The rapid and accurate detection of influenza A virus (FluA), influenza B virus (FluB), and respiratory syncytial virus (RSV) improves patient care. Sample-to-answer (STA) platforms based on nucleic acid amplification and detection of these viruses are simple, automated, and accurate.
KEYWORDS: FluA/B and RSV, sample to answer, clinical microbiology
ABSTRACT
The rapid and accurate detection of influenza A virus (FluA), influenza B virus (FluB), and respiratory syncytial virus (RSV) improves patient care. Sample-to-answer (STA) platforms based on nucleic acid amplification and detection of these viruses are simple, automated, and accurate. We compared six such platforms for the detection of FluA, FluB, and RSV: Cepheid GeneXpert Xpress Flu/RSV (Xpert), Hologic Panther Fusion Flu A/B/RSV (Fusion), Cobas influenza A/B & RSV (Liat), Luminex Aries Flu A/B & RSV (Aries), BioFire FilmArray respiratory panel (RP), and Diasorin Simplexa Flu A/B & RSV (Simplexa). Nasopharyngeal (NP) swab specimens (n = 225) from children previously tested by RP were assessed on these platforms. The results were compared to those of the Centers for Disease Control and Prevention (CDC)-developed real-time reverse transcription-PCR (rRT-PCR) assay for influenza A/B viruses and RSV. Subtyping for FluA and FluB was performed for discrepant analysis where applicable. The percent sensitivities/specificities for FluA detection were 100/100 (Fusion), 98.6/99.3 (Xpert), 100/100 (Liat), 98.6/100 (Aries), 98.6/100 (Simplexa), and 100/100 (RP). The percent sensitivities/specificities for FluB detection were 100/100 (Fusion), 97.9/99.4 (Xpert), 97.9/98.3 (Liat), 93.7/99.4 (Aries), 85.4/99.4 (Simplexa), and 95.8/97.7 (RP); and those for RSV detection were 98.1/99.4 (Xpert), 98.1/99.4 (Liat), 96.3/100 (Fusion), 94.4/100 (Aries), 87/94.4 (Simplexa), and 94.4/100 (RP). The 75 strains confirmed to be FluA included 29 pH1N1, 39 H3N2, 4 sH1N1, and 3 untyped strains. The 48 strains confirmed to be FluB included 33 strains of the Yamagata lineage, 13 of the Victoria lineage, 1 of both the Yamagata and Victoria lineages, and 1 of an unknown lineage. All six STA platforms demonstrated >95% sensitivity for FluA detection, while three platforms (Fusion, Xpert, and Liat) demonstrated >95% sensitivity for FluB and RSV detection.
INTRODUCTION
Acute respiratory tract infections are the most common illnesses affecting individuals of all ages and genders across the globe (1, 2). Among the diverse group of pathogens that cause these illnesses, influenza viruses (influenza A virus [FluA] and influenza B virus [FluB]) and respiratory syncytial virus (RSV) are two of the most prevalent (3, 4). RSV is the leading cause of viral bronchiolitis, resulting in a high rate of emergency department (ED) visits and hospitalization of young children (5, 6). Influenza virus infections, too, result in a high rate of morbidity and mortality in patients, along with the burden of hospitalization, multiple outpatient visits, and workday absenteeism (7–9). The rapid and accurate diagnosis of FluA/B and RSV has the potential to decrease the ED length of stay, reduce the need for additional diagnostic tests with a corresponding reduction in patient charges, and guide the judicious and appropriate use of antibiotics and antiviral agents, thereby resulting in better patient management (10–12).
Traditional laboratory methods, such as culture, serology, and direct immunofluorescence assay (DFA), have been used to identify these viruses. However, the delay in result availability for culture and serology and the increased technical expertise required for DFA have limited their use in recent years. With the shift of emphasis on rapid turnaround time (TAT), lateral flow rapid antigen detection tests (RADTs) were favored over traditional methods due to their ease of use and rapid result availability for both FluA/B and RSV (10, 13). However, clinical experience with RADTs has documented their poor sensitivities compared to those of traditional methods (13, 14). Hence, the majority of the nucleic acid amplification tests (NAATs) designed to amplify and detect viral RNA targets gained prominence due to a TAT shorter than that of culture, their cost-effectiveness, and their superior accuracy (14). Traditional reverse transcription-PCRs (RT-PCRs) require nucleic acid extraction prior to the amplification process, a low- to moderate-complexity (MC) instrument, trained personnel to perform both extraction and PCR, and manual interpretation of results, necessitating a TAT of approximately 3 to 4 h. The recently introduced automated sample-to-answer (STA) NAAT systems have improved the ease of use and provide moderate to highly accurate test results with a faster TAT. At present, a number of STA FluA/B and RSV molecular assays are approved by the Food and Drug Administration (FDA) and available for clinical use: GeneXpert Xpress Flu/RSV (Xpert; Cepheid, Sunnyvale, CA) (15, 16), Diasorin Simplexa Flu A/B & RSV (Simplexa; Diasorin Molecular, Cypress, CA) (17), Alere (Abbott Rapid Diagnostics, IL) (18, 19), Aries Flu A/B & RSV (Aries; Luminex Corporation, Austin, TX) (20), Cobas influenza A/B & RSV (Liat; Roche Diagnostics, Indianapolis, IN) (21), BioFire FilmArray respiratory panel (RP; BioFire Diagnostics, Salt Lake City, UT), and Panther Fusion Flu A/B/RSV (Fusion; Hologic Inc., San Diego, CA), among others (https://www.cdc.gov/flu/professionals/diagnosis/table-nucleic-acid-detection.html). Of these STA assays, Alere, Liat, and Xpert offer Clinical Laboratory Improvement Amendments (CLIA)-waived (CW) assays suitable for point-of-care (POC) testing by nonlaboratory personnel to provide test results within 15 to 30 min in outpatient settings (21, 22).
Our aim was to evaluate the performance of six FDA-cleared STA FluA/B and RSV molecular assays compared to the results of the Centers for Disease Control and Prevention (CDC) influenza virus real-time RT-PCR (rRT-PCR) and RSV rRT-PCR assays as the reference methods (14, 23, 24). Multiple studies have reported the enhanced performance of the CDC Flu A/B rRT-PCR assay over other diagnostic assays (25–27). Moreover, typing of FluA/B into the respective subtypes by the CDC rRT-PCR method provides conclusive evidence for discrepant analysis. The STA FluA/B/RSV assays selected for this comparative study included Fusion, Aries, Xpert, Simplexa, RP, and Liat.
MATERIALS AND METHODS
Samples.
A total of 225 retrospective nasopharyngeal (NP) specimens collected from children between the ages of 2 months and 80 months (median age = 7 months) during the respiratory infection seasons of 2012 to 2017 and previously characterized to be positive (75 specimens positive for FluA, 50 specimens positive for FluB, 51 specimens positive for RSV) or negative (n = 49) by RP were used for the study. The specimens were initially collected in 3 ml universal transport medium (Becton, Dickinson and Company, NJ). Frozen samples were thawed by a deidentifier to distribute appropriate volumes into separate vials on the same day to maintain uniformity, and a single freeze-thaw cycle was performed for all the samples prior to testing across all platforms. The aliquot for Xpert testing was shipped to a neighboring hospital, while the other aliquots were distributed to the designated operators at Children's Mercy Hospital, Kansas City, MO, for Fusion, Aries, Liat, Simplexa, and CDC rRT-PCR testing (23). Test operators were blind to the historical sample results. Appropriate controls were run for each of the assays at the recommended frequencies per manufacturer instructions prior to processing of the samples to ensure the validity of the results. This study was reviewed and approved by the Institutional Review Board at Children's Mercy Hospital, Kansas City, MO.
STA testing.
An allocated amount of the sample was dispensed in the cassette/cartridge for Aries, Liat, and Xpert testing (200 μl) or in the transfer tube for Fusion testing (500 μl) and loaded onto the respective instrument as instructed in the package inserts. For Simplexa testing, 50 μl each of a FluA/FluB/RSV-specific reaction mix and sample was dispensed into the respective wells in the direct assay disc. The disc was then loaded onto the integrated cycler, and the FluA/B- and RSV-specific protocol was run on the instrument. Results were obtained at various time points for the respective assays: 20 min for Liat, 30 min for Xpert, 75 min for Simplexa, 2 h for Aries, and 2.4 h for Fusion. Samples with an initial invalid or error result were repeated once on the respective platforms using an extra frozen aliquot of the specimen.
Reference testing.
Nucleic acid was extracted from 210 μl of specimen (200 μl of clinical specimen plus 10 μl of the MS2 plasmid that served as the internal control) using a NucliSENS easyMag system (bioMérieux) and eluted in 110 μl of elution buffer. Extracted samples were then distributed into 3 aliquots, the first of which was used for a control (as discussed below) and RSV PCR (24), the second of which was used for a FluA/B screening PCR, and the third of which was used for a FluA and -B subtyping PCR (the protocol for FluA/B screening and typing can be obtained by request from https://www.cdc.gov/flu/clsis/index.htm) (23). The same-day aliquoting was done to avoid multiple freeze-thaw cycles of the samples. To determine the quality of the extracted sample and extraction efficiency, we first performed RT-PCR to amplify RNase P (human gene) and MS2 (extraction control). Samples amplifying both targets were evaluated by FluA/B and RSV RT-PCRs. The primers and probes for all targets were designed following CDC guidelines (23, 24). Real-time RT-PCR was performed on an ABI 7500 system. Samples positive by FluA/B screening were subtyped using primers for specific pandemic H1N1 (pH1N1), seasonal H1N1 (sH1N1), and H3N2 strains for influenza A virus and the Yamagata and Victoria lineages for influenza B virus. The FluA/B subtyping test results were used for discrepant analysis of FluA and FluB. RSV subtyping was not performed. The results of all assays and RP (historical results) were compared with those of the reference standard CDC rRT-PCR FluA/B and RSV assays for sensitivity/specificity calculations.
Data analysis.
Reportable data were categorized as true positive (TP), true negative (TN), false positive (FP), and false negative (FN). The following calculations were used to determine the diagnostic yield among the different assays: sensitivity = [number of samples with TP results/(number of samples with TP results + number of samples with FN results)] and specificity = [number of samples with TN results/(number of samples with TN results + number of samples with FP results)]. The 95% confidence intervals were calculated according to the efficient-score method (corrected for continuity). A whisker box plot was used to demonstrate the cycle threshold (CT) values for the three analytes (FluA, FluB, and RSV) on the different platforms with the median and interquartile range (IQR). A two-tailed t test was done to determine the significance between CT values comparing Fusion with the other platforms for each of the targets, FluA, FluB, and RSV.
RESULTS
Of the 225 samples selected on the basis of RP historical results, CDC rRT-PCR results confirmed the following: 75 samples had FluA-positive results, 48 samples had FluB-positive results, 54 samples had RSV-positive results, and 48 samples had negative results. Our final analysis was based on the CDC rRT-PCR FluA/B and RSV assay results.
Influenza virus.
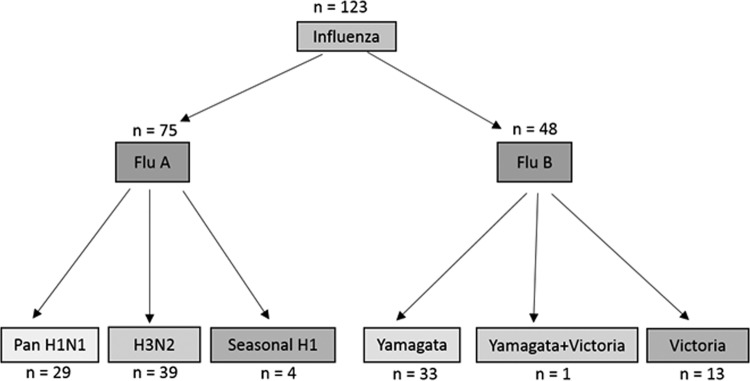
On the basis of the results of the CDC rRT-PCR assay, a total of 123 influenza virus-positive specimens were tested on the 5 STA platforms. As shown in Fig. 1, these 123 specimens comprised 75 FluA-positive and 48 FluB-positive samples. Subtyping with H1N1- and H3N2-specific primers demonstrated 29 pH1N1, 39 H3N2, and 4 sH1N1 strains. However, 3 FluA-positive strains remained untyped, and 2 of these strains were influenza A virus type H3 and 1 was influenza A virus type H1, on the basis of RP historical data. Of the 48 FluB-positive specimens, 33 specimens were of the Yamagata lineage, 13 specimens belonged to the Victoria lineage, and 1 specimen tested positive for both types. The lineage of one FluB-positive specimen remained undetermined (Fig. 1).
FIG 1.
Influenza A and B virus-positive specimens and their subtypes. Of 75 FluA-positive samples, 72 were typed successfully and 3 specimens were nontypeable after two attempts. Of 48 FluB-positive samples, 47 were typed successfully, with 1 sample being nontypeable.
FluA.
Comparing the results across the 6 STA platforms with those of CDC rRT-PCR, Fusion, Liat, and RP were positive for 75 samples, whereas Xpert, Aries, and Simplexa were positive for 74 samples. The sensitivity and specificity of each assay for detecting FluA are shown in Table 1, along with the numbers of samples with FP and FN results. Three different FluA-positive samples were falsely negative on 3 platforms (one each on Aries, Xpert, and Simplexa), and discrepant analysis for these samples demonstrated 2 as H3N2 and 1 as sH1N1 (Table 1). The specificity was 100% for all platforms except Xpert, which had a specificity of 99.3% with 1 FP result.
TABLE 1.
Performance of STA platforms for detection of FluA, FluB, and RSVa
| Virus and parameter | Value(s) for the following platform: |
|||||
|---|---|---|---|---|---|---|
| Fusion | Aries | Liat | Xpert | Simplexa | BioFire RP | |
| FluA | ||||||
| No. of samples with the following result: | ||||||
| TP | 75 | 74 | 75 | 74 | 74 | 75 |
| TN | 150 | 150 | 150 | 149 | 150 | 150 |
| FP | 0 | 0 | 0 | 1 | 0 | 0 |
| FN | 0 | 1 | 0 | 1 | 1 | 0 |
| % sensitivity (95% CI) | 100 (93.9–100) | 98.6 (91.7–99.9) | 100.0 (93.9–100) | 98.6 (91.7–99.9) | 98.63 (91.7–99.9) | 100 (93.9–100) |
| % specificity (95% CI) | 100 (96.8–100) | 100 (96.8–100) | 100 (96.8–100) | 99.3 (95.7 −99.9) | 100 (96.8–100) | 100 (96.8–100) |
| Subtype of sample with FN result | None | pH1N1 | None | H3N2 | H3N2 | None |
| FluB | ||||||
| No. of samples with the following result: | ||||||
| TP | 48 | 45 | 47 | 47 | 41 | 46 |
| TN | 177 | 176 | 174 | 176 | 176 | 173 |
| FP | 0 | 1 | 3 | 1 | 1 | 4 |
| FN | 0 | 3 | 1 | 1 | 7 | 2 |
| % sensitivity (95% CI) | 100 (90.7–100) | 93.7 (81.8–98.3) | 97.9 (87.5–99.8) | 97.9 (87.5–99.8) | 85.4 (71.6–93.5) | 95.8 (84.6–99.2) |
| % specificity (95% CI) | 100 (97.3–100) | 99.4 (96.4–99.9) | 98.3 (94.7–99.6) | 99.4 (96.3–99.9) | 99.4 (96.4–99.9) | 97.7 (93.9–99.2) |
| Subtype of sample with FN result (no. of strains) | None | Yamagata (3) | Yamagata | Yamagata | Yamagata (5), Victoria (2) | Yamagata (2) |
| RSV | ||||||
| No. of samples with the following result: | ||||||
| TP | 52 | 51 | 53 | 53 | 47 | 51 |
| TN | 171 | 171 | 170 | 170 | 170 | 171 |
| FP | 0 | 0 | 1 | 1 | 1 | 0 |
| FN | 2 | 3 | 1 | 1 | 7 | 3 |
| % sensitivity (95% CI) | 96.3 (86.2–99.3) | 94.4 (83.6–98.5) | 98.1 (88.8–99.9) | 98.1 (88.8–99.9) | 87 (74.5–94.2) | 94.4 (83.6–98.5) |
| % specificity (95% CI) | 100 (97.2–100) | 100 (97.2–100) | 99.4 (96.3–99.9) | 99.4 (96.3–99.9) | 99.4 (96.3–99.9) | 100 (97.2–100) |
| Subtype of sample with FN result | Not performed | Not performed | Not performed | Not performed | Not performed | Not performed |
TP, true positive; TN, true negative; FP, false positive; FN, false negative; CI, confidence interval.
FluB.
Of the 48 FluB-positive specimens, Fusion detected all 48, whereas Xpert, Liat, Aries, Simplexa, and RP were positive for 47, 47, 45, 41, and 46 samples, respectively. Table 1 depicts the sensitivity, the specificity, and the overall numbers of samples with FP and FN results for each assay. The highest rate of FN results was observed with the Simplexa assay (7/48), while the highest rate of FP results was observed with RP (4/173). A total of 7 FluB-positive specimens could not be detected by a single STA assay: Simplexa (n = 5), Aries (n = 1), and RP (n = 1). Discrepant analysis of these samples demonstrated 6 infected with strains of the Yamagata lineage and 1 infected with a strain of the Victoria lineage. One FluB specimen tested negative by 3 assays (RP, Aries, Simplexa) and one specimen tested negative by 4 assays (Simplexa, Xpert, Liat, Aries), and both of these belonged to the Yamagata lineage. All STAs except Fusion demonstrated FP results (RP, n = 4; Aries, n = 1; Xpert, n = 1; Simplexa, n = 1; Liat, n = 3).
RSV.
Of the 54 RSV specimens, Xpert and Liat each found that 53 were positive, whereas 52 samples were positive for RSV by Fusion. Aries and RP were each positive for 51 specimens, and Simplexa was positive for 47 specimens. Sensitivity and specificity for RSV detection were the highest in Xpert and Liat and the lowest in Simplexa (Table 1). RSV-positive specimens were undetected six times on a single platform (Fusion, n = 1; Simplexa, n = 5), twice on 2 platforms (Aries and Simplexa, RP and Simplexa), once on 3 platforms (RP, Aries, and Simplexa), and once on all STA platforms. The rate of FN results was the highest for Simplexa, but the rate of FP results was comparable across all assays evaluated (Table 1).
Invalid results.
The result for a sample was deemed invalid when there was an error or a failure to generate a result. On the basis of the initial results, the number of invalid results was the highest on Xpert (n = 7) and Liat and Fusion (n = 5 each), followed by Aries (n = 2) and Simplexa (n = 1). The results for all impacted samples were valid upon retesting on the Fusion, Aries, and Liat platforms. Six samples with invalid results on Xpert had valid results upon repeat testing; however, the result for 1 sample was invalid on repeat testing. The single sample with an invalid result on Simplexa also had an invalid result on repeat testing. The rate of invalid results remained undetermined for RP since we collected only historical data from this platform.
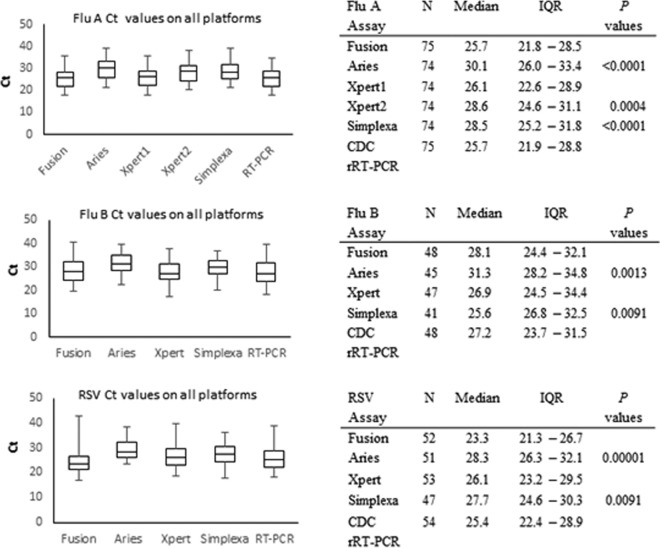
CT.
The median and IQR cycle threshold (CT) values for FluA, FluB, and RSV on the Fusion, Aries, Xpert, and Simplexa platforms and the CDC rRT-PCR are depicted as box plots in Fig. 2. Liat and RP do not report CT values; hence, they could not be incorporated into the analysis. The mean CT ± standard deviation (SD) for the reference method (CDC rRT-PCR) was 25.6 ± 4.31 for FluA, 27.5 ± 5.35 for FluB, and 26.2 ± 5.32 for RSV. The method with the lowest mean CT ± SD was Fusion for FluA (25.4 ± 4.56), with this value being significantly lower than the CT values for all other assays (Fig. 2). Xpert showed the lowest mean CT ± SD for FluB (27.5 ± 5.06). The lowest mean CT ± SD for RSV was demonstrated by Fusion (25.13 ± 6.01), which had a significantly lower CT than Aries and Simplexa (Fig. 2). The highest mean CT ± SD for all three targets was for Aries (for FluA, 30.01 ± 4.54; for FluB, 31.4 ± 4.59; for RSV, 29.4 ± 4), with Aries showing greater than 3 CTs of difference from the CT of the CDC rRT-PCR.
FIG 2.
Box plot diagrams and tables depicting the CT values for FluA (top), FluB (middle), and RSV (bottom) detection on STA platforms and the reference method, rRT-PCR. Significant P values comparing Fusion with the other assays are also reported.
Analysis of the CT values for discrepant samples did not reveal any significant pattern that could be attributed to the lack of detection. CT values for samples in which virus was not detected on the different platforms ranged from 21 to 39 for FluA, 28 to 37 for FluB, and 29 to 42 for RSV. The single specimen that tested negative for RSV with all STA assays had a CT of 33.25 by the CDC rRT-PCR. Thus, we failed to conclude any direct correlation between the incidences of FN and the viral load.
DISCUSSION
It is suggested that although infectious influenza virus is not released from adults after 5 days of illness, virus shedding is typically longer (range, 7 to 10 days) in children, patients with chronic illnesses, and immunocompromised patients (28–30). Current evidence also suggests that the peak viral load is higher in children than in adults (7, 28, 30), which makes detection of pathogens from pediatric respiratory specimens more sensitive. Our study compared 6 different STA assays for FluA/B and RSV detection in a pediatric population. Separate studies with Xpert, Aries, Liat, RP, and Simplexa have established their utility and importance in the rapid diagnosis of these pathogens from respiratory specimens both in children and in adults (15–18, 20, 21, 31–34). Previous studies comparing Aries and Xpert showed 96 to 100% sensitivity and 99.3 to 100% specificity for all three targets between the assays (31) or 96.6% positive percent agreement (PPA) for FluA, 100% PPA for FluB and RSV, and 98.9% negative percent agreement (NPA) (32). In a separate retrospective study evaluating Xpert versus RP, the PPA for FluA and RSV was reported to be 100% and the PPA for FluB was reported to be 92.3% (34). Similarly, a 100% PPA was reported for FluA and FluB and a 97.1% PPA was reported for RSV; an NPA of 95.2%, 99.5%, and 99.6% was reported for FluA, FluB, and RSV, respectively, in a separate study evaluating the performance of the Xpert FluA/B RSV assay (33). Our study showed similar sensitivities and specificities for FluA (98.6% and 99.3%, respectively), FluB (97.9% and 99.4%, respectively), and RSV (99.1% and 98.4%, respectively) by Xpert. A multicenter clinical evaluation of the Aries system reported 95.8%, 93.8%, and 97.1% PPA for FluA, FluB, and RSV, respectively, a 98.4% NPA for FluA and RSV, and a 99.4% NPA for FluB (20). We report a higher specificity for all three targets with Aries and comparable sensitivities in our study. In two studies evaluating the performance of Cobas Liat FluA/B RSV assays, the sensitivity and specificity for all three targets were reported to be 96 to 100% (21, 35), which compare well with findings from our study (sensitivity and specificity, 100% and 100%, respectively, for FluA; 97.9% and 98.3%, respectively, for FluB; and 98.1% and 99.4%, respectively, for RSV). Simplexa showed a significantly lower sensitivity for both FluB (85.4%) and RSV (87%) in our study compared to the results from the earlier study (99.3% for FluB and 96.8% for RSV), but specificity values were similar to those from the earlier study (17). Overall, the assays in our study panel demonstrated greater than 93% sensitivity and specificity for all analytes (FluA, FluB, and RSV), with the exception of Simplexa, which demonstrated 85.3% and 87% sensitivities for FluB and RSV, respectively. The overall highest numbers of FP and FN results were obtained with Liat (n = 4) and Simplexa (n = 16), respectively.
Studies evaluating the detection of influenza A virus have reported a lower sensitivity for some commercial assays for various strains. This finding may be attributed to the assay design, which leads to a variation in the ability to detect circulating strains or emerging strains with antigenic drift or shift (36, 37). Antigenic variability is also seen in influenza B virus and RSV, although at a lower prevalence (38–40). Mutations or polymorphisms in primer or probe binding regions create a probe-target mismatch that may affect the detection of new or unknown variants, resulting in a false-negative result and a missed diagnosis (41–43). Additionally, potentially interfering substances like blood or mucus could also impact the outcome of the assay and its result, especially in the absence of an optimum nucleic acid extraction method. We did not observe significant FN results with FluA in our study. However, a high number of FluB and RSV specimens were undetectable by Simplexa (7/48 and 7/54, respectively), which could be due to the above-mentioned reasons or lower viral loads in the specimens.
Hospitals and clinical labs require the prompt and accurate diagnosis of FluA/B and RSV. Although commonly used STA platforms may have performance trends similar to those depicted in our study, some may offer advantages worth considering. For example, assays with a time to result of less than 30 min are suitable for use in outpatient settings (e.g., ED, urgent care centers) to enable prompt clinical decision making and may be preferred, despite their usually higher cost. Such POC assays are performed on simple, small instruments that are easy to maneuver and do not require specialized training. Since POC testing is performed by clinical staff rather than laboratory-trained individuals, adequate laboratory quality management practices need to be implemented to reduce any errors that could potentially occur due to a lack of training. Therefore, POC testing requires significant support from the laboratory to ensure the integrity of the testing and meet accreditation requirements (44).
Among the STA platforms tested in our study, Liat had the shortest time to a result of 20 min (Table 2), followed by Xpert (30 min), which leads to their popularity for POC testing. These 2 assays are advantageous in settings where continuous testing of single specimens can withstand the steady inflow of patients, providing rapid confirmation of the presence of these 3 pathogens in the outpatient setting.
TABLE 2.
Workflow parameters
| Parameter | Value or result for the following platform: |
|||||
|---|---|---|---|---|---|---|
| Fusion | Aries | Simplexa | Xpert | Liat | RP | |
| Complexitya | HC | MC | MC | CW | CW | MC |
| Sample vol (μl) | 500 | 200 | 50 | 200 | 200 | 200 |
| Assay time (min) | 145 | 120 | 75 | 30 | 20 | 60 |
| Random access | Yes | Yes | No | Yesb | Yesb | No |
| Sample loading capacity per instrument | 120 | 12 | 8 | 1–16 | 1 | 1 |
| Throughput (no. of samples) per instrument in 8 h | 335 | 48 | 50 | 16 | 24 | 8 |
HC, high complexity; MC, moderate complexity; CW, CLIA waived.
Based on the modules (Xpert) or the number of instruments (Liat) implemented.
The Simplexa assay time to result was 75 min, with an additional hands-on time of 10 min for loading the disc to its full capacity. The loading of the sample and the instrument is relatively simple and requires a small cycler that is connected to a computer. Aries and Fusion have a time to result of 2 h and 2.4 h, respectively. The large size of these instruments is not conducive to the ED setting and is better suited to the clinical laboratory.
Additional considerations for the selection of FluA/B and RSV STA assays include throughput, batch mode versus random access ability, the required sample volume, ease of use, invalid result rate, additional test menu, and ability to interface with laboratory information systems (LIS). A random access capability enables testing of individual samples at the precise time that they are most needed. This leads to providing more timely results for physicians and patients and also increases laboratory work flow efficiency. The Liat and Xpert assays process one sample at a time, but samples can also be tested in batches using multiple instruments or modules concurrently (one specimen per instrument). The Simplexa and Aries assays are designed to run in batch modes with a sample size of 8 and 6 to 12, respectively. The Aries M1 system has 1 magazine, and the Aries system has 2 magazines, each of which can hold up to 6 cassettes for testing. Thus, 6 to 12 samples can be tested at a time. On these systems, batching reduces waste and may enable labs managing high volumes of patient specimens (e.g., labs in the inpatient setting or reference labs) to generate mass data in a timely manner. However, testing in batch mode can delay the results while the laboratory waits to accrue sufficient samples to complete the batch. Of the STA methods evaluated, only Fusion's system offers the random access capability to load a significant number of samples on a single instrument (up to 120 controls or specimens) at any time. However, Liat and Xpert allow the opportunity for random access if multiple instruments (Liat) or modules (Xpert) are available.
An invalid or error result, representing a failure to generate a result, requires reanalysis, which is costly and time-consuming and delays the reporting of results. An invalid or error result leading to the failure to report a result could be caused by various events, including but not limited to operator error (e.g., pipetting of an insufficient volume of specimen), system failure (i.e., the pressure limit of a component is exceeded), assay validity criteria (i.e., failure of processing or the internal control), control failure (i.e., criteria for the control are not met), or the clinical specimen (i.e., inhibition or presence of an exogenous interfering substance, such as blood). The number of specimens requiring repeat testing was the highest on Xpert (n = 7), followed by Liat and Fusion (n = 5) and Aries (n = 2). Retesting resolved all impacted samples for Liat, Fusion, and Aries; however, the result for a single sample remained invalid for Xpert and Simplexa.
The volume of specimen required for testing on the different platforms varies from 50 μl for testing on Simplexa to 500 μl for testing on Fusion and 200 μl for testing on the Xpert, Liat, and Aries assays. The low volume of specimen needed for the Simplexa assay makes it an ideal choice in cases where sufficient specimen is unavailable or where repeat testing is required. Specimens already tested by the Fusion assay can be stored in lysis buffer, allowing testing for additional respiratory pathogens in an algorithmic approach to contain costs, in contrast to large multiplex respiratory assays with a one-size-fits-all approach.
Although our study is a comprehensive effort to evaluate and compare the performance of the most popular STA platforms for FluA/B and RSV detection, a few limitations apply. One is the lack of a conclusive discrepant analysis for RSV-positive specimens that were not differentiated further into RSV A or B subtypes. We used aliquots of previously characterized frozen samples instead of fresh specimens for the study. The extended freezer time and freeze-thaw cycle for some of the specimens could affect assay performance. However, to minimize any differential effect on assay performance, we tested the same specimen freeze-thaw cycle on all 5 platforms simultaneously. Another shortcoming of our study is the bias associated with selecting samples that were previously tested with RP, and a future study comparing the performance of RP with that of these assays on prospectively collected specimens would provide more unbiased and conclusive data. However, our approach ensured that we included sufficient FluA/B- and RSV-positive specimens and a sufficient representation of all FluA and FluB subtypes.
Overall, all the assays were easy to perform and required minimum technical skill for operating the instruments. With the presence of equally sensitive and precise diagnostic assays in the field, the decision to select a STA FluA/B and RSV molecular assay depends upon the laboratory setting, testing volume, infrastructure with respect to space and staff, work flow, the cost of the assays, and patient population. Our study evaluated the performance of these platforms in a controlled study design using well-characterized specimens. Data from our study provide important information on the strengths and weaknesses of these assays for the detection of FluA/B and RSV in respiratory specimens collected from children.
ACKNOWLEDGMENTS
We extend our thanks to Brett Whitaker for sharing the protocols for the RSV CDC rRT-PCR.
This work was funded by Hologic Inc., San Diego, CA, USA.
REFERENCES
- 1.Monto AS. 2002. Epidemiology of viral respiratory infections. Am J Med 112(Suppl 6A):4S–12S. [DOI] [PubMed] [Google Scholar]
- 2.Monto AS. 1995. Viral respiratory infections in the community: epidemiology, agents, and interventions. Am J Med 99:24S–27S. doi: 10.1016/S0002-9343(99)80307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair H, Brooks WA, Katz M, Roca A, Berkley JA, Madhi SA, Simmerman JM, Gordon A, Sato M, Howie S, Krishnan A, Ope M, Lindblade KA, Carosone-Link P, Lucero M, Ochieng W, Kamimoto L, Dueger E, Bhat N, Vong S, Theodoratou E, Chittaganpitch M, Chimah O, Balmaseda A, Buchy P, Harris E, Evans V, Katayose M, Gaur B, O'Callaghan-Gordo C, Goswami D, Arvelo W, Venter M, Briese T, Tokarz R, Widdowson MA, Mounts AW, Breiman RF, Feikin DR, Klugman KP, Olsen SJ, Gessner BD, Wright PF, Rudan I, Broor S, Simoes EA, Campbell H. 2011. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet 378:1917–1930. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- 4.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeois FT, Valim C, McAdam AJ, Mandl KD. 2009. Relative impact of influenza and respiratory syncytial virus in young children. Pediatrics 124:e1072–e1080. doi: 10.1542/peds.2008-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller EK, Gebretsadik T, Carroll KN, Dupont WD, Mohamed YA, Morin LL, Heil L, Minton PA, Woodward K, Liu Z, Hartert TV, Williams JV. 2013. Viral etiologies of infant bronchiolitis, croup and upper respiratory illness during 4 consecutive years. Pediatr Infect Dis J 32:950–955. doi: 10.1097/INF.0b013e31829b7e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munywoki PK, Koech DC, Agoti CN, Kibirige N, Kipkoech J, Cane PA, Medley GF, Nokes DJ. 2015. Influence of age, severity of infection, and co-infection on the duration of respiratory syncytial virus (RSV) shedding. Epidemiol Infect 143:804–812. doi: 10.1017/S0950268814001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 9.Dorratoltaj N, Marathe A, Lewis BL, Swarup S, Eubank SG, Abbas KM. 2017. Epidemiological and economic impact of pandemic influenza in Chicago: priorities for vaccine interventions. PLoS Comput Biol 13:e1005521. doi: 10.1371/journal.pcbi.1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chartrand C, Leeflang MM, Minion J, Brewer T, Pai M. 2012. Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann Intern Med 156:500–511. doi: 10.7326/0003-4819-156-7-201204030-00403. [DOI] [PubMed] [Google Scholar]
- 11.Rogan DT, Kochar MS, Yang S, Quinn JV. 2017. Impact of rapid molecular respiratory virus testing on real-time decision making in a pediatric emergency department. J Mol Diagn 19:460–467. doi: 10.1016/j.jmoldx.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonner AB, Monroe KW, Talley LI, Klasner AE, Kimberlin DW. 2003. Impact of the rapid diagnosis of influenza on physician decision-making and patient management in the pediatric emergency department: results of a randomized, prospective, controlled trial. Pediatrics 112:363–367. doi: 10.1542/peds.112.2.363. [DOI] [PubMed] [Google Scholar]
- 13.Chartrand C, Tremblay N, Renaud C, Papenburg J. 2015. Diagnostic accuracy of rapid antigen detection tests for respiratory syncytial virus infection: systematic review and meta-analysis. J Clin Microbiol 53:3738–3749. doi: 10.1128/JCM.01816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merckx J, Wali R, Schiller I, Caya C, Gore GC, Chartrand C, Dendukuri N, Papenburg J. 2017. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann Intern Med 167:394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- 15.Arbefeville S, Thonen-Kerr E, Ferrieri P. 2017. Prospective and retrospective evaluation of the performance of the FDA-approved Cepheid Xpert Flu/RSV XC assay. Lab Med 48:e53–e56. doi: 10.1093/labmed/lmx038. [DOI] [PubMed] [Google Scholar]
- 16.Salez N, Nougairede A, Ninove L, Zandotti C, de Lamballerie X, Charrel RN. 2015. Prospective and retrospective evaluation of the Cepheid Xpert(R) Flu/RSV XC assay for rapid detection of influenza A, influenza B, and respiratory syncytial virus. Diagn Microbiol Infect Dis 81:256–258. doi: 10.1016/j.diagmicrobio.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hindiyeh M, Kolet L, Meningher T, Weil M, Mendelson E, Mandelboim M. 2013. Evaluation of Simplexa Flu A/B & RSV for direct detection of influenza viruses (A and B) and respiratory syncytial virus in patient clinical samples. J Clin Microbiol 51:2421–2424. doi: 10.1128/JCM.00286-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapin KC, Flores-Cortez EJ. 2015. Performance of the molecular Alere I influenza A&B test compared to that of the Xpert flu A/B assay. J Clin Microbiol 53:706–709. doi: 10.1128/JCM.02783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell JJ, Selvarangan R. 2014. Evaluation of the Alere I influenza A&B nucleic acid amplification test by use of respiratory specimens collected in viral transport medium. J Clin Microbiol 52:3992–3995. doi: 10.1128/JCM.01639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juretschko S, Mahony J, Buller RS, Manji R, Dunbar S, Walker K, Rao A. 2017. Multicenter clinical evaluation of the Luminex Aries Flu A/B & RSV assay for pediatric and adult respiratory tract specimens. J Clin Microbiol 55:2431–2438. doi: 10.1128/JCM.00318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binnicker MJ, Espy MJ, Irish CL, Vetter EA. 2015. Direct detection of influenza A and B viruses in less than 20 minutes using a commercially available rapid PCR assay. J Clin Microbiol 53:2353–2354. doi: 10.1128/JCM.00791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trabattoni E, Le V, Pilmis B, Pean de Ponfilly G, Caisso C, Couzigou C, Vidal B, Mizrahi A, Ganansia O, Le Monnier A, Lina B, Nguyen Van JC. 2018. Implementation of Alere i influenza A & B point of care test for the diagnosis of influenza in an emergency department. Am J Emerg Med 36:916–921. doi: 10.1016/j.ajem.2017.10.046. [DOI] [PubMed] [Google Scholar]
- 23.Selvaraju SB, Selvarangan R. 2010. Evaluation of three influenza A and B real-time reverse transcription-PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J Clin Microbiol 48:3870–3875. doi: 10.1128/JCM.02464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fry AM, Chittaganpitch M, Baggett HC, Peret TC, Dare RK, Sawatwong P, Thamthitiwat S, Areerat P, Sanasuttipun W, Fischer J, Maloney SA, Erdman DD, Olsen SJ. 2010. The burden of hospitalized lower respiratory tract infection due to respiratory syncytial virus in rural Thailand. PLoS One 5:e15098. doi: 10.1371/journal.pone.0015098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karre T, Maguire HF, Butcher D, Graepler A, Weed D, Wilson ML. 2010. Comparison of Becton Dickinson Directigen EZ Flu A+B test against the CDC real-time PCR assay for detection of 2009 pandemic influenza A/H1N1 virus. J Clin Microbiol 48:343–344. doi: 10.1128/JCM.02063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbas MG, Gallego SV, Castro GM, Baumeister E, Kademian S, De Leon J, Cudola A. 2012. Performance of a commercial assay for the diagnosis of influenza A (H1N1) infection in comparison to the Centers for Disease Control and Prevention protocol of real time RT-PCR. Rev Argent Microbiol 44:26–29. doi: 10.1590/S0325-75412012000100006. [DOI] [PubMed] [Google Scholar]
- 27.Bosevska G, Panovski N, Janceska E, Mikik V, Topuzovska IK, Milenkovik Z. 2015. Comparison of Directigen Flu A+B with real time PCR in the diagnosis of influenza. Folia Med (Plovdiv) 57:104–110. doi: 10.1515/folmed-2015-0027. [DOI] [PubMed] [Google Scholar]
- 28.Lee N, Chan PK, Hui DS, Rainer TH, Wong E, Choi KW, Lui GC, Wong BC, Wong RY, Lam WY, Chu IM, Lai RW, Cockram CS, Sung JJ. 2009. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis 200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato M, Hosoya M, Kato K, Suzuki H. 2005. Viral shedding in children with influenza virus infections treated with neuraminidase inhibitors. Pediatr Infect Dis J 24:931–932. doi: 10.1097/01.inf.0000180976.81055.ce. [DOI] [PubMed] [Google Scholar]
- 30.Killingley B, Greatorex J, Cauchemez S, Enstone JE, Curran M, Read RC, Lim WS, Hayward A, Nicholson KG, Nguyen-Van-Tam JS. 2010. Virus shedding and environmental deposition of novel A (H1N1) pandemic influenza virus: interim findings. Health Technol Assess 14:237–354. doi: 10.3310/hta14460-04. [DOI] [PubMed] [Google Scholar]
- 31.Ling L, Kaplan SE, Lopez JC, Stiles J, Lu X, Tang YW. 2018. Parallel validation of three molecular devices for simultaneous detection and identification of influenza A and B and respiratory syncytial viruses. J Clin Microbiol 56:e01691-. doi: 10.1128/JCM.01691-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMullen P, Boonlayangoor S, Charnot-Katsikas A, Beavis KG, Tesic V. 2017. The performance of Luminex ARIES((R)) Flu A/B & RSV and Cepheid Xpert((R)) Flu/RSV XC for the detection of influenza A, influenza B, and respiratory syncytial virus in prospective patient samples. J Clin Virol 95:84–85. doi: 10.1016/j.jcv.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Cohen DM, Kline J, May LS, Harnett GE, Gibson J, Liang SY, Rafique Z, Rodriguez CA, McGann KM Sr, Gaydos CA, Mayne D, Phillips D, Cohen J. 2018. Accurate PCR detection of influenza A/B and respiratory syncytial viruses by use of Cepheid Xpert Flu+RSV Xpress assay in point-of-care settings: comparison to Prodesse ProFlu. J Clin Microbiol 56:e01237-. doi: 10.1128/JCM.01237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahrenbrock MG, Matushek S, Boonlayangoor S, Tesic V, Beavis KG, Charnot-Katsikas A. 2016. Comparison of Cepheid Xpert Flu/RSV XC and BioFire FilmArray for detection of influenza A, influenza B, and respiratory syncytial virus. J Clin Microbiol 54:1902–1903. doi: 10.1128/JCM.00084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson J, Schechter-Perkins EM, Mitchell P, Mace S, Tian Y, Williams K, Luo R, Yen-Lieberman B. 2017. Multi-center evaluation of the Cobas((R)) Liat((R)) influenza A/B & RSV assay for rapid point of care diagnosis. J Clin Virol 95:5–9. doi: 10.1016/j.jcv.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Overmeire Y, Vanlaere E, Hombrouck A, De Beenhouwer H, Simons G, Brink A, Van den Abeele AM, Verfaillie C, Van Acker J. 2016. Severe sensitivity loss in an influenza A molecular assay due to antigenic drift variants during the 2014/15 influenza season. Diagn Microbiol Infect Dis 85:42–46. doi: 10.1016/j.diagmicrobio.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Stellrecht KA, Nattanmai SM, Butt J, Maceira VP, Espino AA, Castro AJ, Landes A, Dresser N, Butt SA. 2017. Effect of genomic drift of influenza PCR tests. J Clin Virol 93:25–29. doi: 10.1016/j.jcv.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi XS, Hu A, Bolar TV, Al-Rimawi W, Zhao P, Tam JS, Rappaport R, Cheng SM. 2005. Detection and characterization of new influenza B virus variants in 2002. J Clin Microbiol 43:2345–2349. doi: 10.1128/JCM.43.5.2345-2349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa N, Nukuzuma S, Haratome S, Go S, Nakagawa T, Hayashi K. 2002. Emergence of an influenza B virus with antigenic change. J Clin Microbiol 40:3068–3070. doi: 10.1128/JCM.40.8.3068-3070.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, Storch GA, Zang C, Peret TC, Park CS, Anderson LJ. 1999. Genetic variability in envelope-associated protein genes of closely related group A strains of respiratory syncytial virus. Virus Res 59:89–99. doi: 10.1016/S0168-1702(98)00132-4. [DOI] [PubMed] [Google Scholar]
- 41.Kamau E, Agoti CN, Lewa CS, Oketch J, Owor BE, Otieno GP, Bett A, Cane PA, Nokes DJ. 2017. Recent sequence variation in probe binding site affected detection of respiratory syncytial virus group B by real-time RT-PCR. J Clin Virol 88:21–25. doi: 10.1016/j.jcv.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawkinson D, Abhyankar S, Aljitawi O, Ganguly S, McGuirk JP, Horvat R. 2013. Delayed RSV diagnosis in a stem cell transplant population due to mutations that result in negative polymerase chain reaction. Diagn Microbiol Infect Dis 75:426–430. doi: 10.1016/j.diagmicrobio.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 43.Lee HK, Lee CK, Loh TP, Chiang D, Koay ES, Tang JW. 2011. Missed diagnosis of influenza B virus due to nucleoprotein sequence mutations, Singapore, April 2011. Euro Surveill 16(33):pii=19943 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19943. [PubMed] [Google Scholar]
- 44.Shaw JLV. 2016. Practical challenges related to point of care testing. Pract Lab Med 4:22–29. doi: 10.1016/j.plabm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]