Current tests for the detection of Clostridioides (formerly Clostridium) difficile free toxins in feces lack sensitivity, while nucleic acid amplification tests lack clinical specificity. We have evaluated the Singulex Clarity C.
KEYWORDS: C. difficile, ultrasensitivity, single-molecule counting technology
ABSTRACT
Current tests for the detection of Clostridioides (formerly Clostridium) difficile free toxins in feces lack sensitivity, while nucleic acid amplification tests lack clinical specificity. We have evaluated the Singulex Clarity C. diff toxins A/B assay (currently in development), an automated and rapid ultrasensitive immunoassay powered by single-molecule counting technology, for detection of C. difficile toxin A (TcdA) and toxin B (TcdB) in stool. The analytical sensitivity, analytical specificity, repeatability, and stability of the assay were determined. In a clinical evaluation, frozen stool samples from 311 patients with suspected C. difficile infection were tested with the Clarity C. diff toxins A/B assay, using an established cutoff value. Samples were tested with the Xpert C. difficile/Epi assay, and PCR-positive samples were tested with an enzyme immunoassay (EIA) (C. Diff Quik Chek Complete). EIA-negative samples were further tested with a cell cytotoxicity neutralization assay. The limits of detection for TcdA and TcdB were 0.8 and 0.3 pg/ml in buffer and 2.0 and 0.7 pg/ml in stool, respectively. The assay demonstrated reactivity to common C. difficile strains, did not show cross-reactivity to common gastrointestinal pathogens, was robust against common interferents, allowed detection in fresh and frozen stool samples and in samples after three freeze-thaw cycles, and provided results with high reproducibility. Compared to multistep PCR and toxin-testing procedures, the Singulex Clarity C. diff toxins A/B assay yielded 97.7% sensitivity and 100% specificity. The Singulex Clarity C. diff toxins A/B assay is ultrasensitive and highly specific and may offer a standalone solution for rapid detection and quantitation of free toxins in stool.
INTRODUCTION
Clostridioides (formerly Clostridium) difficile, a spore-forming, anaerobic, Gram-positive bacterium, is the most common cause of antibiotic-associated diarrhea and nosocomial infection in Europe and the United States and places a high financial burden on the health care system (1, 2). The clinical manifestations caused by C. difficile range in severity from asymptomatic colonization to mild diarrhea to fatal pseudomembranous colitis or toxic megacolon (3, 4). C. difficile infection (CDI) is mediated by toxin A (TcdA) and toxin B (TcdB), and detection of either free toxins or toxigenic C. difficile in stool is part of the CDI case definition (4).
Current laboratory tests used in the diagnosis of CDI have crucial limitations. The cell cytotoxicity neutralization assay (CCNA) has relatively high sensitivity and specificity for free TcdB, but its long turnaround time makes it clinically impractical. Nucleic acid amplification tests (NAATs) targeting tcdA and/or tcdB are rapid and sensitive but not clinically specific. Current enzyme immunoassays (EIAs) targeting both toxins, although rapid and specific, are hampered by a lack of sensitivity (4, 5). Although it is highly debated (6), the presence of toxins may better correlate with clinical outcomes than do molecular testing results, and there is a concern that the use of NAATs leads to overdiagnosis of CDI (7, 8). Compared with current EIAs, however, NAATs allow safe exclusion of CDI (6). Many laboratories have now implemented algorithms for CDI testing that combine the results of NAATs and EIAs in multiple steps (4, 5). Thus, there remains a need for a simple standalone test with the specificity of EIAs and the sensitivity of NAATs.
The Singulex Clarity C. diff toxins A/B assay, powered by single-molecule counting technology and in development for use on the Singulex Clarity system (Singulex Inc., Alameda, CA, USA), is an automated and rapid ultrasensitive immunoassay for the detection of C. difficile TcdA and TcdB in stool. The sensitivity of the Singulex single-molecule counting technology has been illustrated previously in numerous studies (9–12). Here, we describe the analytical and clinical performance of the Singulex Clarity C. diff toxins A/B assay.
MATERIALS AND METHODS
Singulex Clarity C. diff toxins A/B assay.
The Singulex Clarity C. diff toxins A/B assay (in development) measures TcdA and TcdB in stool on the Singulex Clarity system, an automated, in vitro diagnostics platform utilizing single-molecule counting technology. The assay is a paramagnetic microparticle-based immunoassay that uses single-photon fluorescence detection for analyte measurement. The Singulex Clarity C. diff toxins A/B assay uses TcdA- and TcdB-specific monoclonal antibodies (BiosPacific, Emeryville, CA, USA; BBI Solutions, Cardiff, United Kingdom). A 100-μl volume of unformed stool sample or 0.1 g of solid stool sample is diluted 1:20 with 1.9 ml of standard buffer (Tris-buffered saline [TBS]-EDTA with 3% bovine serum albumin [BSA]). The sample is centrifuged at 14,000 × g for 10 min, and 300 μl of the supernatant is transferred into a sample tube and loaded onto the Singulex Clarity instrument. The instrument automatically transfers each sample to a reaction vessel, where it is mixed and incubated for 5 min at 37°C with a 1:1 mixture of paramagnetic microparticles precoated with either anti-TcdA or anti-TcdB antibodies (capture reagent) and fluorescently labeled (Alexa Fluor 647; Thermo Fischer Scientific, Waltham, MA, USA) toxin-specific antibodies (detection reagent). During this time, toxins present in the sample are bound by both the capture and detection antibodies, forming an immune complex. Unbound material in the mixture is washed away during subsequent wash steps. Elution buffer is added to cleave the immune complexes from the paramagnetic microparticles, releasing the fluorescently labeled antibodies. The resulting eluate, containing the dissociated, fluorescently labeled detection antibodies, is transferred to a detection vessel, where the labeled molecules are detected and counted. The Singulex Clarity system reader is a confocal fluorescence microscope with an avalanche photodiode detector. A proprietary algorithm counts detected events, and compares the results to a previously established 11-point standard curve for native TcdA and TcdB isolated from strain VPI 10463, toxinotype 0 (Native Antigen Company, Kidlington, United Kingdom). Concentrations of toxins for generation of the standard curve were prepared by diluting toxin in Tris buffer with 3% BSA. The Singulex Clarity software interpolates the data, including the detected events prime signal, into a combined TcdA-TcdB concentration. The total turnaround time (sample in to result out) is 32 min. The system can process 1 to 48 samples in an assay run.
Analytical sensitivity. (i) Limit of detection.
Five frozen, deidentified stool samples obtained from patients with suspected CDI (Bristol scale scores of 4 to 7; TriCore Reference Laboratories, Albuquerque, NM, USA), which tested negative by EIA (C. Diff Quik Chek Complete assay; TechLab Inc., Blacksburg, VA, USA), NAAT (BD MAX Cdiff assay; Becton, Dickinson Inc., Franklin Lakes, NJ, USA), and CCNA (C. difficile TOX-B test; Techlab; tested at ARUP Laboratories, Salt Lake City, UT, USA), were pooled. TcdA (CDA-TNL-100; Native Antigen Company) and TcdB (CDB-TNL-100; Native Antigen Company) were spiked at 12 different concentrations, at a TcdA-TcdB concentration ratio of 1:1, into pooled C. difficile-negative stool (range, 0 to 50,000 pg/ml) and antigen-free sample diluent (TBS-EDTA with 3% BSA) (range, 0 to 10,000 pg/ml). Each point on each curve was run in triplicate, using one reagent lot, on one Clarity instrument. TcdA and TcdB standard curves were interpolated from the TcdA-TcdB combination curve to analyze the concentration of each toxin. The analytical limit of detection (LoD) was calculated as 2 times the standard deviation (SD) of the blank divided by the slope of the standard curve.
(ii) Cutoff value establishment.
A derivation cohort consisting of 103 frozen deidentified stool samples from patients with suspected CDI (74 samples with Bristol scale scores of 5 to 7 and 29 samples with a Bristol scale score of 4; TriCore Reference Laboratories and Discovery Life Sciences Laboratories, Los Osos, CA, USA) was used to establish a preliminary cutoff value for the Singulex Clarity C. diff toxins A/B assay, compared to the CCNA. Of the patients, 63.1% (65 patients) were women and 99.0% (102 patients) were ≥18 years of age. All samples, including 27 CCNA-positive samples (26%) and 76 CCNA-negative samples (74%), were tested in triplicate, and the mean concentration was used for cutoff value establishment. Using the CCNA as the reference method, the area under the receiver operating characteristic curve (AuROC) was calculated. The cutoff value was established as the Clarity C. diff toxins A/B assay concentration that minimized the joint difference between the test sensitivity and the test specificity, in order to apply equal penalties to false-negative and false-positive results (13). The cutoff value was used to classify positive and negative Clarity assay results in the subsequent studies.
(iii) Analytical reactivity.
Two frozen deidentified stool samples from patients with suspected CDI (Bristol scale scores of 5 to 7; TriCore Reference Laboratories) that tested triple positive or triple negative by EIA, NAAT, and CCNA were pooled to make sample sets with TcdA-TcdB concentrations just above (“low positive”; range, 19 to 32 pg/ml) or just below (“high negative”; range, 6 to 13 pg/ml) the cutoff value. The analytical reactivity against 38 toxigenic or nontoxigenic strains of C. difficile with 8 different toxinotypes was determined; strains were selected according to FDA guidelines (14). Toxinotyping was based on restriction fragment length polymorphism analysis of the 19-kb region of C. difficile pathogenicity locus, which encodes TcdA and TcdB, and ribotyping was based on comparisons of the patterns of PCR products in the 16S-23S rRNA intergenic spacer region. The C. difficile strains were cultured for 24 to 48 h in cycloserine-cefoxitin mannitol broth with taurocholate and lysozyme and cycloserine-cefoxitin fructose agar. Colonies were harvested into sterile phosphate-buffered saline (PBS) and diluted with PBS to a turbidity of a 0.5 McFarland standard (calculated based on an optical density at 600 nm of 1.0 equaling 1 × 109 cells/ml) prior to inoculation into sample matrices, to achieve a theoretical bacterial concentration of >106 CFU/ml. Each strain was spiked into stool with high-negative toxin concentrations and was tested in up to 3 replicates using one reagent lot on one Clarity instrument.
Analytical specificity. (i) Cross-reactivity.
Cross-reactivity against other gastrointestinal pathogens (29 aerobic bacteria, 3 microaerophilic bacteria, 18 anaerobic bacteria, 1 fungus, and 8 viruses) at medically relevant concentrations (based on estimations of bacterial concentrations according to the optical density of the inoculum) of >106 CFU/ml was determined in accordance with FDA guidelines (14). Bacteria and fungi were cultured according to ATCC recommendations and spiked into the TcdA-TcdB low-positive and high-negative stool samples. Viruses, which were vendor confirmed to be at medically relevant concentrations of >105 PFU/ml, were inoculated into stool samples directly from frozen-thawed stock (viral culture fluid). Each strain was tested in 3 replicates using one reagent lot on one Clarity instrument.
(ii) Potential interference.
Potential interference by 11 endogenous or exogenous substances at medically relevant concentrations was determined. Sample panels with low-positive or high-negative TcdA-TcdB concentrations were spiked with an interfering substance or with the corresponding solvent (control case). Each sample was tested in 4 replicates using one reagent lot on one Clarity instrument. The potential interference was assessed based on the difference in qualitative results and on the percent difference between the signals obtained with spiked and control samples. The acceptance criterion for determining interference was that the difference, if any, was within ±20%.
Assay repeatability and stability. (i) Reproducibility.
Eighty-five frozen deidentified stool samples from patients with suspected CDI (TriCore Reference Laboratories and Discovery Life Sciences Laboratories) were tested in triplicate using one reagent lot on one Clarity instrument. Of the patients, 63.5% (54 patients) were women and 97.6% (83 patients) were ≥18 years of age.
(ii) Stability.
To determine sample stability, 5 C. difficile-positive stool samples (Bristol scale scores of 5 to 7) and 5 C. difficile-negative stool samples (3 with Bristol scale scores of 5 to 7 and 2 with a Bristol scale score of 4; TriCore Reference Laboratories and Discovery Life Sciences Laboratories), which had been tested with the CCNA, were stored under room temperature (20°C to 25°C), refrigerated (2°C to 8°C), or frozen (−70°C) conditions. Samples stored at room temperature were tested within 4 h after arrival at the laboratory (baseline) and again 4 to 8 h after arrival. Samples stored at 2°C to 8°C were tested at day 2 and at week 1, and samples stored at −70°C were tested at weeks 1, 2, and 3 and at months 3 and 6. As an additional test, samples were subjected to three freeze-thaw cycles. Each sample was tested in 3 replicates at each time point, using two reagent lots on one Clarity instrument. The qualitative result for each time point was used to determine the stability of each sample.
Clinical evaluation.
The study was approved by the institutional review board at Stanford University (Palo Alto, CA, USA) (protocol IRB-43749). Frozen stool samples from 311 patients with suspected CDI, collected at the Stanford Health Care Diagnostic Microbiology Laboratory, were tested with the Singulex Clarity C. diff toxins A/B assay (Fig. 1). The samples were not thawed prior to this study. Of the patients, 46.0% (143 patients) were women and 84.9% (264 patients) were ≥18 years of age. Sample collection and testing for this cohort were described previously for PCR-positive samples (15). In summary, all samples had been previously tested at Stanford Health Care with a PCR assay detecting tcdB (Xpert C. difficile/Epi; Cepheid Inc., Sunnyvale, CA, USA), and PCR-positive samples (n = 211) were tested for the presence of free toxins with a rapid membrane EIA (C. Diff Quik Chek Complete). EIA-negative samples (n = 110) were further tested with a CCNA (C. difficile TOX-B test, yielding 31 CCNA-positive samples and 79 CCNA-negative samples). The study was designed based on a meta-analysis showing that PCR-negative samples are CCNA negative and EIA-positive samples are CCNA positive (5). EIA-positive or CCNA-positive samples were considered true toxin-positive samples, and PCR-negative samples were considered C. difficile-negative samples.
FIG 1.
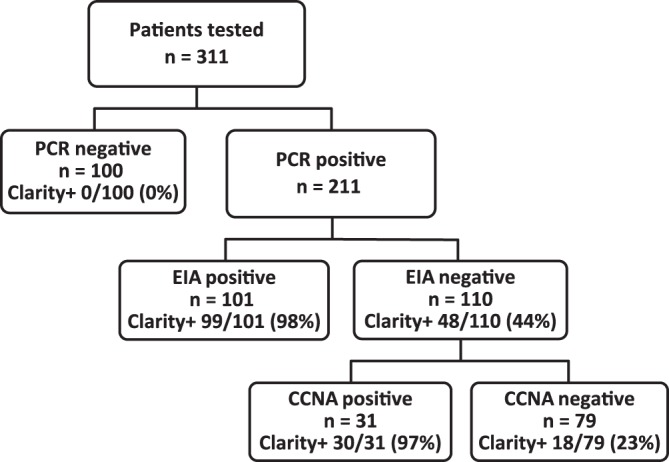
Distribution of patients enrolled in the study and their test results. EIA, C. Diff Quik Chek Complete.
Statistical methods.
Statistical differences in toxin concentrations were assessed by the Mann-Whitney U test if subjects were separated into two groups, by the Kruskal-Wallis test if subjects were separated into more than two unordered groups, and by analysis of variance (ANOVA) contrast models if subjects were separated into more than two ordered groups. Common procedures were used to calculate the sensitivity and specificity, with associated exact Clopper-Pearson 95% confidence intervals (CIs). Spearman's correlation coefficient was used to measure the strength and direction of the linear association between the Clarity C. diff toxins A/B assay and PCR cycle threshold (CT) results. Statistical analyses were performed with SAS 9.4, Analyze-It for MS Excel 4.51, and GraphPad Prism 5.0 software.
RESULTS
Analytical sensitivity. (i) LoD.
The LoDs for TcdA and TcdB were 0.8 and 0.3 pg/ml in buffer and 2.0 and 0.7 pg/ml in stool, respectively.
(ii) Cutoff value establishment.
The AuROC curve demonstrated a C-statistic of 0.99 (95% CI, 0.98 to 1.00). The cutoff value was set at 16.7 pg/ml. In this cohort, the sensitivity was 96.3% (95% CI, 81.0 to 99.9%), compared to the CCNA, and the specificity was 96.1% (95% CI, 88.9 to 99.2%).
(iii) Analytical reactivity.
At an estimated concentration of >1 × 108 CFU/ml for each toxinotype, the Singulex Clarity C. diff toxins A/B assay successfully detected toxins from all tested toxinotypes (see Table S1 in the supplemental material). The analytical reactivity did not diminish when only one toxin was present. In the absence of TcdA and TcdB, the assay correctly yielded a negative result.
Analytical specificity. (i) Cross-reactivity.
When a microorganism was spiked into stool at either a low-positive or high-negative C. difficile toxin concentration, all samples were correctly reported as positive or negative, respectively (Table S1).
(ii) Potential interference.
No interference was detected when common endogenous and exogenous substances were tested with the Singulex Clarity C. diff toxins A/B assay (Table S2).
Assay repeatability and stability. (i) Repeatability.
All 3 replicates gave the same result for 84/85 samples tested (repeatability, 99.0%).
(ii) Stability.
Qualitative analysis at each time point showed no drift, compared to the baseline (<4 h) reading. Samples for C. difficile toxin testing were stable for up to 8 h at room temperature, 1 week at 2°C to 8°C, 3 months at −70°C, and three freeze-thaw cycles (Table S3).
Clinical evaluation.
The median toxin concentrations in the 100 PCR-negative, 79 PCR-positive/EIA-negative/CCNA-negative, 31 PCR-positive/EIA-negative/CCNA-positive, and 101 PCR-positive/EIA-positive samples were 0 pg/ml (interquartile range [IQR], 0 to 1.25 pg/ml), 3.5 pg/ml (IQR, 0.3 to 3.5 pg/ml), 319.3 pg/ml (IQR, 116.3 to 319.3 pg/ml), and 4,334 pg/ml (IQR, 1,030 to 15,487 pg/ml; P < 0.001), respectively (Fig. 2). The Singulex Clarity C. diff toxins A/B assay results were positive for 129 of the 132 PCR-positive/EIA-positive or EIA-negative/CCNA-positive samples (sensitivity, 97.7% [95% CI, 93.0% to 99.4%]), and results for all PCR-negative samples were negative (specificity, 100% [95% CI, 95.4% to 100%]). The 3 false-negative samples included 2 EIA-positive samples and 1 EIA-negative/CCNA-positive sample. Among the 79 PCR-positive/EIA-negative/CCNA-negative samples, 18 (22.8%) were positive by the Singulex Clarity C. diff toxins A/B assay. The median toxin concentrations in toxin-positive samples with the Singulex Clarity C. diff toxins A/B assay were 31.3 pg/ml (IQR, 22.3 to 209.7 pg/ml), 352.2 pg/ml (IQR, 124.6 to 540.2 pg/ml), and 4,680.5 pg/ml (IQR, 1,067.3 to 15,839.2 pg/ml; P < 0.001) for the 18 PCR-positive/EIA-negative/CCNA-negative, 30 PCR-positive/EIA-negative/CCNA-positive, and 99 PCR-positive/EIA-positive samples, respectively. The 2 PCR-positive/EIA-positive but Clarity assay-negative samples were further investigated; both were non-027 strains. One of the samples was EIA negative when retested, while the second sample had insufficient volume remaining for retesting.
FIG 2.
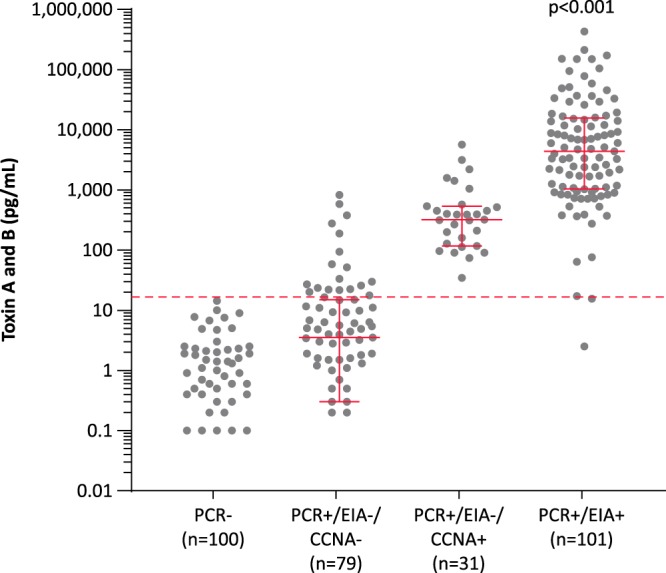
C. difficile toxin concentrations in samples in various result categories. The Singulex Clarity C. diff toxins A/B assay concentrations (combined TcdA and TcdB concentrations) are shown for stool samples with different PCR (Xpert C. difficile/Epi), EIA (C. Diff Quik Chek Complete), and CCNA results. The dashed line shows the preliminary cutoff value for the Singulex Clarity C. diff toxins A/B assay.
The median toxin concentration was higher in samples with 027 (n = 28) versus non-027 (n = 183) strains (2,846 pg/ml [IQR, 96.4 to 7,729 pg/ml] versus 369.8 pg/ml [IQR, 5.0 to 3,124 pg/ml]; P = 0.0058) (Fig. S1). The median CT values determined by PCR for low-negative (0 pg/ml [n = 17]), high-negative (0.1 to 16.7 pg/ml [n = 47]), low-positive (16.8 to 1,000.0 pg/ml [n = 64]), and high-positive (>1,000.0 pg/ml [n = 83]) samples (as determined by the Clarity assay) were 33.0 (IQR, 31.1 to 34.4), 30.2 (IQR, 26.2 to 33.9), 24.1 (IQR, 22.3 to 27.5), and 23.0 (IQR, 21.6 to 24.2; P < 0.001), respectively, and there was a significant continuous correlation between toxin concentrations and CT values (Spearman's correlation coefficient, −0.64; P < 0.001) (Fig. 3). Individual sample results are presented in Fig. 3.
FIG 3.
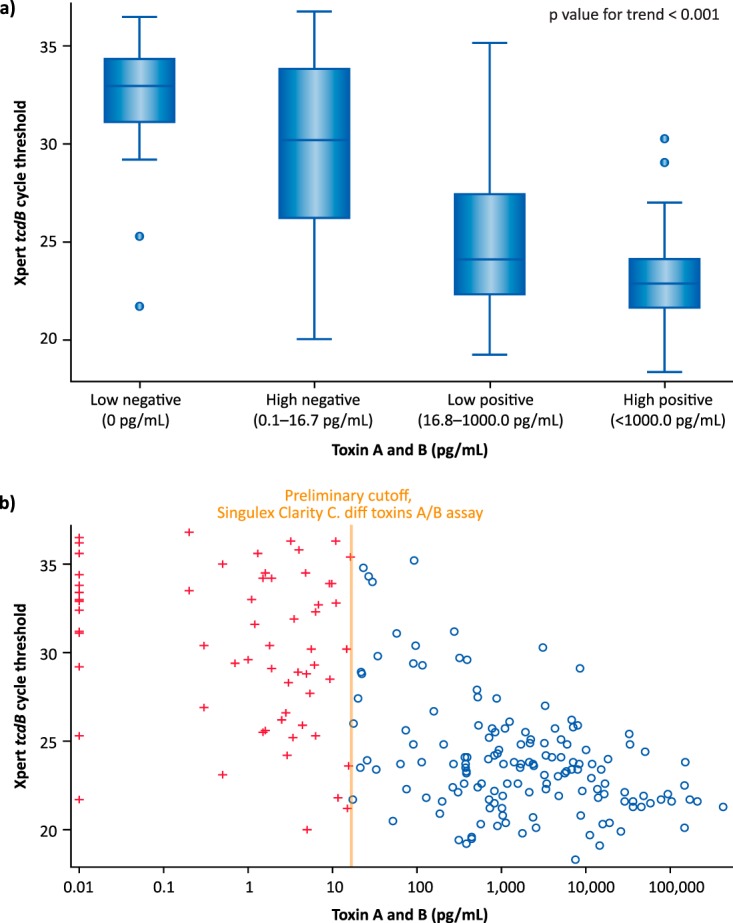
C. difficile toxin concentrations and CT values determined by PCR (Xpert C. difficile/Epi) for samples tested with the Singulex Clarity C. diff toxins A/B assay. (a) CT values for low-negative (0 pg/ml), high-negative (0.1 to 16.7 pg/ml), low-positive (16.8 to 1,000.0 pg/ml), and high-positive (>1,000.0 pg/ml) samples. (b) Distribution of samples and correlation between C. difficile toxin concentrations and CT values. Red plus signs indicate samples negative by the Clarity C. diff toxins A/B assay, and blue circles indicate samples positive by the Clarity C. diff toxins A/B assay.
DISCUSSION
There is a need for a standalone C. difficile diagnostic tool that can detect the C. difficile toxins TcdA and TcdB with the sensitivity of a CCNA, the speed of NAATs, and the automation of random-access NAATs. In this study, we showed that the Singulex Clarity C. diff toxins A/B assay had 97.7% sensitivity and 100% specificity, compared to a multistep toxin-testing procedure that included CCNA testing for sensitive detection of free TcdB. This ultrasensitive toxin assay, which is automated and rapid (i.e., 32 min), has the potential to be a standalone test to replace the multistep testing algorithms currently recommended for C. difficile diagnosis (4, 5). Implementation of this assay in health care systems around the globe may improve antibiotic stewardship, given its high sensitivity and negative predictive value for free fecal toxins.
With LoDs in stool of 2.0 pg/ml (TcdA) and 0.7 pg/ml (TcdB), the analytical sensitivity of the Singulex Clarity C. diff toxins A/B assay is orders of magnitude higher than that of any commercially available assay for the detection of toxins (16). Current EIAs have LoDs ranging from 800 to 2,500 pg/ml, and more complex methods, such as cell-based assays and real-time cellular analysis assays, do not detect concentrations lower than 200 pg/ml and have turnaround times of up to 60 h (17, 18). Several assays have been described for ultrasensitive toxin detection, but none is commercially available for clinical application. A cell-based immunocytotoxicity assay based on a real-time cell electronic sensing system achieved a TcdA LoD of 0.1 to 1 pg/ml in buffer, but the LoD in stool was not reported and the turnaround time was up to 4 h (19). Digital enzyme-linked immunosorbent assays (ELISAs) by Quanterix Inc. (Lexington, MA, USA), detecting molecules on paramagnetic beads, were shown to detect toxins A and B in stool with LoDs of 0.45 and 1.5 pg/ml, respectively, and had a cutoff value in the same range as the Clarity assay in a clinical evaluation (20). The Clarity C. diff toxins A/B assay offers a simple sample-to-answer solution and, unlike the CCNA, can be performed in less than 1 hour.
Animal studies have shown that toxin positivity is necessary for C. difficile virulence (21, 22). Clinical studies have shown that toxin-positive patients have more severe outcomes than toxin-negative patients and that NAAT-positive/toxin-negative and NAAT-negative patients have similar outcomes (7, 8). Most NAAT-positive/toxin-negative patients are currently treated, due to lack of an ultrasensitive toxin assay. The Clarity C. diff toxins A/B assay detected 97.7% of EIA-positive or CCNA-positive samples and, in addition, detected toxins in 23% of PCR-positive/EIA-negative/CCNA-negative samples. The latter finding may be due to the fact that the Clarity assay may have higher analytical sensitivity than the CCNA and the CCNA detects only TcdB, while the Clarity assay detects both TcdA and TcdB. Recent studies reported that some C. difficile strains may produce only TcdA (23). Our findings indicate that the Clarity C. diff toxins A/B assay can refine the selection of patients who are most likely to benefit from treatment. Studies have also shown that toxin concentrations may predict clinical outcomes and identify patients who need more aggressive treatment (16, 17, 24). A clinical study is under way to define the cutoff value for the Clarity C. diff toxins A/B assay based on clinical severity, outcomes, and the need for therapy.
In the absence of a sensitive and rapid test for C. difficile toxins in stool, it has been shown that PCR CT values, which correlate with organism burdens, may be used for prediction of toxin results (25–27). Studies estimating CT values for toxin prediction used either toxin EIA or CCNA as reference methods. With the advent of ultrasensitive toxin assays, the impact of toxin concentrations on CDI pathology can be evaluated; further studies are needed to understand the clinical utility of both ultrasensitive toxin tests and CT values for diagnosis and therapy guidance (28).
The median toxin concentration was higher in samples with ribotype 027 versus non-027 strains. Although outbreaks of 027 strains have been associated with more severe outcomes and such strains are thought to be more virulent due to increased toxin production and other mechanisms (29), there is growing evidence opposing this, at least in nonepidemic settings (15, 30). Toxin concentrations may reflect only one of many 027 virulence factors, while increased sporulation capacity (31) and fluoroquinolone administration (32) may drive separate virulence mechanisms. The need for 027 genotyping is not currently recommended for routine clinical laboratories diagnosing nonepidemic CDI cases (33).
Although the findings of this study are promising, this study has several limitations. First, given the concern that storage conditions can affect toxin stability, testing with fresh samples might have been preferred over frozen samples. However, multiple studies have shown that, although toxins deteriorate at room temperature, they are stable in longer-term storage under refrigerated or frozen conditions (34–36). Second, this study did not correlate toxin findings with clinical data. A follow-up study is under way to investigate clinical outcomes for toxin-positive and toxin-negative patients, including PCR-positive/CCNA-negative/Clarity-positive patients. Lastly, this was a single-center study. Given the geographical variation of C. difficile strains, multicenter studies are preferred and will be performed for regulatory approval purposes.
In summary, the Singulex Clarity C. diff toxins A/B assay described in this study is an ultrasensitive and highly specific assay, compared with multistep toxin-testing algorithms. It may be used as a standalone test for rapid detection of free toxins in stool, to guide treatment for patients with CDI.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Lee, Wendy Trinh, Kent Chau, Orr Hadass, Louisa Luk, Priyanka Lakkaraju, Michael Mateling, Dylan Valdez, Abegail Garcia, Valerie DeGuzman, Genalyn Amond, Samir Khan, Derek Chang, Numan Syed, Garrett Goff, and Michael Daffern, all at Singulex, Inc., for their valuable contributions and hard work completing the studies.
This study was sponsored by Singulex, Inc.
J.S., A.B., A.A., S.T., S.B., S.A., J.J.B., N.N., J.E., and J.T. are employees of Singulex, Inc. N.B. is a member of the Singulex medical advisory board.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00908-18.
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassini A, Plachouras D, Eckmanns T, Abu Sin M, Blank H-P, Ducomble T, Haller S, Harder T, Klingeberg A, Sixtensson M, Velasco E, Weiß B, Kramarz P, Monnet DL, Kretzschmar ME, Suetens C. 2016. Burden of six healthcare-associated infections on European population health: estimating incidence-based disability-adjusted life years through a population prevalence-based modelling study. PLoS Med 13:e1002150. doi: 10.1371/journal.pmed.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. 2013. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 4.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crobach MJT, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ. 2016. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 22(Suppl 4):S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Fang FC, Polage CR, Wilcox MH. 2017. Point-counterpoint: what is the optimal approach for detection of Clostridium difficile infection? J Clin Microbiol 55:670–680. doi: 10.1128/JCM.02463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang Y-W, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH. 2015. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, O'Connor L, Oakley SJ, Pope CF, Wren MW, Shetty NP, Crook DW, Wilcox MH. 2013. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis 13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter JE, Honegger U, Puelacher C, Mueller D, Wagener M, Schaerli N, Strebel I, Twerenbold R, Boeddinghaus J, Nestelberger T, Sazgary L, Marbot S, du Fay de Lavallaz J, Kaiser C, Osswald S, Wild D, Rentsch K, Zellweger M, Reichlin T, Mueller C. 2018. Prospective validation of a biomarker-based rule out strategy for functionally relevant coronary artery disease. Clin Chem 64:386–395. doi: 10.1373/clinchem.2017.277210. [DOI] [PubMed] [Google Scholar]
- 10.Neumann JT, Havulinna AS, Zeller T, Appelbaum S, Kunnas T, Nikkari S, Jousilahti P, Blankenberg S, Sydow K, Salomaa V. 2014. Comparison of three troponins as predictors of future cardiovascular events: prospective results from the FINRISK and BiomaCaRE studies. PLoS One 9:e90063. doi: 10.1371/journal.pone.0090063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apple FS, Steffen LM, Pearce LA, Murakami MM, Luepker RV. 2012. Increased cardiac troponin I as measured by a high-sensitivity assay is associated with high odds of cardiovascular death: the Minnesota Heart Survey. Clin Chem 58:930–935. doi: 10.1373/clinchem.2011.179176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apple FS. 2009. A new season for cardiac troponin assays: it's time to keep a scorecard. Clin Chem 55:1303–1306. doi: 10.1373/clinchem.2009.128363. [DOI] [PubMed] [Google Scholar]
- 13.Habibzadeh F, Habibzadeh P, Yadollahie M. 2016. On determining the most appropriate test cut-off value: the case of tests with continuous results. Biochem Med (Zagreb) 26:297–307. doi: 10.11613/BM.2016.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Food and Drug Administration. 2015. Class II special controls guideline document: toxin gene amplification assays for the detection of Clostridium difficile: guideline for industry and Food and Drug Administration staff. Food and Drug Administration, Rockville, MD: https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM459917.pdf. [Google Scholar]
- 15.Senchyna F, Gaur RL, Gombar S, Truong CY, Schroeder LF, Banaei N. 2017. Clostridium difficile PCR cycle threshold predicts free toxin. J Clin Microbiol 55:2651–2660. doi: 10.1128/JCM.00563-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollock NR. 2016. Ultrasensitive detection and quantification of toxins for optimized diagnosis of Clostridium difficile infection. J Clin Microbiol 54:259–264. doi: 10.1128/JCM.02419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryder AB, Huang Y, Li H, Zheng M, Wang X, Stratton CW, Xu X, Tang Y-W. 2010. Assessment of Clostridium difficile infections by quantitative detection of tcdB toxin by use of a real-time cell analysis system. J Clin Microbiol 48:4129–4134. doi: 10.1128/JCM.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang B, Jin D, Zhang J, Sun JY, Wang X, Stiles J, Xu X, Kamboj M, Babady NE, Tang Y-W. 2014. Real-time cellular analysis coupled with a specimen enrichment accurately detects and quantifies Clostridium difficile toxins in stool. J Clin Microbiol 52:1105–1111. doi: 10.1128/JCM.02601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Wang J, Steele J, Sun X, Nie W, Tzipori S, Feng H. 2009. An ultrasensitive rapid immunocytotoxicity assay for detecting Clostridium difficile toxins. J Microbiol Methods 78:97–100. doi: 10.1016/j.mimet.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song L, Zhao M, Duffy DC, Hansen J, Shields K, Wungjiranirun M, Chen X, Xu H, Leffler DA, Sambol SP, Gerding DN, Kelly CP, Pollock NR. 2015. Development and validation of digital enzyme-linked immunosorbent assays for ultrasensitive detection and quantification of Clostridium difficile toxins in stool. J Clin Microbiol 53:3204–3212. doi: 10.1128/JCM.01334-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 23.Banz A, Lantz A, Riou B, Foussadier A, Miller M, Davies K, Wilcox M. 2018. Sensitivity of single-molecule array assays to detect Clostridium difficile toxins in comparison to conventional laboratory testing algorithms. J Clin Microbiol 56:e00452-. doi: 10.1128/JCM.00452-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akerlund T, Svenungsson B, Lagergren A, Burman LG. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J Clin Microbiol 44:353–358. doi: 10.1128/JCM.44.2.353-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crobach MJT, Duszenko N, Terveer EM, Verduin CM, Kuijper EJ. 2018. Nucleic acid amplification test quantitation as predictor of toxin presence in Clostridium difficile infection. J Clin Microbiol 56:e01316-17. doi: 10.1128/JCM.01316-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamboj M, Brite J, McMillen T, Robilotti E, Herrera A, Sepkowitz K, Babady NE. 2018. Potential of real-time PCR threshold cycle (CT) to predict presence of free toxin and clinically relevant C. difficile infection (CDI) in patients with cancer. J Infect 76:369–375. doi: 10.1016/j.jinf.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie JL, Cohen SH, Solnick JV, Polage CR. 2012. Role of fecal Clostridium difficile load in discrepancies between toxin tests and PCR: is quantitation the next step in C. difficile testing? Eur J Clin Microbiol Infect Dis 31:3295–3299. doi: 10.1007/s10096-012-1695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollock NR, Banz A, Chen X, Williams D, Xu H, Cuddemi CA, Cui AX, Perrotta M, Alhassan E, Riou B, Lantz A, Miller MA, Kelly CP. 2018. Comparison of Clostridioides difficile stool toxin concentrations in adults with symptomatic infection and asymptomatic carriage using an ultrasensitive quantitative immunoassay. Clin Infect Dis ciy415 doi: 10.1093/cid/ciy415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 30.Wilson V, Cheek L, Satta G, Walker-Bone K, Cubbon M, Citron D, Gerding DN, Llewelyn MJ. 2010. Predictors of death after Clostridium difficile infection: a report on 128 strain-typed cases from a teaching hospital in the United Kingdom. Clin Infect Dis 50:e77–e81. doi: 10.1086/653012. [DOI] [PubMed] [Google Scholar]
- 31.Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, Gerding DN, Vedantam G. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol 192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pépin J, Saheb N, Coulombe M-A, Alary M-E, Corriveau M-P, Authier S, Leblanc M, Rivard G, Bettez M, Primeau V, Nguyen M, Jacob C-E, Lanthier L. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis 41:1254–1260. doi: 10.1086/496986. [DOI] [PubMed] [Google Scholar]
- 33.Kociolek LK, Gerding DN. 2016. Clinical utility of laboratory detection of Clostridium difficile strain BI/NAP1/027. J Clin Microbiol 54:19–24. doi: 10.1128/JCM.02340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfa MJ, Olson N, Murray B-L. 2014. Fecal specimens for Clostridium difficile diagnostic testing are stable for up to 72 hours at 4°C. J Med Microbiol Diagn 3:140. doi: 10.4172/2161-0703.1000140. [DOI] [Google Scholar]
- 35.Modi C, DePasquale JR, Nguyen NQ, Malinowski JE, Perez G. 2010. Does the handling time of unrefrigerated human fecal specimens impact the detection of Clostridium difficile toxins in a hospital setting? Indian J Gastroenterol 29:157–161. doi: 10.1007/s12664-010-0040-1. [DOI] [PubMed] [Google Scholar]
- 36.Schora DM, Peterson LR, Usacheva EA. 2018. Immunological stability of Clostridium difficile toxins in clinical specimens. Infect Control Hosp Epidemiol 39:434–438. doi: 10.1017/ice.2018.20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


