1998;27(2):392–401. doi: 10.1002/hep.510270212. [DOI] [PubMed] [Google Scholar]
- 122.Lin T.C., Hou H.A., Chou W.C., Ou D.L., Yu S.L., Tien H.F., Lin L.I. CEBPA methylation as a prognostic biomarker in patients with de novo acute myeloid leukemia. Leukemia. 2011;25(1):32–40. doi: 10.1038/leu.2010.222. [DOI] [PubMed] [Google Scholar]
- 123.Annamaneni S., Kagita S., Gorre M., Digumarti R.R., Satti V., Battini M.R. Methylation status of CEBPA gene promoter in chronic myeloid leukemia. Hematology. 2014;19(1):42–44. doi: 10.1179/1607845413Y.0000000081. [DOI] [PubMed] [Google Scholar]
- 124.Kumagai T., Akagi T., Desmond J.C., Kawamata N., Gery S., Imai Y., Song J.H., Gui D., Said J., Koeffler H.P. Epigenetic regulation and molecular characterization of C/EBPalpha in pancreatic cancer cells. Int. J. Cancer. 2009;124(4):827–833. doi: 10.1002/ijc.23994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lefterova M.I., Zhang Y., Steger D.J., Schupp M., Schug J., Cristancho A., Feng D., Zhuo D., Stoeckert C.J., Jr, Liu X.S., Lazar M.A. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22(21):2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu X., Liu C., Laurini E., Posocco P., Pricl S., Qu F., Rocchi P., Peng L. Efficient delivery of sticky siRNA and potent gene silencing in a prostate cancer model using a generation 5 triethanolamine-core PAMAM dendrimer. Mol. Pharm. 2012;9(3):470–481. doi: 10.1021/mp2006104. [DOI] [PubMed] [Google Scholar]
- 127.Lima W.F., Wu H., Nichols J.G., Sun H., Murray H.M., Crooke S.T. Binding and cleavage specificities of human Argonaute2. J. Biologic. Chem. 2009;284(38):26017–26028. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huan H., Wen X., Chen X., Wu L., Liu W., Habib N.A., Bie P., Xia F. C/EBPalpha short-activating RNA suppresses metastasis of hepatocellular carcinoma through inhibiting EGFR/beta-catenin signaling mediated EMT. PLoS One. 2016;11(4):e0153117. doi: 10.1371/journal.pone.0153117. [DOI] [PMC free article] [PubMed] [Google Scholar]