A mixed infection of a single tick or host by Lyme disease spirochetes is common and a unique challenge for the diagnosis, treatment, and surveillance of Lyme disease. Here, we describe a novel protocol for differentiating Lyme strains on the basis of deep sequencing of the hypervariable outer surface protein C locus (ospC).
KEYWORDS: Lyme disease, Borrelia burgdorferi, Borreliella, Ixodes scapularis, outer surface protein C, next-generation sequencing, frequency-dependent selection
ABSTRACT
A mixed infection of a single tick or host by Lyme disease spirochetes is common and a unique challenge for the diagnosis, treatment, and surveillance of Lyme disease. Here, we describe a novel protocol for differentiating Lyme strains on the basis of deep sequencing of the hypervariable outer surface protein C locus (ospC). Improving upon the traditional DNA-DNA hybridization method, the next-generation sequencing-based protocol is high throughput, quantitative, and able to detect new pathogen strains. We applied the method to more than one hundred infected Ixodes scapularis ticks collected from New York State, USA, in 2015 and 2016. An analysis of strain distributions within individual ticks suggests an overabundance of multiple infections by five or more strains, inhibitory interactions among coinfecting strains, and the presence of a new strain closely related to Borreliella bissettiae. A supporting bioinformatics pipeline has been developed. The newly designed pair of universal ospC primers target intergenic sequences conserved among all known Lyme pathogens. The protocol could be used for culture-free identification and quantification of Lyme pathogens in wildlife and potentially in clinical specimens.
INTRODUCTION
Lyme disease occurs widely in the Northern Hemisphere and is the most prevalent vector-borne disease in the United States (1–3). The causative agents of Lyme disease are obligate bacterial parasites of vertebrates transmitted predominantly by hard-bodied Ixodes ticks. Lyme pathogens and related strains, formerly known as the Borrelia burgdorferi sensu lato species group, have been recently (and controversially) classified as a new spirochetal genus Borreliella (4, 5). In the United States, B. burgdorferi causes the majority of the Lyme disease cases, while new human-pathogenic Borreliella species (e.g., B. mayonii) continue to be discovered (6). Seven additional Borreliella species are formally recognized in North America, including B. americana, B. andersonii, B. bissettiae, B. californiensis, B. carolinensis, B, kurtenbachii, and B. lanei, although to date, there are no confirmed cases of human infection caused by these species (7–13). Borreliella species vary not only in genomic sequences but also in geographic distribution, host preferences, human pathogenicity, and disease manifestations (2, 6, 14–17). In addition, the Ixodes ticks in the United States and elsewhere are frequently coinfected with Borrelia miyamotoi, a member of the redefined Borrelia genus now consisting exclusively of strains grouped with agents of relapsing fever (14, 18).
A hallmark of Lyme disease endemics is the coexistence of multiple spirochete species and strains within local populations and oftentimes within a single vector, host, or patient (19–26). The high genetic diversity within local pathogen populations is to a large extent driven and maintained by frequency-dependent selection under which rare strains gain selective advantage over common ones in establishing superinfection in a host (20, 27–29). In addition, the diversification of local conspecific strains may be driven by host specificity and other phenotypes, including tissue invasiveness (19, 23, 26, 30). Against this backdrop of the vast geographic, genetic, and phenotypic variations of Lyme disease pathogens across the globe and within regions of endemicity, it is essential to develop accurate, sensitive, and scalable technologies for identifying species and strains of Lyme pathogens in order to understand, monitor, and control the range expansion of Lyme disease (16, 31, 32).
Early molecular technologies for identifying Lyme pathogen strains relied on amplifying and detecting genetic variations at a single variable locus, including the outer surface protein A locus (ospA), outer surface protein C locus (ospC), and the intergenic spacer regions of rRNA genes (rrs-rrlA and rrfA-rrlB) (21, 25, 33). The availability of the first Lyme pathogen genome facilitated the development of more sensitive multilocus sequence typing (MLST) technologies targeting genetic variations at a set of single-copy housekeeping genes (22, 30, 34, 35). For the direct identification of Lyme strains in tick and host specimens without first culturing and isolating the organisms, a reverse line blotting (RLB) technology has been developed on the basis of DNA-DNA hybridization (19, 36–38). The RLB technology, while sensitive and able to detect mixed infection in ticks and hosts, is difficult to scale up or to standardize and does not yield quantitative measures of strain diversity. A further limitation of the RLB technology is that it depends on oligonucleotide probes of known ospC major group alleles and is not able to detect strains with novel ospC alleles.
Next-generation sequencing (NGS) technologies circumvent the limitations of traditional methods in scalability, standardization, and ability for de novo strain detection while offering high sensitivity and high-throughput quantification (39). Using the hybridization capture technology to first enrich pathogen genomes from tick extracts and subsequently obtaining genome-wide short-read sequences using the Illumina NGS platform, >70% of field-collected nymphal ticks from the Northeast and Midwest United States were found to be infected with multiple B. burgdorferi strains due to a mixed inoculum (24). In an NGS-based study of European Lyme pathogen populations, a combination of quantitative PCR and high-throughput sequencing on the 454 pyrosequencing platform targeting the ospC locus revealed a similarly high rate (77.1%) of mixed infection of nymphal ticks by Borrelia afzelii and Borrelia garinii (20).
Here, we report an improved NGS technology for identifying Lyme pathogen strains through deep sequencing of ospC sequences amplified from individual ticks. We applied the technology to more than 100 pathogen-infected Ixodes scapularis ticks collected from New York State in a period of 2 years. Our results suggest a new putative Borreliella species, competitive interactions among coinfecting strains, and genetic homogeneity within a region of endemicity.
MATERIALS AND METHODS
Tick collection and DNA extraction.
Adult and nymphal blacklegged ticks (Ixodes scapularis) were captured by dragging canvas sheets across vegetation in 2015 and 2016 during their host-questing seasons from four locations in areas of endemicity of Lyme disease surrounding New York City (Fig. 1). The ticks were stored at −80°C before dissection. Each tick was immersed in a 5% solution of Chelex 100 resin (Sigma-Aldrich, St. Louis, MO, USA) containing 20 mg/ml proteinase K in Milli-Q water (EMD Millipore, Billerica, MA, USA) with total volumes of 30 μl for nymphs, 100 μl for male ticks, and 200 μl for female ticks. The ticks were dissected into four or more pieces using a sterilized scalpel or disposable pipette tips. The dissected mixture was incubated at 56°C overnight, heated to 100°C for 10 min in a dry bath, and then briefly centrifuged to separate the tick debris and Chelex resin from the supernatant. The supernatant containing the extracted DNA was transferred to a fresh tube and stored at 4°C (or frozen at −20°C for long-term storage).
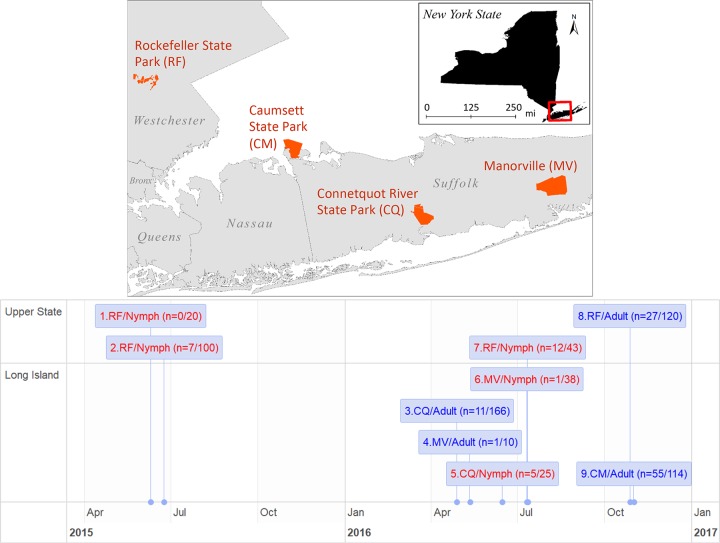
FIG 1.
Study sites and timeline. Nine cohorts of adult and nymphal Ixodes scapularis ticks were collected from four study sites in New York State, USA (top), during their host-questing seasons in a period of 18 months (bottom). Nymphal samples are colored in red and adult samples in blue. Numbers in parenthesis indicate the numbers of ticks infected by Lyme disease spirochetes (numerator) and the total sample size (denominator).
Single-round PCR amplification of full-length ospC.
A pair of new primer sequences targeting full-length ospC was designed according to available Borreliella genomes, with sequences 5′-AATAAAAAGGAGGCACAAATTAATG-3′ (Oc-Fwd, targeting the intergenic spacer between BB_B18 and BB_B19) and 5′-ATATTGACTTTATTTTTCCAGTTAC-3′ (Oc-Rev, targeting the intergenic spacer between BB_B19 and BB_B22). The alignments of primer regions flanking the ospC locus in a diverse set of Lyme pathogen genomes are provided in Section S1 in the supplemental material.
Each 20-μl reaction mixture contained 200 μM each deoxynucleoside triphosphate (dNTP), 1 U Roche FastStart Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany), 2 μl of 10× Roche FastStart buffer (Roche Diagnostics, California, USA), 0.4 μM each primer, and 1 μl DNA extract. The reaction mixture was heated at 95°C for 4 min, amplified for 36 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 60 s, and finally incubated at 72°C for 5 min. The PCR products were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and imaged under a UV light. An Agencourt AMPure XP PCR purification kit (Beckman Coulter, Brea, CA, USA) was used to remove excess primers, dNTPs, and other reagents. The amplicon quantity was measured on the Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) using the companying Qubit dsDNA HS assay kit.
NGS library preparation and short-read sequencing.
The Nextera XT DNA Library Prep (catalog no. FC-131-1024; Illumina, CA, USA) protocol was used to prepare the amplicon libraries for sequencing. First, the PCR products were diluted to 0.2 ng/μl after DNA quantification using a DNA 1000 kit on a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). The samples were tagmented by incubating 5 μl DNA sample at 55°C for 5 min in a solution containing 10 μl Tagment DNA buffer and 5 μl amplicon Tagment mix. The tagmentation reaction was terminated by adding 5 μl Neutralize Tagment buffer. Tagmented samples were amplified and barcoded (with set A and set B) by PCR in a solution containing 5 μl of each of the barcoded primers and the Nextera PCR master mix. The thermal cycling parameters were incubation at 72°C for 3 min, 95°C for 30 s, 12 cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s, and a final incubation at 72°C for 5 min. The indexed amplicon libraries were cleaned using an AMPure XP PCR purification kit and concentrations quantified using the High-Sensitivity DNA 1000 kit on a 2100 Bioanalyzer. The amplicon libraries were diluted to the same concentrations and then combined to a total concentration of 2 nM to 4 nM with a volume of 5 μl or more.
In preparation for loading in the MiSeq sequencer, the pooled library was denatured by mixing 5 μl of 0.2 N NaOH with 5 μl of sample, incubating at room temperature for 5 min, and then adding 990 μl prechilled hybridization buffer, resulting in a total of 1 ml (10 pM concentration) of denatured pooled amplicon library (Illumina denature and dilute libraries guide publication no. 15039740). Furthermore, 5% PhiX sequencing control (catalog no. FC-110-3001; Illumina, CA, USA) was added to the sample pool before loading to the MiSeq sequencer. The sequencing kit used was the MiSeq reagent kit v3 for 150 cycles (catalog no. MS-102-3001; Illumina, CA, USA) for paired-end reads of 75 bases at each end. After sequencing, a total of 4.24 gigabases of sequence were generated by the instrument, corresponding to 57,463,220 reads, with approximately 90% of the reads (52,311,968) passing the built-in filter for quality control. Finally, the samples were automatically demultiplexed to individual FASTQ files after completion of the sequencing run by the MiSeq reporter software on the basis of Nextera XT barcodes corresponding to each sample (the barcodes were trimmed by the software from each read).
Amplicon cloning and Sanger sequencing.
New alleles were identified when the majority of reads were not aligned to reference sequences. For such samples, a de novo assembly of short reads was performed to obtain candidate allele sequences (see below). The novel alleles were subsequently validated by cloning and Sanger sequencing. The cloning of PCR products was performed using the TOPO TA Cloning kit for sequencing (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's protocol. Five bacterial colonies containing plasmids with the PCR amplicons as inserts were selected for further growth in selective liquid medium. Plasmid DNA was extracted and purified using PureLink Quick plasmid miniprep kits (Thermo Fisher Scientific, Waltham, MA, USA). Nucleotide sequences of cloned PCR amplicons were obtained using the Sanger method through commercial sequencing services, including Genewiz (South Plainfield, NJ, USA) and Macrogen (Rockville, MD, USA).
Bioinformatics methods for allele identification and quantification.
Alleles present in tick samples were identified and quantified by aligning the paired-end short reads to a set of reference sequences. These reference sequences consisted of full-length ospC sequences and were obtained from published genome sequences, from Sanger sequencing of cloned or uncloned amplicons (see “Amplicon cloning and Sanger sequencing”), or from de novo assembly of short reads (see below) (the 20 reference sequences are listed in Section S2).
The short reads were indexed and aligned to the reference sequences using software packages bwa (40) and samtools (41). The coverage of reads at each site of each reference sequence was obtained by using bedtools (42) and visualized using ggplot2 in the R statistical computing environment (43, 44). The presence of new alleles was noted when a large number of reads were unmapped. For these samples, the PCR amplicons were cloned and then sequenced using Sanger sequencing (see “Amplicon cloning and Sanger sequencing”). A number of known alleles do not have full-length ospC sequences from sequenced genomes or from GenBank. For these alleles, de novo assembly of reads was performed to obtain the 5′- and 3′-end sequences using the assembler metaSPAdes (45).
To test the accuracy and sensitivity of the bioinformatics pipelines, simulated short reads with known allele identities and known proportions of allele mixtures were generated using wgsim, which is a part of the samtools package (41). Key steps and commands for allele identification, coverage calculation, de novo assembly, and simulated reads are provided in Section S3.
Statistical analysis of genetic diversity.
The relative amounts of spirochete loads in individual infected ticks were estimated by the weights (in ng) of PCR amplicons. The diversity of ospC alleles in individual infected ticks was first measured with multiplicity, i.e., the number of unique alleles present in an infected tick. Allelic diversity was more quantitatively measured with the Shannon diversity index, where pi is the frequency of allele i and n is the number of distinct alleles in an infected tick (allowing α = 0 for n = 1). This Shannon diversity index, also known as the Shannon equitability index, is a normalized measure of biodiversity ranging from 0 (infected with a single strain) to 1 (all strains being equally frequent) (46). Allele frequencies in an infected tick were obtained as, where Ci is the coverage of allele i averaged over all nucleotide positions.
The genetic differentiation between two populations (A and B) was measured with the Fst statistic (47), where HS(A), HS(B), and HT are the heterozygosity of sample A, sample B, and the total sample, respectively, and nA and nB are the sample sizes. The statistical significance of an Fst value was estimated by a randomization procedure by which the two population samples are combined and randomly divided into two pseudosamples with the same sample sizes. An Fst value was calculated between the two pseudosamples. The procedure was repeated 999 times, and a P value was obtained for the proportion of permutated Fst values that was greater than or equal to the observed value. Genetic differentiation was further tested using F statistics implemented in the hierfstat package in the R statistical computing environment (48).
Data availability.
New sequences have been deposited in GenBank with consecutive accessions MH071430 through MH071436. Experimental data have been archived in a custom relational database. An interactive website has been developed using the D3js (http://d3js.org) JavaScript library to visualize allele composition and read depth for the 119 infected ticks and is publicly available at http://diverge.hunter.cuny.edu/~weigang/ospC-sequencing/. Data sets and R scripts are publicly available at Github (https://github.com/weigangq/ocseq).
RESULTS
Tick infection rates, coinfections, specificity, and sensitivity.
Approximately 25% of nymphal ticks and 50% of adult ticks were infected with Borreliella species or Borrelia miyamotoi. For example, the nymphal infection rate for Borreliella burgdorferi was 27.9% (with a 95% confidence interval of 15.3% to 43.7%) in cohort 7, and the adult infection rate for Borreliella burgdorferi was 42.1% (48 of 114 ticks, 32.9% to 51.7% confidence interval) for cohort 9 (Fig. 1). The infection rate for Borrelia miyamotoi in adult ticks was 6.1% (7 of 114 ticks; 2.5% to 12.2% confidence interval) for cohort 9. Four ticks in cohort 9 were infected with both Borreliella burgdorferi and Borrelia miyamotoi (coinfection rate of 3.5%, 0.96% to 8.74% confidence interval). These rates are consistent with results from other studies conducted in the same region and appear to be stable through recent decades (29, 38). The rates from other cohorts were underestimates due to a lack of success in tick processing, DNA amplification, or NGS sequencing during protocol development. For example, ticks preserved in 95% alcohol yielded a lower amplification rate than those from the same cohort that were kept frozen in −80°C.
The sequencing outputs averaged ∼108,000 reads per tick sample. The coverage (i.e., read depth) of an allele depended on the total number of tick samples in a pooled library and the number of alleles present in a tick. Alleles were identified if the reads covered all nucleotide positions of a reference allele and the total read percentage was at least 1% of the most abundant allele. The total sample of 119 successfully sequenced ticks was divided into four subpopulations according to geographic origin and life stage, with allele counts of pathogens in each of the four populations listed in Table 1. The allele counts from adult ticks were much higher from Long Island samples (n = 192 alleles) than from upper state samples (n = 57 alleles). This is mostly due to the difference in the overall numbers of infected ticks (67 versus 27). If corrected for sample size, the relative allele counts were not significantly different (2.9 versus 2.1 alleles per infected adult tick, P = 0.3273 by chi-square test). Therefore, we pooled the numbers from different locations together in subsequent analyses of spirochete loads and strain multiplicity (see below).
TABLE 1.
Allele countsa
| Population | Origin (n) | Tick stage | No. of alleles |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | B3 | C | C14 | D | E | F | G | H | I | J | K | L | M | N | O | T | U | vsp | Total | |||
| 1 | Upper state (27) | Adult | 2 | 4 | 0 | 7 | 0 | 1 | 6 | 0 | 7 | 2 | 3 | 0 | 3 | 4 | 2 | 5 | 0 | 5 | 4 | 2 | 57 |
| 2 | Upper state (27) | Nymph | 3 | 3 | 0 | 1 | 1 | 1 | 4 | 1 | 1 | 3 | 0 | 0 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 3 | 32 |
| 3 | Long Island (67) | Adult | 12 | 8 | 1 | 4 | 0 | 5 | 12 | 18 | 16 | 10 | 14 | 4 | 17 | 0 | 8 | 8 | 8 | 24 | 13 | 10 | 192 |
| 4 | Long Island (6) | Nymph | 3 | 0 | 0 | 1 | 0 | 0 | 3 | 3 | 2 | 2 | 2 | 3 | 1 | 0 | 3 | 3 | 3 | 0 | 1 | 4 | 34 |
| Sum | 20 | 15 | 1 | 13 | 1 | 7 | 25 | 22 | 26 | 17 | 19 | 7 | 23 | 5 | 15 | 18 | 13 | 31 | 18 | 19 | 315 | ||
See Fig. 5 for allele frequency distributions.
The specificity of allele identification was tested by generating simulated reads from a single reference sequence and aligning the simulated reads to all reference sequences. This simulation-based test showed that the bioinformatics protocol for allele identification was highly specific, with only a small fraction of cross-aligned reads at the first ∼200 conserved positions for some ospC alleles (see Section S4 in the supplemental material).
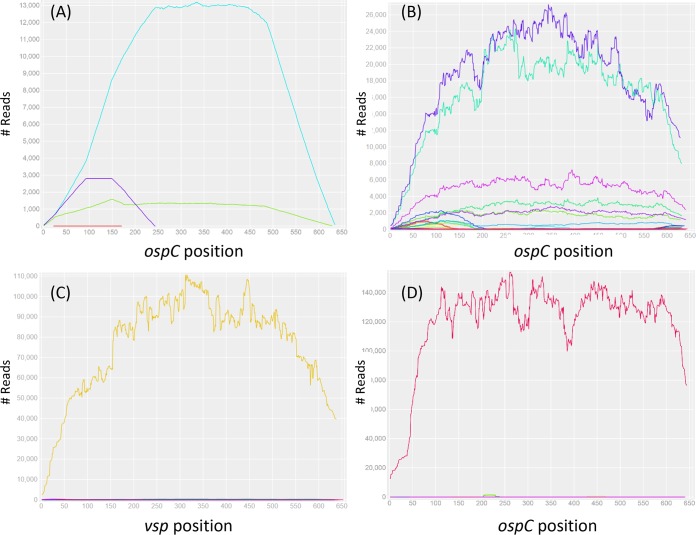
The sensitivity of allele quantification was tested by generating a known proportion of simulated reads from two reference sequences. For example, a 10:1 mixed sample of short reads generated on the basis of the sequences of alleles “J” and “F” was quantified using the bioinformatics protocol, resulting in an ∼13:1 quantification (Fig. 2A).
FIG 2.
Read depths of ospC alleles in simulated and tick samples.(A) Simulated reads were generated on the basis of nucleotide sequences of two ospC alleles (F and J) which were subsequently mixed in a 1F:10J proportion. The reads were aligned to all 20 reference sequences (see Section S2 in the supplemental material), and only the two input alleles (J, cyan; F, green; E, red; K, purple) showed complete full-length coverages and approximately the same input proportions, validating the specificity and sensitivity of the bioinformatics protocol for allele identification (a full test of specificity is shown in Section S4). (B) Seven ospC alleles (O, I, U, H, T, E, and K) were detected in an adult tick (M119, male, Rockefeller State Park [RF], fall of 2016). (C) The universal ospC primer set was able to amplify not only the ospC locus in Borreliella species but also the vsp locus in Borrelia miyamotoi (see alignment in Section S1). Here, the vsp locus was detected in a nymph tick (N030, RF, summer of 2015). (D) A previously unknown ospC allele (C14, GenBank accession MH071431) was detected in a nymph tick (N150, RF, summer of 2016), suggesting the presence of a new B. bissettiae-like species.
New strain, spirochete load, and multiplicity.
The NGS protocol was able to not only detect the presence of multiple strains but also to quantify their relative frequencies in individual ticks infected by multiple strains (Fig. 2B). Two new alleles not in the initial reference ospC sequences were discovered. Our protocol was able to detect infection by Borrelia miyamotoi, as shown by the presence of one of its vsp (variable surface protein gene, locus name AXH25_04790) amplicons in samples (Fig. 2C). The vsp allele was cloned, sequenced with the Sanger method, and assigned a GenBank accession (MH071435). Another allele (labeled C14_N150) had no known high-identity homologs in GenBank, with the top BLASTp hit being the B. bissettiae strain 25015 ospC with 75% identity in protein sequence (AAC45540) (49). This allele likely represents an unidentified Borreliella species (Fig. 2D). This allele was cloned, sequenced with the Sanger method, and assigned a GenBank accession (MH071431). The full-length F allele was cloned, sequenced with the Sanger method, and assigned a GenBank accession (MH071432). The full-length O allele (MH071435) was sequenced with the Sanger method directly from the singly infected tick N045 without cloning of the PCR amplicon. The sequences of full-length alleles “B3,” “N,” and “T” (MH071430, MH071433, and MH071436, respectively) were obtained by de novo assembly of short reads using metaSPAdes (45).
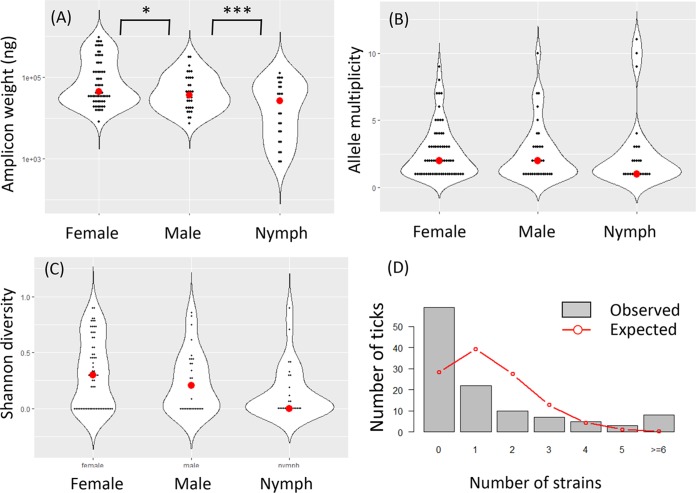
Assuming that the spirochete load is correlated with the total weight of PCR amplicons, we found that female adult ticks carried a significantly higher spirochete load than male adult ticks (P = 0.022 by Student's t test), which in turn carried a higher infection load than nymphal ticks (P = 8.1e−3) (Fig. 3A). There was no significant difference in the average numbers of strains infecting a single tick (P > 0.5 by Mann-Whitney U test), although the median values were two strains per infected adult tick and one strain per nymphal tick (Fig. 3B). Similarly, there were no significant differences in strain diversity measured by the Shannon diversity indexes between male, female, and nymphal ticks (P > 0.5 by Mann-Whitney U test) (Fig. 3C).
FIG 3.
Spirochete load and diversity. (A) Spirochete loads, estimated with the weight of amplicons (y axis, log10 scale), were significantly higher in female ticks than in males ticks, which in turn were higher than in nymphal ticks. (B) There were no significance differences among the three life stages in strain diversity as measured by the numbers of distinct strains within a tick (“multiplicity”). (C) Shannon diversity, which takes allele frequencies into account (see Materials and Methods), was also not significantly different among the tick stages. These results support the notion that strain diversity in individual ticks is contributed more by a mixed inoculum in hosts than by the number of blood meals (24). (D) Observed counts of infected and uninfected adult ticks (n = 144 from cohort 9) (see Fig. 1). Expected counts were based on a Poisson model assuming that strains infect ticks independently. The observed distribution shows an overabundance of uninfected ticks and ticks infected by five or more strains but an underabundance of ticks infected by 1 to 3 strains, suggesting reservoir hosts tend to be either uninfected or repeatedly infected.
Aggregated infection and negative strain interactions.
A previous study of multistrain infection by B. afzelii in Europe found that strains tend to be aggregated in infected ticks, suggesting that the infection of ticks and hosts was more successfully established by multiple spirochete strains than by a single strain alone (50). Our data support their conclusion. In cohort 9, for example, we detected a total of 159 ospC alleles in 55 infected ticks out of a total of 114 processed adult ticks. Assuming a Poisson model of independent infection among strains with an average successful infection rate λ = 159/114 = 1.395 strains per tick, we expected on average 28.2 uninfected ticks and 39.4 ticks infected by a single strain (the observed and expected counts are shown in Fig. 3D). In fact, 59 ticks were uninfected in this cohort, more than twice the expected count. Meanwhile, 22 ticks were infected by a single strain, approximately half of the expected number. It appeared that ticks tend to be either free of infection or infected by multiple spirochete strains, supporting the aggregated infection hypothesis (50).
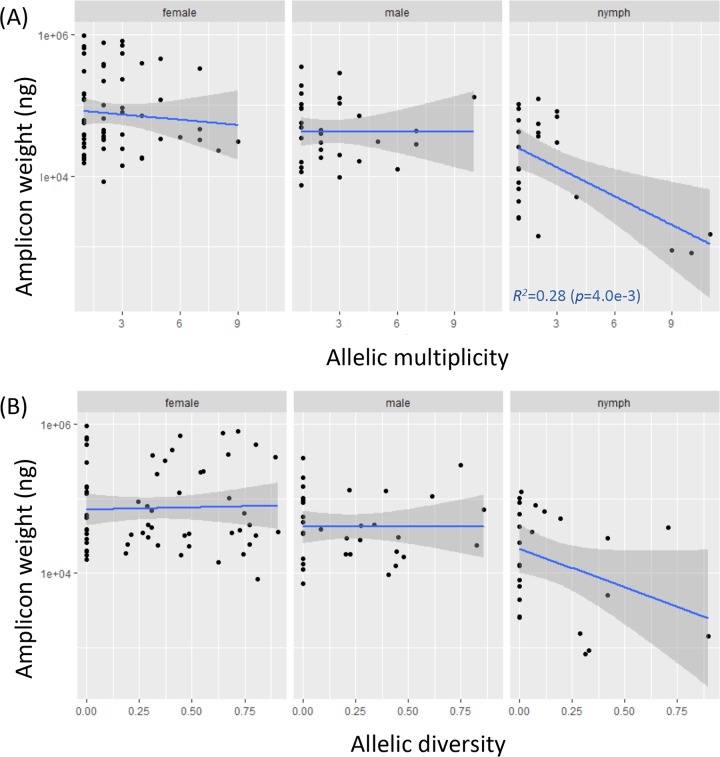
In infected ticks, previous studies concluded either a negative or a lack of interactions among coinfecting strains (20, 24, 50). Our analysis supports the presence of negative or inhibitory interactions among coinfecting strains. First, multiple strains tended to be unevenly distributed in their spirochete loads, with some strains dominating others (e.g., Fig. 2B). This is more generally shown with the Shannon diversity index, which was on average approximately half of the maximum attainable diversity (when all strains are equally abundant) in ticks with mixed infections (see Section S5). There was, however, no evidence that any particular strains were consistently more dominant than another (Section S5). Second, we plotted the total pathogen load with respect to multiplicity or the Shannon diversity index in individual ticks (Fig. 4). For the most part, the regression lines were not significantly different from a slope of zero, except that the spirochete load in nymphs decreased significantly with increasing numbers of strains. The overall flat trend supported a negative rather than facilitating interaction (20, 24). This is because if strains were independent from each other or facilitating each other's growth, one expects ticks infected with multiple strains to have a higher spirochete load than ticks infected with a single strain.
FIG 4.
Negative interactions among coinfecting strains. (A) Spirochete load was either flat or decreasing with increasing strain multiplicity, supporting inhibitory interactions among coinfecting strains (20, 24). (B) The pattern of inhibitory interactions was similar when strain diversity was measured by Shannon diversity.
Similar strain distributions among regions and life stages.
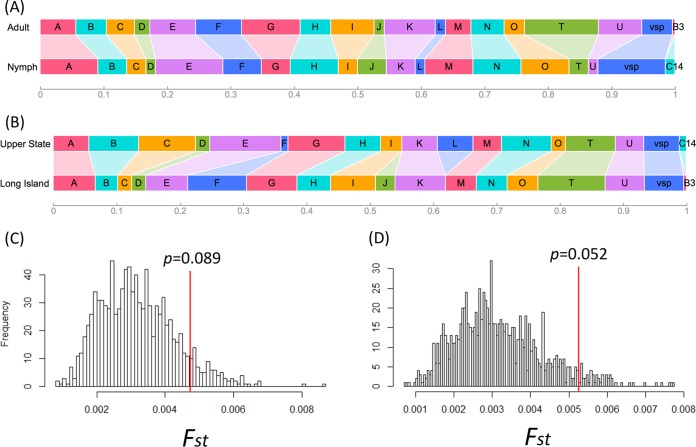
The spirochete populations infecting adult and nymph ticks were similar in strain composition (Fst = 4.7e−3 and P = 0.089 by resampling, P = 0.369 by F test) (Fig. 5A and C). The genetic differentiation between the upper state and Long Island populations was more pronounced but nonetheless lacked statistical significance (Fst = 5.3e−3 and P = 0.052 by resampling, P = 0.245 by F test). The groups F and J strains appeared to be more common on Long Island than in the upper state area, while the group L strain showed the opposite pattern of distribution (Fig. 5B and D).
FIG 5.
Geographic and life-stage differences in pathogen strain compositions. (A) Strain compositions (width of each colored rectangle representing frequency of an allele in a population sample) between those infecting adult ticks (population 1 [Pop1] plus Pop3) (see Table 1) and those infecting nymph ticks (Pop2 plus Pop4). (B) Strain compositions in two regional populations (upper state, Pop1 plus Pop2; Long Island, Pop3 plus Pop4). (C) There was no significant genetic difference in strain compositions between those infecting adult and those infecting nymph ticks (P value obtained by resampling 999 times; see Materials and Methods). (D) Two regional populations showed nonsignificant albeit stronger genetic differentiation.
DISCUSSION
In this report, we describe a new experimental and bioinformatics protocol for detecting and quantifying Lyme disease pathogen strains infecting individual ticks on the basis of next-generation sequencing technology. Improving upon the previous reverse line blotting (RLB) technology, the protocol enabled de novo detection of previously unknown pathogen strains. Indeed, one of the ticks carried a putative new Borreliella species with a novel ospC allele (C14, MH071431). The protocol was highly sensitive and specific, enabling the quantification of genetic diversity within single ticks and rigorously testing ecological hypotheses such as strain interactions (Fig. 4) and genetic differentiation (Fig. 5).
High-throughput sequencing has been used for the quantification of Lyme strains in ticks on the basis of either genome capture or ospC amplicons (20, 24). Our technology is novel for an improved PCR protocol by using a set of universal PCR primers able to amplify full-length ospC in all Borreliella species as well as one of the vsp loci in Borrelia miyamotoi (51) Furthermore, the PCR protocol is simplified from two rounds to a single round of thermal cycling. Due to these critical improvements in the PCR protocol, our method could be used for the detection and quantification of Lyme disease (and possibly relapsing fever) pathogens in clinical and wildlife specimens across their species ranges worldwide.
Genotyping Lyme pathogen strains in clinical samples is desirable for precise diagnosis and treatment. First, Borreliella strains vary greatly in disease symptomology as well as in their ability for within-body dissemination (6, 23, 26, 52). Second, at present, the strains causing Lyme disease in individual patients are rarely known, hindering clinical studies, including epitope identification and vaccine development (53). Third, strain identification in patients could serve as a clinical marker of reinfection or superinfection considering the lack of cross-immunity protection against heterologous strains (27). For disease control and prevention, the high genotypic diversity of Lyme pathogens in local communities is indicative of high Lyme disease risks, while relatively low strain diversity is indicative of leading edges of tick and pathogen range expansion (31, 54).
Our strain identification method is based on the assumption of a strict one-to-one correspondence (i,e., complete genetic linkage) between ospC alleles and B. burgdorferi strains. The ospC locus is the most polymorphic single-copy locus in the Borreliella genome (17). The linkage between the ospC locus and the whole genome is indeed nearly complete for Borreliella populations in the northeast United States (17, 55). In fact, the diversification of strains in local Borreliella populations is likely driven by frequency-dependent selection targeting the ospC locus (28, 32). However, the complete linkage between ospC and other genomic loci is not universal, and exceptions exist in the midwestern and southern United States as well as in Canadian populations due to recombination (30, 54, 56). Cross-species and cross-strain exchange of ospC alleles is also common in European populations. For example, whole-genome sequencing showed that the European B. burgdorferi strain BOL26 obtained its ospC and its flanking genes from a conspecific strain through horizontal gene transfer (32). For population samples elsewhere, therefore, it might be necessary to add a 2nd locus for strain identification (57). One complemental genetic marker could be the rRNA spacer (rrs-rrlA), which is a single-copy and highly variable locus (33). Experimental methods for high-throughput sequencing of the rrs-rrlA locus are yet to be developed.
While we were able to estimate the relative spirochete loads in individual ticks on the basis of the quantification of ospC amplicons (Fig. 2A), we have not attempted to directly quantify the number of spirochetes in infected ticks using methods such as quantitative PCR (20). In the future, we plan to quantify spirochete loads in individual ticks by running our experimental and bioinformatics procedures with known quantities of genomic DNA and generating a standard calibration curve.
Nonetheless, using relative estimates of spirochete loads in individual ticks, we were able to validate a number of hypotheses on multistrain infections. First, the lack of differences in strain diversity between the questing adult ticks, which have taken two blood meals, and the questing nymphal ticks, which have taken one blood meal (Fig. 3), supports the conclusion that strain diversity in individual ticks is for the most part due to a mixed inoculum in infected hosts (24). Second, the ticks were more likely to be infected by five or more strains than expected by chance (Fig. 4D), supporting the aggregated infection hypothesis (50). Rather than strains actively facilitating each other in establishing infections, however, strain aggregation in ticks may be a reflection of reservoir hosts being either free of spirochetes (in the case of resistant and healthy hosts) or infected by multiple strains (in the case of susceptible and weakened hosts). Regardless, it appears that once a host is infected by a strain, it becomes susceptible for superinfection by additional immunologically distinct strains (27). Third, we found an uneven distribution of strains in infected ticks as well as a flat or decreasing spirochete load with increasing strain diversity (Fig. 5), supporting inhibitory interactions among coinfecting strains driven by competitive growth in reservoir hosts (20, 24). Fourth, we found weak genetic differentiation between the eastern and northern suburbs of the New York City (Fig. 5B), suggesting either a recent common origin, similar reservoir hosts, or both. Fifth, we observed cocirculation of B. miyamotoi and other Borreliella species in the same area. The lower prevalence as well as minimal genetic diversity at ospC or vsp loci of these low-prevalence spirochetes relative to those of B. burgdorferi suggests that ospC hypervariability may be a key adaptation underlining the ecological success of B. burgdorferi in this region.
To summarize, we have established a next-generation-sequencing-based taxonomically broad procedure that has the potential to become a standard protocol for genotyping and quantifying Lyme disease pathogens across the globe. The increased sensitivity of high-throughput sequencing technologies employed here and elsewhere highlights the prevalence of multiple infections in wildlife samples and a pressing need for broad-spectrum vaccines for the control and prevention of Lyme disease (58, 59).
Supplementary Material
ACKNOWLEDGMENTS
We thank Li Zhai and Edward Skolnik of the New York University School of Medicine for facilitating cloning experiments and Brian Sulkow for participating in fieldwork.
This work was supported by Public Health Service grants AI107955 and AI139782 (to W.-G.Q.) from the National Institute of Allergy and Infectious Diseases (NIAID) and the grant MD007599 (to Hunter College) from the National Institute on Minority Health and Health Disparities (NIMHD) of the National Institutes of Health (NIH). Roy Nunez is supported by the grant GM60665 (to Hunter College) of the National Institute of General Medical Sciences (NIGMS) of NIH.
The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of NIAID, NIMHD, or NIH.
L.D. contributed to the study methodology, investigation, data curation, software, and visualization; Z.W. contributed to the methodology, investigation, and writing of the original draft; S.A. contributed to the methodology, investigation, and resources; C.Y. and A.L. contributed to the study investigation; L.L. contributed to the resources and investigation; C.D. contributed to the resources and visualization; R.N. and D.M.C. contributed resources; N.L.G. contributed to the supervision and funding acquisition; K.K. contributed to the study methodology, funding acquisition, and writing of the manuscript; W.-G.Q. contributed to the study conceptualization, funding acquisition, supervision, and writing of the original draft.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00940-18.
REFERENCES
- 1.Hengge UR, Tannapfel A, Tyring SK, Erbel R, Arendt G, Ruzicka T. 2003. Lyme borreliosis. Lancet Infect Dis 3:489–500. doi: 10.1016/S1473-3099(03)00722-9. [DOI] [PubMed] [Google Scholar]
- 2.Margos G, Vollmer SA, Ogden NH, Fish D. 2011. Population genetics, taxonomy, phylogeny and evolution of Borrelia burgdorferi sensu lato. Infect Genet Evol 11:1545–1563. doi: 10.1016/j.meegid.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz AM, Hinckley AF, Mead PS, Hook SA, Kugeler KJ. 2017. Surveillance for Lyme disease - United States, 2008–2015. MMWR Surveill Summ 66:1–12. doi: 10.15585/mmwr.ss6622a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adeolu M, Gupta RS. 2014. A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie Van Leeuwenhoek 105:1049–1072. doi: 10.1007/s10482-014-0164-x. [DOI] [PubMed] [Google Scholar]
- 5.Margos G, Marosevic D, Cutler S, Derdakova M, Diuk-Wasser M, Emler S, Fish D, Gray J, Hunfeldt K-P, Jaulhac B, Kahl O, Kovalev S, Kraiczy P, Lane RS, Lienhard R, Lindgren PE, Ogden N, Ornstein K, Rupprecht T, Schwartz I, Sing A, Straubinger RK, Strle F, Voordouw M, Rizzoli A, Stevenson B, Fingerle V. 2017. There is inadequate evidence to support the division of the genus Borrelia. Int J Syst Evol Microbiol 67:1081–1084. doi: 10.1099/ijsem.0.001717. [DOI] [PubMed] [Google Scholar]
- 6.Pritt BS, Mead PS, Johnson DKH, Neitzel DF, Respicio-Kingry LB, Davis JP, Schiffman E, Sloan LM, Schriefer ME, Replogle AJ, Paskewitz SM, Ray JA, Bjork J, Steward CR, Deedon A, Lee X, Kingry LC, Miller TK, Feist MA, Theel ES, Patel R, Irish CL, Petersen JM. 2016. Identification of a novel pathogenic Borrelia species causing Lyme borreliosis with unusually high spirochaetaemia: a descriptive study. Lancet Infect Dis 16:556–564. doi: 10.1016/S1473-3099(15)00464-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolan MC, Hojgaard A, Hoxmeier JC, Replogle AJ, Respicio-Kingry LB, Sexton C, Williams MA, Pritt BS, Schriefer ME, Eisen L. 2016. Vector competence of the blacklegged tick, Ixodes scapularis, for the recently recognized Lyme borreliosis spirochete Candidatus Borrelia mayonii. Ticks Tick Borne Dis 7:665–669. doi: 10.1016/j.ttbdis.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Marconi RT, Liveris D, Schwartz I. 1995. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol 33:2427–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margos G, Fedorova N, Kleinjan JE, Hartberger C, Schwan TG, Sing A, Fingerle V. 2017. Borrelia lanei sp. nov. extends the diversity of Borrelia species in California. Int J Syst Evol Microbiol 67:3872–3876. doi: 10.1099/ijsem.0.002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margos G, Lane RS, Fedorova N, Koloczek J, Piesman J, Hojgaard A, Sing A, Fingerle V. 2016. Borrelia bissettiae sp. nov. and Borrelia californiensis sp. nov. prevail in diverse enzootic transmission cycles. Int J Syst Evol Microbiol 66:1447–1452. doi: 10.1099/ijsem.0.000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margos G, Piesman J, Lane RS, Ogden NH, Sing A, Straubinger RK, Fingerle V. 2013. Borrelia kurtenbachii sp. nov., a widely distributed member of the Borrelia burgdorferi sensu lato species complex in North America. Int J Syst Evol Microbiol 64(Pt 1):128–130. doi: 10.1099/ijs.0.054593-0. [DOI] [PubMed] [Google Scholar]
- 12.Rudenko N, Golovchenko M, Grubhoffer L, Oliver JH. 2011. Borrelia carolinensis sp. nov., a novel species of the Borrelia burgdorferi sensu lato complex isolated from rodents and a tick from the south-eastern USA. Int J Syst Evol Microbiol 61(Pt 2):381–383. doi: 10.1099/ijs.0.021436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudenko N, Golovchenko M, Lin T, Gao L, Grubhoffer L, Oliver JH Jr. 2009. Delineation of a new species of the Borrelia burgdorferi sensu lato complex, Borrelia americana sp. nov. J Clin Microbiol 47:3875–3880. doi: 10.1128/JCM.01050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbour AG, Bunikis J, Travinsky B, Hoen AG, Diuk-Wasser MA, Fish D, Tsao JI. 2009. Niche partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am J Trop Med Hyg 81:1120–1131. doi: 10.4269/ajtmh.2009.09-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casjens SR, Di L, Akther S, Mongodin EF, Luft BJ, Schutzer SE, Fraser CM, Qiu W-G. 2018. Primordial origin and diversification of plasmids in Lyme disease agent bacteria. BMC Genomics 19:218. doi: 10.1186/s12864-018-4597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurtenbach K, Hanincová K, Tsao JI, Margos G, Fish D, Ogden NH. 2006. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol 4:660–669. doi: 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- 17.Mongodin EF, Casjens SR, Bruno JF, Xu Y, Drabek EF, Riley DR, Cantarel BL, Pagan PE, Hernandez YA, Vargas LC, Dunn JJ, Schutzer SE, Fraser CM, Qiu W-G, Luft BJ. 2013. Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: genome stability and adaptive radiation. BMC Genomics 14:693. doi: 10.1186/1471-2164-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagemakers A, Staarink PJ, Sprong H, Hovius JWR. 2015. Borrelia miyamotoi: a widespread tick-borne relapsing fever spirochete. Trends Parasitol 31:260–269. doi: 10.1016/j.pt.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Brisson D, Dykhuizen DE. 2004. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics 168:713–722. doi: 10.1534/genetics.104.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durand J, Herrmann C, Genné D, Sarr A, Gern L, Voordouw MJ. 2017. Multistrain infections with Lyme borreliosis pathogens in the tick vector. Appl Environ Microbiol 83:e02552-16. doi: 10.1128/AEM.02552-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman DS, Wang PW, Wang IN, Bosler EM, Luft BJ, Dykhuizen DE. 1996. Multiple infections of Ixodes scapularis ticks by Borrelia burgdorferi as revealed by single-strand conformation polymorphism analysis. J Clin Microbiol 34:652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu W-G. 2008. Wide distribution of a high-virulence Borrelia burgdorferi clone in Europe and North America. Emerg Infect Dis 14:1097–1104. doi: 10.3201/eid/1407.070880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang IN, Wormser GP, Schriefer ME, Luft BJ. 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun 67:3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter KS, Carpi G, Evans BR, Caccone A, Diuk-Wasser MA. 2016. Vectors as epidemiological sentinels: patterns of within-tick Borrelia burgdorferi diversity. PLoS Pathog 12:e1005759. doi: 10.1371/journal.ppat.1005759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang I-N, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. 1999. Genetic diversity of ospc in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wormser GP, Brisson D, Liveris D, Hanincová K, Sandigursky S, Nowakowski J, Nadelman RB, Ludin S, Schwartz I. 2008. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis 198:1358–1364. doi: 10.1086/592279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatia B, Hillman C, Carracoi V, Cheff BN, Tilly K, Rosa PA. 2018. Infection history of the blood-meal host dictates pathogenic potential of the Lyme disease spirochete within the feeding tick vector. PLoS Pathog 14:e1006959. doi: 10.1371/journal.ppat.1006959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haven J, Vargas LC, Mongodin EF, Xue V, Hernandez Y, Pagan P, Fraser-Liggett CM, Schutzer SE, Luft BJ, Casjens SR, Qiu W-G. 2011. Pervasive recombination and sympatric genome diversification driven by frequency-dependent selection in Borrelia burgdorferi, the Lyme disease bacterium. Genetics 189:951–966. doi: 10.1534/genetics.111.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.States SL, Brinkerhoff RJ, Carpi G, Steeves TK, Folsom-O'Keefe C, DeVeaux M, Diuk-Wasser MA. 2014. Lyme disease risk not amplified in a species-poor vertebrate community: similar Borrelia burgdorferi tick infection prevalence and OspC genotype frequencies. Infect Genet Evol 27:566–575. doi: 10.1016/j.meegid.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanincova K, Mukherjee P, Ogden NH, Margos G, Wormser GP, Reed KD, Meece JK, Vandermause MF, Schwartz I. 2013. Multilocus sequence typing of Borrelia burgdorferi suggests existence of lineages with differential pathogenic properties in humans. PLoS One 8:e73066. doi: 10.1371/journal.pone.0073066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilpatrick AM, Dobson ADM, Levi T, Salkeld DJ, Swei A, Ginsberg HS, Kjemtrup A, Padgett KA, Jensen PM, Fish D, Ogden NH, Diuk-Wasser MA. 2017. Lyme disease ecology in a changing world: consensus, uncertainty and critical gaps for improving control. Philos Trans R Soc Lond B Biol Sci 372:20160117. doi: 10.1098/rstb.2016.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu W-G, Martin CL. 2014. Evolutionary genomics of Borrelia burgdorferi sensu lato: findings, hypotheses, and the rise of hybrids. Infect Genet Evol 27:576–593. doi: 10.1016/j.meegid.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G, Liveris D, Mukherjee P, Jungnick S, Margos G, Schwartz I. 2014. Molecular typing of Borrelia burgdorferi. Curr Protoc Microbiol 34:12C.5.1–12C.5.31. doi: 10.1002/9780471729259.mc12c05s34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, Vugt R van, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 35.Margos G, Gatewood AG, Aanensen DM, Hanincová K, Terekhova D, Vollmer SA, Cornet M, Piesman J, Donaghy M, Bormane A, Hurn MA, Feil EJ, Fish D, Casjens S, Wormser GP, Schwartz I, Kurtenbach K. 2008. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc Natl Acad Sci U S A 105:8730–8735. doi: 10.1073/pnas.0800323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durand J, Jacquet M, Paillard L, Rais O, Gern L, Voordouw MJ. 2015. Cross-immunity and community structure of a multiple-strain pathogen in the tick vector. Appl Environ Microbiol 81:7740–7752. doi: 10.1128/AEM.02296-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morán Cadenas F, Rais O, Humair P-F, Douet V, Moret J, Gern L. 2007. Identification of host bloodmeal source and Borrelia burgdorferi sensu lato in field-collected Ixodes ricinus ticks in Chaumont (Switzerland). J Med Entomol 44:1109–1117. doi: 10.1093/jmedent/44.6.1109. [DOI] [PubMed] [Google Scholar]
- 38.Qiu W-G, Dykhuizen DE, Acosta MS, Luft BJ. 2002. Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the northeastern United States. Genetics 160:833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. 2015. Next-generation sequencing for infectious disease diagnosis and management. J Mol Diagn 17:623–634. doi: 10.1016/j.jmoldx.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 44.Wickham H. 2009. Ggplot2 elegant graphics for data analysis. Springer Science & Business Media, Berlin, Germany. [Google Scholar]
- 45.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidakovic B. 2011. Statistics for bioengineering sciences: with MATLAB and WinBUGS support. Springer Science & Business Media, Berlin, Germany. [Google Scholar]
- 47.Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70:3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goudet J. 2005. hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 49.Tilly K, Casjens S, Stevenson B, Bono JL, Samuels DS, Hogan D, Rosa P. 1997. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol 25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 50.Andersson M, Scherman K, Råberg L. 2013. Multiple-strain infections of Borrelia afzelii: a role for within-host interactions in the maintenance of antigenic diversity? Am Nat 181:545–554. doi: 10.1086/669905. [DOI] [PubMed] [Google Scholar]
- 51.Barbour AG. 2016. Multiple and diverse vsp and vlp sequences in Borrelia miyamotoi, a hard tick-borne zoonotic pathogen. PLoS One 11:e0146283. doi: 10.1371/journal.pone.0146283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallais F, De Martino SJ, Sauleau EA, Hansmann Y, Lipsker D, Lenormand C, Talagrand-Reboul E, Boyer PH, Boulanger N, Jaulhac B, Schramm F. 2018. Multilocus sequence typing of clinical Borreliella afzelii strains: population structure and differential ability to disseminate in humans. Parasit Vectors 11:374. doi: 10.1186/s13071-018-2938-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baum E, Randall AZ, Zeller M, Barbour AG. 2013. Inferring epitopes of a polymorphic antigen amidst broadly cross-reactive antibodies using protein microarrays: a study of OspC proteins of Borrelia burgdorferi. PLoS One 8:e67445. doi: 10.1371/journal.pone.0067445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyler S, Tyson S, Dibernardo A, Drebot M, Feil EJ, Graham M, Knox NC, Lindsay LR, Margos G, Mechai S, Van Domselaar G, Thorpe HA, Ogden NH. 2018. Whole genome sequencing and phylogenetic analysis of strains of the agent of Lyme disease Borrelia burgdorferi from Canadian emergence zones. Sci Rep 8:10552. doi: 10.1038/s41598-018-28908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casjens SR, Gilcrease EB, Vujadinovic M, Mongodin EF, Luft BJ, Schutzer SE, Fraser CM, Qiu W-G. 2017. Plasmid diversity and phylogenetic consistency in the Lyme disease agent Borrelia burgdorferi. BMC Genomics 18:165. doi: 10.1186/s12864-017-3553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mechai S, Margos G, Feil EJ, Lindsay LR, Ogden NH. 2015. Complex population structure of Borrelia burgdorferi in southeastern and south central Canada as revealed by phylogeographic analysis. Appl Environ Microbiol 81:1309–1318. doi: 10.1128/AEM.03730-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbour AG, Cook VJ. 2018. Genotyping strains of Lyme disease agents directly from ticks, blood, or tissue. Methods Mol Biol 1690:1–11. doi: 10.1007/978-1-4939-7383-5_1. [DOI] [PubMed] [Google Scholar]
- 58.Earnhart CG, Buckles EL, Marconi RT. 2007. Development of an OspC-based tetravalent, recombinant, chimeric vaccinogen that elicits bactericidal antibody against diverse Lyme disease spirochete strains. Vaccine 25:466–480. doi: 10.1016/j.vaccine.2006.07.052. [DOI] [PubMed] [Google Scholar]
- 59.Livey I, O'Rourke M, Traweger A, Savidis-Dacho H, Crowe BA, Barrett PN, Yang X, Dunn JJ, Luft BJ. 2011. A new approach to a Lyme disease vaccine. Clin Infect Dis 52 Suppl 3:s266–s270. doi: 10.1093/cid/ciq118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New sequences have been deposited in GenBank with consecutive accessions MH071430 through MH071436. Experimental data have been archived in a custom relational database. An interactive website has been developed using the D3js (http://d3js.org) JavaScript library to visualize allele composition and read depth for the 119 infected ticks and is publicly available at http://diverge.hunter.cuny.edu/~weigang/ospC-sequencing/. Data sets and R scripts are publicly available at Github (https://github.com/weigangq/ocseq).