Campylobacter spp. are foodborne and waterborne pathogens.
KEYWORDS: World Health Organization, Global Foodborne Infections Network, proficiency test, quality assurance, Campylobacter jejuni, Campylobacter coli, identification, antimicrobial susceptibility testing
ABSTRACT
Campylobacter spp. are foodborne and waterborne pathogens. While rather accurate estimates for these pathogens are available in industrialized countries, a lack of diagnostic capacity in developing countries limits accurate assessments of prevalence in many regions. Proficiency in the identification and susceptibility testing of these organisms is critical for surveillance and control efforts. The aim of the study was to assess performance for identification and susceptibility testing of thermotolerant Campylobacter spp. among laboratories participating in the World Health Organization (WHO) Global Foodborne Infections Network (GFN) External Quality Assurance System (EQAS) over a 9-year period. Participants (primarily national-level laboratories) were encouraged to self-evaluate their performance as part of continuous quality improvement. The ability to correctly identify Campylobacter spp. varied by year and ranged from 61.9% (2008) to 90.7% (2012), and the ability to correctly perform antimicrobial susceptibility testing (AST) for Campylobacter spp. appeared to steadily increase from 91.4% to 93.6% in the test period (2009 to 2012). The poorest performance (60.0% correct identification and 86.8% correct AST results) was observed in African laboratories. Overall, approximately 10% of laboratories reported either an incorrect identification or antibiogram. As most participants were supranational reference laboratories, these data raise significant concerns regarding capacity and proficiency at the local clinical level. Addressing these diagnostic challenges is critical for both patient-level management and broader surveillance and control efforts.
INTRODUCTION
Campylobacteriosis in humans typically presents as acute diarrhea with fever. However, more significant sequelae, such as Guillain-Barré syndrome (GBS), reactive arthritis (ReA), and irritable bowel syndrome (IBS), have been reported following Campylobacter gastroenteritis (1).
Most human cases are caused by thermotolerant Campylobacter spp., which are zoonotic bacteria found in animals, such as poultry, cattle, and pigs, as well as contaminants of various foodstuffs and water (2, 3).
Campylobacter jejuni and Campylobacter coli are the most commonly implicated species, and campylobacteriosis is the most frequently reported bacterial foodborne illness in most developed countries. However, data from developing countries are often limited by a lack of diagnostic capacity (1, 2, 4).
Antimicrobials are typically not suggested for treatment of mild/moderate enteritis in otherwise healthy individuals. However, antimicrobial therapy may be warranted for severe or bloody diarrhea. Antimicrobials are also used in the management of extraintestinal (invasive) infections. Macrolides (e.g., erythromycin and azithromycin) are commonly used for empirical treatment of campylobacteriosis, and fluoroquinolones (e.g., ciprofloxacin) may be a second-line therapy for adults. Accurate antimicrobial susceptibility testing is critical for developing empirical therapy guidelines and monitoring emerging resistance. Increasing antimicrobial resistance (AMR), especially multidrug resistance to fluoroquinolones and azithromycin, significantly limits treatment options for severe/invasive disease. Access to last-line drugs, such as carbapenems, is often beyond the reach of many people in the developing world (5).
Since 2000, the World Health Organization (WHO) Global Foodborne Infections Network (GFN) (formerly WHO Global Salm-Surv [WHO GSS]) has functioned as an international platform to enhance the capacity of countries to detect, control, and prevent foodborne and other enteric infections. Part of this capacity-building work has focused on identification and susceptibility testing of Campylobacter spp. Since 2000, the WHO GFN has offered members the opportunity to participate at no cost in an annual External Quality Assurance System (EQAS). Although the primary focus of the EQAS is serotyping and antimicrobial susceptibility testing (AST) of Salmonella spp., identification and AST of Campylobacter spp. are included as a separate module. Laboratories may choose to participate in all or some components. Approximately 200 laboratories participate in one or more components of the EQAS. Of these laboratories, approximately 50% will participate in the Campylobacter module. The WHO GFN program focuses activities mainly at reference-level facilities (supranational, national, or subnational). While some of these facilities may perform clinical testing, clinical diagnostic laboratories typically do not participate in EQAS (6, 7). Participants report results electronically and receive their results immediately. Participants are encouraged to utilize their results as part of continuous quality improvement.
The aim of this paper is to summarize and describe temporal and geographic trends in the performance of the Campylobacter component of the EQAS (identification and AST) observed between 2003 and 2012.
MATERIALS AND METHODS
The Campylobacter identification component has included C. jejuni and C. coli since 2003 and Campylobacter lari from 2003 to 2008. AST of C. jejuni and C. coli has been included since 2009. Due to the limited availability of epidemiological cutoff values (ECOFFs) for other Campylobacter spp., the AST component includes only C. jejuni and C. coli. Since 2003, the Technical University of Denmark, National Food Institute (DTU Food), in collaboration with members of the WHO GFN steering committee, has organized this proficiency test annually (except 2005). DTU Food coordinated the selection of test strains and verified the identification and AST of test strains. The results obtained by DTU Food were reconfirmed in a blind manner by a referee laboratory (U.S. Centers for Disease Control and Prevention [CDC]). Further details on the preparatory work and the EQAS setup are described in the annual EQAS reports available online (http://www.who.int/gfn/activities/eqas/en/).
While the target audience for the EQAS is national public health, food, and veterinary reference laboratories, in special instances (particularly in countries without a designated referral laboratory), the organizers occasionally permit a select number of clinical and/or research laboratories to participate in the EQAS. EQAS participants have the option to participate in all or some of the components. Participants in the WHO GFN proficiency test for identification and/or AST of Campylobacter spp. receive two vials, each containing a lyophilized Campylobacter isolate (challenge strains). In addition, all laboratories participating in the Campylobacter AST component were provided an isolate of C. jejuni ATCC 33560 upon request. Protocols available online (http://www.antimicrobialresistance.dk/233-169-215-eqas.htm) describe how to revive and test the isolates and refer to a manual on subculturing and maintaining quality control (QC) strains.
For the identification component, the protocol specified that the laboratory's routine methods should be applied. Laboratories were free to utilize conventional phenotypic identification, molecular identification, or a combination of methods for identification. Laboratories that participated in only the AST component (i.e., did not perform identification) were provided upon request the identification of the coded Campylobacter strains.
While multiple methods were used for identification, during this period, validated methods for susceptibility testing of Campylobacter spp. by disk diffusion or Etest were not internationally available. As such, only broth or agar dilution methods (measuring the MIC) were accepted for the AST component. Epidemiological cutoff values (ECOFFs) for the interpretation of disk diffusion zones for ciprofloxacin, erythromycin, and tetracycline against C. coli, as well as against ciprofloxacin, erythromycin, tetracycline, and gentamicin for C. jejuni are now incorporated into the EUCAST guidelines. Participants could test and submit results for chloramphenicol (CHL), ciprofloxacin (CIP), erythromycin (ERY), gentamicin (GEN), nalidixic acid (NAL), streptomycin (STR), and tetracycline (TET). The protocol listed the interpretative criteria applied for this EQAS, i.e., ECOFFs according to EUCAST (http://www.eucast.org), which allowed two categories of characterization (resistant [R; non-wild type], or susceptible [S; wild type]) for the C. jejuni and C. coli test strains.
The WHO GFN EQAS was set up as a self-evaluating system in which participants, directly upon submission of results, received a report comparing their obtained results with quality-assured and verified expected results and itemizing the laboratory's eventual deviations. Deviations for the identification component were reported as incorrect results, and no attempt was made to quantify their severity. For the AST component, the acceptance limit was set at 5% deviations, i.e., one deviation would categorize the laboratory's results as unacceptable. The analysis was based on assigning all results the same level of influence, i.e., disregarding the impact of the variation in the selection of test strains from year to year (the susceptibility testing of some strains could be more difficult than others) and the various participation levels (some years, more laboratories participated than in other years).
For each of the world regions, the annual proportions of correctly identified species were analyzed for (i) significant variation between the years 2003 and 2012 using the function Fisher.test in R and (ii) a time trend from 2003 to 2012 by performing a chi-square test for trend in proportions using the function prop.trend.test from the R package stats. Next, for each region, the data were aggregated to the overall proportion of correctly identified Campylobacter species over the whole period from 2003 to 2012. These data were used to test if the proportions of correctly identified species were different between the regions using the prop.test function in the R package stats.
To assess potential differences between regions in AST performance, the proportions of correct AST results for each antibiotic (CHL, CIP, ERY, GEN, NAL, STR, and TET) were analyzed for significant differences between regions using the prop.test function in the stat R package stats.
All presented 95% confidence intervals for the proportions were obtained using the function binconf in the R package Hmisc.
RESULTS
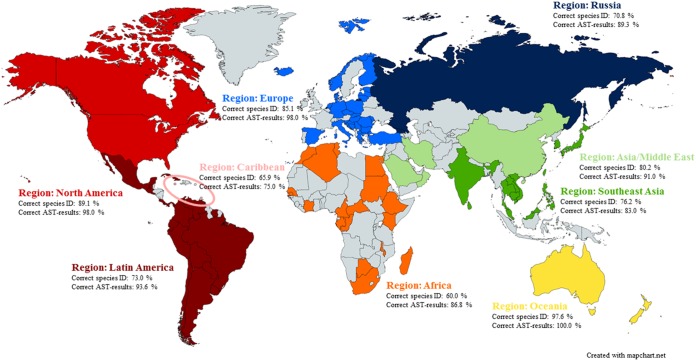
In total, laboratories from 96 countries (Fig. 1) participated in the Campylobacter sp. identification and/or AST component of the EQAS in one or more iterations from 2003 to 2012; the list included national and other reference laboratories from the veterinary, food, and public health sectors.
FIG 1.
Map indicating the participating countries colored with respect to the region to which they belong (Africa, Asia/Middle East, Caribbean, Europe, Latin America, North America, Oceania, Russia, or Southeast Asia). Performance with regard to the species identification and antimicrobial susceptibility testing is indicated as average % of correct results. The 96 participating countries included the following, by region: in Africa, Algeria, Botswana, Cameroon, Central African Republic, Republic of the Congo, Egypt, Ethiopia, Gambia, Gabon, Ivory Coast, Kenya, Madagascar, Malawi, Mauritius, Morocco, Senegal, South Africa, Sudan, and Tunisia; in Asia and the Middle East, China, Islamic Republic of Iran, Israel, Kuwait, Oman, and Saudi Arabia; in the Caribbean, Barbados, Grenada, Jamaica, and Trinidad and Tobago; in Europe, Bosnia and Herzegovina, Bulgaria, Croatia, Cyprus, Czech Republic, Denmark, Estonia, Finland, Germany, Greece, Hungary, Iceland, Italy, Latvia, Lithuania, Luxembourg, Macedonia, Malta, Republic of Moldova, The Netherlands, Norway, Poland, Romania, Serbia, Slovakia, Slovenia, Spain, and Turkey; in Latin America, Argentina, Bolivia, Brazil, Chile, Colombia, Costa Rica, Cuba, Ecuador, El Salvador, Guatemala, Mexico, Panama, Paraguay, Peru, Suriname, Uruguay, and Venezuela; in North America, Canada, and the United States; in Oceania, Australia, New Caledonia, and New Zealand; in the Russian region, Belarus, Georgia, Russian Federation, and Ukraine; and in Southeast Asia, Brunei Darussalam, Cambodia, India, Japan, Republic of Korea, Lao People's Democratic Republic, Malaysia, the Philippines, Singapore, Sri Lanka, Taiwan, Thailand, and Vietnam.
For the identification component of the EQAS, the number of countries participating each year were 53 (2003), 62 (2004), 59 (2006), 59 (2007), 63 (2008), 54 (2009), 62 (2010), 57 (2011), and 69 (2012). In some cases, multiple laboratories from a country participated in the EQAS (e.g., Ministry of Health [MoH] and Ministry of Agriculture [MoA] laboratories). The cumulative numbers of laboratories participating in the Campylobacter module were 97, 111, 100, 104, 112, 92, 100, 82, and 112 in the years 2003 to 2012, respectively.
For the AST of Campylobacter spp., the numbers of participating countries from each region in the years 2009, 2010, 2011, and 2012, respectively, were 2, 2, 7, and 4 for Africa; 1, 1, 1, and 3 for Asia and the Middle East; 0, 0, 0, and 1 for the Caribbean; 7, 9, 7, and 11 for Europe; 4, 7, 6, and 6 for Latin America; 1, 2, 2, and 2 for North America; 0, 0, 1, and 0 for Oceania; 0, 1, 1, and 0 for Russia; and 4, 5, 4, and 6 for Southeast Asia. The cumulative numbers of laboratories participating from each country were 25, 37, 38, and 47 for the years 2009 to 2012, respectively. In all, 18 laboratories participated twice, 14 laboratories participated three times, and 11 laboratories participated in all four Campylobacter AST iterations from 2009 to 2012.
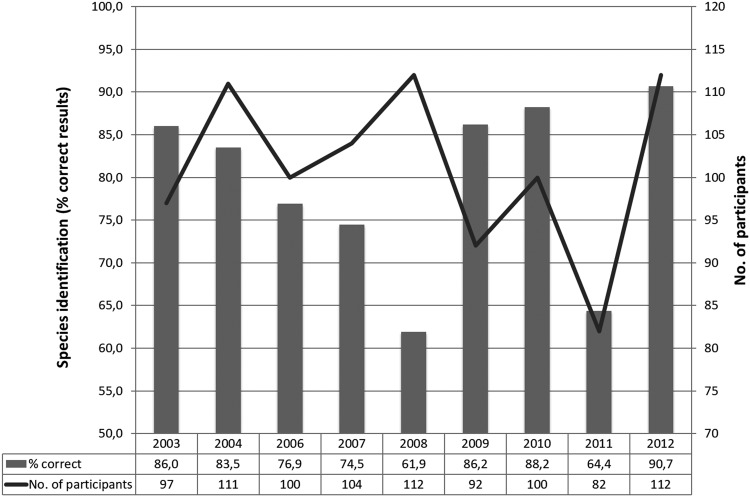
The overall ability to correctly identify Campylobacter spp. fluctuated over the years 2003 to 2012 (Fig. 2), and when focusing on C. coli and C. jejuni bacteria between the regions, a significant difference could be identified in Europe and Latin America in the proportion of correctly identified Campylobacter species between years (data not shown). No time-related trend was identified in Europe or Latin America, and other factors (such as new laboratories joining or previous participants leaving the program) likely contributed to this variation. A significant variation between the years was not found in any other regions.
FIG 2.
Species identification of Campylobacter. Summary of the performance per year of results covering all nine participating regions (Africa, Asia/Middle East, Caribbean, Europe, Latin America, North America, Oceania, Russia, and Southeast Asia).
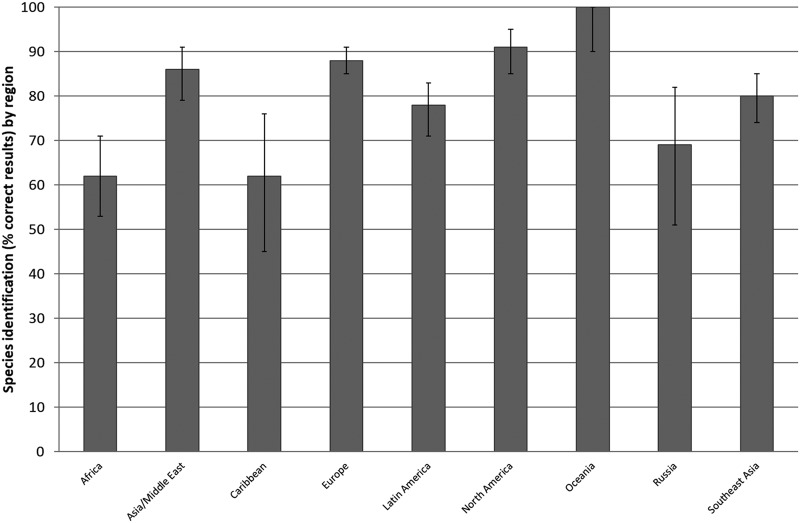
There was a significant difference between the regions as to the proportion of correctly identified Campylobacter species, with Oceania and North America exhibiting the highest and Africa and the Caribbean exhibiting the lowest proportions of correctly identified Campylobacter species (Fig. 3). It is important to consider that in some regions (e.g., Oceania), participation was limited (n = 1) and may fail to reflect true regional capacity.
FIG 3.
Species identification of Campylobacter coli and C. jejuni. Summary of the performance per region of results over the years 2003 to 2012 (excluding 2005).
In the iterations from 2003 to 2012, identifications were correctly performed between 59.3% (2011, C. coli, n = 81) and 96.4% (2012, C. jejuni, n = 112), with an average over the years of 79.3% (based on the result of two isolates per year, i.e., 18 isolates in total of C. jejuni, C. coli, and C. lari; total number of observations, n = 1,736). Comparing the species, it appears that participants were more able to correctly identify C. jejuni (90.4% correct) than C. coli and C. lari (74.1% and 68.8% correct identifications, respectively). This result was not unexpected, as, typically, C. jejuni is readily identified by hippurate hydrolysis, whereas other species require additional more complex tests.
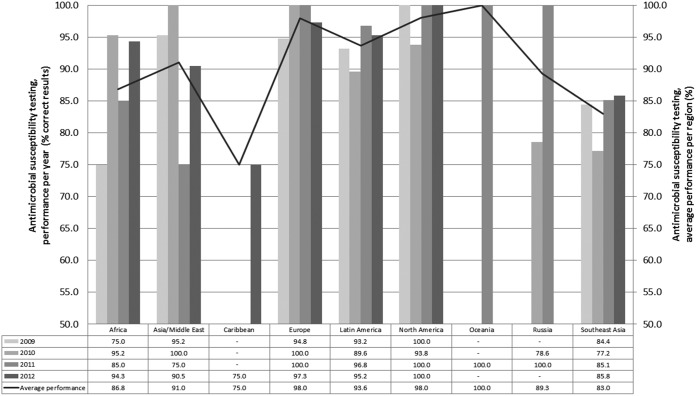
Subsequent to the validation of the submitted data, 1,565 AST results were included for analysis for the eight Campylobacter isolates in the four iterations from 2009 to 2012. Of these results, 7.3% deviated from the expected. For all antimicrobials included in the antimicrobial susceptibility performance test, there was a significant difference between the performances of the regions, where Europe and North America exhibited the highest correspondence with the expected results and Southeast Asia the lowest (Fig. 4). Additionally, results from Oceania presented a high performance percentage (100%), and results from the Caribbean region presented a low percentage (75%); however, these results should be interpreted with care, as they represent results from a limited number of laboratories per region (n = 1).
FIG 4.
Antimicrobial susceptibility testing of Campylobacter spp. showing the performance per year and region.
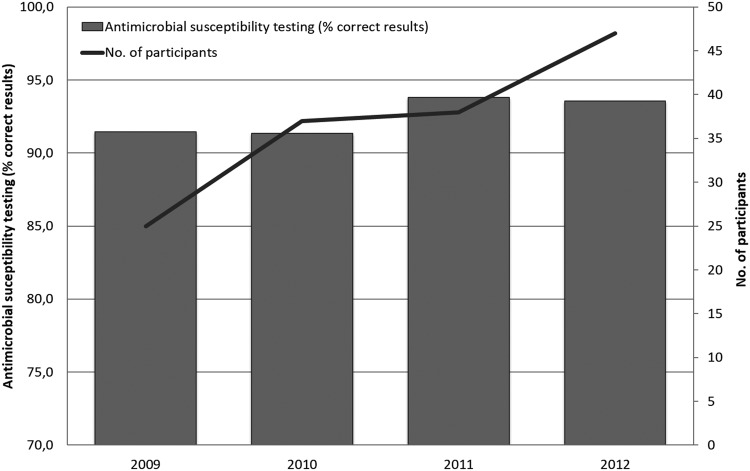
Deviations from the expected results appeared to be particularly related to streptomycin, with a deviation level at 11.5%; however, fairly high deviation levels were also seen for erythromycin, tetracycline, and ciprofloxacin, (8.5%, 8.1%, and 6.4%, respectively). In particular, one bacterial strain, WHO 2009 C-9.2 (resistant [non-wild type] to CIP, NAL, and TET), caused a high level of deviation, with 11.3% of the submitted results deviating from the expected. A low deviation level (4.2%) was obtained for chloramphenicol, to which all the test strains were susceptible (wild type). When the obtained results from all laboratories that participated in the AST component in one or more of the four iterations were compared, a slight increase in performance was observed across the years from 2009 to 2012 (Fig. 5). Disregarding results from the Caribbean and Oceania regions due to the limited number of submitted results, the summary of the 4 years of results per region provided an indication of a generally low performance of the participants in the Southeast Asian region and the African region, with deviation levels at 17.0% and 13.2%, respectively. In 2012, however, all regions except Southeast Asia (deviation level, 14.2%) exhibited deviation levels lower than 10% (the Russian region did not participate in the 2012 iteration).
FIG 5.
Antimicrobial susceptibility testing of Campylobacter spp. Summary of the performance per year of results covering all nine participating regions (Africa, Asia/Middle East, Caribbean, Europe, Latin America, North America, Oceania, Russia, and Southeast Asia).
In the four iterations, 51 (75%) laboratories uploaded one or more values for the QC reference strain C. jejuni ATCC 33560, suggesting that 25% of the labs did not test the QC strain for reference. Of the submitted values for the QC reference strain, an average of 17.2% were out of the QC range when evaluated against one of the validated methods described by the CLSI (e.g., VET01-A4) (8). An analysis of regional differences in this context revealed Africa to be the region with the highest level of laboratories submitting AST results for the Campylobacter test strains without submitting results for the C. jejuni ATCC 33560 reference strain.
DISCUSSION
The proficiency test results are intended to be utilized for continuous quality improvement. However, some participating laboratories did not demonstrate improvement in identification or AST over time. Information about corrective actions implemented in the individual laboratories based on the deviations in their evaluation report could have added to the analysis but unfortunately was not available. This proficiency test shows a worrisome tendency, where even national reference laboratories in regions normally anticipated to perform flawlessly have approximately 10% incorrect results in both tests. Given the complex microbiology of Campylobacteraceae and the fastidious nature of these organisms, when these results are extrapolated to first-line facilities, such as clinical laboratories, the ability to correctly identify and perform antimicrobial susceptibility tests on Campylobacter spp. is likely substantially higher than the 10% error observed among participating laboratories. In addition, results for Campylobacter spp. were received only from ∼50% of the total number of participating laboratories in the WHO GFN EQAS. This suggests that nearly 50% of participants lack a basic capacity for testing Campylobacter spp. These findings are worrying in light of the importance of the Campylobacter spp. as a foodborne pathogen. Thus, actions are still required to improve the performance of laboratories and report correct data on Campylobacter spp. worldwide.
Performance (pass/fail) criteria were not specified for the Campylobacter species identification module, although the average of 79.3% correctly identified strains indicates that some laboratories would need to assess their routine to improve their performance. Assessing the general performance of the AST component of all participating laboratories over the 4 years, the average deviation level at 7.3% exceeds the defined acceptance limit of 5%. The development in the annual deviation levels, however, could not indicate a trend with statistical significance. For the AST, the high level of incorrectly reported results is critical, especially for macrolides (8.5%) and fluoroquinolones (6.4%), since these two antimicrobial classes are the preferred choice for treatment.
The frequent incidence of Campylobacter diarrhea, increasing drug resistance, and the potential for long-term sequelae highlight the importance of accurately understanding the socioeconomic burden of campylobacteriosis (1). Increased competence of reference laboratories for identification and AST of C. jejuni and C. coli therefore supports disease surveillance and control programs. However, the ability of a referral center to impact surveillance is contingent upon an isolate/specimen flow from first-line laboratories. On average, only 50% of EQAS participants reported results for Campylobacter spp., suggesting widespread gaps in capacity. The advancement in whole-genome sequencing and in silico bioinformatics tools combined with lower prices is a promising development in enhancing the ability of national reference laboratories to correctly identify and perform susceptibility testing of Campylobacter spp. in the future. Similarly, molecular assays and other culture-independent tests may increase surveillance capacity at the clinical level. While these technologies currently are not a substitute for culture, they may provide estimates of burden and help determine which specimens should be subjected to conventional culture.
The self-evaluation design of this proficiency test was intended to challenge the participating laboratories to assess their current identification and AST methods for Campylobacter spp., while allowing them to include the proficiency test outcome as an external quality assessment of the relevant methods. Self-evaluation is a concept well known to laboratories following a quality assurance standard requiring quality control procedures, e.g., ISO/IEC 17025:2005 (9), and might include monitoring the validity of test results by the regular use of an internal quality control using reference materials or participation in proficiency-testing programs. Apart from the obvious connection to the WHO GFN EQAS as a proficiency test, laboratories participating in the program are offered material for internal control in the form of both the certified ATCC reference strain, C. jejuni ATCC 33560, and the test strains, which can be regarded as internal control strains and, consequently, should be stored and maintained.
In addition to self-evaluation, the possibility of introducing approaches, such as mentoring of participants, training courses, and E-learning, could be explored and suggested to the participants. The question of resources must, however, be considered; for example, mentoring of participants appears to be a rewarding but also a resource-demanding approach of capacity building. Regional follow-up is likely to be a rewarding approach and should be based on an evaluation of regional needs and challenges. Especially for the African and Southeast Asia regions, it appears that specific follow-up is required. For example, the submitted results for the AST component indicate that many laboratories did not perform adequate internal quality control (17.2% of submitted results for the ATCC reference strain were out of range), which is why WHO GFN capacity-building efforts focus on encouraging the maintenance of relevant quality assurance as part of the laboratory routines. Internal laboratory quality assurance (QA) ensures the minimization of variable factors influencing the obtained result for a test strain. These factors include the medium content, the activity of the antimicrobial, and the testing of a QC reference strain according to an internationally recognized standard (e.g., one by the CLSI). Laboratories that have introduced relevant quality assurance of the variable factors facilitate good performance. For all laboratories performing AST of Campylobacter species, testing of the C. jejuni reference strain (ATCC 33560) should be a routine QA measure providing quality control for both the method and the reagents. Moreover, results outside the quality control ranges should always induce an appropriate follow-up.
In conclusion, this annually provided proficiency test supports the identification and AST of Campylobacter spp. and allows national reference laboratories, free of charge, to evaluate their obtained results by comparison to quality-assured and verified expected results. Overall, we found that global ability to correctly identify Campylobacter spp. fluctuated over the years up to 90.7%, and the ability to correctly perform AST appeared to steadily increase to 93.6%. African laboratories, followed by Southeast Asian laboratories, had the lowest performance in both identifying the Campylobacter spp. and conducting AST. Our results reveal a worrisome tendency where approximately 10% of laboratories reported either an incorrect diagnosis or antimicrobial susceptibility profile for treatment. This issue will compromise the ability to correctly diagnose illness and effectively treat patients, and it will provide unreliable data for pathogen and AST surveillance systems if not addressed.
ACKNOWLEDGMENTS
This work was funded by the World Health Organization Global Foodborne Infections Network (http://www.who.int/gfn) and by COMPARE, which has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement 643476.
We thank Arne B. Jensen for setting up the submission database for capturing and evaluating the results of the EQAS participants.
REFERENCES
- 1.World Health Organization.2013. The global view of campylobacteriosis: report of an expert consultation, Utrecht, Netherlands, 9–11 July 2012. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/handle/10665/80751/9789241564601_eng.pdf?sequence=1&isAllowed=y. [Google Scholar]
- 2.European Food Safety Authority (EFSA), European Centre for Disease Prevention and Control (ECDC). 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J 10:2597 doi: 10.2903/j.efsa.2012.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs-Reitsma W, Lyhs U, Wagenaar J. 2008. Campylobacter in the food supply, p 427–444. In Nachamkin I, Szymanski CM, Blaser MJ (ed), Campylobacter, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 4.Food and Agriculture Organization of the United Nations/World Health Organization (FAO/WHO). 2009. Risk assessment of Campylobacter spp. in broiler chickens: interpretative summary. Microbiological risk assessment series no. 11. Food and Agriculture Organization of the United Nations/World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis 7:24–34. doi: 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendriksen RS, Mikoleit M, Carlson VP, Karlsmose S, Vieira AR, Jensen AB, Seyfarth AM, DeLong SM, Weill F-X, Lo Fo Wong DM, Angulo FJ, Wegener HC, Aarestrup FM. 2009. WHO Global Salm-Surv External Quality Assurance System for serotyping of Salmonella isolates from 2000 to 2007. J Clin Microbiol 47:2729–2736. doi: 10.1128/JCM.02437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendriksen RS, Seyfarth AM, Jensen AB, Whichard J, Karlsmose S, Joyce K, Mikoleit M, Delong SM, Weill F-X, Aidara-Kane A, Lo Fo Wong DMA, Angulo FJ, Wegener HC, Aarestrup FM. 2009. Results of use of WHO Global Salm-Surv External Quality Assurance System for antimicrobial susceptibility testing of Salmonella isolates from 2000 to 2007. J Clin Microbiol 47:79–85. doi: 10.1128/JCM.00894-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CLSI. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 4th ed Approved standard CLSI document VET01-A4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.ISO/IEC. 2005. ISO/IEC 17025:2005 General requirements for the competence of testing and calibration laboratories. International Organization for Standardization, Geneva, Switzerland: https://www.iso.org/standard/39883.html. [Google Scholar]