Mycoplasma gallisepticum, the primary etiologic agent of chronic respiratory disease, is a significant poultry pathogen, causing severe inflammation and leading to economic losses worldwide. Immunodominant proteins encoded by the variable lipoprotein and hemagglutinin (vlhA) gene family are thought to be important for M. gallisepticum-host interaction, pathogenesis, and immune evasion, but their exact role remains unknown.
KEYWORDS: Mycoplasma gallisepticum, RNA-seq, phase variable, vlhA
ABSTRACT
Mycoplasma gallisepticum, the primary etiologic agent of chronic respiratory disease, is a significant poultry pathogen, causing severe inflammation and leading to economic losses worldwide. Immunodominant proteins encoded by the variable lipoprotein and hemagglutinin (vlhA) gene family are thought to be important for M. gallisepticum-host interaction, pathogenesis, and immune evasion, but their exact role remains unknown. Previous work has demonstrated that vlhA phase variation is dynamic throughout the earliest stages of infection, with vlhA 3.03 being the predominant vlhA expressed during the initial infection, and that the pattern of dominant vlhA expression may be nonrandom and regulated by previously unrecognized mechanisms. To further investigate this gene family, we assessed the vlhA profile of two well-characterized vaccine strains, GT5 and Mg7, a vlhA 3.03 mutant strain, and an M. gallisepticum population expressing an alternative immunodominant vlhA. Here, we report that two M. gallisepticum vaccine strains show different vlhA profiles over the first 2 days of infection compared to that of wild-type Rlow, while the population expressing an alternative immunodominant vlhA gene reverted to a profile indistinguishable from that of wild-type Rlow. Additionally, we observed a slight shift in the vlhA gene expression profile but no reduction in virulence in a vlhA 3.03 mutant. Taken together, these data further support the hypothesis that M. gallisepticum vlhA genes change in a nonstochastic temporal progression of expression and that vlhA 3.03, while preferred, is not required for virulence. Collectively, these data may be important in elucidating mechanisms of colonization and overall pathogenesis of M. gallisepticum.
INTRODUCTION
Mycoplasma gallisepticum, the primary etiologic agent of chronic respiratory disease (CRD), causes significant disease and monetary loss throughout the poultry industry worldwide. This highly transmissible pathogen affects the respiratory tract, causing significant inflammation of the air sacs, lungs, and trachea, as well as the reproductive tract, resulting in decreased weight gain and egg production. Mycoplasma gallisepticum is also pathogenic in other avian species, causing infectious sinusitis in turkeys and severe conjunctivitis in house finch (1, 2).
Despite much effort, little is understood about the mechanisms of survival and persistence employed by M. gallisepticum. It has been established that the primary attachment proteins, GapA and CrmA (3), fibronectin binding proteins, PlpA and Hlp3 (4), sugar transport permease, MalF (5), and dihydrolipoamide dehydrogenase, Lpd (6), all play key roles in the survival and persistence of M. gallisepticum in the host. Additionally, Ron et al. identified 13 proteins preferentially expressed during in vivo infection, including GapA, PlpA, and Hlp3 (7).
Likely important in the virulence of M. gallisepticum are the members of the variable lipoprotein and hemagglutinin (vlhA) gene family, consisting of 43 closely related genes distributed across 5 loci and comprising just over 10% of the entire genome in strain Rlow (8–10). While the function of these gene products remains unknown, it has been speculated to be related to evasion of the host adaptive immune response during infection (11), and variation in vlhA gene complement has been observed among M. gallisepticum strains differing in virulence (12). These gene products have been hypothesized to be involved in attachment and have been shown to bind host red blood cells (13) and host apolipoprotein A1 (14).
Previous work from our laboratory has demonstrated that M. gallisepticum strain Rlow vlhA gene expression changes in a nonstochastic temporal progression in the initial stages of infection in the natural host and may be regulated, at least in part, by previously unrecognized mechanisms (15). M. gallisepticum vlhA 3.03 gene expression is dominant during the very initial stages of infection and increases in expression through 1 day postinfection before rapidly decreasing in expression during the remaining infection.
Here, we expand on this work, examining the early vlhA gene expression profiles from four different M. gallisepticum strains from experimentally inoculated chickens over the earliest stages of infection. We hypothesize that perturbation to the established model will result in changes in the ordered pattern of M. gallisepticum vlhA gene expression, providing valuable information about what may be contributing to the changes in vlhA gene expression.
We report the vlhA profiles over 2 days for two well-characterized vaccine strains, GT5 (16) and Mg7 (6), that produce no significant tracheal lesions or changes to the tracheal architecture, in addition to an M. gallisepticum population stably expressing an alternative dominant vlhA (15). Additionally, we report the virulence and vlhA gene expression profile of an M. gallisepticum strain harboring a transposon insertion mutation in the primary immunodominant vlhA 3.03 gene.
RESULTS
Mg7 vlhA gene expression.
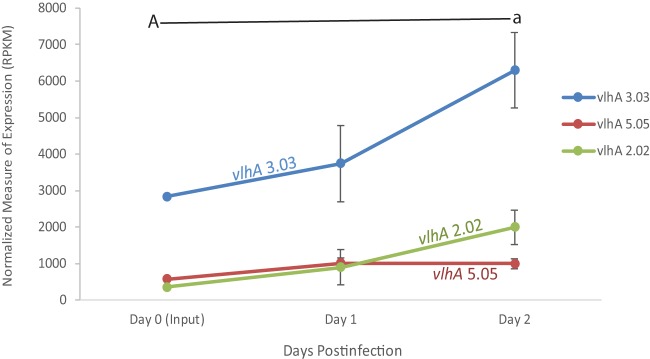
The pattern of the M. gallisepticum vaccine strain, Mg7, over the course of the 2-day infection showed that the expression of vlhA 3.03 increased (Fig. 1; see also Fig. S1 in the supplemental material), with a 1.31-fold increase at day one, compared to that of the broth-grown input culture used for inoculation. Even more dramatic was the increase in expression of vlhA 3.03 (1.68-fold) between days one and two postinfection. This pattern of vlhA gene expression observed in Mg7 is strikingly different from that of wild-type Rlow in vivo, as reported by Pflaum et al. (15), where a dramatic drop in the expression of initially dominant vlhA 3.03 is observed to begin between days one and two postinfection.
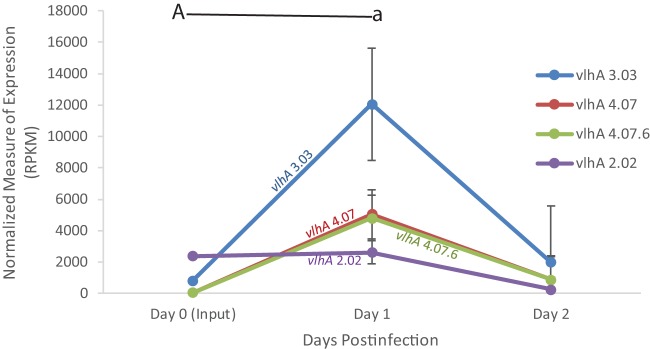
FIG 1.
vlhA expression profile of M. gallisepticum Mg7 extracted directly from tracheas of experimentally inoculated birds over the course of the 2-day infection, as determined by RNA sequencing. Only vlhA genes discussed are displayed. Each data point represents an average RPKM value from the results determined for five animals (with the exception of broth). Error bars show standard errors of the means (SEM). Key statistically significant changes between two time points are indicated by paired upper- and lowercase letters for vlhA 3.03 (A/a).
Interestingly, an increase in vlhA 2.02 was observed at days one and two postinfection, an observation that was not seen with the wild-type Rlow strain. Furthermore, we did not see an increase in the expression of vlhA 4.07 and 4.07.6 at days one and two postinfection, as was observed previously in wild-type virulent Rlow.
Most notable is the continued increase in the expression of vlhA 3.03 over the earliest stages of infection in this live attenuated vaccine strain, which does not result in any significant tracheal lesions at 2 weeks postinfection (6) and which is in contrast to the previous vlhA pattern observed with virulent strain Rlow (15).
GT5 vlhA gene expression.
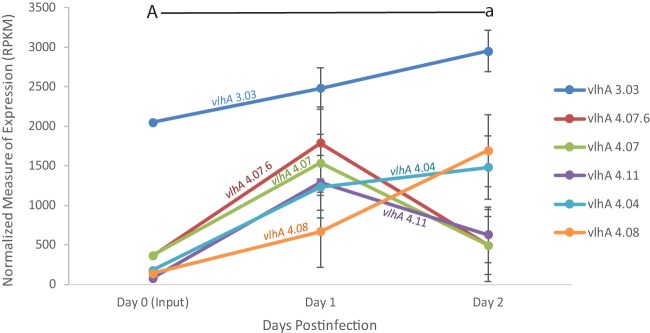
Similar to the vlhA gene expression pattern observed in the Mg7 vaccine strain, M. gallisepticum recovered from chickens inoculated with GT5 showed a continued increase in expression of vlhA 3.03 over the course of the 2-day infection compared to the broth-grown inoculum, with an increase at day one postinfection continuing through day two postinfection (Fig. 2 and Fig. S2).
FIG 2.
vlhA expression profile of M. gallisepticum GT5 extracted directly from tracheas of experimentally inoculated birds over the course of the 2-day infection, as determined through RNA sequencing. Only vlhA genes discussed are displayed. Each data point represents an average RPKM value from the results determined for five animals (with the exception of broth). Error bars show SEM. Key statistically significant changes between two time points are indicated by paired upper- and lowercase letters for vlhA 3.03 (A/a).
We observed an increase in the expression of vlhA 4.07 and 4.07.6 at day one postinfection, as was previously observed in the wild-type Rlow strain. Interestingly, we saw an increase in the expression of vlhA 4.08 at days one and two postinfection, a pattern we did not previously observe until much later time points with wild-type virulent Rlow. Like Mg7, the vlhA expression pattern of this vaccine strain, which does not result in any significant tracheal lesions at 2 weeks postinfection, differs from that of virulent Rlow, in that the expression of vlhA 3.03 increased significantly over the earliest days of infection.
vlhA 3.03 mutant induced host pathology and recovery.
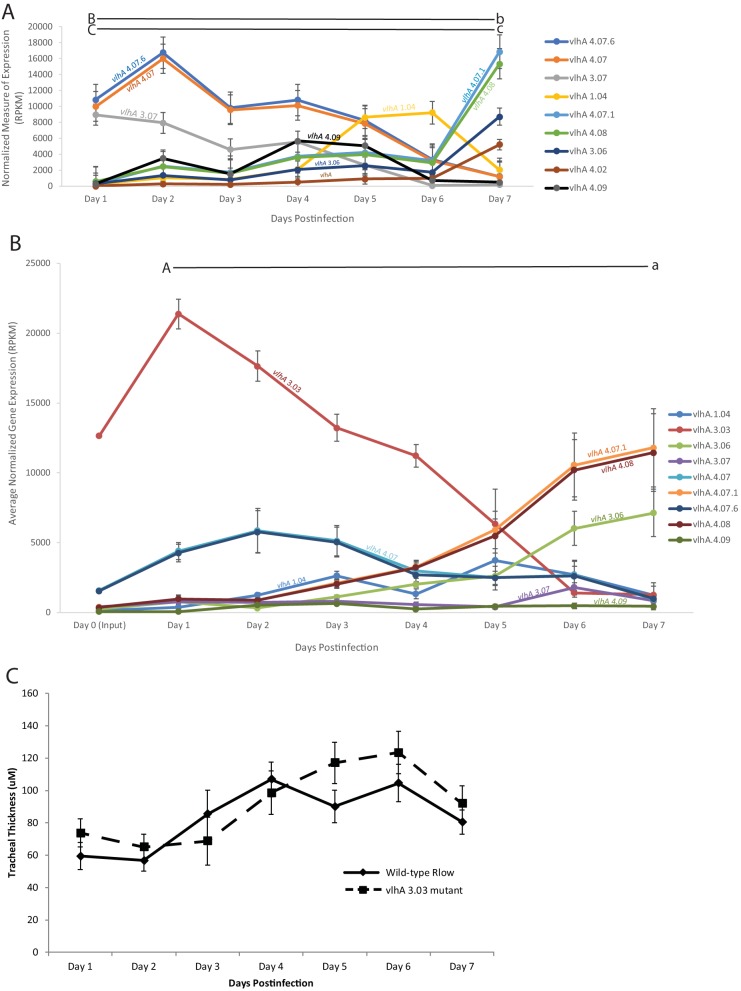
Surprisingly, the transposon insertion in the vlhA 3.03 gene did not cause a significant reduction in virulence. There was a statistically indistinguishable difference in tracheal thickness measurements between chickens challenged with wild-type Rlow and the vlhA 3.03 mutant, both demonstrating moderate mucosal thickening (Fig. 3C), suggesting that this mutant displayed virulence at a level comparable to that of wild-type Rlow in the chicken respiratory tract. The tracheal thicknesses induced by both wild-type Rlow and vlhA 3.03, while indistinguishable from each other, were significantly higher than those previously observed from a medium control-inoculated chicken (17).
FIG 3.
vlhA expression profile of the M. gallisepticum vlhA 3.03 mutant (A) or wild-type M. gallisepticum Rlow (B) extracted directly from tracheas of experimentally inoculated birds over the course of the 7-day infection, as determined by RNA sequencing. Only vlhA genes discussed are shown. Each data point represents an average RPKM value from the results determined for five animals. Error bars show SEM. Key statistically significant changes between two time points are indicated by paired upper- and lowercase letters for the genes vlhA 3.03 (A/a), vlhA 4.07.6 and vlhA 4.07 (B/b), and vlhA 4.07.1 and vlhA 4.08 (C/c). (C) Tracheal thickness of chickens challenged with wild-type M. gallisepticum (solid line) or the M. gallisepticum 3.03 mutant (dashed line). Each data point represents an average thickness measure of 4 measurements from 5 animals each. Error bars show SEM.
M. gallisepticum was recovered from the trachea of chickens challenged with the vlhA 3.03 mutant in at least 4 out of 5 birds at each day throughout the 7-day infection time course (data not shown). This rate of M. gallisepticum recovery was indistinguishable from that of chickens challenged with the virulent wild-type Rlow strain, suggesting that the mutation in the immunodominant vlhA 3.03 does not negatively impact the ability of M. gallisepticum to colonize and survive in the respiratory tract of chickens.
Taken together, these data suggest that the M. gallisepticum vlhA 3.03 mutation did not have a negative effect on the colonization or virulence of the pathogen in the chicken respiratory tract.
vlhA 3.03 mutant vlhA gene expression.
The vlhA 3.03 mutant of Rlow demonstrated an in vivo vlhA gene expression pattern that was similar to that previously demonstrated for wild-type Rlow. In both strains, vlhA 4.07 and 4.07.6 peaked at day two, albeit at higher expression levels and as the predominant vlhA in the vlhA 3.03 mutant and as subdominant to vlhA 3.03 in wild-type Rlow. Both the vlhA 3.03 mutant and wild-type Rlow demonstrated peak expression of vlhA 1.04 at days 5 and 6 (Fig. 3A and B and Fig. S3). Notably, the vlhA 3.03 mutant demonstrated increased expression of vlhA genes that were not expressed above baseline in wild-type Rlow, specifically vlhA 3.07, which peaked at day 1, and vlhA 4.09, which peaked at day 4. Overall, vlhA gene expression in the vlhA 3.03 mutant resembled that of wild-type M. gallisepticum Rlow temporally and in genes expressed, save the lack of vlhA 3.03 and the additional genes expressed above background.
vlhA expression pattern of a population with an alternative dominant vlhA gene.
We observed an immediate switch back to a vlhA expression pattern highly similar to that of wild-type Rlow, with a 25-fold increase of vlhA 3.03 at day one postinfection (Fig. 4A and Fig. S4). vlhA 4.07.6 and 4.07 showed a 65-fold increase in expression by day one postinfection, a pattern similar to that for wild-type Rlow. By day 1 postinfection the vlhA pattern was statistically indistinguishable (with the exception of minor vlhA genes) from previous wild-type Rlow vlhA profile patterns (data not shown). These data demonstrate that challenging chickens with an M. gallisepticum population that is expressing a non-vlhA 3.03 dominant vlhA gene does not affect the overall progression of vlhA expression and that vlhA 3.03 expression immediately and dramatically increases on day one.
FIG 4.
vlhA expression profile of the M. gallisepticum population expressing vlhA 2.02 as the predominant vlhA extracted directly from tracheas of experimentally inoculated birds over the course of the 2-day infection, as determined by RNA sequencing. Only vlhA genes discussed are displayed. Each data point represents an average RPKM value from the results determined for five animals (with the exception of broth). Error bars show SEM. Key statistically significant changes between two time points are indicated by paired upper- and lowercase letters for vlhA 3.03 (A/a).
DISCUSSION
This study has compared the vlhA gene expression profiles of populations of four different strains of M. gallisepticum when RNA is collected directly from the tracheal mucosa of experimentally inoculated chickens over the earliest stages of infection. We hypothesize that vlhA switching within the first week of infection may be driven, at least in part, by changes in the host cellular architecture as the disease pathology progresses over time. To test this hypothesis, we exposed chickens intratracheally to live attenuated vaccine strains GT5 and Mg7. Since these two vaccine strains do not cause the tracheal changes that are traditionally associated with wild-type virulent Rlow, such as deciliation of host cells and squamous cell metaplasia, we expect to see a dramatically different vlhA gene expression profile over the earliest stages of infection.
The vaccine strain Mg7 showed very dramatic changes in vlhA expression compared to that of wild-type virulent Rlow. The predominant vlhA expressed in the broth (vlhA 3.03) continues to increase in expression at day one and day two postinfection, in contrast to wild-type Rlow. Several other vlhA genes, such as vlhA 4.07 and vlhA 4.07.6, did not show any significant increase or change in expression over the 2-day time course. This finding was particularly significant, as the Mg7 vaccine strain does not cause significant pathology (6), and supports our hypothesis. Additionally, the vaccine strain GT5 showed a pattern of vlhA gene expression different from that of wild-type Rlow. An increase in the expression of vlhA 3.03 at days one and two postinfection similar to that of Mg7 was observed and was consistent with our hypothesis.
Since we see a different vlhA gene expression profile in the populations of M. gallisepticum strains that do not cause significant pathology in the host, it is possible that the changes in tracheal architecture associated with disease pathology could play a role, at least in part, in driving the changes in M. gallisepticum vlhA gene expression. The previously reported nonstochastic patterned progression of vlhA expression (15) is seen when there are significant pathological changes in the trachea associated with M. gallisepticum infection. This same ordered pattern of vlhA expression is not seen when there are not significant cellular changes in the trachea (e.g., inoculation with live attenuated vaccine strains); the vlhA gene expression pattern appears to be altered where vlhA 3.03 increases in expression at day one postinfection and continues to increase through day two postinfection.
Pathogenesis of the vlhA 3.03 negative mutant was not reduced relative to that of Rlow in the chicken respiratory tract during the first week of infection, indicating that vlhA 3.03 is not essential for the early stages of infection and colonization. However, it is possible that another vlhA gene, such as vlhA 4.07, which shows an increase in expression at day one postinfection, could be similar enough in function to serve the same role as vlhA 3.03 in vivo, highlighting the potential importance of the redundancy within this unique gene family.
While vlhA 3.03 may be dispensable for initial colonization and virulence of M. gallisepticum in the chicken trachea, vlhA 3.03 may play an essential role in a niche within the host that has not yet been explored.
The vlhA 3.03 mutant showed a slight shift in the patterned progression of vlhA gene expression over the 7-day time course, with some vlhA genes (vlhA 4.07 and vlhA 4.07.6) peaking in expression later than was observed with wild-type virulent Rlow. This demonstrates resilience and plasticity in the ordered expression pattern among vlhA gene paralogs. When there is a mutation in the preferred dominant vlhA gene (vlhA 3.03), the progression of vlhA gene expression is slightly shifted, but the overall pattern was still maintained over the course of a 7-day infection.
Challenging chickens with a population of M. gallisepticum expressing an alternative dominant vlhA (vlhA 2.02) does not alter the pattern progression of vlhA expression. In fact, by day one postinfection, the vlhA expression pattern is virtually indistinguishable from that of the wild-type virulent Rlow strain predominantly expressing vlhA 3.03, suggesting that this progression of vlhA expression is important during the initial stages of the infection process. These data, along with the vlhA 3.03 mutant data, also suggest that while the dominant expression of vlhA 3.03 at the very earliest stages of the infection process is favored, it may not be essential for the colonization and virulence of M. gallisepticum in the chicken trachea.
Additionally, these data further support the hypothesis that vlhA gene expression is changing, at least in part, as a response to the changing cellular environment of the trachea. The vaccine strains Mg7 and GT5 show a significant reduction in virulence (6, 16) in the host and do not show the same initial progression of vlhA gene expression as that shown by wild-type virulent Rlow. Additionally, M. gallisepticum GT5 and Mg7 do not cause the same host cell stress response or immune dysregulation (J. Beaudet, unpublished data) that is observed in chickens experimentally challenged with virulent wild-type Rlow (18). It is possible that since the trachea is not being disrupted or damaged by the vaccine strains, the vlhA genes are not forced to change in response. These data also suggest that M. gallisepticum colonizing other organs, such as the lungs and air sacs, expresses different suites of vlhA genes best suited for M. gallisepticum survival in their current environments.
Collectively, these data further support the hypothesis that the vlhA genes change in a nonstochastic temporal progression of expression and that vlhA 3.03, while preferred, is not required to be the initially predominant vlhA expressed at the time of infection for colonization and virulence. These results, revealing global vlhA expression changes over the course of early infection in different strains of M. gallisepticum, are important in elucidating mechanisms of colonization, persistence, and overall pathogenesis of M. gallisepticum in the natural host. While these data provide detailed insights into the vlhA gene family of M. gallisepticum, it is not clear, at this time, exactly what molecules or mechanisms are driving the change in vlhA gene expression seen here. It is also important to note that all vlhA expression observations were made on a population level and do not exclude the possibility that the changes observed were due to the survival of members of the population expressing the suite of vlhA genes best suited for the current microenvironment in the host airway. Full and complete understanding of the phase variation of M. gallisepticum vlhA genes may play a pivotal role in the understanding of the earliest stages of the infection process of M. gallisepticum and how the pathogen responds to changes of the microenvironment within the host. To our knowledge, the current study, combined with our previous work, is the first time phase variation has been assessed in a bacterial pathogen at the earliest stages of infection. These findings may be directly relevant to other important bacterial pathogens that possess phase-variable gene families expressing variable surface proteins.
MATERIALS AND METHODS
Animals.
Four-week-old female specific-pathogen-free White Leghorn chickens (SPAFAS, North Franklin, CT, USA) were received and divided randomly into groups, placed in HEPA-filtered isolators, and allowed to acclimate for 1 week prior to the start of the study. Nonmedicated feed and water were provided ad libitum throughout the experiment. All animal studies were performed in accordance with approved UConn IACUC protocol number A15-056.
Chicken study.
Stocks of M. gallisepticum strain Rlow (passage 17) were grown overnight at 37°C in Hayflick's complete medium until mid-log phase was reached, as indicated by a color shift from red to orange. Bacterial concentrations were determined by the optical density at 620 nm, and 10-fold serial dilutions were conducted to confirm viable color-changing unit titers as previously described (4, 6, 16). Bacteria were pelleted by centrifugation at 10,000 × g for 10 min and resuspended in Hayflick's complete medium. A previously identified population of M. gallisepticum stably expressing vlhA 2.02 as the predominant vlhA (15) was verified for predominant vlhA 2.02 expression by RNA sequencing (RNA-seq), quantitated, and frozen at −80°C. At the time of infection, the stocks were thawed and immediately used for infection to ensure there were no changes in vlhA expression.
Previously characterized M. gallisepticum vaccine strains GT5 (16) and Mg7 (6), in addition to the vlhA 3.03 mutant (described in detail below), were grown as described above in Hayflick's medium supplemented with 150 μg/ml gentamicin to maintain the transposon insertions. Chickens were inoculated intratracheally as previously described (6) with 1 × 108 CFU/200 μl of the respective cultures, and RNA was collected directly from the tracheal mucosa daily, as described below.
Two-day time course studies were conducted with M. gallisepticum strains GT5 and Mg7 to assess the very early changes in vlhA expression, as the virulence of these strains has been previously reported (6, 16), in addition to the population expressing vlhA 2.02 as the predominant vlhA. A 7-day time course study was conducted on the vlhA 3.03 mutant to assess the virulence of the mutant in addition to the very early changes in the vlhA gene expression profile.
RNA extraction.
Five infected chickens per group were humanely sacrificed by cervical dislocation daily for a total of 2 (GT5, Mg7, and the vlhA 2.02 predominant population) or 7 (vlhA 3.03 mutant) days. After sacrifice, tracheas were excised and total RNA was extracted from each individual trachea by washing the lumen with 1 ml TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was then purified using the Zymo Direct-zol RNA miniprep kit (Zymo Research Corporation, Irvine, CA, USA), and standard PCR was conducted to ensure that the RNA preparations were free of any DNA. RNA was quality checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and high-quality samples with RNA integrity numbers (RIN) of >8 were utilized to construct cDNA libraries.
To enrich for bacterial RNA, total RNAs were subjected to a poly(A) depletion step to remove the eukaryotic mRNA using the NEBNext poly(A) mRNA magnetic isolation module (New England BioLabs, Ispwich, MA, USA) as previously described (15). Briefly, 5 μg of total RNA combined with an equal volume of bead binding buffer was bound to the poly(T) oligonucleotide-attached magnetic beads at 65°C for 5 min. The remaining supernatant was collected, cleaned, and concentrated using the Zymo DNA Clean & Concentrator 25 kit (Zymo Research Corporation) and eluted in 25 μl RNase-free water.
Both prokaryotic and eukaryotic rRNAs were removed from 2.5 μg of poly(A)-depleted RNA using the RiboZero magnetic gold kit (Epidemiology) (Illumina Inc., San Diego, CA, USA) by following the manufacturer's instructions. Each rRNA-depleted RNA sample obtained after cleaning and concentrating with the Zymo DNA Clean & Concentrator 25 kit (Zymo Research Corporation) was eluted in 25 μl of RNase-free water and used to create a cDNA library.
Illumina sequencing and RNA-seq analysis.
The cDNA libraries were created using the Illumina TruSeq stranded mRNA library preparation kit (Illumina Inc., San Diego, CA, USA) and sequenced on a NextSeq500 sequencing platform (Illumina Inc.) as previously described (15).
RNA-seq analysis was performed as previously described (15). Briefly, Fastq data were assembled and mapped, and differential gene expression was assessed using Rockhopper with Bowtie2 parameters allowing zero mismatches (19, 20). The data were normalized by the standard method of determining the ratio of reads per kilobase per million (RPKM) mapped, allowing for comparisons both within and between samples. The fold change data were determined from the log2 transformation of the RPKM data between two samples. The differential levels of gene expression were determined by pairwise comparisons between the normalized values of expression of a given vlhA gene from two different days. The program-generated P value was used to determine the significance of the differential gene expression by calculating q values based on the Benjamini-Hochberg correction with a false discovery rate of <1%. Differences in expression values were considered significant when the q value was <0.02 (20).
Identification of the vlhA 3.03 mutant.
A library of 3,600 Rlow transposon (Tn) mutants was generated via electroporation using plasmid pMT85, as described previously (6). The plasmid carries mini-Tn4001-gentamicin, which encodes a gentamicin resistance gene. Pools of 30 mutants were grown, and genomic DNAs were extracted. PCR screening of mutant pools for transposon insertions in the vlhA 3.03 gene was performed in 96-well plates by using a gene-specific primer in conjunction with a 5′ or 3′ transposon-specific primer. PCR was run on positive pools in the reverse orientation (reverse gene primer with the opposite-end transposon primer to ensure the transposon was located within the gene). Pools with positive PCR products that exhibited the correct gene size for a transposon insertion were selected for a second round of screening. DNA was extracted from each individual pool member and screened again as described above. Sanger sequencing was used to identify the Tn insertion site and confirm there was only a single Tn insertion, as described previously (6, 21).
A reverse transcription-quantitative PCR (RT-qPCR) assay was used to verify that the transposon insertion was sufficient to disrupt the expression of vlhA 3.03 using the QuantiTect reverse transcription kit (Qiagen) with the following PCR conditions: 50°C for 30 min, 95°C for 15 min, and then 35 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 31 s with 20 pmol of each forward primer 5′-ACGACCAAGCAAAACCTAATGA-3′ and reverse primer 5′-ACTCCTGAAGCGAACACTCC-3′. The Sanger sequencing analysis of the vlhA 3.03 mutant showed that the transposon was inserted at base pair 75. RT-qPCR analysis showed a 100-fold reduction in the expression of the gene, which was further confirmed by our RNA-seq gene expression data (data not shown).
Tracheal thickness measurements and bacterial recovery.
As an established objective measure of virulence, tracheal thickness was assessed for the M. gallisepticum vlhA 3.03 mutant daily over the course of 7 days. Tracheal thickness measurements on a distal ring of the trachea were determined for all chickens, as previously described (16). Histological data were subjected to nonparametric Kruskal-Wallis one-way analysis of variance (ANOVA) in the ranks test, in which all pairwise multiple-comparison procedures were performed using the Student-Newman-Keuls method for groups of equal sizes. Statistical tests were conducted using SigmaPlot 11.0 (Systat Software, San Jose, CA). To assess the recovery of viable M. gallisepticum from the trachea of infected animals, a ring from the distal portion of the trachea was collected directly into Hayflick's complete medium (with gentamicin for the GT5, Mg7, and vlhA 3.03 mutant strains) and incubated for 5 h at 37°C. After the incubation period, cultures were passed through 0.45-μm-pore-size filters to remove non-Mycoplasma contaminates, adjusted to pH 7.4, and reincubated at 37°C. Samples were considered positive for M. gallisepticum recovery if the color shifted to yellow within 30 days.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kirklyn Kerr for assistance with necropsy. We also thank the Center of Excellence for Vaccine Research for support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00524-18.
REFERENCES
- 1.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev 62:1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley DH, Berkhoff JE, McLaren JM. 1996. Mycoplasma gallisepticum isolated from house finches (Carpodacus mexicanus) with conjunctivitis. Avian Dis 40:480–483. doi: 10.2307/1592250. [DOI] [PubMed] [Google Scholar]
- 3.Papazisi L, Frasca S, Gladd M, Liao X, Yogev D, Geary SJ. 2002. GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence. Infect Immun 70:6839–6845. doi: 10.1128/IAI.70.12.6839-6845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.May M, Papazisi L, Gorton TS, Geary SJ. 2006. Identification of fibronectin-binding proteins in Mycoplasma gallisepticum strain R. Infect Immun 74:1777–1785. doi: 10.1128/IAI.74.3.1777-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng CW, Kanci A, Citti C, Rosengarten R, Chiu CJ, Chen ZH, Geary SJ, Browning GF, Markham PF. 2013. MalF is essential for persistence of Mycoplasma gallisepticum in vivo. Microbiology (United Kingdom) 159:1459–1470. doi: 10.1099/mic.0.067553-0. [DOI] [PubMed] [Google Scholar]
- 6.Hudson P, Gorton TS, Papazisi L, Cecchini K, Frasca S, Geary SJ. 2006. Identification of a virulence-associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect Immun 74:931–939. doi: 10.1128/IAI.74.2.931-939.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ron M, Gorelick-Ashkenazi A, Levisohn S, Nir-Paz R, Geary SJ, Tulman E, Lysnyansky I, Yogev D. 2015. Mycoplasma gallisepticum in vivo induced antigens expressed during infection in chickens. Vet Microbiol 175:265–274. doi: 10.1016/j.vetmic.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Glew MD, Baseggio N, Markham PF, Browning GF, Walker ID. 1998. Expression of the pMGA genes of Mycoplasma gallisepticum is controlled by variation in the GAA trinucleotide repeat lengths within the 5′ noncoding regions. Infect Immun 66:5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glew MD, Browning GF, Markham PF, Walker ID. 2000. pMGA phenotypic variation in Mycoplasma gallisepticum occurs in vivo and is mediated by trinucleotide repeat length variation. Infect Immun 68:6027–6033. doi: 10.1128/IAI.68.10.6027-6033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papazisi L, Gorton TS, Kutish G, Markham PF, Browning GF, Nguyen DK, Swartzell S, Madan A, Mahairas G, Geary SJ. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain Rlow. Microbiology 149:2307–2316. doi: 10.1099/mic.0.26427-0. [DOI] [PubMed] [Google Scholar]
- 11.Noormohammadi AH. 2007. Role of phenotypic diversity in pathogenesis of avian mycoplasmosis. Avian Pathol 36:439–444. doi: 10.1080/03079450701687078. [DOI] [PubMed] [Google Scholar]
- 12.Tulman ER, Liao X, Szczepanek SM, Ley DH, Kutish GF, Geary SJ. 2012. Extensive variation in surface lipoprotein gene content and genomic changes associated with virulence during evolution of a novel North American house finch epizootic strain of Mycoplasma gallisepticum. Microbiology (United Kingdom) 158:2073–2088. doi: 10.1099/mic.0.058560-0. [DOI] [PubMed] [Google Scholar]
- 13.Vogl G, Plaickner A, Szathmary S, Stipkovits L, Rosengarten R, Szostak MP. 2008. Mycoplasma gallisepticum invades chicken erythrocytes during infection. Infect Immun 76:71–77. doi: 10.1128/IAI.00871-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu F, Zhao C, Bi D, Tian W, Chen J, Sun J, Peng X. 2016. Mycoplasma gallisepticum (HS strain) surface lipoprotein pMGA interacts with host apolipoprotein A-I during infection in chicken. Appl Microbiol Biotechnol 100:1343–1354. doi: 10.1007/s00253-015-7117-9. [DOI] [PubMed] [Google Scholar]
- 15.Pflaum K, Tulman ER, Beaudet J, Liao X, Geary SJ. 2016. Global changes in Mycoplasma gallisepticum phase-variable lipoprotein gene vlhA expression during in vivo infection of the natural chicken host. Infect Immun 84:351–355. doi: 10.1128/IAI.01092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gates AE, Frasca S, Nyaoke A, Gorton TS, Silbart LK, Geary SJ. 2008. Comparative assessment of a metabolically attenuated Mycoplasma gallisepticum mutant as a live vaccine for the prevention of avian respiratory mycoplasmosis. Vaccine 26:2010–2019. doi: 10.1016/j.vaccine.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Pflaum K, Tulman ER, Beaudet J, Liao X, Dhondt KV, Dhondt AA, Hawley DM, Ley DH, Kerr KM, Geary SJ. 2017. Attenuated phenotype of a recent house finch-associated Mycoplasma gallisepticum isolate in domestic poultry. Infect Immun 85:e00185-17. doi: 10.1128/IAI.00185-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaudet J, Tulman ER, Pflaum K, Liao X, Kutish GF, Szczepanek SM, Silbart LK, Geary SJ. 2017. Transcriptional profiling of the chicken tracheal response to virulent Mycoplasma gallisepticum strain Rlow. Infect Immun 85:e00343-17. doi: 10.1128/IAI.00343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szczepanek SM, Boccaccio M, Pflaum K, Liao X, Geary SJ. 2014. Hydrogen peroxide production from glycerol metabolism is dispensable for virulence of Mycoplasma gallisepticum in the tracheas of chickens. Infect Immun 82:4915–4920. doi: 10.1128/IAI.02208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.