In 2012, VALidation of human papillomavirus (HPV) GENotyping Tests (VALGENT) was initiated to provide a formalized and uniform study framework for comparison and validation of HPV assays with genotyping capability. In VALGENT-3, the clinical and analytical performance of Anyplex II HPV HR detection (Anyplex) was compared to that of the Hybrid Capture 2 HPV DNA test (hc2) and the cobas 4800 HPV test (cobas).
KEYWORDS: cervical cancer screening, human papillomavirus, VALGENT, Anyplex, Hybrid Capture 2, cobas, clinical validation, analytical evaluation
ABSTRACT
In 2012, VALidation of human papillomavirus (HPV) GENotyping Tests (VALGENT) was initiated to provide a formalized and uniform study framework for comparison and validation of HPV assays with genotyping capability. In VALGENT-3, the clinical and analytical performance of Anyplex II HPV HR detection (Anyplex) was compared to that of the Hybrid Capture 2 HPV DNA test (hc2) and the cobas 4800 HPV test (cobas). The panel comprises 1,300 stored samples that were obtained from women 25 to 64 years old who participated in the Slovenian cancer screening program, enriched with 300 samples from women with abnormal cervical cytology. The sensitivity and specificity of Anyplex were noninferior to those of hc2, with a relative sensitivity of 1.01 (95% confidence interval [CI], 0.97 to 1.04) for cervical intraepithelial neoplasia grade 2 or worse (CIN2+) and 1.01 (95% CI, 0.97 to 1.06) for CIN3+ and relative specificity of 1.02 (95% CI, 1.00 to 1.03) for a CIN grade of ≤1. The clinical sensitivity of Anyplex for CIN2+ and CIN3+ was comparable to that of hc2 (P values for McNemar test [pMcN] of 0.655 and 0.564, respectively), but its specificity was significantly higher (pMcN = 0.008). The sensitivity and specificity of Anyplex were also noninferior to those of cobas, with relative sensitivity of 1.01 (95% CI, 0.98 to 1.04) for CIN2+ and 1.01 (95% CI, 0.99 to 1.04) for CIN3+ and relative specificity of 1.00 (95% CI, 0.99 to 1.01) (pMcN value of >0.05 in all cases). Regardless of the clinical outcome (CIN2+ or CIN3+), age restriction (women ≥30 years old), or comparator test used, Anyplex consistently showed excellent clinical performance and can be considered validated for primary cervical cancer screening.
INTRODUCTION
The development of cervical cancer and its immediate precursors is inevitably associated with persistent infection with high-risk human papillomaviruses (hrHPV). Several large randomized trials and screening cohort studies in the United States (1, 2) and in Europe (3–8) have shown that HPV testing as a stand-alone test or in combination with cytology (cotesting) is superior to cytology alone for detection of underlying cervical intraepithelial neoplasia grade 2 or worse (CIN2+) (3, 9), provides better protection against invasive cervical cancer, and allows prolonged intervals between screening rounds (5, 10–12). These findings led to the implementation of HPV testing in organized cervical cancer screening programs (9, 13–18) and simultaneously to the dramatic increase in the number of commercially available HPV tests during the last decade (19). However, only clinically validated HPV tests that fulfill the minimum requirements as set in the international consensus guidelines should be used in primary cervical cancer screening (20, 21). Thus, VALGENT (VALidation of HPV GENotyping tests) was initiated in 2012 to provide a formalized and uniform study framework for comparison and validation of HPV assays with genotyping capability. Thus far, three VALGENT panels (VALGENT-1 to -3) were completed, and samples were provided from Belgium, Scotland, and Slovenia, respectively, with eight HPV assays evaluated through this framework thus far (22–31).
This VALGENT-3 study evaluated both clinical and analytical performance of the Anyplex II HPV HR detection test (Anyplex; Seegene, Seoul, South Korea). In addition to a standard comparator test, the Hybrid Capture 2 HPV DNA test (hc2; Qiagen, Gaithersburg MD, USA), the performance of Anyplex was evaluated against the cobas 4800 HPV test (cobas; Roche Molecular Systems, Alameda, CA, USA), because cobas has been extensively evaluated, is FDA approved, and has consistently shown good clinical performance in cohort studies as well as in longitudinal studies (21, 32–37). Finally, analytical agreement between Anyplex and cobas was assessed.
MATERIALS AND METHODS
VALGENT-3 and study population.
VALGENT-3 comprises a total of 1,600 samples. Of them, 1,300 were samples from the Slovenian HPV prevalence study, collected between December 2009 and August 2010, as described in detail previously (30, 31, 38). Briefly, cervical specimens were collected from women 25 to 64 years old who participated in the organized cancer screening program in Slovenia, which had more than 73% national coverage of the target population (39). In accordance with the VALGENT protocol, the general screening population was enriched with a total of 300 consecutive samples obtained from women referred to the largest colposcopy clinic in the country (University Medical Centre Ljubljana) until 100 women with atypical squamous cervical cells of undetermined significance (ASC-US), 100 women with low-grade squamous intraepithelial lesion (LSIL), and 100 women with high-grade squamous intraepithelial lesion (HSIL) were enrolled (27). All women were referred to colposcopy according to the criteria of the Slovenian National Cervical Cancer Screening Program (atypical squamous cells that cannot exclude HSIL [ASC-H] or worse) or due to HPV16/18 positivity, irrespective of cytological findings. Colposcopically directed punch biopsy specimens were obtained from the suspicious areas and histopathologically assessed by certified pathologists.
Two cervical specimens were obtained from each woman, one for traditional cytological examination and the second for HPV testing. A sample for HPV testing was placed into ThinPrep PreservCyt solution (Hologic, Marlborough, MA), transported to the laboratory, labeled with an anonymous study number upon arrival, and split into several aliquots. Samples were stored at −70°C until testing.
This study was approved by the Medical Ethics Committee of the Republic of Slovenia (consent numbers 83/11/09 and 109/08/12).
HPV testing.
All samples were tested with Anyplex, cobas, and hc2. hc2 is a semiquantitative test designed to detect 13 hrHPV genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) and is one of the two standard comparator tests recommended by the Meijer guidelines for clinical evaluation of new HPV tests for use in primary cervical cancer screening (20). Results of hc2 testing were described previously (30, 31).
Anyplex is designed to simultaneously detect and quantify 14 hrHPV genotypes (all 13 genotypes included in hc2 as well as HPV66). It is a fully automated real-time PCR that includes three basic steps: DNA extraction, PCR amplification, and target detection. DNA extraction was performed from 400 μl of original ThinPrep sample using an automated purification system, NIMBUS IVD (Hamilton, Reno, NV, USA), with subsequent PCR amplification on a CFX96 real-time thermocycler (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. The Anyplex assay utilizes TOCE technology (tagging oligonucleotide cleavage and extension), with key components named DPO (dual priming oligonucleotide primer), Pitcher (a tagging oligonucleotide), and Catcher (a fluorescently labeled artificial template with a sequence complementary to the tagging portion of Pitcher). The DPO and Pitcher hybridize specifically on opposite sides of the target sequence of the HPV nucleic acid. The tagging portion of Pitcher is released during the DPO primer extension with Taq polymerase, which enables its hybridization to the capturing portion of Catcher. When “Duplex Catcher” (the tagging portion of Pitcher and the complementary Catcher sequence) is fully extended, it separates the reporter molecule from the quencher molecule, which results in a fluorescent signal. As an internal control, the human housekeeping gene is coamplified simultaneously with the L1 gene of the targeted HPV types. Data analysis of the test result is generated automatically using Anyplex software (40–42). Up to 40 clinical samples per run could be processed by the automated system in less than 6 h: DNA extraction takes 2 h and PCR amplification 3 h and 40 min. Less than 30 min of hands-on time is required for specimen and PCR machine handling (42).
cobas is an automated multiplex real-time PCR-based test that allows detection of 14 hrHPV types with concurrent distinction of HPV16 and HPV18 from 12 other hrHPV genotypes (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68). Cobas was launched in 2009 and has been thoroughly evaluated in several clinical trials during the last decade (21, 32–37).
All assays were performed according to the manufacturer's instructions at the Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana. Testing of all samples with Anyplex was performed between July 2016 and September 2016 and with cobas between January 2015 and July 2015. hc2 testing was performed, as described previously (30, 31), within 2 weeks after the sample was obtained; thus, the testing of the screening set was performed between December 2009 and September 2010, and testing of the enrichment set was between January 2014 and June 2015.
Clinical outcomes and statistical analysis.
For sensitivity estimates, we included all women with histologically confirmed high-grade lesions (CIN2 or worse) and CIN3+. Women that had two consecutive normal cytology results (at enrollment and after at least 12 months within the 48 months of follow-up) were considered a control group to assess the clinical specificity for ≤CIN1. The clinical performance of hc2, Anyplex, and cobas was evaluated separately for women <30 years old, women ≥30 years old, and the total study population. The clinical accuracy of Anyplex was compared to that of (i) a standard comparator assay (hc2) and (ii) cobas using a noninferiority statistic with a relative sensitivity and specificity threshold of 0.90 and 0.98, respectively (20). The level of statistical significance was set at a value of 0.05, with a P value for McNemar test above 0.05 (pMcN > 0.05) indicating that the observed difference in sensitivity and specificity is not statistically significant, whereas the P value in noninferiority testing below 0.05 (pNi < 0.05) indicates that the observed sensitivity or specificity of the evaluated test is not lower than that of the comparator test. In addition, the overall and HPV type-specific agreement between Anyplex and cobas was assessed using Cohen's kappa statistic with corresponding 95% confidence intervals (CIs) (43). Analysis and evaluation of the data were performed at the Unit of Cancer Epidemiology, Scientific Institute of Public Health (Brussels, Belgium), using STATA version 14 (College Station, TX, USA).
RESULTS
Out of 1,600 samples, two and nine samples had invalid internal control amplification when tested with Anyplex and cobas, respectively. All nine samples with cobas invalid internal control amplification were from the screening population and tested negative for the presence of HPV using hc2 (however, hc2 has no internal control). Thus, 1,598 samples were included in the clinical comparison between Anyplex and hc2 and 1,591 samples in the analytical and clinical comparison between Anyplex and cobas. Out of 1,598 samples included in the clinical comparison between Anyplex and hc2, 1,011 women ≥30 years old had two consecutive normal cytology results and were included in the clinical specificity calculation, and 98 women ≥30 years old were identified with CIN2+ and used to calculate clinical sensitivity. Similarly, out of 1,591 samples included in the clinical comparison between Anyplex and cobas, 1,005 women ≥30 years old had two consecutive normal cytology results and were included in the clinical specificity calculation, and 98 women ≥30 years old were identified with CIN2+ and used to calculate clinical sensitivity. In the total study population, 1,214 and 1,207 women had two consecutive normal cytology results and were included in the clinical specificity calculation when comparing Anyplex to hc2 and Anyplex to cobas, respectively, and 127 women with CIN2+ were used in clinical sensitivity calculation.
Type-specific HPV prevalence using Anyplex by cytology results.
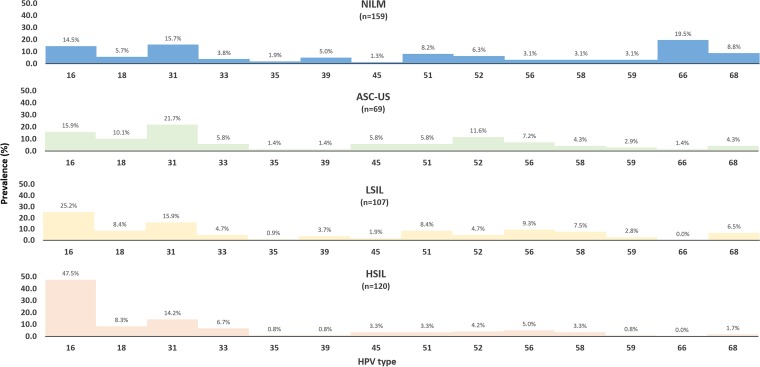
The overall prevalence of hrHPV types as measured by Anyplex, cobas, and hc2 in the screening population were 11.1% (144/1,298; 95% CI, 9.4 to 12.9), 11.0% (142/1,291; 95% CI, 9.3 to 12.8), and 12.2% (159/1,300; 95% CI, 10.5 to 14.1). The type-specific prevalence of HPV type according to the Anyplex result by cytology result was assessed for the total study population (Fig. 1), with an observed positive trend for HPV positivity according to the severity of the cytological result.
FIG 1.
Type-specific prevalence according to the cytology results by Anyplex in the total study population. Abbreviations: NILM, negative for intraepithelial lesion or malignancy; ASC-US, atypical squamous cervical cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion.
Clinical evaluation of Anyplex.
The absolute clinical sensitivity for detecting CIN2+ and CIN3+ and clinical specificity for ≤CIN1 of Anyplex, hc2, and cobas are shown in Table 1, separately for women ≥30 years old and for the total study population, and in Table S1 in the supplemental material for women <30 years old. In women ≥30 years old, Anyplex identified 95/98 CIN2+ and 65/66 of CIN3+ (number of positive sample out of the total number of samples), which corresponds to a clinical sensitivity of 96.9% (95% CI, 91.3 to 99.4) for CIN2+ and 98.5% (95% CI, 91.8 to 100.0) for CIN3+. In the total study population, clinical sensitivity for CIN2+ and CIN3+ was 96.9% (123/127; 95% CI, 92.1 to 99.1) and 98.8% (81/82; 95% CI, 93.4 to 100.0), respectively. Anyplex was negative for hrHPV in 951/1,011 and 1,111/1,214 samples of women with two consecutive negative cytology results, corresponding to a clinical specificity of 94.1% (95% CI, 92.4 to 95.4) and 91.5% (95% CI, 89.8 to 93.0) for women ≥30 years old and the total study population, respectively.
TABLE 1.
Comparison of clinical sensitivity for detection of CIN2+ and CIN3+ with clinical specificity for ≤CIN1 of Anyplex, hc2, and cobasa
| Study group and test | Clinical sensitivity (%) for: |
Clinical specificity (%) | |
|---|---|---|---|
| CIN2+ | CIN3+ | ||
| Women ≥30 years old | |||
| Anyplex | 96.9 (95/98; 91.3–99.4) | 98.5 (65/66; 91.8–100.0) | 94.1 (951/1,011; 92.4–95.4) |
| hc2 | 95.9 (94/98; 89.9–98.9) | 97.0 (64/66; 89.5–99.6) | 92.7 (937/1,011; 90.9–94.2) |
| cobas | 96.9 (95/98; 91.3–99.4) | 97.0 (64/66; 89.5–99.6) | 93.9 (944/1,005; 92.2–95.3) |
| Total study population | |||
| Anyplex | 96.9 (123/127; 92.1–99.1) | 98.8 (81/82; 93.4–100.0) | 91.5 (1,111/1,214; 89.8–93.0) |
| hc2 | 96.1 (122/127; 91.1–98.7) | 97.6 (80/82; 91.5–99.7) | 90.1 (1,094/1,214; 88.3–91.8) |
| cobas | 96.1 (122/127; 91.1–98.7) | 97.6 (80/82; 91.5–99.7) | 91.4 (1,103/1,207; 89.7–92.9) |
Testing was performed on the total study population and on women ≥30 years old. Data in parentheses are total positive for sensitivity or specificity out of the total number of samples and 95% CI.
The performance of Anyplex relative to hc2 and cobas separately for women ≥30 years old and for the total study population is shown in Table 2 and for women <30 years old in Table S2. The sensitivity and specificity of Anyplex were noninferior to those of hc2, with a relative sensitivity of 1.01 (95% CI, 0.97 to 1.04) for CIN2+ and 1.01 (95% CI, 0.97 to 1.06) for CIN3+ and a relative specificity of 1.02 (95% CI, 1.00 to 1.03) for ≤CIN1. Whereas the clinical sensitivity of Anyplex for CIN2+ and CIN3+ was comparable to that of hc2 (pMcN = 0.655 and pMcN = 0.564, respectively), its specificity was significantly higher (pMcN = 0.008). These observations did not differ if the analysis was confined to women ≥30 years old. If cobas was used as a comparator test, the sensitivity and specificity of Anyplex remained noninferior, as shown in Table 2, with a relative sensitivity of 1.01 (95% CI, 0.98 to 1.04) for CIN2+ and 1.01 (95% CI, 0.99 to 1.04) for CIN3+ and relative specificity of 1.00 (95% CI, 0.99 to 1.01) (pMcN value of >0.05 in all cases). Similar results were obtained when the analysis was restricted to women ≥30 years old.
TABLE 2.
Relative sensitivity and specificity of Anyplex compared to those of hc2 and cobas in women ≥30 years old and in the total study populationa
| Comparison by group and clinical outcome | Relative sensitivity | Relative specificity | pMcN | pNi |
|---|---|---|---|---|
| Anyplex vs hc2 | ||||
| Women ≥30 old | ||||
| CIN2+ | 1.01 (0.96–1.06) | 0.655 | 0.001 | |
| CIN3+ | 1.02 (0.96–1.07) | 0.564 | 0.003 | |
| ≤CIN1 | 1.01 (1.00–1.03) | 0.013 | 0.000 | |
| Total study population | ||||
| CIN2+ | 1.01 (0.97–1.04) | 0.655 | <0.001 | |
| CIN3+ | 1.01 (0.97–1.06) | 0.564 | 0.001 | |
| ≤CIN1 | 1.02 (1.00–1.03) | 0.008 | <0.001 | |
| Anyplex vs cobas | ||||
| Women ≥30 old | ||||
| CIN2+ | 1.00 (0.97–1.03) | 1.000 | 0.002 | |
| CIN3+ | 1.02 (0.99–1.05) | 0.317 | 0.003 | |
| ≤CIN1 | 1.00 (0.99–1.01) | 0.827 | 0.002 | |
| Total study population | ||||
| CIN2+ | 1.01 (0.98–1.04) | 0.564 | <0.001 | |
| CIN3+ | 1.01 (0.99–1.04) | 0.317 | 0.001 | |
| ≤CIN1 | 1.00 (0.99–1.01) | 0.858 | <0.001 |
pMcN, P value for McNemar test; pNi, P value in noninferiority testing.
Analytical agreement for hrHPV between Anyplex and cobas.
The overall agreement for hrHPV positivity (97.4% [1,549/1,591; 95% CI, 96.4 to 98.1]) and type-specific agreement for HPV16 (99.4% [1,582/1,591; 95% CI, 98.9 to 99.7]), HPV18 (99.7% [1,587/1,591; 95% CI, 99.3 to 99.9]), and other hrHPV (97.0% [1,543/1,591; 95% CI, 96.0 to 97.7]) between Anyplex and cobas was consistently high (Table 3). These values correspond to kappa values of 0.923 (95% CI, 0.900 to 0.946), 0.959 (95% CI, 0.932 to 0.986), 0.942 (95% CI, 0.884 to 0.999), and 0.885 (95% CI, 0.853 to 0.917), indicating excellent agreement throughout all categories.
TABLE 3.
Analytical agreement between Anyplex and cobas in the total study population stratified by all possible HPV genotyping results
| Anyplex result | cobas result |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HPV16 | HPV18 | Other hrHPV | HPV16, HPV18 | HPV16, other hrHPV | HPV18, other hrHPV | HPV16, HPV18, other hrHPV | Negative | Total | |
| HPV16 | 75 | 4 | 4 | 83 | |||||
| HPV18 | 13 | 2 | 1 | 16 | |||||
| Other hrHPV | 180 | 2 | 2 | 15 | 199 | ||||
| HPV16, HPV18 | 3 | 1 | 1 | 5 | |||||
| HPV16, other hrHPV | 2 | 2 | 26 | 1 | 31 | ||||
| HPV18, other hrHPV | 1 | 12 | 13 | ||||||
| HPV16, HPV18, other hrHPV | 1 | 1 | |||||||
| Negative | 21 | 1,222 | 1,243 | ||||||
| Total | 77 | 14 | 203 | 3 | 33 | 16 | 2 | 1,243 | 1,591 |
DISCUSSION
Anyplex is an automated system that allows simultaneous detection and provides genotype information for 14 hrHPV types in a single reaction. Because Anyplex utilizes relatively new technology, data on clinical performance are scant; however, all previous studies indicate excellent clinical performance. The first study to assess the clinical performance of Anyplex was published in 2016, in which Anyplex was compared to hc2 on a total of 1,137 samples derived from a Korean screening population. In that study, Jung et al. reported a clinical sensitivity of 92.5% (95% CI, 84.3 to 100.0) for CIN2+ and clinical specificity of 87.5% (95% CI, 77.3 to 99.7) and noninferiority compared to hc2 (41). In the following year, Hesselink et al. further confirmed previous findings in a study in which Anyplex was compared to the other standard comparator according to international guidelines: GP5+/6+ PCR enzyme immunoassay (EIA). Anyplex had a clinical sensitivity for CIN2+ of 98.3% (95% CI, 89.1 to 99.8) and a clinical specificity of 93.6 (95% CI, 90.3 to 96.5), and it was shown to be noninferior to the standard comparator test (42). Similar to previous studies, Anyplex demonstrated clinical sensitivity comparable to that of the standard comparator (hc2) as well as to cobas both in women ≥30 years old and in the total study population (pNi of <0.001 in all cases) in our study. Although the clinical specificity of Anyplex assessed for women with two consecutive negative cytology results in our study was similar to that of cobas, the difference was significantly higher than that of hc2 in both age groups (pMcN value of <0.05 for both). Thus, regardless of the comparator test (hc2, GP5+/6+ EIA PCR, or cobas) or medium (ThinPrep or HuroPath) used, clinical outcome (CIN2+ or CIN3+), and age restriction (women ≥30 years old), Anyplex consistently showed noninferior clinical sensitivity and specificity.
Another requirement for the use of an hrHPV test in primary cervical cancer screening is high intra- and interlaboratory reproducibility of the newly developed HPV test compared to the standard comparator. This has also been demonstrated in both of the studies mentioned above (41, 42), in which intralaboratory agreement (96.3% and 94.3%, respectively) and interlaboratory agreement (95.5% and 95.3%, respectively) of Anyplex were far above the given lower confidence bound (higher than 87.0%), with kappa values varying between 0.910 and 0.953 (well above the cutoff of 0.5), confirming excellent reliability and reproducibility of Anyplex (41, 42).
In 2016, analytical comparison between Anyplex, hc2, and cobas was assessed on 400 prospectively collected samples. The level of agreement between Anyplex and cobas was 99.5% for HPV16, 99.8% for HPV18, and 98.8% for other hrHPV, which corresponds to kappa values of 0.98, 0.96, and 0.97, respectively. These results are very similar to those obtained in our study, in which the overall and type-specific agreement were above 97% with kappa values above 0.880 throughout all categories, indicating very good agreement between the two tests. Our results further support previous observations that Anyplex is a robust test with performance comparable to that of other already established HPV tests currently used in clinical practice (44). Furthermore, Cornall et al. compared the clinical performance of four different HPV tests (hc2, cobas, Amplicor HPV test [Roche], and Anyplex) in a cohort of 404 Australian women undergoing management because of high-grade lesions. The clinical sensitivity and specificity of Anyplex were not significantly different from those of any other HPV test, with the exception of Anyplex compared to hc2, which showed significantly higher clinical sensitivity (P < 0.0001) (45).
The strength of this study is direct clinical comparison of Anyplex with the two HPV tests clinically validated in large prospective and/or randomized trials with an abundance of published performance data (hc2 and cobas). Furthermore, the VALGENT-3 framework (once completed) will allow unique comparison between 12 different HPV tests currently used worldwide on a standardized set of cervical specimens using international consensus methodology. An additional strength of our study is that we have calculated clinical specificity for ≤CIN1 using a very strict approach, where only women that had two consecutive normal cytology results (at enrollment and after at least 12 months within the 48 months of follow-up) were considered a control group. However, since the screening population (1,300 women) was artificially enriched with 300 samples from women with abnormal cervical cytology, the calculated clinical specificity may not be directly transposed to the general first-line screening population. One of the potential limitations of the study is that cobas and Anyplex testing was performed 2 and 7 years subsequent to hc2 testing, respectively. However, prolonged specimen storage at −70°C did not influence performance of either evaluated HPV DNA test, since they were both clinically noninferior to hc2. Moreover, several other HPV DNA tests evaluated through the VALGENT-3 framework using stored specimens all exhibit noninferiority to hc2 (30, 31). The VALGENT-3 initiative clearly showed that archived ThinPrep samples can be safely used several years after initial collection to assess clinical performance of HPV DNA tests if appropriately aliquoted, handled, and stored.
In contrast to tests that allow partial genotyping for HPV16 and HPV18, which is increasingly used in the clinical practice in many countries and represents an important method of triage for HPV-positive women, full HPV genotyping tests (tests that provide genotype information for at least 12 hrHPV types) have been used mainly as research tools in basic and epidemiological studies, for surveillance of HPV immunization, and in discordant analysis of samples with different HPV results of two or more HPV tests. However, recent findings suggest that genotyping beyond HPV16 and HPV18 will play an important role in future clinical practice (46), especially in populations with high vaccine coverage. Because Anyplex demonstrated very good performance in all published clinical and analytical evaluations to date and provides individual typing information for 14 hrHPV types, it could be considered a valuable tool in potential novel management algorithms in hrHPV-based primary cervical cancer screening. To conclude, based on the results of current clinical and analytical evaluation and previous studies, Anyplex fulfills all of the requirements for its use in primary cervical cancer screening.
Supplementary Material
ACKNOWLEDGMENTS
M.P., A.O., L.X., and M.A. were supported by the COHEAHR Network (grant no. 603019), which was funded by the 7th Framework Programme of DG Research and Innovation of the European Commission. In VALGENT-3, commercial assays can participate under the condition of a financial contribution, which means that the Scientific Institute of Public Health (Brussels, Belgium) has received funding to perform statistical analyses of VALGENT-3 data, and the Institute of Microbiology and Immunology–Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia, was provided with the Anyplex II HPV HR kits free of charge.
The source of funding did not have any influence on the study design or the analysis of the results.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01176-18.
REFERENCES
- 1.Wright TC, Stoler MH, Behrens CM, Sharma A, Zhang G, Wright TL. 2015. Primary cervical cancer screening with human papillomavirus: end of study results from the ATHENA study using HPV as the first-line screening test. Gynecol Oncol 136:189–197. doi: 10.1016/j.ygyno.2014.11.076. [DOI] [PubMed] [Google Scholar]
- 2.Reid JL, Wright TC Jr, Stoler MH, Cuzick J, Castle PE, Dockter J, Getman D, Giachetti C. 2015. Human papillomavirus oncogenic mRNA testing for cervical cancer screening: baseline and longitudinal results from the CLEAR study. Am J Clin Pathol 144:473–483. doi: 10.1309/AJCPHVD7MIP3FYVV. [DOI] [PubMed] [Google Scholar]
- 3.Ronco G, Dillner J, Elfström KM, Tunesi S, Snijders PJ, Arbyn M, Kitchener H, Segnan N, Gilham C, Giorgi-Rossi P, Berkhof J, Peto J, Meijer CJ, International HPV Screening Working Group. 2014. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet 383:524–532. doi: 10.1016/S0140-6736(13)62218-7. [DOI] [PubMed] [Google Scholar]
- 4.Naucler P, Ryd W, Törnberg S, Strand A, Wadell G, Elfgren K, Rådberg T, Strander B, Johansson B, Forslund O, Hansson BG, Rylander E, Dillner J. 2007. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N Engl J Med 357:1589–1597. doi: 10.1056/NEJMoa073204. [DOI] [PubMed] [Google Scholar]
- 5.Bulkmans NW, Berkhof J, Rozendaal L, van Kemenade FJ, Boeke AJ, Bulk S, Voorhorst FJ, Verheijen RH, van Groningen K, Boon ME, Ruitinga W, van Ballegooijen M, Snijders PJ, Meijer CJ. 2007. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet 370:1764–1772. doi: 10.1016/S0140-6736(07)61450-0. [DOI] [PubMed] [Google Scholar]
- 6.Rijkaart DC, Berkhof J, van Kemenade FJ, Coupe VM, Hesselink AT, Rozendaal L, Heideman DA, Verheijen RH, Bulk S, Verweij WM, Snijders PJ, Meijer CJ. 2012. Evaluation of 14 triage strategies for HPV DNA-positive women in population-based cervical screening. Int J Cancer 130:602–610. doi: 10.1002/ijc.26056. [DOI] [PubMed] [Google Scholar]
- 7.Kitchener HC, Almonte M, Thomson C, Wheeler P, Sargent A, Stoykova B, Gilham C, Baysson H, Roberts C, Dowie R, Desai M, Mather J, Bailey A, Turner A, Moss S, Peto J. 2009. HPV testing in combination with liquid-based cytology in primary cervical screening (ARTISTIC): a randomised controlled trial. Lancet Oncol 10:672–682. doi: 10.1016/S1470-2045(09)70156-1. [DOI] [PubMed] [Google Scholar]
- 8.Ronco G, Giorgi-Rossi P, Carozzi F, Confortini M, Dalla Palma P, Del Mistro A, Ghiringhello B, Girlando S, Gillio-Tos A, De Marco L, Naldoni C, Pierotti P, Rizzolo R, Schincaglia P, Zorzi M, Zappa M, Segnan N, Cuzick J, New Technologies for Cervical Cancer Screening (NTCC) Working Group. 2010. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol 11:249–257. doi: 10.1016/S1470-2045(09)70360-2. [DOI] [PubMed] [Google Scholar]
- 9.Arbyn M, Ronco G, Anttila A, Meijer CJ, Poljak M, Ogilvie G, Koliopoulos G, Naucler P, Sankaranarayanan R, Peto J. 2012. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine 30(Suppl 5):F88–F99. doi: 10.1016/j.vaccine.2012.06.095. [DOI] [PubMed] [Google Scholar]
- 10.Roland KB, Soman A, Benard VB, Saraiya M. 2011. Human papillomavirus and Papanicolaou tests screening interval recommendations in the United States. Am J Obstet Gynecol 205:447.e1–447.e8. doi: 10.1016/j.ajog.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Gage JC, Schiffman M, Katki HA, Castle PE, Fetterman B, Wentzensen N, Poitras NE, Lorey T, Cheung LC, Kinney WK. 2014. Reassurance against future risk of precancer and cancer conferred by a negative human papillomavirus test. J Natl Cancer Inst 106:dju153. doi: 10.1093/jnci/dju153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vink MA, Bogaards JA, Meijer CJ, Berkhof J. 2015. Primary human papillomavirus DNA screening for cervical cancer prevention: can the screening interval be safely extended? Int J Cancer 137:420–427. doi: 10.1002/ijc.29381. [DOI] [PubMed] [Google Scholar]
- 13.Arbyn M, Sasieni P, Meijer CJ, Clavel C, Koliopoulos G, Dillner J. 2006. Chapter 9: clinical applications of HPV testing: a summary of meta-analyses. Vaccine 24(Suppl 3):S3/78–89. doi: 10.1016/j.vaccine.2005.01.101. [DOI] [PubMed] [Google Scholar]
- 14.Barzon L, Giorgi C, Buonaguro FM, Palu G, the Italian Society for Virology. 2008. Guidelines of the Italian Society for Virology on HPV testing and vaccination for cervical cancer prevention. Infect Agents Cancer 3:14. doi: 10.1186/1750-9378-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox JT. 2009. History of the use of HPV testing in cervical screening and in the management of abnormal cervical screening results. J Clin Virol 45(Suppl 1):S3–S12. doi: 10.1016/S1386-6532(09)70002-2. [DOI] [PubMed] [Google Scholar]
- 16.Cuschieri KS, Cubie HA. 2005. The role of human papillomavirus testing in cervical screening. J Clin Virol 32(Suppl 1):S34–S42. doi: 10.1016/j.jcv.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 17.Cuzick J, Arbyn M, Sankaranarayanan R, Tsu V, Ronco G, Mayrand MH, Dillner J, Meijer CJ. 2008. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine 26(Suppl 10):K29–K41. doi: 10.1016/j.vaccine.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Desai MS, Cubie HA. 2005. The HPV test in cervical screening: a brave new world? Cytopathology 16:3–6. doi: 10.1111/j.1365-2303.2005.00237.x. [DOI] [PubMed] [Google Scholar]
- 19.Poljak M, Kocjan BJ, Ostrbenk A, Seme K. 2016. Commercially available molecular tests for human papillomaviruses (HPV): 2015 update. J Clin Virol 76(Suppl 1):S3–S9. doi: 10.1016/j.jcv.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Meijer CJ, Berkhof J, Castle PE, Hesselink AT, Franco EL, Ronco G, Arbyn M, Bosch FX, Cuzick J, Dillner J, Heideman DA, Snijders PJ. 2009. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int J Cancer 124:516–520. doi: 10.1002/ijc.24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbyn M, Snijders PJ, Meijer CJ, Berkhof J, Cuschieri K, Kocjan BJ, Poljak M. 2015. Which high-risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect 21:817–826. doi: 10.1016/j.cmi.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt M, Depuydt C, Benoy I, Bogers J, Antoine J, Arbyn M, Pawlita M, VALGENT Study Group. 2013. Multiple HPV infections with high viral loads are associated with cervical lesions but do not differentiate grades of cervical abnormalities. J Clin Microbiol 51:1458–1464. doi: 10.1128/JCM.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt M, Depuydt C, Benoy I, Bogers J, Antoine J, Arbyn M, Pawlita M, VALGENT Study Group. 2013. Prevalence and viral load of 51 genital human papillomavirus types and 3 subtypes. Int J Cancer 132:2395–2403. doi: 10.1002/ijc.27891. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt M, Depuydt C, Benoy I, Bogers J, Antoine J, Pawlita M, Arbyn M, VALGENT Study Group. 2013. Viral load of high-risk human papillomaviruses as reliable clinical predictor for the presence of cervical lesions. Cancer Epidemiol Biomarkers Prev 22:406–414. doi: 10.1158/1055-9965.EPI-12-1067. [DOI] [PubMed] [Google Scholar]
- 25.Geraets D, Cuschieri K, Koning M, van Doorn L, Snijders P, Meijer CJ, Quint WG, Arbyn M. 2014. Clinical evaluation of a GP5+/6+-based Luminex assay having full high-risk HPV genotyping capability and an internal control. J Clin Microbiol 52:3996–4002. doi: 10.1128/JCM.01962-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuschieri K, Geraets DT, Moore C, Quint W, Duvall E, Arbyn M. 2015. Clinical and analytical performance of the Onclarity HPV assay using the VALGENT framework. J Clin Microbiol 53:3272–3279. doi: 10.1128/JCM.01366-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arbyn M, Depuydt C, Benoy I, Bogers J, Cuschieri K, Schmitt M, Pawlita M, Geraets D, Heard I, Gheit T, Tommasino M, Poljak M, Bonde J, Quint W. 2016. VALGENT: a protocol for clinical validation of human papillomavirus assays. J Clin Virol 76(Suppl 1):S14–S21. doi: 10.1016/j.jcv.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Heard I, Cuschieri K, Geraets DT, Quint W, Arbyn M. 2016. Clinical and analytical performance of the PapilloCheck HPV-screening assay using the VALGENT framework. J Clin Virol 81:6–11. doi: 10.1016/j.jcv.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Cuschieri K, Geraets D, Cuzick J, Cadman L, Moore C, Vanden Broeck D, Padalko E, Quint W, Arbyn M. 2016. Performance of a cartridge based assay for the detection of clinically significant HPV infection–lessons from VALGENT (validation of HPV genotyping tests). J Clin Microbiol 54:2337–2347. doi: 10.1128/JCM.00897-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polman NJ, Ostrbenk A, Xu L, Snijders P, Meijer CJLM, Poljak M, Heideman DAM, Arbyn M. 2017. Evaluation of the clinical performance of the HPV-risk assay using the VALGENT-3 panel. J Clin Microbiol 55:3544–3551. doi: 10.1128/JCM.01282-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L, Oštrbenk A, Poljak M, Arbyn M. 2018. Assessment of the Roche linear array HPV genotyping test within the VALGENT framework. J Clin Virol 98:37–42. doi: 10.1016/j.jcv.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Stoler MH, Wright TC Jr, Sharma A, Apple R, Gutekunst K, Wright TL, ATHENA (Addressing THENeed for Advanced HPVDiagnostics) Study Group. 2011. High-risk human papillomavirus testing in women with ASC-US cytology: results from the ATHENA HPV study. Am J Clin Pathol 135:468–475. doi: 10.1309/AJCPZ5JY6FCVNMOT. [DOI] [PubMed] [Google Scholar]
- 33.Castle PE, Stoler MH, Wright TC Jr, Sharma A, Wright TL, Behrens CM. 2011. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol 12:880–890. doi: 10.1016/S1470-2045(11)70188-7. [DOI] [PubMed] [Google Scholar]
- 34.Wright TC Jr, Stoler MH, Sharma A, Zhang G, Behrens C, Wright TL, ATHENA (Addressing THENeed for Advanced HPVDiagnostics) Study Group. 2011. Evaluation of HPV-16 and HPV-18 genotyping for the triage of women with high-risk HPV+ cytology-negative results. Am J Clin Pathol 136:578–586. doi: 10.1309/AJCPTUS5EXAS6DKZ. [DOI] [PubMed] [Google Scholar]
- 35.Heideman DA, Hesselink AT, Berkhof J, van Kemenade F, Melchers WJ, Daalmeijer NF, Verkuijten M, Meijer CJ, Snijders PJ. 2011. Clinical validation of the cobas 4800 HPV test for cervical screening purposes. J Clin Microbiol 49:3983–3985. doi: 10.1128/JCM.05552-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lloveras B, Gomez S, Alameda F, Bellosillo B, Mojal S, Muset M, Parra M, Palomares JC, Serrano S. 2013. HPV testing by cobas HPV test in a population from Catalonia. PLoS One 8:e58153. doi: 10.1371/journal.pone.0058153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuzick J, Cadman L, Mesher D, Austin J, Ashdown-Barr L, Ho L, Terry G, Liddle S, Wright C, Lyons D, Szarewski A. 2013. Comparing the performance of six human papillomavirus tests in a screening population. Br J Cancer 108:908–913. doi: 10.1038/bjc.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poljak M, Ostrbenk A, Seme K, Ucakar V, Hillemanns P, Bokal EV, Jancar N, Klavs I. 2011. Comparison of clinical and analytical performance of the Abbott Realtime high risk HPV test to the performance of hybrid capture 2 in population-based cervical cancer screening. J Clin Microbiol 49:1721–1729. doi: 10.1128/JCM.00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basu P, Ponti A, Anttila A, Ronco G, Senore C, Bhadra Vale D, Segnan N, Tomatis M, Soerjomataram I, Primic Zakelj M, Dillner J, Elfstrom KM, Lonnberg S, Sankaranarayanan R. 2018. Status of implementation and organization of cancer screening in the European Union member states–summary results from the second European screening report. Int J Cancer 142:44–56. doi: 10.1002/ijc.31043. [DOI] [PubMed] [Google Scholar]
- 40.Kwon MJ, Roh KH, Park H, Woo HY. 2014. Comparison of the Anyplex II HPV28 assay with the Hybrid Capture 2 assay for the detection of HPV infection. J Clin Virol 59:246–249. doi: 10.1016/j.jcv.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Jung S, Lee B, Lee KN, Kim Y, Oh EJ. 2016. Clinical validation of Anyplex II HPV HR detection test for cervical cancer screening in Korea. Arch Pathol Lab Med 140:276–280. doi: 10.5858/arpa.2015-0117-OA. [DOI] [PubMed] [Google Scholar]
- 42.Hesselink AT, Sahli R, Berkhof J, Snijders PJ, van der Salm ML, Agard D, Bleeker MC, Heideman DA. 2016. Clinical validation of Anyplex II HPV HR detection according to the guidelines for HPV test requirements for cervical cancer screening. J Clin Virol 76:36–39. doi: 10.1016/j.jcv.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Fleiss JL, Levin B, Paik MC. 2003. Statistical methods for rates and proportions, 3rd ed Wiley, New York, NY. [Google Scholar]
- 44.Lee DH, Hwang NR, Lim MC, Yoo CW, Joo J, Kim JY, Park SY, Hwang SH. 2016. Comparison of the performance of Anyplex II HPV HR, the cobas 4800 human papillomavirus test and Hybrid Capture 2. Ann Clin Biochem 53:561–567. doi: 10.1177/0004563215614036. [DOI] [PubMed] [Google Scholar]
- 45.Cornall AM, Poljak M, Garland SM, Phillips S, Tan JH, Machalek DA, Quinn MA, Tabrizi SN. 2017. Anyplex II HPV28 detection and Anyplex II HPV HR detection assays are highly concordant with other commercial assays for detection of high-risk HPV genotypes in women with high grade cervical abnormalities. Eur J Clin Microbiol Infect Dis 36:545–551. doi: 10.1007/s10096-016-2831-5. [DOI] [PubMed] [Google Scholar]
- 46.Cuzick J, Wheeler C. 2016. Need for expanded HPV genotyping for cervical screening. Papillomavirus Res 2:112–115. doi: 10.1016/j.pvr.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.