Abstract
Background:
Potassium bromate (KBrO3), a food additive, has been used in many bakery products as an oxidizing agent. It has been shown to induce renal cancer in many in-vitro and in-vivo experimental models
Objectives:
This study evaluated the carcinogenic potential of potassium bromate (KBrO3) and the chemopreventive mechanisms of the anti-oxidant and anti-inflammatory phytochemical, curcumin against KBrO3-induced carcinogenicity.
Method:
Lactate dehydrogenase (LDH) cytotoxicity assay and morphological characteristics were used to assess curcumin's cytoprotective potential against KBrO3 toxicity. To assess the chemopreventive potential of curcumin against KBrO3-induced oxidative insult, intracellular H2O2 and the nuclear concen-tration of the DNA adduct 8-OHdG were measured. PCR array, qRT-PCR, and western blot analysis were used to identify dysregulated genes by KBrO3 exposure. Furthermore, immunofluorescence was used to evaluate the ciliary loss and the disturbance of cellular tight junction induced by KBrO3.
Results:
Oxidative stress assays showed that KBrO3 increased the levels of intracellular H2O2 and the DNA adduct 8-OHdG. Combination of curcumin with KBrO3 efficiently reduced the level of H2O2 and 8-OHdG while up-regulating the expression of catalase. PCR array, qRT-PCR, and western blot analysis revealed that KBrO3 dysregulated multiple genes involved in inflammation, proliferation, and apoptosis, namely CTGF, IL-1, and TRAF3. Moreover, qRT-PCR and immunofluorescence studies showed that KBrO3 negatively affected the tight junctional protein (ZO-1) and induced a degeneration of primary ciliary proteins. The negative impact of KBrO3 on cilia was markedly repressed by curcumin.
Conclusion:
Curcumin could potentially be used as a protective agent against carcinogenicity of KBrO3.
Keywords: Potassium bromate (KBrO3), chemoprevention, curcumin, primary cilia, kidney cancer, inflammation
1. INTRODUCTION
Potassium bromate (KBrO3), a food additive, has been used in different bakery products as an oxidizing agent. In 2005, the use of KBrO3 as a food additive was banned by countries of the European Union, Canada, Japan, China, India, and South America. Surprisingly, it has not been banned by the FDA in the US and still can be found in some bakery products [1, 2]. KBrO3 was shown to induce renal cancer in many in-vitro and in-vivo experimental models [3-5]. Shiao et al. found that rats exposed to KBrO3 in drinking water caused a mutation in the Von Hippel-Lindau tumor (VHL) gene, a crucial tumor suppressor that has been found to be mutated in renal cell carcinomas [6]. KBrO3, was found to induce oxidative DNA damage and DNA adduct formation that lead to various gene mutations. For instance, the formation of the mutagenic 8-hydroxy deoxyguanosine (8-OHdG) was detected following the exposure of porcine kidney cells to KBrO3 [7]. Interestingly, KBrO3 was found to interact with GSH to form the highly reactive 8-oxodeoxy-guanosine (8-oxodG), thus causing DNA double strand break [8, 9].
Following DNA damage, many genes including those controlling apoptosis and inflammation are dysregulated. For instance, Bader and Hsu mentioned that many pro-inflammatory genes such as HIF1a, HIF2a, TNF-α, TGF-β, and NF-KB were found to be up-regulated in response to DNA damage that eventually induced VHL mutation [10].
Primary cilia are immotile sensory organelles that play an important role in cell differentiation, polarity, and quiescence. They can receive mechanical and chemical signals from other cells as well as, from the surrounding environment [11]. Normally, cells assemble cilia on their membranes when they “stop” dividing, exit the cell cycle, and start to differentiate [12]. However, cells tend to lose cilia when they re-enter the cell cycle and mitosis [13]. In most renal cell carcinomas where the suppressor VHL gene is inactivated, Schraml et al. found that ciliary loss in clear cell renal cell carcinoma (ccRCC) was strongly associated with the mutation of this tumor suppression gene. In contrast, the author found that in papillary renal cell carcinomas, the frequency of cilia was higher than ccRCC and the ciliary loss was VHL independent, which points to the differences in the biological pattern between these types of cancer cells [14].
Chemoprevention is a novel aspect in cancer development and treatment referring to the use of natural, semi-synthetic or synthetic compounds to halt, stop, or reverse tumor formation and progression [15]. Chemopreventives can block tumor initiation; therefore, they are termed blocking agents. For instance, anti-oxidants, free radical scavengers, phase I drug-metabolizing enzymes inhibitors, and phase II drug-metabolizing enzymes inducers are deemed cancer blocking agents. Whereas compounds that halt the stages of tumor promotion and progression are referred to as tumor suppressors. Induction of apoptosis, terminal cell differentiation, inhibition of cell proliferation and clonal expansion, and alteration of gene expression of preneoplastic tumors are examples of tumor suppression [15-17].
Curcumin, also known as diferuloylmethane, is a polyphenolic compound derived from the root and rhizome of the plant Curcuma longa [18, 19]. Curcumin has a chemoprevention potential owing to its anti-oxidant, anti-inflammatory, immunomodulatory, and pro-apoptotic potential [20]. While curcumin abrogates several oncogenic pathways such as NF-KB, Akt/PI3K and MAPK, it has recently been found to induce anti-tumor potential via epigenetic modulations of critical genes. For instance, it dysregulates several oncogenic and tumor suppressor miRNAs, namely, miR-21, miR-17-5p, miR-22, miR-15a, miR-20a, and miR-27a [21, 22].
This study aimed at investigating the carcinogenic potential of KBrO3 and the mechanisms by which curcumin can prevent the carcinogenic insults of KBrO3 on the renal epithelial cells (RPTEC/TERT1). Moreover, our group has reported that targeting RPTEC/TERT1 with a subtoxic concentration of KBrO3 was associated with loss of primary cilia [23], therefore this study also investigated the preventive potential of curcumin against KBrO3 induced deciliation, and thus inhibited proliferation and dedifferentiation.
2. MATERIALS AND METHOD
2.1. Cell Culture and Treatment
The human renal proximal tubular epithelial (RPTEC/TERT1) [24] and ACHN cell lines were obtained from the American Tissue Culture Collection (ATCC). Cells were maintained in low glucose (5 mM) Dulbecco’s Modified Eagle / Nutrient Mix F-12 medium supplemented with 5μg/ml insulin, 5μg/ml transferrin, 5μg/ml selenite (ITS), 36ng/ml hydrocortisone, 10ng/ml epidermal growth factor (EGF) (Sigma‐Aldrich) 50 U/ml penicillin, 50μg/ml streptomycin (P/S), and 2mM L-glutamine (Gibco, Life Technologies). ACHN cells were maintained in Minimum Eagle Medium (Sigma‐Aldrich) with 10% FBS and P/S. Both cell lines were incubated at 37o C in a 5% CO2 humidified atmosphere.
RPTEC/TERT1 were seeded at a density of 1x106 cells/ml. Cells were maintained for 10 days after reaching 100% confluency to allow stabilization of the monolayer [23]. While ACHN cells were seeded at the same density one day in advance before the treatment. KBrO3 and curcumin were purchased from (Sigma-Aldrich, Taufkirchen, Germany). KBrO3 was dissolved in water to prepare 100mM stock solution which was then further diluted with culture medium to get the indicated concentrations. Curcumin and silymarin were dissolved in DMSO to prepare stock solutions of 250 mM and 50mM, respectively. They were then further diluted with culture medium to final concentration 200 µM and 25 µM, respectively. In most experiments, where curcumin or silymarin was used, control cells were exposed to a maximum 0.1% DMSO.
2.2. Lactate Dehydrogenase (LDH) Release Assay
The LDH assay was performed using a Roche kit (Roche Diagnostics, Mannheim, Germany). Briefly, RPTEC/TERT1 cells were cultured at density of 1x106 cells/ ml in 24-well plates and allowed to form a fully confluent monolayer. 10 days post confluency, cells were treated with six different concentrations of KBrO3 (0.5,1,2,3,5, and 10 mM) only or in a combination with 25µM curcumin for 24h. The positive control for maximum LDH release or 100% cell death was prepared by treating cells with 2% Triton-X 100 for 5 min at 37°C. Following 15min incubation of cell supernatants with the reaction mixture (provided by the kit), the absorbance was measured at 490nm using Spectamax2 plate reader (Molecular devices, Winnersh/ UK). LDH release was expressed as a percentage of the maximum LDH activity.
2.3. Phase Contrast Microscopy
RPTEC/TERT1 cells at 1x106 cells/ ml were cultured in 24- well plates and allowed to form a fully confluent monolayer. 10 days post-confluency, cells were treated with KBrO3 (10mM) only or in a combination with 25µM curcumin for 24h at 37°C. Cellular morphology was observed by phase contrast microscopy using a JVC high-resolution digital camera (KY-F55BE) attached to a Nikon TMS phase contrast microscope. Micrographs were processed using ImageJ v.1.49.
2.4. Determination of Intracellular H2O2 Concentration
Intracellular hydrogen peroxide (H2O2) concentrations were measured using Amplex red assay kit (Thermo Fisher scientific, Carlsbad, CA) following the manufacturer instructions. Briefly, cells were cultured in 12 well culture plates. Following treatment, cells were lysed in 200 µl ice-cold lysis buffer (0.1% Triton X-100 in 0.05 M sodium phosphate buffer, pH 7.4). Following this, lysates were transferred to pre-chilled microfuge tubes and vortexed every 3 min for 15 min for complete cell homogenization. Tubes were centrifuged at 14,000 g at 4°C for 15 min. A volume of 50 µl of supernatants and standard H2O2 solutions (0, 0.05, 0.1, 0.25, 0.5, 0.75, 1, 1.25, 2, and 5 µM) were loaded in 96-well opaque black microplate. An equal volume of Amplex Red reaction mixture (0.1 mM Amplex red reagent and 0.2 U/ml horseradish peroxidase in 1X reaction buffer) was added to the pre-loaded wells to initiate the reaction. The fluorescence was measured kinetically every 30 sec for 30 min at excitation and emission wavelengths of 530 and 590 nm, respectively using a scanning microplate reader (Molecular Devices Inc, Sunnyvale, CA, USA). Intracellular H2O2 concentration was calculated using the standard curve and normalized to the total protein content.
2.5. Determination of Intra-nuclear Concentration of 8-OHdG
This assay consists of 4 stages: DNA extraction, determination of DNA concentration, DNA digestion, and measuring the concentration of the DNA adduct 8-OHdG.
DNA was extracted from cells using WAKO DNA Extractor WB Kit (Wako, Osaka, Japan) which contains sodium iodide (NaI) as chaotropic agent to minimize the oxidation of DNA during the extraction. The concentrations of DNA solutions were calculated using the Thermo Fisher Scientific NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA). The Wako 8-OHdG Assay Preparation Reagent kit was used exclusively to digest DNA and release 8-OHdG. Determination of the intranuclear 8-OHdG concentration was performed using an enzyme-linked immunosorbent assay (ELISA) kit (Highly Sensitive 8-OHdG Check ELISA kit, Japan Institute for the Control of Aging, Fukuroi, Japan). Briefly, a volume of 50 µl of digested DNA samples and standard concentrations were loaded to the provided ELISA plate. A volume of 50 µl primary antibody per a well was added and incubated overnight at 4°C. Next day, the well contents were poured off and washed three times using 250μl/well washing solution. The wells were incubated with100 μl/ well of secondary antibody for 1h at room temperature. The plate was washed before adding a chromatic solution and incubated for 15 min at room temperature in a dark place. Finally, the reaction was terminated by adding 100μl of the reaction terminating solution. The absorbance was read at 450 nm.
2.6. Immunofluorescent Labeling
For immunofluorescent labeling experiments, cells were cultured in 8-well chamber slides (Millipore, USAand allowed to form a fully confluent monolayer. 10 days post-confluency, cells were treated with DMSO-containing medium, 5.5mM KBrO3, 25µM curcumin, or a combination of both for 24h at 37 °C. Following treatment, cells were washed with PBS three times and fixed with 3.7% formaldehyde for 20 min at room temperature. Cells were then washed three times with PBS and permeabilized with 0.2%(v/v) Triton-X 100 in PBS. Background was reduced by blocking nonspecific signals with 0.5% (w/v) BSA in PBS. The ciliary markers Acetylated α-tubulin and Arl 13B were labeled using a mouse anti-human antibody (1:400) (Sigma-Aldrich, Taufkirchen, Germany) and a rabbit anti-human antibody, respectively. ZO-1 was labeled using a rabbit anti-human antibody (1:300) (Zymed, Invitrogen, South San Francisco, CA). Nuclei were stained with Hoechst 33342 (1:1000) (Sigma-Aldrich, Taufkirchen, Germany). Slides were imaged using a Zeiss A-Plan 40X/0.65 objective and a Zeiss M1 Upright AxioImager with CoolLED P3000 light source or a Zeiss C-Plan-Apochromat 40X/1.3 objective and a Zeiss LSM510 UVMETA confocal microscope. Images were deconvolved using Auto Quant-X3 deconvolution software (Version 3.0.3) (Media Cybernetics Inc.) at 5 iterations. Images were adjusted for brightness and contrast using Fiji/ImageJ (Fiji.sc/).
2.7. RNA Extraction and Preparation of cDNA
RPTEC/TERT1 cells were seeded and treated as described earlier using 6-well plate format. TRIzol method was used to extract total RNA. The same method was applied to extract the total RNA from the ACHN cells after 24h of growth. Briefly, TRIzol (Life Technologies, USA) was used to lyse cells (1ml per a well). Cells were then homogenized by passing through pipette tip several times. Following the homogenization, 200µl of 1–bromo–3–chloropropane (BCP), a chloroform derivative, was added to the homogenate, mixed by vertexing, incubated for 3 min at room temperature, then centrifuged at 12,000 × g at 4°C for 15min. The mixture was separated into three phases: aqueous (RNA), interphase and a red lower organic layer (DNA and protein). The aqueous phase was transferred into a clean microfuge tubes then 500µl isopropanol was added to precipitate RNA. Following centrifugation at 12,000 × g at 4°C for 15 min, the supernatant was discarded and the pellets were washed with 500µl of 75% ethanol to remove further impurities. The microfuge tubes were finally centrifuged at 7500g at 4°C for 5 min. The supernatant was discarded and RNA pellets were dissolved in ddH2O. For further RNA purification and removing of genomic DNA contamination, mRNeasy and DNase Max kit (Qiagen, UK) were used according to the manufacturer protocol. The yield of the RNA was assessed by measuring the optical density at 260 nm and 280nm using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The purity of RNA samples was assessed based on the absorbance ratio of 260:280 which should be ≥1.8. A total RNA of 1µg was reversed transcribed to cDNA using a RevertAid H Minus First Strand cDNA synthesis Kit (Fermentas GmbH, St. Leon-Rot, Germany) according to the manufacturer’s instructions.
2.8. Quantitative Real-Time PCR Analysis
Quantitative expression of a total of 192 genes (list of genes provided in the supplementary Table 1) involved in inflammation, angiogenesis, and apoptosis were evaluated using real-time PCR ABI PRISM® 7900HT (Foster City, California USA). These were customized PCR arrays designed by (Sigma-Aldrich, Taufkirchen, Germany) Corp. For the PCR array experiment, 20μL cDNA of each individual treatment was diluted up to 170μL using distilled water. Three biological replicates of a single treatment group were pooled and analyzed. The real-time PCR mix (20μL/well) contained 1μL of pooled cDNA, 3μL water, 6μL primer mix, and 10μL SYBR green master mix. The thermal cycle conditions were 95°C for 15min followed by 45 cycles of 94°C for 15 sec, 55 °C for 30 sec, and 70 °C for 30 sec. The dissociation stage was set at 95 °C for 15 sec, 60°C for 15 sec, and 95°C for 15 sec, as previously described [25] The mRNA abundances were expressed in “cycle threshold” (Ct) values, which represents the number of PCR cycles after which the PCR product crosses a threshold value. A Ct value of 35 was used as the cut-off limit. The normalization of gene expression was carried out based on the abundance of the house-keeping genes βactin (ACTB), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and beta-glucuronidase (GUSB).
Table 1.
List of genes dysregulated following the exposure of RPTEC/TERT1 to KBrO3 for 24h compared with untreated RPTEC/TERT1 cells.
| Genes with Markedly Decreased Expression | Genes with Markedly Increased Expression | ||||
|---|---|---|---|---|---|
|
Fold
Decrease |
Gene Name | Gene |
Fold
Increase |
Gene Name | Gene |
| -57.7 | Interleukin (IL)1-receptor 1 | IL1R1 | 197.55 | Connective tissue growth factor | CTGF |
| -30.89 | Toll-like receptor 3 | TLR3 | 37.53 | Plasminogen activator inhibitor-1 | PAI1 |
| -26.57 | Chemokine (C-X-C Motif) Ligand 1 | CXCL1 | 29.92 | Resistin | RETN |
| -25 | Interleukin 8 | IL8 | 28.21 | Proto-oncogene c-Fos | FOS |
| -23.08 | Chemokine (C-X-C Motif) Ligand 2 | CXCL2 | 15.40 | Pim-3 Proto-Oncogene, Serine/Threonine Kinase | PIM3 |
| -22.29 | TNF receptor-associated factor 5 | TRAF5 | 16.24 | Suppressor of cytokine signaling 1 | SOCS1 |
| -20.26 | Signal Transducer and Activator of Transcription 1 | STAT1 | 11.66 | Tumor necrosis factor | TNFA |
| -17.5 | TNF receptor-associated factor 3 | TRAF3 | 11.28 | Toll-like receptor 4 | TLR4 |
| -17.49 | Myeloid differentiation primary response | MYD88 | 9.76 | Epidermal growth factor | EGF |
| -13.3 | TNF receptor-associated factor 6 | TRAF6 | 9.13 | Lymphotoxin-alpha | LTA |
| -12.9 | Chemokine (C-C motif) ligand 20 | CCL20 | 8.50 | Nuclear receptor related 1 protein | NR4A2 |
| -11.75 | Interleukin 6 | IL6 | 7.02 | Ubiquitin C | UBC |
| -11.59 | Caspase 1, Apoptosis-Related Cysteine Peptidase |
CASP1 | 6.18 | Adrenoceptor Beta 2, Surface | ADRB2 |
| -9.95 | B-Cell CLL/Lymphoma 3 | BCL3 | 6.55 | Interleukin-2 receptor alpha | IL2RA |
| -8.75 | Prostaglandin E Receptor 2 | PTGER2 | 4.42 | Sex Determining Region Y)-Box 9 | SOX9 |
| -8.73 | inhibitor of nuclear factor kappa-B kinase | IKBKB | 3.16 | Tumor Necrosis Factor Receptor Superfamily, Member 10b | TNFRSF10B |
| -8.35 | C-reactive protein | CRP | 3.99 | Superoxide dismutase | SOD1 |
| -7.41 | Plasminogen Activator | PLAT | 3.86 | (Early Growth Response 2 | EGR2 |
| -6.09 | Toll-like receptor 2 | TLR2 | 3.78 | C-C chemokine receptor type 10 | CCR10 |
| -5.87 | Janus Kinase 2 | JAK2 | 3.65 | Vascular cell adhesion protein 1 | VCAM1 |
| -5.57 | Mitogen-Activated Protein Kinase Kinase Kinase 1 | MAP3K1 | 3.63 | Endothelin 1 | EDN1 |
| -5.55 | Intercellular Adhesion Molecule 1 | ICAM1 | 3.54 | Jun Proto-Oncogene | JUN |
| -4.78 | Complement Component 5 | C5 | 3.38 | 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 |
| -4.33 | Leukotriene B4 Receptor 2 | LTB4R2 | 3.31 | Nuclear Receptor Subfamily 4 | NR4A1 |
| -4.18 | Nuclear factor NF-kappa-B p105 | NFKB1 | 2.91 | Insulin-Like Growth Factor Binding Protein 2 | IGFBP2 |
| -4.17 | Insulin-Like Growth Factor Binding Protein 1 | IGFBP1 | 2.48 | Retinoic acid receptor alpha | RARA |
| -4.09 | Phospholipase C, Beta 4 | PLCB4 | 2.46 | Transforming growth factor beta | TGFB1 |
| -3.99 | Chemokine (C-C Motif) Ligand 2 | CCL2 | |||
| -3.98 | TNF receptor-associated factor 2 | TRAF2 | |||
| -3.83 | Signal Transducer And Activator Of Transcription 3 |
STAT3 | |||
| Genes with Markedly Decreased Expression | Genes with Markedly Increased Expression | ||||
|
Fold Decrease |
Gene Name | Gene |
Fold Increase |
Gene Name | Gene |
| -3.7 | Colony Stimulating Factor 2 Receptor | CSF2 | |||
| -3.54 | Tumor Necrosis Factor Receptor Superfamily, Member 1A | TNFRSF1A | |||
| -3.32 | CAMP Responsive Element Binding Protein 1 | CREB1 | |||
| -3.32 | Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 2 | NFKB2 | |||
| -3.16 | Insulin-Like Growth Factor Binding Protein 3 | IGFBP3 | |||
| -2.95 | Epidermal Growth Factor Receptor | EGFR | |||
| -2.84 | v-rel avian reticuloendotheliosis viral oncogene homolog | REL | |||
| -2.78 | Nuclear factor NF-kappa-B p105 subunit | NFKB1 | |||
| -2.77 | Transforming Growth Factor, Beta Receptor 1 | TGFBR1 | |||
| -2.74 | nterleukin 1 Receptor Antagonist | IL1RN | |||
| -2.43 | tumor protein p53 | TP53 | |||
| -2.35 | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | MALT1 | |||
| -2.34 | V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog B | RELB | |||
| -2.09 | Mitogen-Activated Protein Kinase 8 | MAPK8 | |||
Validation of the PCR array was performed using single tube TaqMan-probe based gene expression assays (Applied Biosystems, Foster City, CA, USA). Normalization of genes expression was carried out using the house-keeping gene β-actin. The PCR reaction mixture (10μL) consisted of 0.5μL cDNA, 3.5μL nuclease free water, 0.5μL primer mix, 0.5μL loading control (β-actin), and 5μL TaqMan master mix. The thermal cycle conditions were as follows: 50°C for 2 min, 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min. Samples were loaded in an optical 384 well plate in duplicates (10μL/ well) and Ct values <35 were used as the cut-off limit. For the analysis of both PCR array and qRT-PCR, the 2−ΔΔCt method was applied. Briefly, average ΔCt was calculated as the difference of Ct values of any target gene from the average of the Ct value of the reference gene (s). Then, fold change was calculated as 2(−average ΔCt target gene)/2(−average ΔCt reference gene). A fold difference cut-off point was set at ≥2.0.
2.9. Western Blot Analysis
Western blot analysis was carried out according to the standard method by Buchmann [26]. Following cell treatment, the media was removed, and the cells were lysed by using RIPA buffer (Sigma-Aldrich, Taufkirchen, Germany). Total protein concentration was determined by Bradford method using BCA protein assay kit (Pierce, Rockford, IL, USA) according to the manufacturer protocol. Equal amounts of 20µg of whole cell lysates were placed in each lane and subject to SDS-PAGE electrophoresis, then transferred to a 0.2µm pore size Whatman Protran® nitrocellulose membrane (Thermo Fisher Scientific, Wilmington, DE, USA) using a semi-dry transfer system. After the transfer, the membranes were blocked by incubation with TBS-T buffer (50 mM tris-HCl, pH 7.4, 150 mM NaCl, and 0.05% Tween 20) containing 5% non-fat milk or BSA for 1hr at room temperature. The membranes were then incubated overnight at 4°C with CTGF primary antibody (1:1000) (Santa Cruz, USA) and GAPDH (1:10,000) (Cell Signaling Technology Inc, Danvers, MA, USA). Next day, the blots were washed with T-BST then incubated with TBS-T/ 5% non-fat milk containing an appropriate secondary antibodies coupled with horse radish peroxidase (HRP) (Cell Signaling Technology Inc, Danvers, MA, USA) for 1h at room temperature. Immunodetection was performed using the SuperSignal West Pico Substrate (Thermo Scientific, Rockford, IL, USA).
2.10. Statistical Analysis
All experiments were repeated at least three times. Statistical analyses were performed using Graph Pad Prism 5.0. Data was analyzed using one-way analysis of variance (ANOVA). Comparisons between different treatment groups were made by Newman-Keuls Multiple Comparison post-test. Results were expressed as the mean +/- standard error of the mean (SEM). A probability of 0.05 of less was deemed statistically significant.
3. Results
3.1. Curcumin Protected RPTEC/TERT1 Against KBrO3 - Induced Cytotoxicity
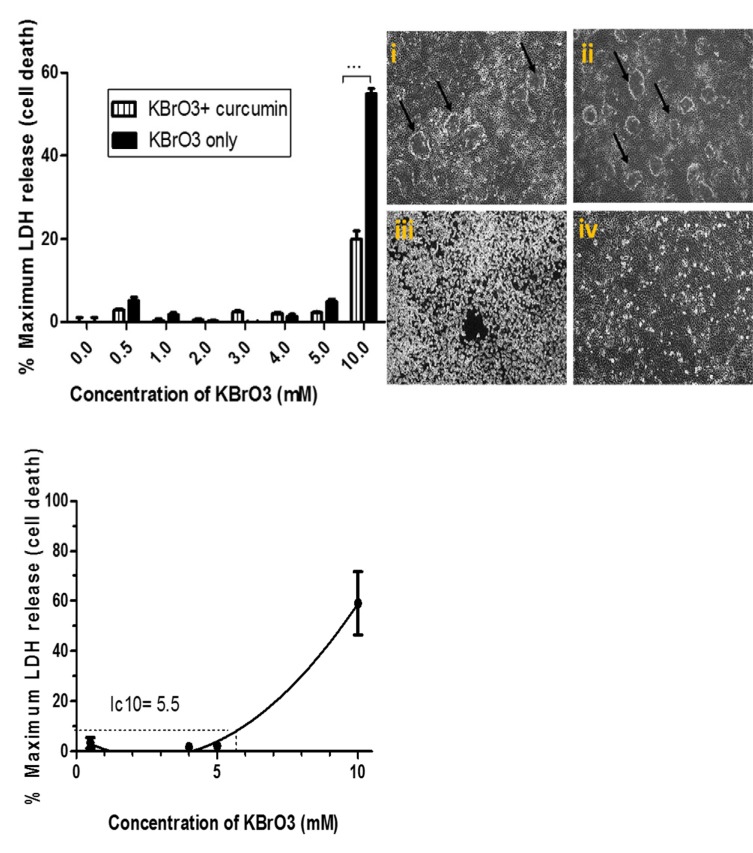
The potential cytoprotective effect of curcumin was investigated by comparing the activity of the released LDH following the exposure to KBrO3 alone and KBrO3/ curcumin combination. The morphological pattern of cells was assessed using phase contrast microscopy. As shown in Fig. (1a), KBrO3 sharply increased the relative activity of LDH at 10mM KBrO3 concentration. The combination of KBrO3 with curcumin exhibited an extremely significant (p< 0.001) lowering of LDH release compared to KBrO3 treatment only. These findings revealed that curcumin exhibited a cytoprotective potential against KBrO3 induced cell death
Fig. (1).
Examination of the cytoprotective effects of curcumin measured by cytotoxicity assay and assessed by morphological characteristics.
Fig. (1a) : The cytoprotective potential of curcumin was assessed using the LDH cytotoxicity assay. RPTEC/TERT1 cells were treated either with the indicated concentrations of KBrO3 only or a combination of KBrO3 concentrations with 25µM curcumin. The figure represents mean± SEM of six independent experiments. (*** = p<0.001)
Fig. (1b) Analysis of the morphological characteristics of RPTEC/TERT1 cells. Cells were seeded in 24 well plates, 10 days post 100% confluency, the cells formed fluid filled dooms which reflect a well-functioning transport system. (i) RPTEC/TERT1 were treated with 0.1% DMSO (control), or (ii) with 25µM curcumin for 24h. The domes were maintained and cells were morphologically unaffected by the treatment. (iii) Cells were treated with 10mM KBrO3;. (iv) Cells were treated with the combination of 10mM KBrO3+ 25µM curcumin for 24 hr.
Fig. (1c) Estimation of subtoxic IC10 and the toxic IC50 concentrations of KBrO3. RPTEC/TERT1 cells were seeded in 24 well plates. 10 days post 100% confluency, they were treated with the indicated concentrations of KBrO3 to estimate subtoxic IC10 and the toxic IC50 concentrations. 2% Triton-TX100 was used as a positive control of cell death. The figure represents mean± SEM of six independent experiments.
The effect of each treatment on cellular morphology was assessed using phase contrast microscopy (Fig. 1b). When RPTEC/TERT1 cells were treated with DMSO (i) or with curcumin (ii), the cells showed normal cobblestone appearance, tight interconnections, and formed characteristic domes. Such morphological characteristics reflect an intact cellular transport system and indicated that the cells were “happy”, “healthy”, and fully differentiated. Under toxic conditions (iii), KBrO3 caused severe cellular damage with a complete loss of domes and loss of tight junctions between cells. Curcumin clearly minimized KBrO3-induced cellular damage and loss of tight junctions. However, it only partially re-established the characteristic domes of RPTEC/TERT1 cells (Fig. 1b).
Following 24h exposure, the toxic (IC50) and the subtoxic (IC10) concentrations of KBrO3 were estimated to be 5.5 and 7.5mM, respectively using an LDH release assay (Fig. 1c).
3.2. Curcumin Suppressed KBrO3 Induced Oxidative Stress and DNA Damage
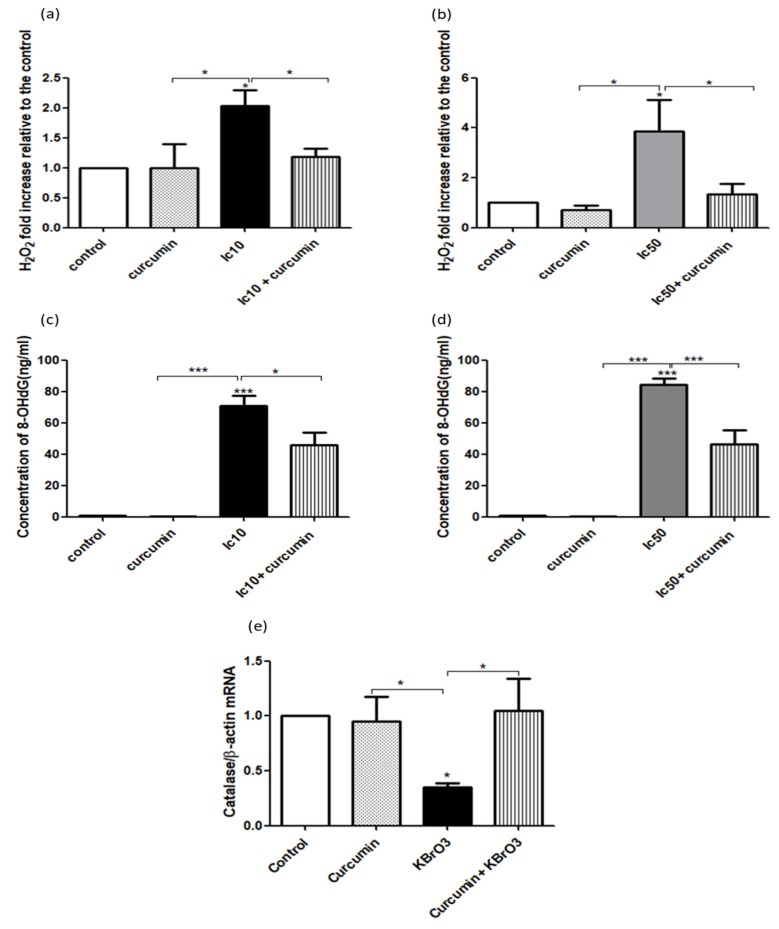
The potential chemopreventive activity of curcumin was further assessed by comparing the level of oxidative stress following the exposure of RPTEC/TERT1 cells to KBrO3 alone or in combination with curcumin. In this context, intracellular H2O2 and 8-OHdG concentrations were measured. Of note, at both IC10 and IC50 concentrations, KBrO3 significantly increased the level of H2O2 (p<0.05) (Fig. 2a, b), and 8-OHdG (p< 0.001) (Fig. 2c, d) compared to the control or curcumin-only treatment. The combination of curcumin with IC10 or IC50 KBrO3 concentrations significantly (p<0.05) inhibited the levels of both H2O2 and 8-OHdG compared to KBrO3 treatment only (Fig. 2a-d). In addition, KBrO3 significantly (p<0.05) reduced catalase gene expression compared to the control or curcumin only treatment. The combination of curcumin with KBrO3, significantly (p<0.05) reversed the negative effect of KBrO3 by upregulating catalase gene expression (Fig. 2e).
Fig. (2).
Analysis of curcumin protection on RPTEC/TERT1 against KBrO3 induced oxidative stress and DNA adduct formation.
Fig. (2a, b) Cells were seeded in 24 well plates until fully confluent. After 10 days, cells were treated with 0.1% DMSO (control), curcumin, KBrO3 and a combination of KBrO3 + curcumin. Intracellular concentration of H2O2 was detected using the Amplex Red Hydrogen Peroxide Assay Kit. The data represents three independent experiments, *= p< 0.05.
Fig. (2c, d) DNA adduct formation was investigated by measuring the intracellular concentration of the DNA adduct 8-OHdG, using the Highly Sensitive 8-OHdG Check ELISA kit following the manufacturer kit instructions. The data represents three independent experiments. *= p< 0.05, **= p<0.01, and ***=p <0.001.
Fig. (2e) Catalase gene expression was examined in KBrO3 (5.5mM) treated RPTEC/TERT1 cells after 24h treatment by RT-PCR analysis (* = P < 0.05).
3.3. KBrO3 Induced Dysregulation of Target Genes
The effects of KBrO3 on a panel of 192 genes, was assessed using SYBR green based PCR array technology. These genes are involved in the regulation of inflammation, oxidative stress, angiogenesis, epithelial-mesenchymal transition (EMT), ciliary formation, and apoptosis (supplementary Table 1).
Following the exposure of RPTEC/TERT1 cells to 5.5mM KBrO3, many genes were dysregulated, as shown in Table 1. Namely, connective tissue growth factor (CTGF) was the first most overexpressed gene, while interleukin (IL)1-receptor 1 (IL-1R1) was the first most downregulated compared to the untreated RPTEC/TERT1 cells. Genes that were differentially dysregulated in renal cancerous ACHN cells compared to normal RPTEC/TERT1 cells are shown in Table 2. In this regard, CTGF was one of the top three most overexpressed genes, while IL-1R1 was the most down-regulated gene. The status of genes, that were up-/down-regulated following the exposure of RPTEC/TERT1 cells to KBrO3, was compared to the congruent genes in ACHN cells. ACHN cell line was used as a positive control of carcinogenesis.
Table 2.
List of genes that were differentially dysregulated in cancerous ACHN cells compared to untreated RPTEC/TERT1 cells.
| Genes with Decreased Expression | Genes with Increased Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold Decrease | Gene Name | Gene | Fold Increase | Gene Name | Gene | |||||
| -53.30 | Interleukin (IL)1-receptor 1 | IL1R1 | 185.52 | Vascular cell adhesion molecule 1 | VCAM1 | |||||
| -51.65 | major histocompatibility complex, class I, A | HLAA | 23.24 | Chemokine (C-C Motif) Receptor 10 | CCR10 | |||||
| -46.38 | interleukin (IL)1-receptor 1 | IL1RN | 20.88 | Connective Tissue Growth Factor | CTGF | |||||
| -41.10 | Superoxide dismutase 2 | SOD2 | 18.96 | Insulin-Like Growth Factor Binding Protein 2 | IGFBP2 | |||||
| -39.13 | Nuclear receptor related 1 protein | NR4A2 | 5.86 | Resistin | RETN | |||||
| -29.53 | TNF receptor-associated factor 3 | TRAF3 | 3.99 | Matrix Metallopeptidase 2 | MMP2 | |||||
| -27.81 | tumor necrosis factor a (TNF superfamily, member 2 | TNFA | 3.53 | Adrenoceptor Beta 2, Surface | ADRB2 | |||||
| -24.24 | Interleukin 6 | IL6 | 3.09 | Signal Transducer And Activator Of Transcription 1 | STAT1 | |||||
| -21.16 | Interleukin 6 receptor | IL6R | 3.08 | Phospholipase C, Beta 4 | PLCB4 | |||||
| -20.59 | TNF receptor-associated factor 5 | TRAF5 | 2.87 | Early Growth Response-2 | EGR2 | |||||
| Genes with Decreased Expression | Genes with Increased Expression | |||||||||
| Fold Decrease | Gene Name | Gene | Fold Increase | Gene Name | Gene | |||||
| -18.65 | C-reactive protein | CRP | 2.08 | Plasminogen activator inhibitor-1 | PAI1 | |||||
| -17.67 | Chemokine (C-X-C Motif) Ligand 1 | CXCL1 | 6.11 | (Interleukin 2 Receptor, Alpha | IL2RA | |||||
| -16.81 | Intercellular Adhesion Molecule 1 | ICAM1 | 3.58 | TATA Box Binding Protein (TBP)-Associated Factor | TRAF1 | |||||
| -16.79 | Janus Kinase 2 | JAK2 | 2.55 | Toll-Like Receptor 4 | TLR4 | |||||
| -15.28 | Insulin-Like Growth Factor Binding Protein 3 |
IGFBP3 | ||||||||
| -14.84 | Colony Stimulating Factor 2 Receptor, Alpha |
CSF2 | ||||||||
| -13.78 | Transcription factor SOX-9 | SOX9 | ||||||||
| -13.60 | Interleukin-23 | IL23 | ||||||||
| -12.45 | Mitogen-Activated Protein Kinase Kinase Kinase 1 |
MAP3K1 | ||||||||
| -10.66 | Colony Stimulating Factor 2 Receptor, Alpha |
CSF1 | ||||||||
| -9.45 | Chemokine (C-X-C Motif) Ligand 2 | CXCL2 | ||||||||
| -8.77 | Transforming Growth Factor, Beta Receptor 1 |
TGFBR1 | ||||||||
| -8.62 | Vascular Endothelial Growth Factor A | VEGFA | ||||||||
| -8.50 | Prostaglandin E Receptor 2 (Subtype EP2) | PTGER2 | ||||||||
| -8.10 | Basal Cell Adhesion Molecule | BCAM | ||||||||
| -8.07 | Caspase 1, Apoptosis-Related Cysteine Peptidase | CASP1 | ||||||||
| -7.80 | B-cell lymphoma 6 protein | BCL6 | ||||||||
| -7.22 | liases for IKBKB Gene | IKBKB | ||||||||
| -6.30 | Tumor Necrosis Factor Receptor Superfamily, Member 10 | TNFRSF10A | ||||||||
| -6.07 | Inhibitor Of Kappa Light Polypeptide Gene Enhancer In B-Cells | SCOS3 | ||||||||
Interestingly, we found that a total of 47 genes were differentially dysregulated in the same manner in both KBrO3 treated RPTEC/TERT1 and in the cancerous ACHN cell lines (Table 3).
Table 3.
Summary of genes that were dysregulated in both carcinogen (KBrO3) treated RPTEC/TERT1 and in cancerous ACHN cells compared to the untreated RPTEC/TERT1 cells.
| Downregulated | Upregulated |
|---|---|
| BCL3 | CTGF |
| CASP1 | PAI1 |
| CCL2 | VCAM1 |
| CCL20 | RETN |
| CREB1 | CCR10 |
| CRP | IGFBP2 |
| CSF2 | ADRB2 |
| CXCL1 | EGR2 |
| CXCL2 | PAI1 |
| ICAM1 | IL2RA |
| IGFBP1 | |
| IGFBP3 | |
| IKBKB | |
| IL1R1 | |
| IL1RN | |
| IL6 | |
| IL8 | |
| JAK2 | |
| LTB4R2 | |
| MAP3K1 | |
| MYD88 | |
| NFKB1 | |
| NFKB1 | |
| NFKB2 | |
| PTGER2 | |
| REL | |
| RELB | |
| STAT3 | |
| TGFBR1 | |
| TLR2 | |
| TLR3 | |
| TNFRSF1A | |
| TP53 | |
| TRAF2 | |
| TRAF3 | |
| TRAF5 | |
| TRAF6 |
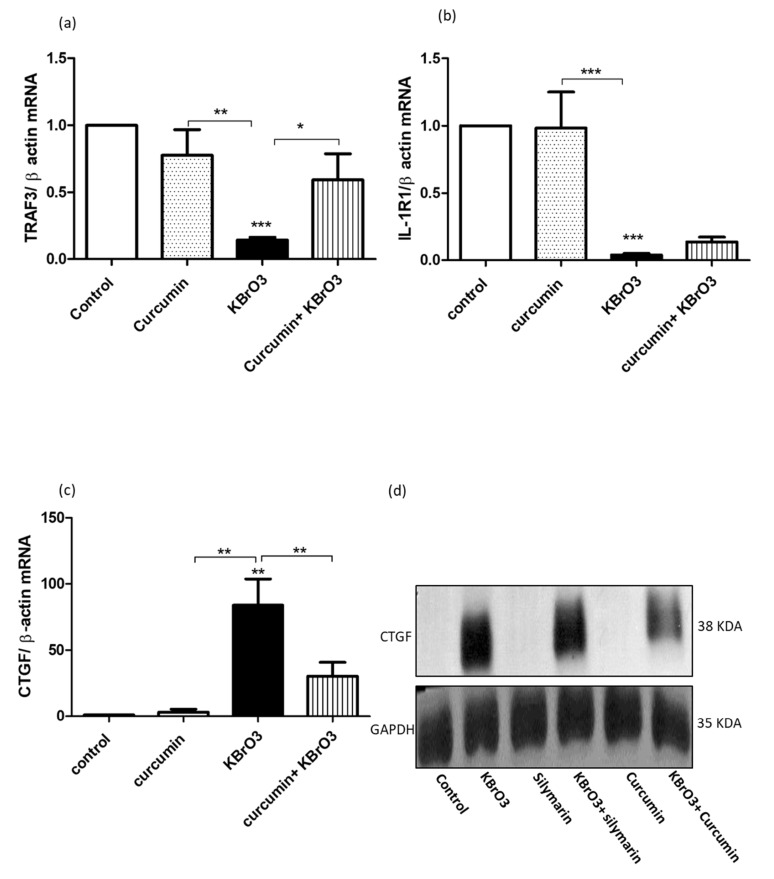
3.4. Curcumin Effectively Reduced KBrO3 Induced TRAF3 and IL1-R1 Downregulation and CTGF Upregulation
To validate the results of the PCR array, individual TaqMan- based probe assays were used to measure the expression of TRAF3, IL1-R1, and CTGF quantitatively using qRT-PCR. The exposure of RPTEC/TERT1 to 5.5mM KBrO3 significantly (p<0.001) inhibited TRAF3 and IL-1R1 gene expression compared to untreated or curcumin-treated cells (Fig. 3a and 3b). In contrast, KBrO3 treatment significantly (p<0.01) upregulated CTGF gene expression in KBrO3-treated RPTEC/TERT1 compared with untreated or curcumin-treated cells (Fig. 3c). More importantly, the co-treatment of 25µM curcumin with KBrO3 significantly (p<0.05) diminished the negative effect of KBrO3 on TRAF3 expression (Fig. 3a), and induced a significant (p<0.01) reduction of the overexpressed CTGF level when combined with KBrO3 compared with KBrO3 alone (Fig. 3c). However, the protective effect of curcumin on KBrO3 induced IL-1R1 suppression wasn’t significant (p>0.05), (Fig. 3b).
Fig. (3).
Analysis of the effects of KBrO3 on TRAF3, IL-1R1, and CTGF gene expression, and the protective potential of curcumin.
Fig. (3a, b) KBrO3 (5.5mM) treatment of RPTEC/TERT1 for 24h onTRAF3 and IL-1 gene expression, respectively compared with the untreated or curcumin-treated cells. Tthe data represents three independent experiments. (*= P> 0.05).
Fig. (3c, d) KBrO3 (5.5mM)- treatment of RPTEC/TERT1 for 24h on CTGF at both mRNA and protein levels in comparison the untreated or curcumin-treated cells. The data represents three independent experiments (**=P < 0.01).
For further validation, we investigated the expression of CTGF at the protein level. In this case, we compared the CTGF repressor activity of curcumin with silymarin, a natural CTGF repressor [27, 28]. KBrO3 clearly up-regulated CTGF protein expression, which was markedly repressed by curcumin. Strikingly, curcumin showed a higher CTGF repressor activity than silymarin when combined with KBrO3 compared with KBrO3-only treatment (Fig. 3d).
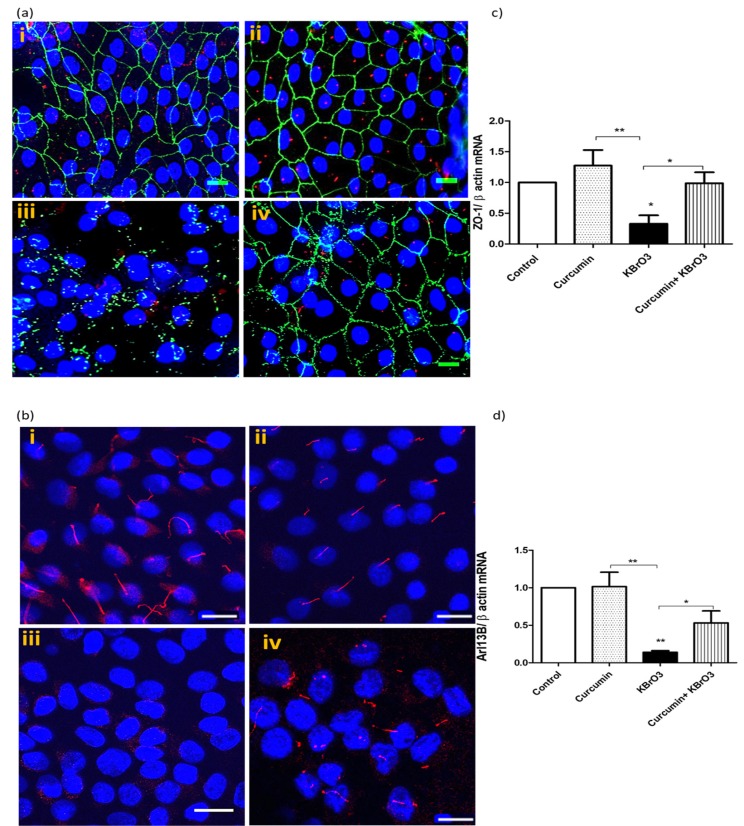
3.5. Curcumin Diminishes the Deciliating Effects of the Genotoxic Carcinogen KBrO3
It has been reported that some renal carcinogens including KBrO3 induce ciliary loss [23]. For this reason, we investigated whether curcumin has a protective potential against KBrO3-induced
Values are expressed as fold change in gene expression relative to vehicle treated control cells. Positive and negative values indicate up- and down-regulation of gene expression, respectively. A fold difference cut-off point was set at ≥2.5.
Values are expressed as fold change in gene expression relative to vehicle treated control cells. Positive and negative values indicate up- and down-regulation of gene expression, respectively. A fold difference cut-off point was set at ≥2.5. deciliation. As shown in Fig. (4a) (i) and (ii), RPTEC/TERT1 cells were treated with 0.1% DMSO (control) or with 25µM curcumin. Under both conditions, RPTEC/TERT1 exhibited well-defined tight junctional barriers between cells (green), and prominent cilia (red). Treating cells with 5.5mM KBrO3 (iii) resulted in disruption of the tight junctional protein ZO-1 and a complete loss of the ciliary marker protein, acetylated α-tubulin. The co-treatment of curcumin with KBrO3 (iv) minimized ciliary loss and junctional protein damage.
Fig. (4).
Effect of Curcumin on KBrO3 induced loss of primary cilia and disruption of tight junction proteins.
Fig. (4a) KBrO3 (5.5mM) treatment of RPTEC/TERT1 for 24h on ciliary and junctional proteins in comparison with untreated or curcumin-treated cells. RPTEC/TERT1 cells were cultured in 8-well chamber slides and treated with after 10 days of 100% confluency. Confocal images (40x) show the primary cilia with red labelled, α-acetylated tubulin, while the green staining represents the tight junctional protein ZO-1. Hoechst 33342 was used to stain the nuclei (blue), scale 50µm. (i) 0.1% DMSO (control), (ii) curcumin 25 µM, (iii) 5.5 mM KBrO3 (ic10), (iv) Co-treatment of curcumin + KBrO3. The images are representative of 3 independent experiments. Scale =50 µm.
Fig. (4b) KBrO3 (5.5mM) treatment of RPTEC/TERT1 for 24h on a ciliary protein in comparison with untreated or curcumin-treated cells. Confocal images (40x) show the primary cilia with red labelled Arl13b. Hoechst 33342 was used to stain the nuclei (blue), scale 50µm. (i) 0.1% DMSO (control), (ii) curcumin 25 µM, (iii) 5.5 mM KBrO3 (ic10), (iv) Co-treatment of curcumin + KBrO3. The images are representative of 3 independent experiments.
Fig. (4c, d) KBrO3 (5.5mM) treatment of RPTEC/TERT1 for 24h on ZO-1 and Arl13b gene expression, respectively compared with the untreated or curcumin-treated cells. The data represents three independent experiments. *= p> 0.05, ** = p>0.01. (The color version of the figure is available in the electronic copy of the article).
To confirm the results, we investigated the effect of KBrO3 on Arl13b, a specific ciliary protein, using confocal microscopy. Very prominent and intact cilia were observed in both untreated and curcumin-treated cells, as shown in Fig. (4b) (i) and (ii), respectively. The exposure of RPTEC/TERT1 cells to 5.5mM KBrO3 for 24h caused a significant loss of cilia. In contrast, the co-treatment of 25µM curcumin with KBrO3 clearly reduced the deciliating effect of KBrO3 on RPTEC/TERT1 cells (Fig. 4b iv).
We also examined gene expression levels of ZO-1 and Arl13b using TaqMan probe-based gene assays, Fig. 4c and 4d. As seen in both figures, KBrO3 at 5.5 mM significantly down-regulated the expression of both ZO-1 (p<0.05) and Arl13b (p<0.01) compared with untreated or curcumin-treated cells. However, the co-treatment of 25µM curcumin with KBrO3 significantly (p<0.01) reversed the negative effect of KBrO3 on ZO-1 and Arl13b gene expression.
4. Discussion
Potassium bromate, KBrO3, a salt of the bromate ion, is a nephrotoxic and neurotoxic agent in humans and a proven carcinogen in animals [29, 30]. KBrO3 is considered a possible human carcinogen (IIb) according to the International Agency for Research on Cancer (IARC) [31]. It was used as a model of cancers in many in vivo studies [4, 32-34].
The cytotoxic effects of KBrO3 were previously assessed by measuring the activity of the LDH enzyme. Akanji et al. found that after administration of a single dose of KBrO3, LDH activity was significantly increased in rat renal and intestinal tissues compared to other tissues [35]. In addition, Ahmed et al. reported that the release of LDH from KBrO3- treated cells was linked to KBrO3-induced oxidative stress [36]. In our model, we found that the co-treatment of KBrO3 with curcumin resulted in decreased LDH release which reflects the protective potential of curcumin against KBrO3-induced cytotoxicity. Curcumin has antioxidant, anti-inflammatory, and free radical scavenging activity. Its structure provides two methoxyphenyl groups and an enol form of β-diketone which illicit typical radical trapping activity [37].
In this study, treating human renal RPTEC/TERT1 cells with IC10 or IC50 concentrations of KBrO3 caused an increase of H2O2 and 8-OHdG levels suggesting that the induction of oxidative stress is one of the mechanisms by which KBrO3 induces its toxic and carcinogenic effects. We measured H2O2 levels as it is more stable than other oxidative species; also, H2O2 represents the precursor molecule of the hydroxyl radical that directly targets DNA and forms DNA adducts [38]. The co-treatment of KBrO3 with curcumin suppressed KBrO3 induced elevation of H2O2 and 8-OHdG levels at both toxic and subtoxic concentrations. Our findings are in line with a number of previous studies [39-42]. 8-OHdG is a biomarker for oxidative stress and carcinogenicity [43, 44] and has been proven to be a factor in initiating and promoting the process of carcinogenesis [45]. It has been reported that reactive species such as Reactive Oxygen Species (ROS) can attack nucleotide pools such as dGTP forming 8-OHdG. The adduct formed can bind adenine and cytosine nucleotide bases causing A:G mismatch. If the mismatch is irreparable, G:C to A:T transversion, a mutation will form. This mutation is mainly detected in many proto-oncogenes and tumor suppressor genes. In addition, the formation of DNA adducts such as 8-OHdG can cause steric hindrance which affect the fidelity of DNA replication [46]. 8-OHdG is considered as an important biomarker to measure the extent of DNA damage following the exposure to cancer initiating agents. Furthermore, it is also considered as a cofactor of cancer initiation and promotion [47].
Curcumin's chemopreventive potential has been proven by many in vitro and in vivo studies. Much research has shown that curcumin can efficiently protect cells from H2O2 -induced oxidative cell injury [38, 48]. Due to its antioxidant potential, curcumin was shown to have the ability to reduce lipid peroxidation and DNA damage, while increasing the level of vitamin C, vitamin E, and total anti-oxidant capacity [49, 50]. Furthermore, curcumin has been shown to induce phase II metabolism while suppressing phase I metabolizing enzymes such as renal ornithine decarboxylase [51]. Because the catalase enzyme potentially detoxifies and decomposes H2O2 to H2O [52], the activation of catalase by curcumin is considered another effective way to counteract oxidative stress. In this study, KBrO3 was shown to suppress the anti-oxidant catalase enzyme which represents one mechanism by which KBrO3 increases oxidative stress in cells. Our finding is in agreement with a previous study [53]. Interestingly, curcumin effectively reversed KBrO3 induced catalase suppression, which suggests that this may be an important mechanism by which curcumin mediates its chemopreventive effects. Taken together, we can conclude that curcumin blocked the carcinogenic potential of KBrO3 by increasing catalase enzyme activity thus reducing H2O2 and 8-OHdG levels.
Previous studies have shown that oxidative DNA damage causes activation of many inflammatory genes which creates a positive feedback loop leading to increased DNA damage, thus promoting cellular transformation and tumor progression [54-56]. Therefore to determine the role of inflammatory genes in our model, we measured a total of 192 target genes following the treatment of RPTEC/TERT1 cells with a subtoxic concentration of KBrO3 and compared the dysregulation status of the genes with the congruent genes in a human renal cancerous ACHN cell line. We found that CTGF was the most overexpressed gene following KBrO3 treatment and the third most overexpressed gene in the cancerous ACHN cell line. To our knowledge, this is the first study to provide evidence of the increased expression of CTGF following KBrO3 treatment at both transcriptional and translational levels. There is abundant evidence from previous studies showing that CTGF can be overexpressed by oxidative stress conditions [57-60], and it has also been shown that CTGF is up-regulated in many cancers [61-64] including renal cell carcinomas [65]. Taken together, we propose that the carcinogenic potential of KBrO3 might be through DNA adduct formation and the dysregulation of several inflammatory-regulating genes including CTGF. We also compared the potential CTGF repressor activity of curcumin with silymarin, another chemopreventive agent with a well-known CTGF repressor activity [27, 66-68]. Co-treatment of curcumin with KBrO3 significantly reduced the expression of CTGF at both RNA and protein levels. Our results are consistent with Zheng and Chen who found that curcumin reduced CTGF overexpression by interfering with upstream TGF-β signaling and TGF-β receptor activation. In addition to this, curcumin was found to induce GSH antioxidant synthesis and peroxisome proliferator-activated receptor (PPAR)-γ activation. Furthermore, curcumin was shown to modulate extracellular matrix gene expression such as α1(I)-collagen fibronectin and α-smooth muscle actin (α-SMA) by interrupting TGF-β signaling including its downstream effector, CTGF [69]. Curcumin halted the CTGF pathway via inhibition of ERK, P38 MAPK, and NF-KB pathways [70-72].
We have also shown that treating human renal RPTEC/TERT1 cells with KBrO3 negatively affected ciliary and junctional protein expression. KBrO3 caused a reduction in the number of ciliated cells and disturbed cellular borders. Our group has previously reported that KBrO3 induced RPTEC/TERT1 ciliary loss was accompanied by an increased proportion of cells at G2/S phase, and thus caused activation of cell cycle progression and proliferation [23]. We have found that curcumin protected RPTEC/TERT1 cells against KBrO3 induced deciliation. Another study showed that curcumin exerted an anti-proliferative effect via inhibition of cell cycle regulators of hepatic cells when they were exposed to diethylnitrosamine, a genotoxic carcinogen [73]. Blackmore et al. found that curcumin targeted colorectal cancer cells by arresting the G2/M phase of the cell cycle through disruption of microtubular orientation and assembly, as well as chromosomal condensation and congression [74]. Taken together, these studies show that KBrO3 ’s deciliating effect caused loss of cellular differentiation and promoted cellular proliferation. Strikingly, such negative effects of KBrO3 were found to be significantly minimized by co-treatment with curcumin in our system.
We have also shown that treating cells with a subtoxic concentration of KBrO3 caused inhibition of IL-1 and TRAF3 gene expression. However the impact of KBrO3 was reduced when treated in combination with curcumin.. Il-1 and TRAF3 were selected for further analysis due to their direct link with apoptosis-mediated cell death [75, 76]. Furthermore, it has been found that IL-1 can regulate ciliary length. For instance, a study by Wann and Knight showed that ciliary length increased by approximately 50% following only 3 hours of exposure to IL-1. The mechanism of ciliary elongation is through a protein kinase A (PKA) dependent mechanism [77]. Therefore, by inhibiting IL-1 gene expression, KBrO3 can counteract apoptosis and induce cellular dedifferentiation. TRAF3 is a cytoplasmic component with E3 ubiquitin ligase activity; following the binding to CD40, TRAF3 activates B-lymphocyte cells. It has been found that mice homozygous for a null allele of TRAF3 develop B-cell lymphoma. It also participates in the inhibition of NF-KB signaling pathway, namely interacting with Act 1 in cancer cells. Cells with mutant TRAF3 become cancerous due to the activation of NF-KB [78]. Generally, TRAF3 controls an alternative pathway of NF-KB activation with no effects on the classical pathway. TRAF3 negatively regulates NF-κB inducing kinase (NIK) levels, therefore the mutation of TRAF3 genes or activation of receptor-mediated proteasomal degradation is associated with the accumulation and auto-phosphorylation of NIK and activation of IKKα, which leads to the activation of NF-κB [79]. TRAF3 is sequestered to the cell cytoskeleton via TRAF3 interacting protein 1 (TRAF3ip1) which is localized to the cilium and is necessary for ciliogenesis. TRAF3ip1 mutant cells have been shown to be incapable of generating a primary cilium [80].
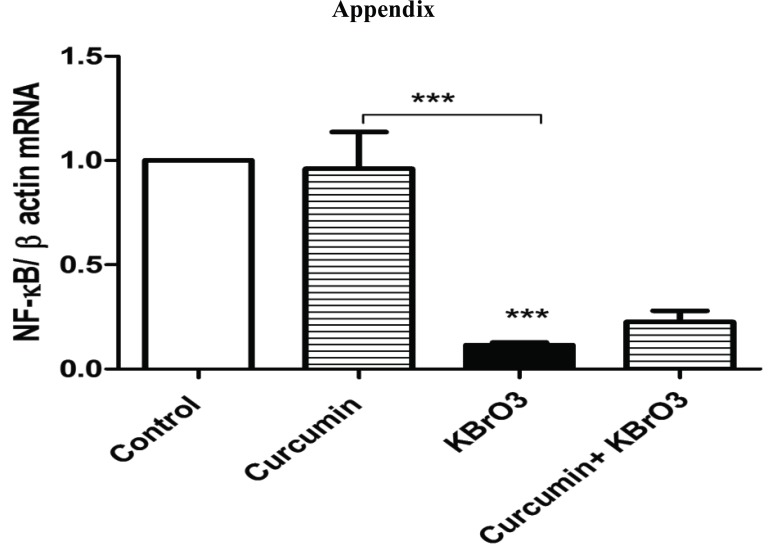
We have shown that KBrO3 induced downregulation of NF-KB gene expression at the mRNA level. However, no rescue effect was observed of curcumin against KBrO3-induced NF-KB downregulation (supplementary, Fig. 1).
A study by Wann et al. suggested that primary cilia are considered an important influence on NF-KB activation, thus loss of cilia was associated with deregulation of NF-KB activation [81]. Furthermore, Sinha et al. found that TRAF3 can participate in NF-KB activation through the x-linked ectodermal dysplasia receptor [82]. Therefore downregulation of TRAF3, NF-KB, and IL-1 gene expression and disruption of cilia by KBrO3 might be important mechanisms by which to counteract apoptosis and induce cellular dedifferentiation in carcinogenesis. However, further studies are required to investigate the effect of ciliary loss on the expression of inflammatory genes.
A significant number of publications have described curcumin’s anti-inflammatory potential as an important property to curtail the progression of several diseases including cancers. Curcumin shows potent anti-inflammatory potential due to its direct regulatory effects on several transcription factors such as STAT, MAPK, and NF-KB, interleukins (ILs) such as TNF-a, IL-1,2,6,8 and 12. Furthermore, it has shown to inhibit lipoxygenase, cyclooxygenase-2 (COX-2), and inducible nitric oxide synthase (iNOS) enzymes activities [83-85].
Despite the broad spectrum of curcumin’s health benefits and the wide range of biological significance, its clinical applications are potentially limited due to poor bioavailability. The slight aqueous solubility, instability, and photo degradation of curcumin represent major obstacles that limit its use in different therapeutic applications. Therefore, more than 1500 papers had been published by 2015 to find suitable solutions for such fundamental dilemmas in the clinical use of curcumin [86]. Perhaps, the incorporation of curcumin in a phospholipid system to form a liposomal-curcumin complex is one of the successful trials to improve curcumin’s bioavailability. Conjugation of liposomal curcumin with different molecules such as polyethylene glycol and folic acid has been reported to highly extend the biological half-life and targetability of the liposomal curcumin. Therefore, a substantial improvement of both pharmacokinetics and pharmacodynamics of curcumin molecules was recorded following the administration of liposomal curcumin in vivo [87]. Other strategies include induction of structural modifications to improve curcumin;s in vivo stability and effectiveness. For instance, a recent study has shown that dimethoxy curcumin has a unique anti-tumor activity due to its ability to suppress the transcription factor activator protein-1 (AP-1) and induce degradation of androgen receptors (AR). At the same time, dimethoxy curcumin showed a high metabolic stability compared to curcumin itself [88]. While another study found that a modification in curcumin’s aromatic ring resulted in the formation of 10 different curcumin analogues with more potent in vitro and in vivo anti-tumor activities than curcumin [89]. In this study we provide significant evidence that the food additive KBrO3 induced carcinogenic alteration via induction of ROS and activation of several oncogenic genes, counteracting tumor suppressor genes and favouring cell dedifferentiation. To our knowledge, this is the first study to identify the dysregulated genes of KBrO3-treated normal kidney cells that might be implicated in KBrO3-induced carcinogenesis via compar them with the genes of renal cancerous cells. Furthermore, this is the first study to report another protective mechanism of curcumin via inhibition of KBrO3-induced cell dedifferentiation.
Conclusion
In conclusion, the molecular mechanisms of curcumin chemoprevention have been investigated and are due in particular to its potential to block KBrO3 induced intracellular oxidative stress, DNA damage, and the dysregulation of signaling hubs that control inflammation, apoptosis, and cell differentiation which contribute to the pathogenesis and progression of KBrO3-induced renal cancer. Although challenges such as poor absorption and rapid elimination represent the major roadblocks in curcumin’s clinical applications, research is still ongoing to tackle these problems. However, the future seems bright particularly with the development of new curcumin analogues that are more potent, with higher bioavailability. Therefore, curcumin has considerable scope as a novel cancer prevention agent, due to its pleiotropic actions and in particular its cyto-protective and chemopreventive potential against chemical carcinogenesis.
Ethics Approval and Consent to Participate
Not applicable.
Human and Animal Rights
No Animals/Humans were used for studies that are base of this research.
Exposing RPTEC/TERT1 to5.5mM KBrO3 for 24h caused a significant downregulation of NF-KB gene expression compared with control. The combination of curcumin with KBrO3 slightly but non-significantly reversed the negative effect of KBrO3 on NF-KB gene expression. The data represents three independent experiments.
Fig. (S1).
Effect of KBrO3 on NF-KB gene expression.
Table S1.
Symbols and names of 187 inflammation-associated genes screened in gene expression array.
| Gene Symbols | Gene Name |
|---|---|
| ACTB | Beta actin |
| ADRB2 | ADRB2 adrenoceptor beta 2(ADRB2) Homo sapiens |
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 |
| BCAM | BCAM basal cell adhesion molecule |
| BCL3 | BCL3 B-cell CLL/lymphoma 3(BCL3) Homo sapiens |
| BCL6 | BCL6 B-cell CLL/lymphoma 6(BCL6) Homo sapiens |
| BCMA | B-Cell Maturation Protein |
| C3 | Complement component 3 |
| C5 | C5 complement C5(C5) Homo sapiens |
| CASP1 | CASP1 caspase 1(CASP1) Homo sapiens |
| CCL11 | CCL11 C-C motif chemokine ligand 11(CCL11) Homo sapiens |
| CCL13 | CCL13 C-C motif chemokine ligand 13(CCL13) Homo sapiens |
| CCL15 | Chemokine (C-C motif) ligand 15 |
| CCL16 | Chemokine (C-C motif) ligand 16 |
| CCL17 | Chemokine (C-C motif) ligand 17 |
| CCL18 | CCL18 C-C motif chemokine ligand 18(CCL18) Homo sapiens |
| CCL19 | CCL19 C-C motif chemokine ligand 19(CCL19) Homo sapiens |
| CCL2 | CCL2 C-C motif chemokine ligand 2(CCL2) Homo sapiens |
| CCL20 | Chemokine (C-C motif) ligand 20 |
| Gene Symbols | Gene Name |
| CCL21 | CCL21 C-C motif chemokine ligand 21(CCL21) Homo sapiens |
| CCL22 | CCL22 C-C motif chemokine ligand 22(CCL22) Homo sapiens |
| CCL23 | CCL23 C-C motif chemokine ligand 23(CCL23) Homo sapiens |
| CCL25 | Chemokine (C-C motif) ligand 25 |
| CCL3 | CCL3 C-C motif chemokine ligand 3(CCL3) Homo sapiens |
| CCL4 | CCL4 C-C motif chemokine ligand 4(CCL4) Homo sapiens |
| CCL5 | CCL5 C-C motif chemokine ligand 5(CCL5) Homo sapiens |
| CCL7 | CCL7 C-C motif chemokine ligand 7(CCL7) Homo sapiens |
| CCL8 | CCL8 C-C motif chemokine ligand 8(CCL8) Homo sapiens |
| CCR1 | Chemokine (C-C motif) receptor 1 |
| CCR10 | CCR10 C-C motif chemokine receptor 10(CCR10) Homo sapiens |
| CCR2 | CCR2 C-C motif chemokine receptor 2(CCR2) Homo sapiens |
| CCR3 | CCR3 C-C motif chemokine receptor 3(CCR3) Homo sapiens |
| CCR4 | Chemokine (C-C motif) receptor 4 |
| CCR5 | Chemokine (C-C motif) receptor 5 |
| CCR6 | Chemokine (C-C motif) receptor 6 |
| CCR7 | CCR7 C-C motif chemokine receptor 7(CCR7) Homo sapiens |
| CCR8 | Chemokine (C-C motif) receptor 8 |
| CCR9 | CCR9 C-C motif chemokine receptor 9(CCR9) Homo sapiens |
| CD14 | CD14 molecule |
| CD4 | CD4 CD4 molecule(CD4) Homo sapiens |
| CD40 | CD40 molecule, TNF receptor superfamily member 5 |
| CD83 | CD83 molecule |
| CD88 | Complement C5a receptor 1 |
| CD8A | CD8A CD8a molecule(CD8A) Homo sapiens |
| CD8B | CD8B CD8b molecule(CD8B) Homo sapiens |
| CLEC7A | C-type lectin domain family 7, member A |
| CREB1 | cAMP responsive element binding protein 1 |
| CREB1 | CREB1 cAMP responsive element binding protein 1(CREB1) |
| CRP | C-reactive protein, pentraxin-related |
| CSF1 | CSF1 colony stimulating factor 1(CSF1) Homo sapiens |
| CSF1R | CSF1R colony stimulating factor 1 receptor(CSF1R) Homo sapiens |
| CSF2 | CSF2 colony stimulating factor 2(CSF2) Homo sapiens |
| CTGF | CTGF connective tissue growth factor(CTGF) Homo sapiens |
| CXCL1 | CXCL1 C-X-C motif chemokine ligand 1(CXCL1) Homo sapiens |
| CXCL13 | CXCL13 C-X-C motif chemokine ligand 13(CXCL13) Homo sapiens |
| CXCL2 | CXCL2 C-X-C motif chemokine ligand 2(CXCL2) Homo sapiens |
| Gene Symbols | Gene Name |
| CXCL3 | CXCL3 C-X-C motif chemokine ligand 3(CXCL3) Homo sapiens |
| CXCL5 | CXCL5 C-X-C motif chemokine ligand 5(CXCL5) Homo sapiens |
| CYSLTR1 | Cysteinyl leukotriene receptor 1 |
| EDN1 | EDN1 endothelin 1(EDN1) Homo sapiens |
| EGF | EGF epidermal growth factor(EGF) Homo sapiens |
| EGFR | EGFR epidermal growth factor receptor(EGFR) Homo sapiens |
| EGR2 | EGR2 early growth response 2(EGR2) Homo sapiens |
| F2 | F2 coagulation factor II, thrombin(F2) Homo sapiens |
| FOS | FOS Fos proto-oncogene, AP-1 transcription factor subunit |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GSTP1 | GSTP1 glutathione S-transferase pi 1(GSTP1) Homo sapiens |
| GUSB | Glucuronidase, beta |
| HIF1A | HIF1A hypoxia inducible factor 1 alpha subunit(HIF1A) Homo sapiens |
| HLAA | Major histocompatibility complex, class I, A |
| HLAB | Major histocompatibility complex, class I, B [Homo sapiens |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 |
| HPRT1 | HPRT1 hypoxanthine phosphoribosyltransferase 1(HPRT1) |
| HRH3 | Histamine receptor H3 |
| ICAM1 | Intercellular adhesion molecule 1 |
| IFNG | Interferon gamma |
| IGFBP1 | IGFBP1 insulin like growth factor binding protein 1(IGFBP1) |
| IGFBP2 | IGFBP2 insulin like growth factor binding protein 2(IGFBP2) |
| IGFBP3 | IGFBP3 insulin like growth factor binding protein 3(IGFBP3) |
| IKBKB | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| IL10 | Interleukin 10 |
| IL12B | Interleukin 12b |
| IL15 | IL15 interleukin 15(IL15) Homo sapiens |
| IL17A | Interleukin 17A |
| IL1B | Interleukin 1, beta |
| IL1R1 | Interleukin 1 receptor, type I |
| IL1RN | Interleukin 1 receptor antagonist |
| IL2 | Interleukin 2 |
| IL23 | Interleukin 23 |
| IL24 | Interleukin 24(IL24) |
| IL2RA | Interleukin 2 receptor, alpha |
| IL4 | Interleukin 4 |
| IL5 | Interleukin 5 |
| Gene Symbols | Gene Name |
| IL6 | Interleukin 6 |
| IL6R | Interleukin 6 receptor |
| IL6ST | Interleukin 6 signal transducer |
| IL8 | Interleukin 8 |
| INSR | Insulin receptor |
| IRAK1 | Interleukin 1 receptor associated kinase 1 |
| IRAK2 | Interleukin 1 receptor associated kinase 2 |
| IRF1 | Interferon regulatory factor 1 |
| JAK2 | Janus kinase 2 |
| JUN | Jun proto-oncogene, AP-1 transcription factor subunit(JUN) |
| LCN2 | Lipocalin 2 |
| LTA | Lymphotoxin alpha |
| LTB4R2 | Leukotriene B4 receptor 2 |
| LTC4S | Leukotriene C4 synthase |
| MALT1 | Mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 |
| MAPK1 | Mitogen-activated protein kinase 1 |
| MAPK8 | Mitogen-activated protein kinase 8 |
| MMP1 | Matrix metallopeptidase 1 (interstitial collagenase) |
| MMP13 | Matrix metallopeptidase 13 (collagenase 3) |
| MMP2 | MMP2 matrix metallopeptidase 2(MMP2) Homo sapiens |
| MMP3 | MMP3 matrix metallopeptidase 3(MMP3) Homo sapiens |
| MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) |
| MTHFR | Methylenetetrahydrofolate reductase (NAD(P)H) |
| MUC5AC | MUC5AC mucin 5AC, oligomeric mucus/gel-forming(MUC5AC) |
| MYD88 | Myeloid differentiation primary response gene 88 |
| NFKB1 | NFKB1 nuclear factor kappa B subunit 1(NFKB1) Homo sapiens |
| NFKB1A | Nuclear factor kappa B 1 A |
| NFKB2 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| NOD2 | NOD2 nucleotide binding oligomerization domain containing 2(NOD2) |
| NOS2 | Nitric oxide synthase 2, inducible |
| NR2C2 | Nuclear receptor subfamily 2, group C, member 2 |
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| NR4A1 | Nuclear receptor subfamily 4, group A, member 1 |
| NR4A1 | Nuclear receptor subfamily 4 group A member 1 |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 |
| PAI1 | Plasminogen activator inhibitor-1 |
| Gene Symbols | Gene Name |
| PCK1 | PCK1 phosphoenolpyruvate carboxykinase 1 |
| PIM1 | PIM1 Pim-1 proto-oncogene, serine/threonine kinase |
| PIM2 | Pim-2 proto-oncogene, serine/threonine kinase |
| PIM3 | Pim-3 proto-oncogene, serine/threonine kinase |
| PLA2G2D | Phospholipase A2, group IID |
| PLAT | Plasminogen activator, tissue type |
| PLCB4 | Phospholipase C beta 4(PLCB4) Homo sapiens |
| PPARG | Peroxisome proliferator-activated receptor gamma |
| PSMD3 | Proteasome 26S subunit, non-ATPase 3 |
| PTGER1 | Prostaglandin E receptor 1 (subtype EP1) |
| PTGER2 | Prostaglandin E receptor 2 (subtype EP2) |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 |
| RARA | Retinoic acid receptor alpha |
| REL | v-rel avian reticuloendotheliosis viral oncogene homolog |
| RELA | v-rel avian reticuloendotheliosis viral oncogene homolog A |
| RELB | RELB proto-oncogene, NF-kB subunit |
| RELMB | Resistin-Like Molecule-beta |
| RETN | RETN resistin(RETN) Homo sapiens |
| RIPK1 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| SELE | Selectin E |
| SOCS1 | Suppressor of cytokine signaling 1 |
| SOCS3 | Suppressor of cytokine signaling 3 |
| SOD1 | SOD1 superoxide dismutase 1, soluble(SOD1) |
| SOD2 | SOD2 superoxide dismutase 2, mitochondrial(SOD2) |
| SOX9 | SRY (sex determining region Y)-box 9 |
| STAT1 | Signal transducer and activator of transcription 1(STAT1) |
| STAT3 | Signal transducer and activator of transcription 3 |
| TBP | TATA-box binding protein |
| TGFB1 | Transforming growth factor, beta 1 |
| TGFB1 | Transforming growth factor beta 1 |
| TGFB2 | Transforming growth factor beta 2 |
| TGFBR1 | TGFBR1 transforming growth factor beta receptor 1 |
| TIRAP | Toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| TIRAP | TIR domain containing adaptor protein |
| TLR1 | Toll-like receptor 1 |
| TLR2 | Toll-like receptor 2 |
| TLR3 | Toll-like receptor 3 |
| Gene Symbols | Gene Name |
| TLR4 | Toll-like receptor 4 |
| TLR6 | Toll-like receptor 6 |
| TLR7 | Toll-like receptor 7 |
| TLR8 | Toll-like receptor 8 |
| TNFA | Tumor necrosis factor a (TNF superfamily, member 2) |
| TNFRSF10A | TNF receptor superfamily member 10a |
| TNFRSF10B | Tumor necrosis factor receptor superfamily, member 10b |
| TNFRSF1A | Tumor necrosis factor receptor superfamily, member 1A |
| TNFSF18 | Tumor necrosis factor (ligand) superfamily, member 18 |
| TP53 | Tumor protein p53 |
| TRAF1 | TNF receptor-associated factor 1 |
| TRAF2 | TNF receptor-associated factor 2 |
| TRAF3 | TNF receptor-associated factor 3 |
| TRAF5 | TNF receptor-associated factor 5 |
| TRAF6 | TNF receptor-associated factor 6 |
| UBC | UBC ubiquitin C(UBC) |
| VCAM | Vascular cell adhesion molecule |
| VCAM1 | Vascular cell adhesion molecule 1 |
| VEGF | Vascular endothelial growth factor |
| VEGFA | Vascular endothelial growth factor A |
ACKNOWLEDGEMENTS
We are grateful to the Iraqi government (MHESR) for fully funding this study, and providing the financial support to undertake all experiments, and to Science Foundation Ireland for funding the UCD Centre for Toxicology, where this project was undertaken.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Radford R., Frain H., Ryan M.P., Slattery C., McMorrow T. Mechanisms of chemical carcinogenesis in the kidneys. Int. J. Mol. Sci. 2013;14(10):19416–19433. doi: 10.3390/ijms141019416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.2017 https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/cfrsearch.cfm?fr=136.110
- 3.Ishidate M., Yoshikawa K. 1979. [Google Scholar]
- 4.Kasai H., Nishimura S., Kurokawa Y., Hayashi Y. Oral administration of the renal carcinogen, potassium bromate, specifically produces 8-hydroxydeoxyguanosine in rat target organ DNA. Carcinogenesis. 1987;8(12):1959–1961. doi: 10.1093/carcin/8.12.1959. [DOI] [PubMed] [Google Scholar]
- 5.DeAngelo A.B., George M.H., Kilburn S.R., Moore T.M., Wolf D.C. Carcinogenicity of potassium bromate administered in the drinking water to male B6C3F1 mice and F344/N rats. Toxicol. Pathol. 1998;26(5):587–594. doi: 10.1177/019262339802600501. [DOI] [PubMed] [Google Scholar]
- 6.Shiao Y.H., Kamata S.I., Li L.M., et al. Mutations in the VHL gene from potassium bromate-induced rat clear cell renal tumors. Cancer Lett. 2002;187(1-2):207–214. doi: 10.1016/s0304-3835(02)00407-x. [DOI] [PubMed] [Google Scholar]
- 7.Ballmaier D., Epe B. Oxidative DNA damage induced by potassium bromate under cell-free conditions and in mammalian cells. Carcinogenesis. 1995;16(2):335–342. doi: 10.1093/carcin/16.2.335. [DOI] [PubMed] [Google Scholar]
- 8.Parsons J.L., Chipman J.K. The role of glutathione in DNA damage by potassium bromate in vitro. Mutagenesis. 2000;15(4):311–316. doi: 10.1093/mutage/15.4.311. [DOI] [PubMed] [Google Scholar]
- 9.Ballmaier D., Epe B. DNA damage by bromate: Mechanism and consequences. Toxicology. 2006;221(2-3):166–171. doi: 10.1016/j.tox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Bader H.L., Hsu T. Systemic VHL gene functions and the VHL disease. FEBS Lett. 2012;586(11):1562–1569. doi: 10.1016/j.febslet.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaud E.J., Yoder B.K. The Primary Cilium in Cell Signaling and Cancer. Cancer Res. 2006;66(13):6463–6467. doi: 10.1158/0008-5472.CAN-06-0462. [DOI] [PubMed] [Google Scholar]
- 12.Pan J., Seeger-Nukpezah T., Golemis E.A. The role of the cilium in normal and abnormal cell cycles: Emphasis on renal cystic pathologies. Cell. Mol. Life Sci. 2013;70(11):1849–1874. doi: 10.1007/s00018-012-1052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quarmby L.M., Parker J.D.K. Cilia and the cell cycle? J. Cell Biol. 2005;169(5):707–710. doi: 10.1083/jcb.200503053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schraml P., Frew I.J., Thoma C.R., et al. Sporadic clear cell renal cell carcinoma but not the papillary type is characterized by severely reduced frequency of primary cilia. Mod. Pathol. 2009;22(1):31–36. doi: 10.1038/modpathol.2008.132. [DOI] [PubMed] [Google Scholar]
- 15.Mehta R.G., Murillo G., Naithani R., Peng X. Cancer chemoprevention by natural products: how far have we come? Pharm. Res. 2010;27(6):950–961. doi: 10.1007/s11095-010-0085-y. [DOI] [PubMed] [Google Scholar]
- 16.Aggarwal B.B., Sundaram C., Malani N., Ichikawa H. Curcumin: the Indian solid gold. The molecular targets and therapeutic uses of curcumin in health and disease. Springer; 2007. pp. 1–75. [DOI] [PubMed] [Google Scholar]
- 17.Steward W.P., Brown K. Cancer chemoprevention: a rapidly evolving field. Br. J. Cancer. 2013;109(1):1–7. doi: 10.1038/bjc.2013.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chendil D., Ranga R.S., Meigooni D., Sathishkumar S., Ahmed M.M. Curcumin confers radiosensitizing effect in prostate cancer cell line PC-3. Oncogene. 2004;23(8):1599–1607. doi: 10.1038/sj.onc.1207284. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S.C., Patchva S., Koh W., Aggarwal B.B. Discovery of curcumin, a component of golden spice, and its miraculous biological activities. Clin. Exp. Pharmacol. Physiol. 2012;39(3):283–299. doi: 10.1111/j.1440-1681.2011.05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzzan B., Benamouzig R. Is Curcumin a Chemopreventive Agent for Colorectal Cancer? Curr. Colorectal Cancer Rep. 2016;12(1):35–41. [Google Scholar]
- 21.Momtazi A.A., Shahabipour F., Khatibi S., Johnston T.P., Pirro M., Sahebkar A. Curcumin as a MicroRNA Regulator in Cancer: A Review. Rev. Physiol. Biochem. Pharmacol. 2016;171:1–38. doi: 10.1007/112_2016_3. [DOI] [PubMed] [Google Scholar]
- 22.Banikazemi Z., Haji H.A., Mohammadi M., et al. Diet and cancer prevention: Dietary compounds, dietary MicroRNAs, and dietary exosomes. J. Cell. Biochem. 2018;119(1):185–196. doi: 10.1002/jcb.26244. [DOI] [PubMed] [Google Scholar]
- 23.Radford R., Slattery C., Jennings P., et al. Carcinogens induce loss of the primary cilium in human renal proximal tubular epithelial cells independently of effects on the cell cycle. Am. J. Physiol. Renal Physiol. 2012;302(8):F905–F916. doi: 10.1152/ajprenal.00427.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieser M., Stadler G., Jennings P., et al. hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Renal Physiol. 2008;295(5):F1365–F1375. doi: 10.1152/ajprenal.90405.2008. [DOI] [PubMed] [Google Scholar]
- 25.Robertson R., Guihéneuf F., Bahar B., et al. The Anti-Inflammatory Effect of Algae-Derived Lipid Extracts on Lipopolysaccharide (LPS)-Stimulated Human THP-1 Macrophages. Mar. Drugs. 2015;13(8):5402. doi: 10.3390/md13085402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchmann K., Pedersen L., Glamann J. Humoral immune response of European eel Anguilla anguilla to a major antigen in Anguillicola crassus (Nematoda). Dis. Aquat. Organ. 1991;12(1):55–57. [Google Scholar]
- 27.Tzeng J.I., Chen M.F., Chung H.H., Cheng J.T. Silymarin decreases connective tissue growth factor to improve liver fibrosis in rats treated with carbon tetrachloride. Phytother. Res. 2013;27(7):1023–1028. doi: 10.1002/ptr.4829. [DOI] [PubMed] [Google Scholar]
- 28.Duval F., Moreno-Cuevas J.E., González-Garza M.T., Rodríguez-Montalvo C., Cruz-Vega D.E. Protective mechanisms of medicinal plants targeting hepatic stellate cell activation and extracellular matrix deposition in liver fibrosis. Chin. Med. 2014;9(1):27. doi: 10.1186/s13020-014-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.IARC 1986.
- 30.Geter D.R., Ward W.O., Knapp G.W., et al. Kidney Toxicogenomics of Chronic Potassium Bromate Exposure in F344 Male Rats. Transl. Oncogenomics. 2006;1:33–52. [PMC free article] [PubMed] [Google Scholar]
- 31.IARC Evaluation of carcinogenic risks to humans. 1987.
- 32.Kurokawa Y., Takayama S., Konishi Y., et al. Long-term in vivo carcinogenicity tests of potassium bromate, sodium hypochlorite, and sodium chlorite conducted in Japan. Environ. Health Perspect. 1986;69:221–235. doi: 10.1289/ehp.8669221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa Y., Aoki S., Matsushima Y., Takamura N., Imazawa T., Hayashi Y. Dose-Response Studies on the Carcinogenicity of Potassium Bromate in F344 Rats After Long-Term Oral Administration. J. Natl. Cancer Inst. 1986;77(4):977–982. [PubMed] [Google Scholar]
- 34.Sakamoto K., Tominaga Y., Yamauchi K., et al. MUTYH-Null Mice Are Susceptible to Spontaneous and Oxidative Stress–Induced Intestinal Tumorigenesis. Cancer Res. 2007;67(14):6599–6604. doi: 10.1158/0008-5472.CAN-06-4802. [DOI] [PubMed] [Google Scholar]
- 35.Akanji M., Nafiu M., Yakubu M. Enzyme activities and histopathology of selected tissues in rats treated with potassium bromate. Afr. J. Biomed. Res. 2008;11(1) [Google Scholar]
- 36.Ahmad M.K., Khan A.A., Ali S.N., Mahmood R. Chemoprotective effect of taurine on potassium bromate-induced DNA damage, DNA-protein cross-linking and oxidative stress in rat intestine. PLoS One. 2015;10(3):e0119137. doi: 10.1371/journal.pone.0119137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuda T., Maekawa T., Hidaka K., Bando H., Takeda Y., Yamaguchi H. Chemical Studies on Antioxidant Mechanism of Curcumin: Analysis of Oxidative Coupling Products from Curcumin and Linoleate. J. Agric. Food Chem. 2001;49(5):2539–2547. doi: 10.1021/jf001442x. [DOI] [PubMed] [Google Scholar]
- 38.Mahakunakorn P., Tohda M., Murakami Y., Matsumoto K., Watanabe H., Vajaragupta O. Cytoprotective and cytotoxic effects of curcumin: dual action on H2O2-induced oxidative cell damage in NG108-15 cells. Biol. Pharm. Bull. 2003;26(5):725–728. doi: 10.1248/bpb.26.725. [DOI] [PubMed] [Google Scholar]
- 39.Kurokawa Y., Maekawa A., Takahashi M., Hayashi Y. Toxicity and carcinogenicity of potassium bromate--a new renal carcinogen. Environ. Health Perspect. 1990;87:309–335. doi: 10.1289/ehp.9087309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giri U., Iqbal M., Athar M. Potassium bromate (KBrO3) induces renal proliferative response and damage by elaborating oxidative stress. Cancer Lett. 1999;135(2):181–188. doi: 10.1016/s0304-3835(98)00290-0. [DOI] [PubMed] [Google Scholar]
- 41.Khan R.A., Khan M.R., Sahreen S., et al. Potassium bromate (KBrO3) induced nephrotoxicity: protective effects of n-hexane extract of Sonchus asper. J. Med. Plants Res. 2011;5(25):6017–6023. [Google Scholar]
- 42.Khan R.A., Khan M.R., Sahreen S. Protective effects of rutin against potassium bromate induced nephrotoxicity in rats. BMC Complement. Altern. Med. 2012;12(1):204. doi: 10.1186/1472-6882-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasai H. Analysis of a form of oxidative DNA damage, 8-hydroxy-2′-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat. Res. 1997;387(3):147–163. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- 44.Nakae D., Umemura T., Kurokawa Y. Reactive oxygen and nitrogen oxide species-induced stress, a major intrinsic factor involved in carcinogenic processes and a possible target for cancer prevention. Asian Pac. J. Cancer Prev. 2002;3:313–318. [PubMed] [Google Scholar]
- 45.Valavanidis A., Vlachogianni T., Fiotakis C. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): A Critical Biomarker of Oxidative Stress and Carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2009;27(2):120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 46.Klaunig J.E., Kamendulis L.M., Hocevar B.A. Oxidative stress and oxidative damage in carcinogenesis. Toxicol. Pathol. 2010;38(1):96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 47.Valavanidis A., Vlachogianni T. Fau - Fiotakis C, Fiotakis C. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. Electronic; 2009. pp. 1532–4095. [DOI] [PubMed] [Google Scholar]
- 48.Cohly H.H.P., Taylor A., Angel M.F., Salahudeen A.K. Effect of Turmeric, Turmerin and Curcumin on H2O2-Induced Renal Epithelial (LLC-PK1) Cell Injury. Free Radic. Biol. Med. 1998;24(1):49–54. doi: 10.1016/s0891-5849(97)00140-8. [DOI] [PubMed] [Google Scholar]
- 49.Rai B., Kaur J., Jacobs R., Singh J. Possible action mechanism for curcumin in pre-cancerous lesions based on serum and salivary markers of oxidative stress. J. Oral Sci. 2010;52(2):251–256. doi: 10.2334/josnusd.52.251. [DOI] [PubMed] [Google Scholar]
- 50.Ciftci G., Aksoy A., Cenesiz S., et al. Therapeutic role of curcumin in oxidative DNA damage caused by formaldehyde. Microsc. Res. Tech. 2015;78(5):391–395. doi: 10.1002/jemt.22485. [DOI] [PubMed] [Google Scholar]
- 51.Iqbal M., Okazaki Y., Okada S. Curcumin attenuates oxidative damage in animals treated with a renal carcinogen, ferric nitrilotriacetate (Fe-NTA): implications for cancer prevention. Mol. Cell. Biochem. 2009;324(1):157–164. doi: 10.1007/s11010-008-9994-z. [DOI] [PubMed] [Google Scholar]
- 52.Petrucci R.H. General Chemistry: Principles & Modern Applications. 9th ed. Prentice Hall; 2007. [Google Scholar]
- 53.Watanabe S., Tajima Y., Yamaguchi T., Fukui T. Potassium bromate-induced hyperuricemia stimulates acute kidney damage and oxidative stress. J. Health Sci. 2004;50(6):647–653. [Google Scholar]
- 54.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reuter S., Gupta S.C., Chaturvedi M.M., Aggarwal B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halliday GM. 2005.
- 57.Ohshiro Y., Ma R.C., Yasuda Y., et al. Reduction of diabetes-induced oxidative stress, fibrotic cytokine expression, and renal dysfunction in protein kinase Cβ–null mice. Diabetes. 2006;55(11):3112–3120. doi: 10.2337/db06-0895. [DOI] [PubMed] [Google Scholar]
- 58.Park S.K., Kim J., Seomun Y., et al. Hydrogen peroxide is a novel inducer of connective tissue growth factor. Biochem. Biophys. Res. Commun. 2001;284(4):966–971. doi: 10.1006/bbrc.2001.5058. [DOI] [PubMed] [Google Scholar]
- 59.Branchetti E., Poggio P., Sainger R., et al. Oxidative stress modulates vascular smooth muscle cell phenotype via CTGF in thoracic aortic aneurysm. Cardiovasc. Res. 2013;100(2):316–324. doi: 10.1093/cvr/cvt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuda S., Gomi F., Katayama T., Koyama Y., Tohyama M., Tano Y. Induction of connective tissue growth factor in retinal pigment epithelium cells by oxidative stress. Jpn. J. Ophthalmol. 2006;50(3):229–234. doi: 10.1007/s10384-005-0317-6. [DOI] [PubMed] [Google Scholar]
- 61.Wenger C., Ellenrieder V., Alber B., et al. Expression and differential regulation of connective tissue growth factor in pancreatic cancer cells. Oncogene. 1999;18(4):1073–1080. doi: 10.1038/sj.onc.1202395. [DOI] [PubMed] [Google Scholar]
- 62.Jiang W.G., Watkins G., Fodstad O., Douglas-Jones A., Mokbel K., Mansel R.E. Differential expression of the CCN family members Cyr61, CTGF and Nov in human breast cancer. Endocr. Relat. Cancer. 2004;11(4):781–791. doi: 10.1677/erc.1.00825. [DOI] [PubMed] [Google Scholar]
- 63.Chen P-P., Li W-J., Wang Y., et al. Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS One. 2007;2(6):e534. doi: 10.1371/journal.pone.0000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu C-Y., Chang C-C., Prakash E., Kuo M-L. Connective tissue growth factor (CTGF) and cancer progression. J. Biomed. Sci. 2008;15(6):675–685. doi: 10.1007/s11373-008-9264-9. [DOI] [PubMed] [Google Scholar]
- 65.Chintalapudi M.R., Markiewicz M., Kose N., et al. Cyr61/CCN1 and CTGF/CCN2 mediate the proangiogenic activity of VHL-mutant renal carcinoma cells. Carcinogenesis. 2008;29(4):696–703. doi: 10.1093/carcin/bgn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.ZHAO Y-z 2010.
- 67.LIU Z-g 2012.
- 68.Yang C.H., Ting W.J., Shen C.Y., et al. SHSST-cyclodextrin complex inhibits TGF-β/Smad3/CTGF to a greater extent than silymarin in a rat model of carbon tetrachloride-induced liver injury. Mol. Med. Rep. 2015;12(4):6053–6059. doi: 10.3892/mmr.2015.4190. [DOI] [PubMed] [Google Scholar]
- 69.Zheng S., Chen A. Curcumin suppresses the expression of extracellular matrix genes in activated hepatic stellate cells by inhibiting gene expression of connective tissue growth factor. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290(5):G883–G93. doi: 10.1152/ajpgi.00450.2005. [DOI] [PubMed] [Google Scholar]
- 70.O’Connell M., Rushworth S. Curcumin: potential for hepatic fibrosis therapy? Br. J. Pharmacol. 2008;153(3):403–405. doi: 10.1038/sj.bjp.0707580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen A., Zheng S. Curcumin inhibits connective tissue growth factor gene expression in activated hepatic stellate cells in vitro by blocking NF-κB and ERK signalling. Br. J. Pharmacol. 2008;153(3):557–567. doi: 10.1038/sj.bjp.0707542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng Y-T., Chen H-M., Cheng S-J., Chiang C-P., Kuo M.Y-P. Arecoline-stimulated connective tissue growth factor production in human buccal mucosal fibroblasts: Modulation by curcumin. Oral Oncol. 2009;45(9):e99–e105. doi: 10.1016/j.oraloncology.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 73.Chuang S-E., Cheng A-L., Lin J-K., Kuo M-L. Inhibition by curcumin of diethylnitrosamine-induced hepatic hyperplasia, inflammation, cellular gene products and cell-cycle-related proteins in rats. Food Chem. Toxicol. 2000;38(11):991–995. doi: 10.1016/s0278-6915(00)00101-0. [DOI] [PubMed] [Google Scholar]
- 74.Blakemore L.M., Boes C., Cordell R., Manson M.M. Curcumin-induced mitotic arrest is characterized by spindle abnormalities, defects in chromosomal congression and DNA damage. Carcinogenesis. 2012 doi: 10.1093/carcin/bgs345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Georgopoulos N.T., Steele L.P., Thomson M.J., Selby P.J., Southgate J., Trejdosiewicz L.K. A novel mechanism of CD40-induced apoptosis of carcinoma cells involving TRAF3 and JNK/AP-1 activation. Cell Death Differ. 2006;13(10):1789–1801. doi: 10.1038/sj.cdd.4401859. [DOI] [PubMed] [Google Scholar]
- 76.England H., Summersgill H.R., Edye M.E., Rothwell N.J., Brough D. Release of interleukin-1alpha or interleukin-1beta depends on mechanism of cell death. J. Biol. Chem. 2014;289(23):15942–15950. doi: 10.1074/jbc.M114.557561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wann A.K., Knight M.M. Primary cilia elongation in response to interleukin-1 mediates the inflammatory response. Cell. Mol. Life Sci. 2012;69(17):2967–2977. doi: 10.1007/s00018-012-0980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edwards S.K., Moore C.R., Liu Y., Grewal S., Covey L.R., Xie P. N-benzyladriamycin-14-valerate (AD 198) exhibits potent anti-tumor activity on TRAF3-deficient mouse B lymphoma and human multiple myeloma. BMC Cancer. 2013;13(1):481. doi: 10.1186/1471-2407-13-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hacker H., Tseng P.H., Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011;11(7):457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 80.Berbari N.F., Kin N.W., Sharma N., Michaud E.J., Kesterson R.A., Yoder B.K. Mutations in Traf3ip1 reveal defects in ciliogenesis, embryonic development, and altered cell size regulation. Dev. Biol. 2011;360(1):66–76. doi: 10.1016/j.ydbio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wann A.K., Chapple J.P., Knight M.M. The primary cilium influences interleukin-1beta-induced NFkappaB signalling by regulating IKK activity. Cell. Signal. 2014;26(8):1735–1742. doi: 10.1016/j.cellsig.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sinha S.K., Zachariah S., Quinones H.I., Shindo M., Chaudhary P.M. Role of TRAF3 and -6 in the activation of the NF-kappa B and JNK pathways by X-linked ectodermal dysplasia receptor. J. Biol. Chem. 2002;277(47):44953–44961. doi: 10.1074/jbc.M207923200. [DOI] [PubMed] [Google Scholar]
- 83.Deguchi A. Curcumin targets in inflammation and cancer. Endocr. Metab. Immune Disord. Drug Targets. 2015;15(2):88–96. doi: 10.2174/1871530315666150316120458. [DOI] [PubMed] [Google Scholar]
- 84.Karimian M.S., Pirro M., Majeed M., Sahebkar A. Curcumin as a natural regulator of monocyte chemoattractant protein-1. Cytokine Growth Factor Rev. 2017;33(Suppl. C):55–63. doi: 10.1016/j.cytogfr.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 85.Shehzad A., Qureshi M., Anwar M.N., Lee Y.S. Multifunctional Curcumin Mediate Multitherapeutic Effects. J. Food Sci. 2017;82(9):2006–2015. doi: 10.1111/1750-3841.13793. [DOI] [PubMed] [Google Scholar]
- 86.Kasi P.D., Tamilselvam R., Skalicka-Woźniak K., et al. Molecular targets of curcumin for cancer therapy: an updated review. Tumour Biol. 2016;37(10):13017–13028. doi: 10.1007/s13277-016-5183-y. [DOI] [PubMed] [Google Scholar]
- 87.Feng T., Wei Y., Lee R.J., Zhao L. Liposomal curcumin and its application in cancer. Int. J. Nanomedicine. 2017;12:6027–6044. doi: 10.2147/IJN.S132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teymouri M., Barati N., Pirro M., Sahebkar A. Biological and pharmacological evaluation of dimethoxycurcumin: A metabolically stable curcumin analogue with a promising therapeutic potential. J. Cell. Physiol. 2018;233(1):124–140. doi: 10.1002/jcp.25749. [DOI] [PubMed] [Google Scholar]
- 89.Zhang L., Zong H., Lu H., Gong J., Ma F. Discovery of novel anti-tumor curcumin analogues from the optimization of curcumin scaffold. Med. Chem. Res. 2017;26(10):2468–2476. [Google Scholar]