The rapid spread of multidrug-resistant Gram-negative organisms constitutes one of the greatest challenges to global health. While Gram-negative organisms have developed several mechanisms to avert the bactericidal effects of commonly prescribed antibiotic agents, the increasing prevalence of carbapenemase-producing organisms (CPO) is particularly concerning given the rapid spread of mobile genetic elements containing carbapenemase genes, the limited treatment options for infections caused by these organisms, and the high mortality rates associated with CPO infections.
KEYWORDS: carbapenemase-producing Enterobacteriaceae, modified Hodge test, Carba NP test, modified carbapenem inactivation method, lateral flow assay, Carba NP, carbapenemase, carbapenemase-producing organism, lateral flow antigen, phenotypic
ABSTRACT
The rapid spread of multidrug-resistant Gram-negative organisms constitutes one of the greatest challenges to global health. While Gram-negative organisms have developed several mechanisms to avert the bactericidal effects of commonly prescribed antibiotic agents, the increasing prevalence of carbapenemase-producing organisms (CPO) is particularly concerning given the rapid spread of mobile genetic elements containing carbapenemase genes, the limited treatment options for infections caused by these organisms, and the high mortality rates associated with CPO infections. Understanding if an organism is carbapenemase producing and, if so, the class of carbapenemase(s) produced has treatment implications, as some agents preferentially have activity against specific carbapenemases. Furthermore, CPO disseminate between patients with greater ease than non-CP-carbapenem-resistant organisms and warrant more intensive infection control measures than would be employed in the absence of carbapenemase production. Phenotypic assays currently used in clinical practice to detect CPO consist of the following: (i) growth-based assays which measure carbapenem resistance based on organism growth in the presence of a carbapenem antibiotic (e.g., modified Hodge test and modified carbapenem inactivation method), (ii) hydrolysis methods which detect carbapenem degradation products (e.g., Carba NP test and matrix-assisted laser desorption–ionization time of flight mass spectrometry), and (iii) lateral flow immunoassays which detect carbapenemase enzymes through the use of specific antibodies. Although there is no single phenotypic test that meets all specifications of the ideal test, as we describe in this review, there are a number of tests that are user-friendly, affordable, accurate, and feasible for implementation in clinical microbiology laboratories of all sizes.
INTRODUCTION
Infections due to carbapenemase-producing organisms (CPO) are associated with alarming rates of mortality (1). Carbapenemase genes offer a stable and transferable form of resistance, enabling spread via clonal expansion or by horizontal transfer of genes to naive bacteria (2). Carbapenemases defy geographic boundaries, making the prevention of CPO a significant public health concern that requires international coordination to contain (1). Distinguishing CPO from Gram-negative organisms that are carbapenem resistant due to non-carbapenemase-mediated mechanisms is important, as CPO disseminate between patients more readily than non-CPO and warrant implementation of more intensive infection control measures than would be employed in the absence of carbapenemase production (2). The U.S. Centers for Disease Control and Prevention (CDC) recommends that clinical laboratories consider actively screening isolates for carbapenemase production that meet the CDC surveillance definition for carbapenem-resistant Enterobacteriaceae (http://www.cdc.gov/hai/organisms/cre/definition.html).
Currently, characterization of the underlying mechanism of carbapenem resistance is not undertaken by most clinical microbiology laboratories for therapeutic decision-making. However, understanding if an organism is carbapenemase producing and, if so, the class of carbapenemase(s) produced has treatment implications. Although antimicrobial susceptibility testing (AST) results alone are frequently sufficient for the selection of appropriate antibiotic therapy, the availability of antibiotics, like ceftazidime-avibactam or meropenem-vaborbactam, which have activity against some carbapenemases (e.g., Klebsiella pneumoniae carbapenemases [KPCs]) but not others (e.g., metallo-β-lactamases [MBLs], such as New Delhi metallo-β-lactamases [NDMs]), makes carbapenemase mechanism identification important, particularly when access to susceptibility testing for newer agents is limited. Unfortunately, there is no constellation of AST results that reliably distinguishes carbapenemase producers from noncarbapenemase producers.
CLASSIFICATION OF CARBAPENEMASES
Carbapenemases are commonly categorized using the Ambler classification scheme (3). KPCs are the most common carbapenemases identified in the United States and are members of the class A carbapenemases. Although commonly found in Enterobacteriaceae, they are not exclusive to Enterobacteriaceae and can be occasionally identified in Pseudomonas aeruginosa or Acinetobacter baumannii (4). Class B β-lactamases (MBLs) are characterized by the requirement for zinc ions at their active site, which can be useful diagnostically, as chelators, like EDTA or dipicolinic acid (DPA) inhibit MBL activity by binding zinc (5). Common MBLs include NDM, Verona integron-encoded metallo-β-lactamase (VIM), and IMP (named for its imipenem-resistant phenotype) enzymes. Common class D carbapenemases include OXA-48-like enzymes generally produced by Enterobacteriaceae (6) and OXA-23-like, OXA-40-like, OXA-58-like, and OXA-143-like enzymes, which are frequently produced by A. baumannii (4). Other carbapenemases that belong to a variety of molecular classes (e.g., Serratia marcescens enzymes [SME] and Guiana extended-spectrum β-lactamases [GES]) are reported sporadically (1). These are generally species specific, likely because their corresponding genes are either chromosomal or located on narrow-host-range plasmids, limiting large-scale dissemination (1). The diverse range of carbapenemase enzymes contributes to difficulty in their detection.
DEFINING CARBAPENEM-RESISTANT GRAM-NEGATIVE ORGANISMS
The Clinical and Laboratory Standards Institute (CLSI) lowered the carbapenem breakpoints for the Enterobacteriaceae in 2010 (7). The revised breakpoints recommend meropenem or imipenem susceptibility with MICs of ≤1 μg/ml and ertapenem susceptibility with MICs of ≤0.5 μg/ml (7). Furthermore, updated CLSI recommendations remove the recommendation for the routine identification of carbapenemase production among isolates with elevated carbapenem MICs (7). Therefore, providing that the 2010 carbapenem breakpoints have been implemented, it is recommended that AST results should be reported as tested, regardless of any mechanisms of resistance that have been identified. However, laboratories may choose to continue to screen for carbapenemase production for epidemiologic, infection control, or antibiotic stewardship initiatives. Over the past 10 years, a number of phenotypic methods for the detection of carbapenemase producers among CRO have been developed.
OVERVIEW OF PHENOTYPIC ASSAYS
Both phenotypic and molecular-based assays are available for the detection of carbapenemase producers from cultured isolates. Phenotypic assays currently used in clinical practice consist of the following: (i) growth-based assays which measure resistance based on growth in the presence of an antibiotic (e.g., modified Hodge test [MHT]) and modified carbapenem inactivation method [mCIM]); (ii) hydrolysis methods which detect the product of hydrolysis that is catalyzed by carbapenemase enzymes (e.g., Carba NP and matrix-assisted laser desorption–ionization time of flight mass spectrometry [MALDI-TOF MS] methods); and (iii) lateral flow immunoassays which detect carbapenemase enzymes through the use of specific antibodies. Nucleic acid-based carbapenemase detection directly identifies the molecular determinants of carbapenemase production. This review is limited to phenotypic methods of carbapenemase detection and will not address nucleic acid-based approaches, including PCR, microarray, and whole-genome sequencing. We refer the reader to existing comprehensive reviews to learn more about the utility of nucleic acid-based techniques for carbapenemase gene identification (8, 9).
The selection of a carbapenemase detection test is contingent upon several factors, including local carbapenemase prevalence, regional molecular epidemiology, diagnostic performance characteristics, labor intensity, cost, and turnaround time (TAT) of the test (10). The TAT is important both for therapeutic decision-making and infection control purposes, with same-day results being ideal. Other considerations include the organisms to be tested (i.e., Enterobacteriaceae and/or glucose-nonfermenting Gram negatives), ease of use, workflow, regulatory status, necessary equipment, and reagent preparation requirements. Unfortunately, no currently available assay has a favorable profile for all of the criteria listed above. However, several options are available, allowing labs to choose a method that best suits their needs.
Common phenotypic tests employed by clinical microbiology laboratories include the MHT, the Carba NP test and its variants, and most recently, the mCIM. These tests identify carbapenemase production but in their traditional forms lack guidance regarding the specific carbapenemase being produced. Some modifications to these assays can provide more information on the specific carbapenemase groups being produced. For example, although the mCIM is indifferent to the carbapenemase class being expressed, setting up the test in parallel with the addition of the divalent cation chelator EDTA-mCIM (eCIM) enables the differentiation of serine and metallo-carbapenemases.
Multiplex lateral flow immunoassays (LFIAs) are actively being investigated as an approach to identify the specific carbapenemase group being produced, even when multiple carbapenemases are present simultaneously (11–17). Inhibitor-based methods are alternative phenotypic approaches that can be used to differentiate between carbapenemases by applying class-specific inhibitors. For example, the MBL Etest (AB Biodisk, Solna, Sweden) and the combined EDTA disk test rely on decreased MBL activity as a consequence of EDTA binding to zinc. Although MALDI-TOF MS is commonly used by clinical microbiology laboratories for bacterial and fungal genus and species identification, its role for carbapenemase detection is still evolving. The relative pros and cons of each of these phenotypic approaches for carbapenemase detection are described in more detail below and in Table 1.
TABLE 1.
Select phenotypic tests for carbapenemase detection in clinical isolatesa
| Test parameter | Modified Hodge test (10,18–24) | Carba NP test and variants (10, 24–29, 32) | mCIM (7, 10, 32, 43) | Lateral flow immunoassays (11–16) | Targeted carbapenemase assays (47–53) | MALDI-TOF MS hydrolysis approach (28, 54–61) |
|---|---|---|---|---|---|---|
| Carbapenem-resistant organisms | Enterobacteriaceae | Enterobacteriaceae, Pseudomonas aeruginosa | Enterobacteriaceae, P. aeruginosa | Enterobacteriaceae | Generally Enterobacteriaceae | Enterobacteriaceae, P. aeruginosa, A. baumannii |
| Accuracy | For Enterobacteriaceae, sensitivity, ∼95% and specificity, ∼91%; false-positive results with ESBL- or AmpC-positive isolates with porin defects; false-negative results with MBLs | For Enterobacteriaceae, sensitivity, ∼84% and specificity, ∼100%; for P. aeruginosa, sensitivity, 98% and specificity, 98%; with change in lysis buffer and starting pH, sensitivity increases to close to 99%; false-negative results with OXA-48-like producers or mucoid isolates | For Enterobacteriaceae, sensitivity, 97% and specificity, 99%; for P. aeruginosa, sensitivity, 98% and specificity, 95%; mCIM improves detection of OXA-48-like producers over CIM; false-positive results may occur with AmpC-hyperproducing E. cloacae; addition of eCIM identifies MBLs in Enterobacteriaceae | For Enterobacteriaceae, sensitivity, 100% and specificity, ≥95%; false-positive results with some noncarbapenemase OXA-enzymes | Accuracy varies by test, e.g., OXA-48 disk test sensitivity, 96% and specificity, 98%; KPC gradient strips, sensitivity, 92% and specificity, 100%; MBL gradient strips, sensitivity, 94% and specificity, 95%; BD Phoenix CPO Detect, sensitivity, 97% and specificity, 69% | For Enterobacteriaceae, sensitivities, 77%–100% and specificities, 94%–100% |
| Ease of use | Easy to perform, no special reagents or media necessary | For manual versions, fresh reagents need to be prepared frequently due to limited shelf-life of imipenem solution; pH meter needed; commercially available tests remove the requirements for reagent preparation and need for a pH meter | Easy to perform, no special reagents or media necessary | Easy to perform, no special reagents or media necessary besides the test kit | All supplies generally readily available except gradient strips or disk | Complexity varies based on approach |
| Interpretation of results | Indentation of zone diam of E. coli toward carbapenem disk along streaked isolate; reading of zone indentation can be subjective | Color change from red to yellow or blue to yellow depending on the pH indicator; color change can be subjective for intermediate results | Positive, zone diam of 6–15 mm; indeterminate, 16–18 mm; negative, ≥19 mm; requires initial setup and then plating of disk onto lawn of E. coli following 4 h of incubation | Positive results based on the presence of visible lines specific for carbapenemase-type | Varies by test; for MBL Etest, only used for MBL detection; ratio of the IP/IPI MIC of ≥8 or presence of phantom zone or deformation of ellipse | Complex to interpret; requires MALDI-TOF instrument settings different from those traditionally used for FDA-approved microbial identification |
| Total time to perform test | 15 min for setup and 2 min to read the following day | 40 to 170 min (varies based on test) | 5 min for initial setup, 10 min to inoculate plate, and 2 min to read the following day | 5 min for initial setup and 2 min to read | Varies by test; for MBL Etest, 10-min setup and <5 min to read the following day | 3–4 h |
| Turnaround time | 18–24 h | Same-day result; 30 min to 2 h | 18–24 h; shorter TAT has been described with shorter incubation of the disk with the indicator strain | Same-day result; 15 min | 18–24 h | Same-day result; 4 h |
| Estimated cost | <$1.00 | For manual versions, ≥$2.00; cost increases if imipenem-based reagents are not used within 3 days; commercial versions are more expensive, at ∼$2.50–$15.00 (varies based on test) | <$1.00 | Prices have not yet been established | Varies by test; ranges from <$1.00–$6.00; MBL gradient strips, ∼$6.00 | <$1.00 if MALDI-TOF MS instrument already available |
| Regulatory status | LDT; no longer CLSI endorsed | LDT; except the Rapidec Carba NP is FDA cleared for use with Enterobacteriaceae and P. aeruginosa; manual Carba NP is CLSI endorsed | LDT; CLSI endorsed | RUO | RUO | RUO |
IP, imipenem; IPI, imipenem inhibitor (imipenem plus EDTA); RUO, research use only; LDT, laboratory-developed test. Table modified from reference 10.
Modified Hodge test.
The MHT is probably the most well-known approach for carbapenemase detection. It involves the streaking of a clinical isolate in a line away from a disk impregnated with either ertapenem or meropenem which was previously placed on an agar plate inoculated with a lawn of a carbapenem-susceptible Escherichia coli strain. The MHT relies on the ability of carbapenemase producers to decrease the local concentration of carbapenem antibiotics, enabling the carbapenem-susceptible E. coli isolate to grow uninhibited around the streak line near the carbapenem disk, producing a cloverleaf appearance (Fig. 1A). This assay demonstrates acceptable sensitivity for most carbapenemases, particularly KPC enzymes, but low sensitivity for MBLs (18–20). In one study, the MHT was able to correctly detect carbapenemase activity in only 50% of NDM-producing isolates (18). For U.S. collections of Enterobacteriaceae, where KPC producers comprise greater than 95% of carbapenemases (21, 22), the sensitivity of the MHT has been reported to be between 93% and 98% (23, 24). However, with the rapid spread of NDM-producing Enterobacteriaceae, limitations in detecting this epidemiologically important resistance mechanism may have significant consequences. As isolates producing extended-spectrum β-lactamases (ESBLs) or AmpC cephalosporinases in conjunction with porin mutations often yield false-positive MHT results, the MHT has limited specificity, reported at approximately 91% (10). Although the MHT is inexpensive, relatively straightforward to perform, and uses reagents readily available in most clinical laboratories, it is occasionally challenging to interpret and time-consuming, as it requires an additional 24-h growth step after AST results become available (20). Due to these limitations, the MHT was removed from the CLSI M100 document in 2018, as newer phenotypic approaches with improved accuracy have become available (7).
FIG 1.
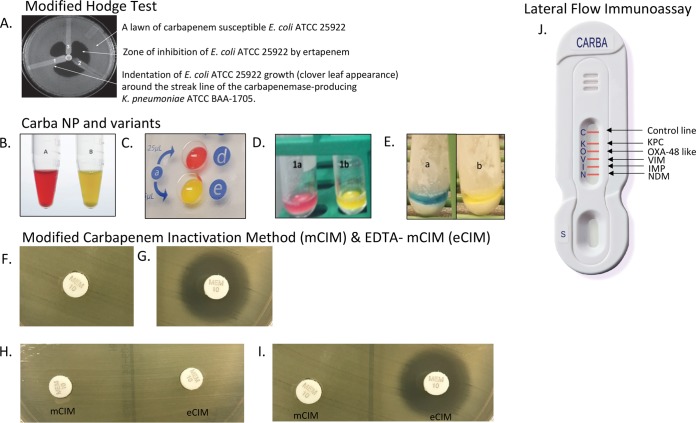
(A) Modified Hodge test. 1, Klebsiella pneumoniae ATCC BAA-1705, positive result; 2, K. pneumoniae ATCC BAA-1706, negative result; 3, clinical isolate, positive result. (B) CLSI Carba NP positive result. Tube A, no imipenem added, red; tube B, imipenem added, yellow. (C) Rapidec Carba NP (bioMérieux, Inc.) positive result. Well d, no imipenem added, red; well e, imipenem added, yellow. (D) Neo-Rapid Carba screen (Rosco Diagnostica) positive result. Tube 1a, no imipenem added, red; tube 1b, imipenem added, yellow (67). (E) Rapid Carb Blue Screen (Rosco Diagnostica) positive result. Tube a, no imipenem added, blue; tube b, imipenem added, yellow. (F) Modified carbapenem inactivation method (mCIM) positive result. (G) mCIM negative result. (H) mCIM and EDTA-mCIM (eCIM) results that are positive for a serine carbapenemase producer, as there is no inhibition of carbapenemase activity in the presence of EDTA. (I) mCIM and eCIM results that are positive for a metallo-beta-lactamase producer, as there is inhibition of carbapenemase activity in the presence of EDTA. (J) NG-Test Carba 5 (NG Biotech) lateral flow immunoassay results for the different carbapenemases detected. Panel A is republished from reference 45 with permission from the publisher. Panels B and F to I are republished from reference 7 with permission from the publisher. The image from panel J was provided by NG Biotech.
Carba NP test and variants.
The Carba NP test detects carbapenemases by measuring the in vitro hydrolysis of imipenem in bacterial extracts and produces color changes within approximately 2 h (24). Imipenem hydrolysis results in a carboxylic derivative, which in turn decreases the pH, producing a resultant color shift of a phenol red indicator from red to yellow (Fig. 1B) (25). It has a high sensitivity for detecting most carbapenemases, with a reported range of 73 to 100% (10, 24–27). Its sensitivity for the OXA-48-like carbapenemases, however, is significantly lower and in one study was only 6% (28). Furthermore, the Carba NP test requires the preparation of reagents with a short shelf-life, and there may be various interpretations of the results (10). Many variants of the Carba NP test have since been described with modifications to the inoculum, extraction regents, starting pH, pH indicators, and reading times, such as the Blue Carba test (29), which utilizes the pH indicator bromothymol blue. The modified Carba NP test is another variant that uses 0.02% cetyltrimethylammonium bromide lysis buffer and a starting pH of 7.5 instead of 7.8, enabling the improved identification of carbapenemase production (30). In an investigation comparing 191 retrospective Enterobacteriaceae isolates, the CLSI Carba NP method had a sensitivity of 84% (95% confidence interval [CI], 76 to 90%) and a specificity of 100% (95% CI, 93 to 100%), whereas the modified Carba NP test had an increased sensitivity of 99% (95% CI, 95 to 100), with the specificity remaining at 100% (95% CI, 93 to 100%) (10). Others have obtained more modest results with the modified Carba NP test. Tijet and colleagues demonstrated that the sensitivity of OXA-48-like detection increased from 56% to 71% with the original Carba NP versus the modified Carba NP test and found that mucoid isolates were particularly prone to false-negative results (26, 31). The Carba NP test II was developed to both identify carbapenemase production and also to discriminate carbapenemase classes (32). It relies on pH changes due to imipenem hydrolysis in conjunction with specific enzyme inhibition through the use of tazobactam for KPC detection and EDTA for MBL detection.
The Carba NP test was also investigated for identifying carbapenemase production in non-glucose-fermenting organisms. The CLSI Carba NP method was used to evaluate 30 P. aeruginosa isolates and 30 A. baumannii isolates at three sites and included representation from Ambler class A, B, and D carbapenemases (33). The mean sensitivity and specificity for carbapenemase detection in P. aeruginosa were 98% (range, 93% to 100%) and 98% (range, 93% to 100%), respectively, whereas the mean sensitivity and specificity for carbapenemase detection in A. baumannii were 19% (range, 9% to 26%) and 100% (range, 100% to 100%), respectively (33). The modified Carba NP test increases the ability to detect carbapenemase production in A. baumannii but remains suboptimal. In one study, the sensitivities of the CLSI Carba NP and modified Carba NP test were 21% (95% CI, 6 to 51%) and 79% (95% CI, 49 to 94%), respectively (34).
The poor sensitivity of the Carba NP for carbapenemase detection in A. baumannii is concerning but not entirely surprising. OXA-type carbapenemases common to carbapenem-resistant A. baumannii strains are known to be inefficient at hydrolyzing the β-lactam ring of carbapenem antibiotics. The elevated carbapenem MICs observed in carbapenem-resistant A. baumannii strains are largely attributable to additional manifestations of resistance, such as reduced porin expression or upregulation of efflux pumps (4). Furthermore, the low-level outer membrane permeability of these organisms encumbers the activity of the Carba NP test that relies on cell lysis and the subsequent release of carbapenemases to detect color changes (4).
The CarbAcineto NP test was designed to overcome some of the impediments associated with detecting carbapenemases produced by A. baumannii (35). The CarbAcineto NP test requires modified lysis conditions and an increased bacterial inoculum compared to the original Carba NP test. More specifically, the lysis buffer (bacterial protein extraction reagent [B-PER II]; Thermo Scientific Pierce, Villebon-sur-Yvette, France) was replaced by a hyperosmotic solution of 5 M NaCl solution in response to the low permeability of the outer membrane, and the inoculum was increased from approximately half a 10-μl loop to a full 10-μl loop (35). The sensitivity of the Carba NP test in detecting acquired carbapenemase subgroups common to A. baumannii, most notably OXA-23-like, OXA-40-like, OXA-58-like, and OXA-143-like subgroups, was 12% in a collection of 151 carbapenemase-producing A. baumannii isolates (35), whereas the CarbAcineto test was able to recognize all of these acquired carbapenemases. Inconsistencies were observed with the CarbAcineto assay's ability to detect OXA-51-like carbapenemases. blaOXA-51-like genes are chromosomally integrated, and OXA-51-like enzymes are either inactive or weakly active at hydrolyzing carbapenem antibiotics in the absence of an ISAba1 insertion sequence at the 5′ end of the blaOXA-51-like gene (36). Overall, the CarbAcineto test had a sensitivity of 95% and a specificity of 100% for detecting carbapenemases commonly associated with A. baumannii (35).
The manual versions of the Carba NP test and its variants require frequent reagent preparation because the imipenem-containing solution has a maximum shelf-life of 72 h. Other commercially available assays using principles similar to those of the Carba NP test have been developed that generally simplify testing, including the Rapidec Carba NP test (37) (bioMérieux, Marcy-l'Etoile, France; Fig. 1C), the Neo-Rapid Carb screen (38) (Rosco Diagnostica, Taastrup, Denmark; Fig. 1D), the β-Carba NP test (Bio-Rad, Marnes-la-Coquette, France), and the Rapid Carb Blue screen (39) (Rosco Diagnostica; Fig. 1E). These commercial tests simplify testing by including all reagents required for testing without the need for reagent preparation. Overall, these tests (not including the β-Carba NP) have sensitivities ranging from 89% to 98% and specificities approaching 100% (10). A recent comparison of the Rapidec Carba NP and the β-Carba NP tests demonstrated that the Rapidec Carba NP test exhibited better performance characteristics (sensitivity, 94%; specificity, 100%) than the β-Carba NP test (sensitivity, 65%; specificity, 90%) (40). Similar to the Carba NP test, the same limitations exist for these commercial assays, as false-negative results occur with OXA-48-like enzymes, and the interpretation of results can be subjective due to minor color changes. More detailed characteristics specific to these assays are presented in Table 1.
Carbapenem inactivation method.
The CIM was first described in 2015 (41). This test is based on the premise that when a 10-μg meropenem disk is incubated for 2 h in water with a 10-μl loop of carbapenemase-producing isolate, the meropenem will be hydrolyzed. Alternatively, if the organism does not produce a carbapenemase, the meropenem disk retains its activity. The meropenem disk is then removed and placed on a Mueller-Hinton agar plate streaked with a susceptible laboratory strain of E. coli. Following overnight incubation, the zone of inhibition is measured. The absence of an inhibition zone indicates the presence of a carbapenemase. In contrast, a zone of inhibition indicates that the meropenem in the disk has preserved its activity and the isolate is not producing a carbapenemase. The CIM yields high sensitivities and specificities, at approximately 91% to 94% and 99% to 100%, respectively (10, 42). It is straightforward both to perform and interpret (10, 41). It requires inexpensive materials readily available in clinical microbiology laboratories at a cost of less than $1 per test, similar to the MHT (10). Both the MHT and CIM are less costly than the Carba NP test ($2 to $10 per test based on the variant), and all of these phenotypic tests are substantially more affordable than molecular assays.
Some concerns based on initial investigations suggested that the CIM may have limitations with the detection of carbapenemases with reduced hydrolytic activity (e.g., OXA-type carbapenemases), MBL enzymes that require divalent cations for activity, or in settings of lower levels of carbapenemase expression (43). Data from a CLSI working group suggested that modifying the carbapenem inactivation step by preparing the bacterial suspension (1-μl loopful of Enterobacteriaceae) in tryptic soy broth (TSB) and extending the time of incubation to 4 h could further improve carbapenemase detection (43). They conducted a multicenter study using the modified CIM (mCIM) approach for 61 isolates provided by the CDC-FDA Antimicrobial Resistance Isolate Bank and identified the mean sensitivity and specificity of the mCIM across the nine testing sites to be 97% (range, 93% to 100%) and 99% (range, 97% to 100%), respectively (43). Excellent reproducibility was shown across laboratories for representative carbapenemases from Ambler classes A, B, and D (43). One of the participating sites compared the CIM to the mCIM using the same set of isolates and demonstrated that the sensitivity increased from 82% to 93% and the specificity remained at 100% (43). Furthermore, at test sites where microbiologists had experience performing the MHT and/or the Carba NP test, interpretation of mCIM results was regarded as less subjective and easier to perform. A major drawback with the mCIM, similar to the MHT, is that it requires overnight incubation, so results will not be available within a single work shift, as is the case with the Carba NP test. Some investigators have implemented shorter incubation periods of the disk with the carbapenem-susceptible indicator strain (6 h) to report CIM or mCIM results within the same day (44). Based on our experience from unpublished work, the specificity of the mCIM might be slightly lower than initially reported, as we have encountered false-positive results with Enterobacter cloacae isolates lacking known carbapenemases.
The CLSI working group recommended a zone diameter of 6 to 15 mm for a result to be identified as positive (Fig. 1F), including when multiple small bacterial colonies are observed within the zone of inhibition around the disk, and a zone diameter of ≥19 mm for a result to be categorized as negative (Fig. 1G). Zone diameters between 16 and 18 mm should be considered indeterminate and require additional testing to establish the presence or absence of carbapenemase production. In 2017, the mCIM was added to the 27th edition of the CLSI M100 supplement document as a reliable method for the detection of carbapenemase production in Enterobacteriaceae (45).
As the mCIM can accurately identify the presence of carbapenemases but cannot distinguish between serine and metallo-β-lactamases, a further modification to this assay with the addition of EDTA has been proposed. The basic premise of this assay is that if the test isolate produces an MBL, EDTA will inhibit carbapenemase production, resulting in the meropenem disk not being hydrolyzed as efficiently as expected in the absence of EDTA (7). Briefly, this method involves adding 20 μl of 0.5 M EDTA to a second 2-ml TSB tube. Then, the mCIM and eCIM tubes are processed in parallel (7). The meropenem disks from the mCIM and eCIM tubes are placed on the same Mueller-Hinton agar plates, inoculated with the E. coli ATCC 25922 indicator strain. The eCIM results should only be interpreted if the mCIM result indicates the presence of a carbapenemase. A ≥5-mm increase in the zone diameter for the eCIM compared to the zone diameter for the mCIM suggests the likelihood of an MBL-producing strain (Fig. 1H and I) (7). Unlike the mCIM, pinpoint colonies surrounding the meropenem disk within the zone of inhibition should be ignored when interpreting eCIM results (7). The eCIM is endorsed in the CLSI M100 supplement as an approach to distinguish MBL carbapenemases from serine carbapenemases, as a CLSI working group determined the eCIM's sensitivity and specificity to be greater than 95% and greater than 92%, respectively (7). Of note, if both serine and metallo-carbapenemases are coproduced by a single isolate, false-negative results may occur. This is important, as the coproduction of both OXA-48-like and NDM enzymes have been reported (10).
The mCIM has also been investigated for the detection of carbapenemases in P. aeruginosa and A. baumannii, using criteria similar to those established with the mCIM for Enterobacteriaceae. A 10-site study evaluating the performance of the mCIM against 30 P. aeruginosa and 30 A. baumannii isolates from the CDC-FDA Antibiotic Resistance Isolate Bank demonstrated a mean sensitivity and specificity of the mCIM for the detection of carbapenemase-producing P. aeruginosa of 98% (range, 87% to 100%) and 95% (range, 93% to 100%) (33). The accuracy of the mCIM for carbapenemase production by A. baumannii proved to be much less reliable. The mean sensitivity and specificity for A. baumannii were 80% (range, 36% to 96%) and 53% (range, 29% to 100%), respectively (33). Significant site-to-site variability in results was identified. Moreover, it was recognized that a larger inoculum (i.e., a 10-μl loopful of organism) was required for reliable carbapenemase detection for non-glucose-fermenting organisms, compared with the Enterobacteriaceae. This was most apparent with VIM-producing P. aeruginosa and OXA-producing A. baumannii isolates. However, the increased sensitivity (60% to 93%) was at the expense of specificity, as was observed with A. baumannii isolates, where the specificity decreased from 100% using a 1-μl loop to 63% using a 10-μl loop (33). Others have found similar limitations with the performance of the mCIM in detecting carbapenemase-producing A. baumannii strains (34). The 28th edition of the CLSI M100 supplement document endorses the use of the mCIM using a 10-μl inoculum for the detection of carbapenemase production among carbapenem-resistant P. aeruginosa strains (7). The mCIM is not currently recommended for the detection of carbapenemase activity in A. baumannii. A modification to the mCIM using 0.5 M Tris-HCl buffer for extraction (CIMTris) detected carbapenemase production in Acinetobacter and Pseudomonas species with a sensitivity of 98% and specificity of 93% (46).
Lateral flow immunoassays.
LFIAs are antibody-based methods to identify the presence of carbapenemases. A number of LFIAs have been recently developed but generally enable the detection of one or a few of the most epidemiologically important carbapenemases. LFIAs have been developed for NDM production (14), IMP-production (15), OXA-48-like production (14), KPC and OXA-48-like production (16), and KPC, NDM, and OXA-48-like production (12). Available data suggest that LFIAs produce accurate results from cultured isolates within 15 min. Recently, an LFIA targeting the five main carbapenemase families (i.e., KPC, NDM, VIM, IMP, and OXA-48-like carbapenemases) was evaluated (11). Investigators both retrospectively and prospectively evaluated this LFIA, named the NG-Test Carba 5, using 296 Enterobacteriaceae isolates (11). Briefly, they collected a single bacterial colony from a Mueller-Hinton agar plate and suspended it in 150 μl of extraction buffer. Subsequently, 100 μl of this extract was loaded on a cassette, and results were read within 15 min of migration, based on the presence of visible lines indicating a positive test, not unlike the interpretation of a home pregnancy test (Fig. 1J). In both the retrospective and prospective studies, the sensitivity of the NG-Test Carba 5 was 100% (11). The specificity was 95% for the retrospective cohort and 100% for the prospective cohort, with two noncarbapenemase OXA enzymes (an OXA-405 and two OXA-163 enzymes) misclassified as OXA-48-like enzymes (11). Importantly, the Carba5 was accurately able to detect isolates producing two carbapenemases (e.g., NDM-type and OXA-48-like carbapenemases), and cross-reactivity was not observed with nontargeted carbapenemases (e.g., GES, SME, etc.). In the future, it is plausible that the NG-Test Carba 5 or similar LFIAs could offer easy-to-use, accurate, rapid, and cost-efficient approaches for clinical microbiology laboratories to identify the presence of specific carbapenemases similar to molecular-based approaches.
Targeted carbapenemase assays.
Targeted phenotypic carbapenemase assays compare carbapenem activity with and without the presence of inhibitors (e.g., phenylboronic acid [PBA] for KPC and EDTA for MBL). There are both commercially available and laboratory-developed options to evaluate for multiple carbapenemase resistance mechanisms (e.g., KPC + MBL Confirm ID kit [Rosco Diagnostica] and Mastdiscs combi Carba Plus [MAST Diagnostics] to evaluate for KPC, MBL, and OXA-48-like producers) (47–49). As an example, a laboratory-developed OXA-48 disk test was developed relying on an imipenem disk, a disk impregnated with EDTA, and another disk containing EDTA and PBA. Evaluating 254 isolates, the OXA-48 disk test had a sensitivity of 96% and a specificity of 98% (48). Targeted carbapenemase tests offer straightforward, affordable, and accurate options for the detection of specific carbapenemases. When used more broadly to identify the production of multiple carbapenemases, as can be commonly observed in some regions of the world (1), however, algorithms and workflows can become complex. To simplify the manual interpretation of these algorithms, the BD Phoenix CPO Detect test was designed to be incorporated into automated AST panels with computer-assisted algorithm-based detection (50). The test consists of nine wells on the BD Phoenix AST panel, each containing a β-lactam antibiotic, alone and in combination with various β-lactamase inhibitors for the detection and classification of carbapenemases, analogous to ESBL tests on automated AST instruments. One high-stringency evaluation of the assay reported a sensitivity of 97% and specificity of 69% to detect carbapenemase producers. Furthermore, it classified carbapenemases appropriately for 85% of class A, 72% of class B, and 89% of class D carbapenemases (50).
Gradient diffusion strips can also be reliable easy-to-use options for identifying specific resistance mechanisms and are growth-based approaches for carbapenemase detection. They rely on a combination of a β-lactam and a β-lactam–β-lactamase inhibitor (generally either boronic acid for KPC or EDTA for MBL) to detect these two carbapenemase types. KPC gradient MIC strips have a reported sensitivity of 92% and specificity of 100% (51). MBL gradient MIC strips have a reported sensitivity of 94% and a specificity of 95% (52).
Notably, there have been reports of false-positive results using MBL Etest strips (AB Biodisk, Solna, Sweden) in the presence of select OXA-type carbapenemases. Certain OXA-type carbapenemases (e.g., OXA-10 and OXA-14 in Enterobacteriaceae or OXA-23-like in A. baumannii) exist in highly active dimeric forms and less-active monomeric forms (53, 54). Metal chelators stabilize the active dimeric forms, converting the enzymes to their less-active monomeric states, reducing carbapenemase activity, yielding false-positive MBL Etest results. Targeted carbapenemase assays can be valuable during outbreaks or when considering treatment options with activity against select carbapenemases.
MALDI-TOF MS.
MALDI-TOF MS platforms are well established for microbial genus and species identification and are increasingly commonplace in clinical microbiology laboratories. Two major applications of MALDI-TOF MS for the rapid identification of carbapenemase production are being pursued. The first (“hydrolysis approach”) detects carbapenem degradation products when bacterial protein extracts are incubated with a carbapenem substrate. The second approach (“plasmid-associated peak approach”) involves the detection of a known carbapenemase-bearing plasmid-associated protein peak.
Regarding the hydrolysis method, a number of protocols have been described; however, a standardized approach is not yet available (28, 55–62). In short, carbapenem antibiotics are incubated with bacterial cultures, the cultures are centrifuged, and the supernatants are analyzed for β-lactam hydrolysis by MALDI-TOF MS. Reported sensitivities and specificities range from 77% to 100% and 94% to 100%, respectively, with a turnaround time of within 4 h. Similar to the Carba NP test, MALDI-TOF MS has difficulty detecting OXA-48-like enzymes. One study reported a sensitivity of 77% and a specificity of 100%, with the lower sensitivity due to missing 16 of 19 OXA-48 producers (28). However, this report suggested that the addition of bicarbonate to the reaction buffer could enhance the detection of OXA-48-like producers, increasing the overall sensitivity to 98%, without compromising the detection of other enzymes (28). Similarly, this modified approach was reported to have both a sensitivity and specificity of 100% in identifying carbapenemase activity in A. baumannii (63). Recent investigations suggest that the inclusion of temocillin, for which high-level resistance is suggestive of OXA-48 activity, may effectively detect this specific resistance mechanism (64). More recently, the role of carbapenem inhibitors was explored. The addition of PBA resulted in the reappearance of MS peaks corresponding to ertapenem for class A carbapenemases, whereas MS peaks associated with the degradation products of ertapenem remain in the case of MBL producers. Conversely, when adding the chelator DPA, MS peaks representing the hydrolyzed form of ertapenem remain in settings with class A carbapenemases, but peaks associated with ertapenem reappear in the case of MBL producers (64).
The plasmid-associated peak approach functions quite differently. Investigators at the National Institutes of Health retrospectively evaluated protein profiles from the pKpQIL plasmid carrying a blaKPC-3 gene involved in a K. pneumoniae outbreak at the NIH clinical center that led to 18 affected patients and 6 deaths (65). An approximately 11,109-Da MS peak corresponding to a gene product of the blaKPC-3-carrying pKpQIL plasmid was identified and was common to all K. pneumoniae isolates and absent from a diverse set of controls (65). Remarkably, plasmid identification using this MALDI-TOF MS method was accomplished in as little as 10 min from isolated colonies. Further work by the same investigators including 140 characterized Enterobacteriaceae isolates showed both sensitivity and specificity over 95% (66). The investigators propose that signature MS peaks may be useful in tracking other plasmids conferring carbapenem resistance.
MALDI-TOF-MS for carbapenemase detection can be a cost-effective approach, if the instrument is already present in a microbiology laboratory, for genus and species identification. Importantly, the use of MALDI-TOF MS for carbapenemase detection by the hydrolysis method requires MALDI-TOF instrument settings different from those traditionally used for FDA-approved microbial identification and requires in-house validation. Currently, the MALDI-TOF MS hydrolysis approach for carbapenemase detection requires reagent preparation and advanced knowledge of the MALDI-TOF MS instrument settings. It can be viewed as being relatively complex to perform and interpret. Although the plasmid-associated peak approach is promising, it has only been evaluated for a single plasmid-associated carbapenemase gene, and more work in developing a more comprehensive database of commonly circulating plasmids and associated carbapenemase genes is needed before it becomes a feasible approach for clinical microbiology laboratories.
CONCLUSION
In summary, the spread of CPO remains a significant clinical and public health concern. Reliable detection of carbapenemase production is an essential first step in combating this problem. In an era of international travel and medical tourism, the association between specific resistance mechanisms and a given region will become less important, making both routine surveillance and further evaluation of carbapenem-resistant Gram-negative clinical isolates imperative. While there is no single phenotypic test that meets all specifications of the “perfect” test, there are a number of options that are user-friendly, accurate, and feasible for implementation in clinical microbiology laboratories of all sizes.
ACKNOWLEDGMENTS
The work was supported by funding from the National Institutes of Health grant K23-AI127935 awarded to P.D.T. and grant R21-AI130608 awarded to P.J.S.
We report no conflicts of interest.
REFERENCES
- 1.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman KE, Simner PJ, Tamma PD, Milstone AM. 2016. Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev Anti Infect Ther 14:95–108. doi: 10.1586/14787210.2016.1106940. [DOI] [PubMed] [Google Scholar]
- 3.Ambler RP. 1980. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 4.Gniadek TJ, Carroll KC, Simner PJ. 2016. Carbapenem-resistant non-glucose-fermenting Gram-negative bacilli: the missing piece to the puzzle. J Clin Microbiol 54:1700–1710. doi: 10.1128/JCM.03264-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logan LK, Bonomo RA. 2016. Metallo-beta-lactamase (MBL)-producing Enterobacteriaceae in United States children. Open Forum Infect Dis 3:ofw090. doi: 10.1093/ofid/ofw090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Lupo A, Papp-Wallace KM, Sendi P, Bonomo RA, Endimiani A. 2013. Non-phenotypic tests to detect and characterize antibiotic resistance mechanisms in Enterobacteriaceae. Diagn Microbiol Infect Dis 77:179–194. doi: 10.1016/j.diagmicrobio.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rood IGH, Li Q. 2017. Review: molecular detection of extended spectrum-beta-lactamase- and carbapenemase-producing Enterobacteriaceae in a clinical setting. Diagn Microbiol Infect Dis 89:245–250. doi: 10.1016/j.diagmicrobio.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Tamma PD, Opene BN, Gluck A, Chambers KK, Carroll KC, Simner PJ. 2017. Comparison of 11 phenotypic assays for accurate detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 55:1046–1055. doi: 10.1128/JCM.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, Cotellon G, Plaisance M, Oueslati S, Dortet L, Jousset A, Simon S, Naas T, Volland H. 2018. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 73:909–915. doi: 10.1093/jac/dkx521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saleh A, Gottig S, Hamprecht AG. 2018. Multiplex immunochromatographic detection of OXA-48, KPC, and NDM carbapenemases: impact of inoculum, antibiotics, and agar. J Clin Microbiol 56:e00050-. doi: 10.1128/JCM.00050-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glupczynski Y, Jousset A, Evrard S, Bonnin RA, Huang TD, Dortet L, Bogaerts P, Naas T. 2017. Prospective evaluation of the OKN K-SeT assay, a new multiplex immunochromatographic test for the rapid detection of OXA-48-like, KPC and NDM carbapenemases. J Antimicrob Chemother 72:1955–1960. doi: 10.1093/jac/dkx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dortet L, Jousset A, Sainte-Rose V, Cuzon G, Naas T. 2016. Prospective evaluation of the OXA-48 K-SeT assay, an immunochromatographic test for the rapid detection of OXA-48-type carbapenemases. J Antimicrob Chemother 71:1834–1840. doi: 10.1093/jac/dkw058. [DOI] [PubMed] [Google Scholar]
- 15.Notake S, Matsuda M, Tamai K, Yanagisawa H, Hiramatsu K, Kikuchi K. 2013. Detection of IMP metallo-beta-lactamase in carbapenem-nonsusceptible Enterobacteriaceae and non-glucose-fermenting Gram-negative rods by immunochromatography assay. J Clin Microbiol 51:1762–1768. doi: 10.1128/JCM.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glupczynski Y, Evrard S, Ote I, Mertens P, Huang TD, Leclipteux T, Bogaerts P. 2016. Evaluation of two new commercial immunochromatographic assays for the rapid detection of OXA-48 and KPC carbapenemases from cultured bacteria. J Antimicrob Chemother 71:1217–1222. doi: 10.1093/jac/dkv472. [DOI] [PubMed] [Google Scholar]
- 17.Boutal H, Naas T, Devilliers K, Oueslati S, Dortet L, Bernabeu S, Simon S, Volland H. 2017. Development and validation of a lateral flow immunoassay for rapid detection of NDM-producing Enterobacteriaceae. J Clin Microbiol 55:2018–2029. doi: 10.1128/JCM.00248-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol 50:477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, Carey RB, Thompson A, Stocker S, Limbago B, Patel JB. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol 45:2723–2725. doi: 10.1128/JCM.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalhaes CG, Picao RC, Nicoletti AG, Xavier DE, Gales AC. 2010. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother 65:249–251. doi: 10.1093/jac/dkp431. [DOI] [PubMed] [Google Scholar]
- 21.Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, Wilson LE, Vaeth E, Lynfield R, Shaw KM, Vagnone PM, Bamberg WM, Janelle SJ, Dumyati G, Concannon C, Beldavs Z, Cunningham M, Cassidy PM, Phipps EC, Kenslow N, Travis T, Lonsway D, Rasheed JK, Limbago BM, Kallen AJ. 2015. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 314:1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamma PD, Goodman KE, Harris AD, Tekle T, Roberts A, Taiwo A, Simner PJ. 2017. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathers AJ, Carroll J, Sifri CD, Hazen KC. 2013. Modified Hodge test versus indirect carbapenemase test: prospective evaluation of a phenotypic assay for detection of Klebsiella pneumoniae carbapenemase (KPC) in Enterobacteriaceae. J Clin Microbiol 51:1291–1293. doi: 10.1128/JCM.03240-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasoo S, Cunningham SA, Kohner PC, Simner PJ, Mandrekar JN, Lolans K, Hayden MK, Patel R. 2013. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J Clin Microbiol 51:3097–3101. doi: 10.1128/JCM.00965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. 2013. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yusuf E, Van Der Meeren S, Schallier A, Pierard D. 2014. Comparison of the Carba NP test with the Rapid Carb screen kit for the detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 33:2237–2240. doi: 10.1007/s10096-014-2199-3. [DOI] [PubMed] [Google Scholar]
- 28.Papagiannitsis CC, Studentova V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabak J. 2015. Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol 53:1731–1735. doi: 10.1128/JCM.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pires J, Novais A, Peixe L. 2013. Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakour S, Garcia V, Loucif L, Brunel JM, Gharout-Sait A, Touati A, Rolain JM. 2015. Rapid identification of carbapenemase-producing Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii using a modified Carba NP test. New Microbes New Infect 7:89–93. doi: 10.1016/j.nmni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. 2014. Reply to “further proofs of concept for the Carba NP test.” Antimicrob Agents Chemother 58:1270. doi: 10.1128/AAC.02285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simner PJ, Johnson JK, Brasso WB, Anderson K, Lonsway DR, Pierce VM, Bobenchik AM, Lockett ZC, Charnot-Katsikas A, Westblade LF, Yoo BB, Jenkins SG, Limbago BM, Das S, Roe-Carpenter DE. 2018. Multicenter evaluation of the modified carbapenem inactivation method and the Carba NP for detection of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii. J Clin Microbiol 56:e1369-. doi: 10.1128/JCM.00472-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simner PJ, Opene BNA, Chambers KK, Naumann ME, Carroll KC, Tamma PD. 2017. Carbapenemase detection among carbapenem-resistant glucose-nonfermenting Gram-negative bacilli. J Clin Microbiol 55:2858–2864. doi: 10.1128/JCM.00775-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dortet L, Poirel L, Errera C, Nordmann P. 2014. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J Clin Microbiol 52:2359–2364. doi: 10.1128/JCM.00594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 37.Poirel L, Nordmann P. 2015. Rapidec Carba NP test for rapid detection of carbapenemase producers. J Clin Microbiol 53:3003–3008. doi: 10.1128/JCM.00977-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dortet L, Agathine A, Naas T, Cuzon G, Poirel L, Nordmann P. 2015. Evaluation of the Rapidec Carba NP, the Rapid Carb screen and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 70:3014–3022. doi: 10.1093/jac/dkv213. [DOI] [PubMed] [Google Scholar]
- 39.Novais A, Brilhante M, Pires J, Peixe L. 2015. Evaluation of the recently launched rapid Carb Blue kit for detection of carbapenemase-producing Gram-negative bacteria. J Clin Microbiol 53:3105–3107. doi: 10.1128/JCM.01170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancini S, Kieffer N, Poirel L, Nordmann P. 2017. Evaluation of the Rapidec Carba NP and beta-Carba tests for rapid detection of carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 88:293–297. doi: 10.1016/j.diagmicrobio.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 41.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in Gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun K, Xu X, Yan J, Zhang L. 2017. Evaluation of six phenotypic methods for the detection of carbapenemases in Gram-negative bacteria with characterized resistance mechanisms. Ann Lab Med 37:305–312. doi: 10.3343/alm.2017.37.4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, Bobenchik AM, Lockett ZC, Charnot-Katsikas A, Ferraro MJ, Thomson RB Jr, Jenkins SG, Limbago BM, Das S. 2017. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol 55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renvoise A, Decre D, Amarsy-Guerle R, Huang TD, Jost C, Podglajen I, Raskine L, Genel N, Bogaerts P, Jarlier V, Arlet G. 2013. Evaluation of the βLacta test, a rapid test detecting resistance to third-generation cephalosporins in clinical strains of Enterobacteriaceae. J Clin Microbiol 51:4012–4017. doi: 10.1128/JCM.01936-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 46.Uechi K, Tada T, Shimada K, Kuwahara-Arai K, Arakaki M, Tome T, Nakasone I, Maeda S, Kirikae T, Fujita J. 2017. A modified carbapenem inactivation method, CIMTris, for carbapenemase production in Acinetobacter and Pseudomonas species. J Clin Microbiol 55:3405–3410. doi: 10.1128/JCM.00893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang TD, Berhin C, Bogaerts P, Glupczynski Y. 2014. Evaluation of avibactam-supplemented combination disk tests for the detection of OXA-48 carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 79:252–254. doi: 10.1016/j.diagmicrobio.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Tsakris A, Poulou A, Bogaerts P, Dimitroulia E, Pournaras S, Glupczynski Y. 2015. Evaluation of a new phenotypic OXA-48 disk test for differentiation of OXA-48 carbapenemase-producing Enterobacteriaceae clinical isolates. J Clin Microbiol 53:1245–1251. doi: 10.1128/JCM.03318-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsakris A, Poulou A, Pournaras S, Voulgari E, Vrioni G, Themeli-Digalaki K, Petropoulou D, Sofianou D. 2010. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrob Chemother 65:1664–1671. doi: 10.1093/jac/dkq210. [DOI] [PubMed] [Google Scholar]
- 50.Thomson G, Turner D, Brasso W, Kircher S, Guillet T, Thomson K. 2017. High-stringency evaluation of the automated BD Phoenix CPO detect and Rapidec Carba NP tests for detection and classification of carbapenemases. J Clin Microbiol 55:3437–3443. doi: 10.1128/JCM.01215-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girlich D, Halimi D, Zambardi G, Nordmann P. 2013. Evaluation of Etest strips for detection of KPC and metallo-carbapenemases in Enterobacteriaceae. Diagn Microbiol Infect Dis 77:200–201. doi: 10.1016/j.diagmicrobio.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Walsh TR, Bolmstrom A, Qwarnstrom A, Gales A. 2002. Evaluation of a new Etest for detecting metallo-beta-lactamases in routine clinical testing. J Clin Microbiol 40:2755–2759. doi: 10.1128/JCM.40.8.2755-2759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danel F, Paetzel M, Strynadka NC, Page MG. 2001. Effect of divalent metal cations on the dimerization of OXA-10 and -14 class D beta-lactamases from Pseudomonas aeruginosa. Biochemistry 40:9412–9420. doi: 10.1021/bi0025969. [DOI] [PubMed] [Google Scholar]
- 54.Segal H, Elisha BG. 2005. Use of Etest MBL strips for the detection of carbapenemases in Acinetobacter baumannii. J Antimicrob Chemother 56:598. doi: 10.1093/jac/dki265. [DOI] [PubMed] [Google Scholar]
- 55.Hrabák J, Chudackova E, Walkova R. 2013. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev 26:103–114. doi: 10.1128/CMR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hrabák J, Studentova V, Walkova R, Zemlickova H, Jakubu V, Chudackova E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerova T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hrabák J, Walkova R, Studentova V, Chudackova E, Bergerova T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 49:3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol 49:3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knox J, Jadhav S, Sevior D, Agyekum A, Whipp M, Waring L, Iredell J, Palombo E. 2014. Phenotypic detection of carbapenemase-producing Enterobacteriaceae by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and the Carba NP test. J Clin Microbiol 52:4075–4077. doi: 10.1128/JCM.02121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirande C, Canard I, Buffet Croix Blanche S, Charrier JP, van Belkum A, Welker M, Chatellier S. 2015. Rapid detection of carbapenemase activity: benefits and weaknesses of MALDI-TOF MS. Eur J Clin Microbiol Infect Dis 34:2225–2234. doi: 10.1007/s10096-015-2473-z. [DOI] [PubMed] [Google Scholar]
- 61.Lasserre C, De Saint Martin L, Cuzon G, Bogaerts P, Lamar E, Glupczynski Y, Naas T, Tande D. 2015. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization–time of flight mass spectrometry in less than 30 minutes. J Clin Microbiol 53:2163–2171. doi: 10.1128/JCM.03467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carvalhaes CG, Cayo R, Visconde MF, Barone T, Frigatto EA, Okamoto D, Assis DM, Juliano L, Machado AM, Gales AC. 2014. Detection of carbapenemase activity directly from blood culture vials using MALDI-TOF MS: a quick answer for the right decision. J Antimicrob Chemother 69:2132–2136. doi: 10.1093/jac/dku094. [DOI] [PubMed] [Google Scholar]
- 63.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization–time of flight mass spectrometry. PLoS One 7:e31676. doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oviaño M, Barba MJ, Fernandez B, Ortega A, Aracil B, Oteo J, Campos J, Bou G. 2016. Rapid detection of OXA-48-producing Enterobacteriaceae by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 54:754–759. doi: 10.1128/JCM.02496-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn JH, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. 2014. A rapid matrix-assisted laser desorption ionization–time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol 52:2804–2812. doi: 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youn JH, Drake SK, Weingarten RA, Frank KM, Dekker JP, Lau AF. 2016. Clinical performance of a matrix-assisted laser desorption ionization–time of flight mass spectrometry method for detection of certain blaKPC-containing plasmids. J Clin Microbiol 54:35–42. doi: 10.1128/JCM.01643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simner PJ, Gilmour MW, DeGagne P, Nichol K, Karlowsky JA. 2015. Evaluation of five chromogenic agar media and the Rosco Rapid Carb screen kit for detection and confirmation of carbapenemase production in Gram-negative bacilli. Clin Microbiol 53:105–112. doi: 10.1128/JCM.02068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]