Conjugate vaccines against Streptococcus pneumoniae have significantly reduced the incidence of diseases caused by the serotypes included in those vaccines; however, there is still a need for vaccines that confer serotype-independent protection. In the current study, we have constructed a library of conserved surface proteins from S. pneumoniae and have screened for IL-17A and IL-22 production in human immune cells obtained from adenoidal/tonsillar tissues of children and IL-17A production in splenocytes from mice that had been immunized with a killed whole-cell vaccine or previously exposed to pneumococcus.
KEYWORDS: Streptococcus pneumoniae, colonization, antigen discovery, Th17
ABSTRACT
Conjugate vaccines against Streptococcus pneumoniae have significantly reduced the incidence of diseases caused by the serotypes included in those vaccines; however, there is still a need for vaccines that confer serotype-independent protection. In the current study, we have constructed a library of conserved surface proteins from S. pneumoniae and have screened for IL-17A and IL-22 production in human immune cells obtained from adenoidal/tonsillar tissues of children and IL-17A production in splenocytes from mice that had been immunized with a killed whole-cell vaccine or previously exposed to pneumococcus. A positive correlation was found between the rankings of proteins from human IL-17A and IL-22 screens, but not between those from human and mouse screens. All proteins were tested for protection against colonization, and we identified protective antigens that are IL-17A dependent. We found that the likelihood of finding a protective antigen is significantly higher for groups of proteins ranked in the top 50% of all three screens than for groups of proteins ranked in the bottom 50% of all three. The results thus confirmed the value of such screens for identifying Th17 antigens. Further, these experiments have evaluated and compared the breadth of human and mouse Th17 responses to pneumococcal colonization and have enabled the identification of potential vaccine candidates based on immunological responses in mouse and human cells.
INTRODUCTION
Streptococcus pneumoniae, or pneumococcus, is a Gram-positive bacterium that is a major cause of morbidity and mortality in infants, toddlers, and the elderly in both developed and developing countries. The introduction of the first conjugate vaccine, PCV7, followed by PCV10 and PCV13, has greatly reduced the incidences of invasive disease and colonization caused by the serotypes included in these vaccines (1). Despite the success of these vaccines, serotypes that are not included in these vaccines pose an emerging threat. Indeed, there are at least 97 identified pneumococcal serotypes, with variability in the regional distribution of predominant serotypes; furthermore, the phenomenon of serotype replacement has reduced the impact of these vaccines in many settings (2).
While it is well established that anticapsular antibodies are sufficient to prevent invasive diseases such as pneumonia, meningitis, and sepsis, other mechanisms of immunity may also play important roles. Either live exposure to pneumococcus or immunization with a killed whole-cell vaccine (S. pneumoniae whole-cell vaccine [SPWCV]) can induce CD4+ Th17-dependent protection against nasopharyngeal colonization in mice (3–5). The reduction in the incidence of colonization in mice is dependent on the generation of pneumococcus-specific Th17 immunity and the recruitment of neutrophils to the mucosal site (3).
Evidence is accumulating to suggest that Th17-based immunity is also important for protection against pneumococcus in humans. The existence of pneumococcus-specific Th17-type T cells has been demonstrated in both children and adults by analysis of Th17 responses to a killed preparation of pneumococcus (Streptococcus pneumoniae whole-cell antigen [WCA]) or individual pneumococcal antigens (6). Furthermore, the frequency of pneumococcus-specific Th1 and Th17 CD4+ T cells within mucosal lymphoid tissue demonstrates age-dependent increases, likely due to cumulative exposure to pneumococcus (7). In an experimental human challenge model, lung interleukin 17A (IL-17A)-secreting CD4+ memory T cells were detected following intentional pneumococcal carriage (8). Furthermore, patients with autosomal dominant hyper-IgE syndrome (Job's syndrome), who lack the ability to generate memory Th17 cells due to mutations in the STAT3 gene, are highly susceptible to recurrent pneumococcal infections (9, 10). More recently, two studies provided support for a role of IL-17A in pneumococcal carriage in children: a specific polymorphism of the IL-17 gene (G152A) in Finnish children (11) and decreased IL-17A secretion in Fijian children (12) were associated with increased risks of pneumococcal carriage.
In addition to Th17 cells, antibodies are likely to play an important role in protection against pneumococcal disease. An SPWCV consisting of WCA and aluminum adjuvant given subcutaneously or intramuscularly elicits anti-protein antibody-mediated protection against pneumococcal pneumonia and sepsis in mice, and this protection can also be reproduced by passive transfer of antibodies obtained from rabbits immunized with SPWCV (13). Naturally acquired protection against pneumococcal disease in humans has recently been shown to depend largely on antibodies to protein antigens rather than to capsular polysaccharides (14).
As a consequence, there has been a concerted effort over many years to identify protective protein antigens against pneumococcal disease (15, 16). Traditional methods, such as protein separation by 2-dimensional gel electrophoresis and identification by mass spectroscopy, have been used to identify proteins from the cell wall fraction of pneumococcus that interact with convalescent-phase sera from patients (17). As another approach, libraries of purified surface proteins have been used to identify antigens that may induce protection against invasive pneumococcal disease (18). A display library expressing 15 to 150 amino acid fragments of the pneumococcal proteome was used to identify proteins that interact with sera from infected individuals, leading to the selection of StkP and PcsB as candidate antigens (19).
Using similar approaches, other investigators have used pneumococcal antigens to identify potentially protective T-cell antigens. Putative Th17-eliciting antigens were identified from the soluble fraction of the WCA using preparative SDS gel electrophoresis followed by mass spectroscopy (20). In a more comprehensive approach using the atlas system, an expression library containing >96% of predicted pneumococcal proteins was used to identify antigens recognized by Th17 cells from SPWCV-immunized mice (21) and from human volunteers naturally exposed to pneumococcus (22).
The studies cited above used either mouse splenocytes or human peripheral blood mononuclear cells (PBMCs) as a source of immune cells. Some studies using an arguably more relevant source of immune cells, human mucosal lymphoid tissue, probed for Th17 responses following stimulation with a small number of pneumococcal proteins (23–25), but a more comprehensive analysis of the range of Th17 responses to the pneumococcal proteome has not been performed. In this study, we constructed a protein library consisting of genetically conserved pneumococcal surface proteins. We used this library to screen human adenoidal cells for Th17 and Th22 cytokine-inducing antigens and compared these human responses to those observed in splenocytes from mice immunized with SPWCV or exposed to pneumococcus. Positive correlations between two human screens, but no correlations between human and murine screens, were observed. Several antigens that subsequently showed IL-17A-dependent protection against pneumococcal colonization in mice were identified.
RESULTS
Age-dependent immune response to WCA.
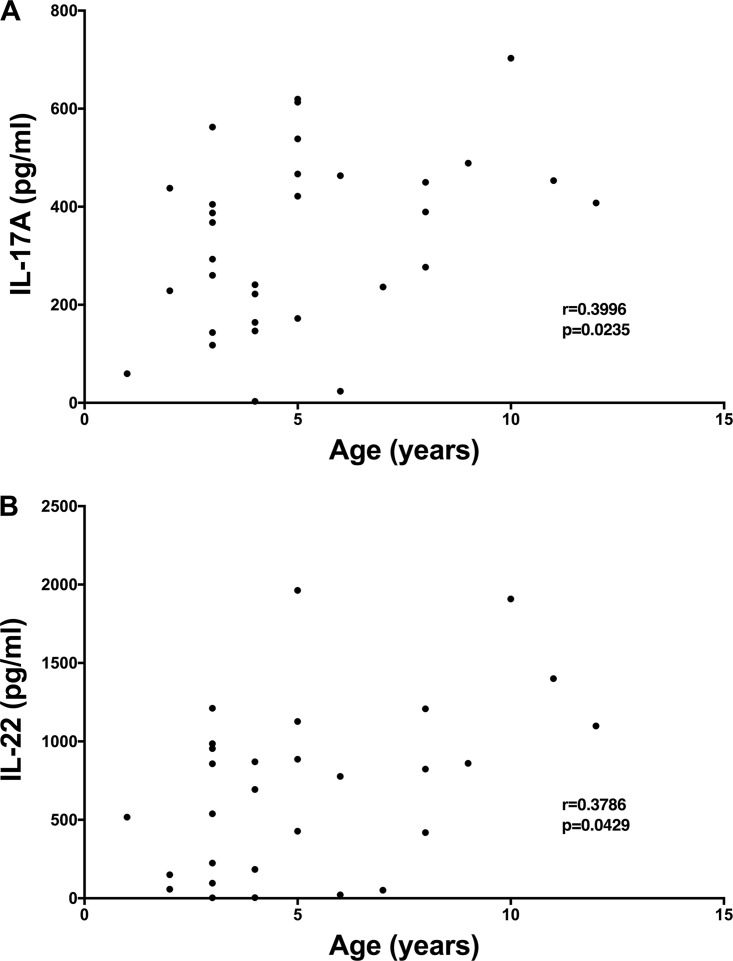
We obtained 35 adenoidal samples from children undergoing adenoidectomy and analyzed IL-17A and IL-22 production in response to stimulation with WCA in 33 of these samples. As shown in Fig. 1, there were significant age-dependent increases in IL-17A (P = 0.0235; r = 0.3996) and IL-22 (P = 0.0429; r = 0.3786) responses. The finding that immune responses to pneumococcus in adenoids in children tend to increase with age supports the hypothesis that these adenoidal cells represent a good source of responsive immune cells for antigen discovery.
FIG 1.
IL-17A and IL-22 responses to SPWCV are positively correlated with donors' ages. Adenoidal cells were stimulated with SPWCV, and IL-17A (A) and IL-22 (B) concentrations in the cell supernatants were measured. The correlations were evaluated using the Spearman test.
Screening antigens using human adenoidal cells.
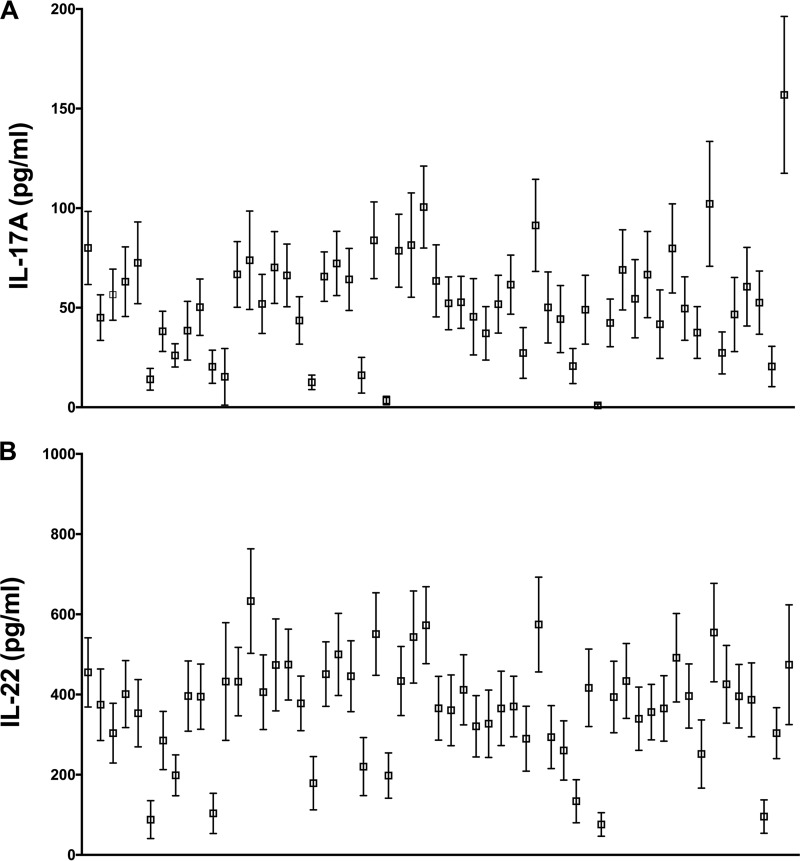
Among 81 genetic constructs, 56 recombinant proteins were successfully expressed in Escherichia coli and were purified using nickel-nitrilotriacetic acid (Ni-NTA) chromatography. These proteins were further purified by gel filtration before being used in stimulation experiments with adenoidal cells. Due to the limited number of cells obtained from each adenoid, we were able to screen all 56 proteins in 13 samples and only a subgroup of these proteins with the other 22 human samples. Both we and others have shown that IL-17A production in humans in response to pneumococcal whole-cell antigen or purified pneumococcal proteins is due mostly to the stimulation of memory T cells (26, 27). The results for IL-17A and IL-22 production in these 13 samples after subtraction of the baseline responses following incubation with medium alone are shown in Fig. 2. IL-17A responses from each protein were generally in the range of 50 to 100 pg/ml (mean, 157 pg/ml), and WCA exhibited the highest responses (Fig. 2A, far right). IL-22 responses were generally higher than IL-17A responses, mostly ranging from 200 to 600 pg/ml (Fig. 2B).
FIG 2.
Responses of human adenoidal cells to stimulation with each protein or with the whole-cell vaccine. Human adenoidal mononuclear cells were stimulated with 8 μg/ml of recombinant proteins or SPWCA at a concentration representing 1 × 106 CFU of killed bacteria/ml for 7 days, and cytokines IL-17A (A) and IL-22 (B) were measured by ELISA. Data are presented as means ± standard errors of the means. The antigens used for screening are as follows, from left to right: SP0079, SP0084, SP0092, SP0098, SP0127, SP0149, SP0191, SP0198, SP0249, SP0346, SP0435, SP0453, SP0564, SP0582, SP0601, SP0604, SP0617, SP0620, SP0629, SP0648-1, SP0648-2, SP0648-3, SP0659, SP0662-1, SP0662-2, SP0678, SP0724, SP0742, SP0757, SP0785, SP0787, SP0878, SP0899, SP1002, SP1032, SP1069, SP1154-2, SP1386, SP1404, SP1479, SP1500, SP1534, SP1545, SP1560, SP1652, SP1683, SP1826, SP1872, SP1942, SP2070, SP2083, SP2145, SP2192, SP2197, SP2207, SP2218, and SPWCA. SP0321 was toxic to human cells and was not used in human screens.
Screening antigens using murine immune cells.
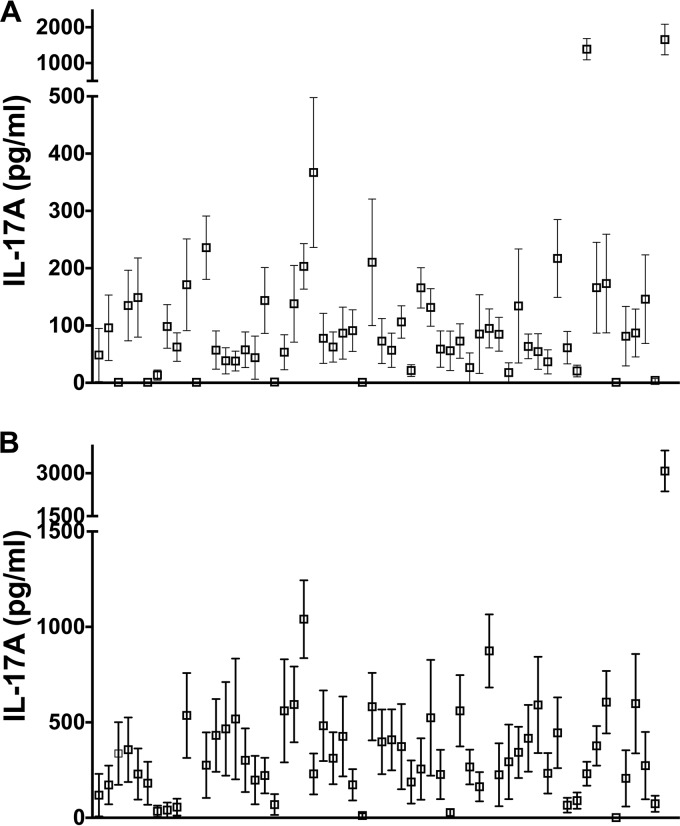
While colonization and infection models in mice are widely used to test candidate pneumococcal vaccines, the extent to which murine immune responses mimic human responses is still unclear. A comparison of the responses of murine and human immune cells following stimulation with proteins from the same pneumococcal library may improve our understanding of the differences between these two species as well as their common features. To this end, murine splenocytes were obtained from mice either immunized with SPWCV or previously colonized with a serotype 6B pneumococcal strain for 4 weeks (the latter model may more closely mimic pneumococcal exposure in humans). Both methods result in the generation of protective and antigen-specific memory CD4+ T cell responses to nasopharyngeal colonization in mice (3, 13, 28, 29), and we have confirmed that these mice were protected from nasal colonization (data not shown). IL-17A responses following the stimulation of splenocytes from these two mouse models with the library of proteins are shown in Fig. 3. Stimulation with SPWCA and proteins in splenocytes from naïve mice did not generate any IL-17A (data not shown). Responses in SPWCV-immunized mice, most of which were between 50 and 200 pg/ml (Fig. 3A), were generally lower than those in pneumococcus-exposed mice, most of which were between 100 and 500 pg/ml (Fig. 3B). SPWCA induced the largest responses in both mouse models (Fig. 3, far-right data points).
FIG 3.
Responses of murine splenocytes to stimulation with each protein or with the whole-cell vaccine. Murine splenocytes were stimulated with 5 μg/ml of recombinant proteins or with SPWCA at a concentration representing 1 × 106 CFU of killed bacteria/ml for 3 days, and IL-17A was measured by ELISA. Data in the graphs are presented as means ± standard errors of the means. The antigens used for screening are as follows, from left to right: SP0079, SP0084, SP0092, SP0098, SP0127, SP0149, SP0191, SP0198, SP0249, SP0321, SP0346, SP0435, SP0453, SP0564, SP0582, SP0601, SP0604, SP0617, SP0620, SP0629, SP0648-1, SP0648-2, SP0648-3, SP0659, SP0662-1, SP0662-2, SP0678, SP0724, SP0742, SP0757, SP0785, SP0787, SP0878, SP0899, SP1002, SP1032, SP1069, SP1154-2, SP1386, SP1404, SP1479, SP1500, SP1534, SP1545, SP1560, SP1652, SP1683, SP1826, SP1872, SP1942, SP2070, SP2083, SP2145, SP2192, SP2197, SP2207, SP2218, and SPWCA. Shown are IL-17A levels following stimulation of splenocytes from SPWCV-immunized mice (A) and from pneumococcus-exposed mice (B).
Evaluation of antigens in colonization models.
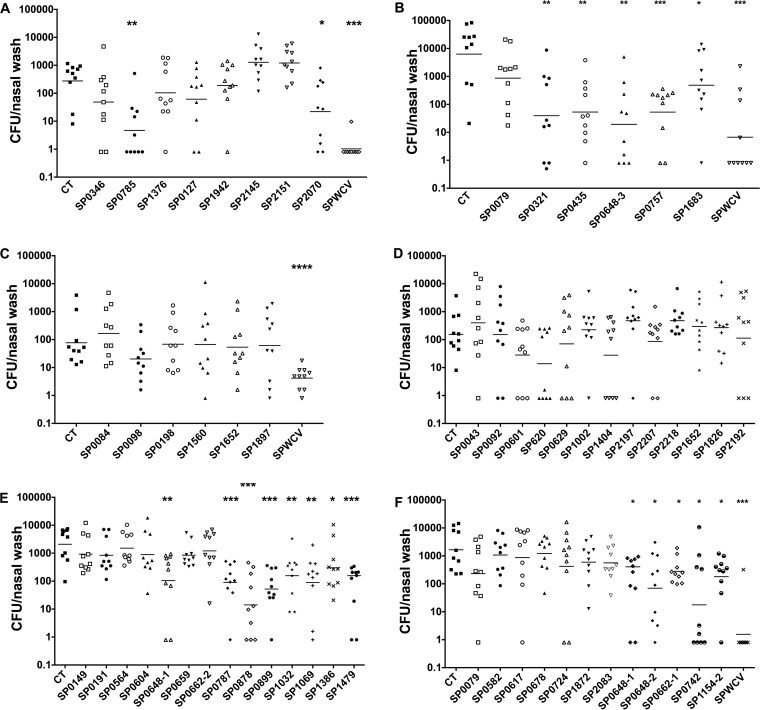
Next, we tested the ability of the library of proteins to provide protection against colonization in mice when used as immunogens. Mice were immunized intranasally (i.n.) twice, at a 1-week interval, with one of the top five murine proteins (10 μg/dose) and 1 μg of cholera toxin (CT); control mice received 1 μg of CT alone. The whole library was tested in six different experiments, as shown in Fig. 4. A total of 18 constructs (16 antigens) were found to be protective in this model; their identities and predicted functions are listed in Table 1.
FIG 4.
Protection against colonization by antigens from the surface protein library. Mice were immunized with 10 μg of each protein and 1 μg of cholera toxin twice (1 week apart) and were challenged 4 weeks after the last immunization with a serotype 6B clinical strain. Nasal wash specimens were collected 7 days later, and bacterial CFU counts were determined by plating. As shown in panels A to F, six different challenge experiments were performed. Each data point represents the CFU recovered from one mouse, and the horizontal lines represent geometric means. Colonization rates for mice immunized with an antigen plus CT adjuvant were compared to those for control mice (receiving CT alone) by use of the unpaired nonparametric Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TABLE 1.
Antigens protective against pneumococcal colonization
| Gene name | Annotation |
|---|---|
| SP0321b | Phosphotransferase system, IIA component |
| SP0648a,b,c,e | β-Galactosidase |
| SP0662a,b,c | Sensor histidine kinase |
| SP0742a | Hypothetical protein |
| SP0757a,b,c | Cell division protein FtsX |
| SP0785a,b,c | Hypothetical protein |
| SP0787a,c | Hypothetical protein |
| SP0878b | SpoE family protein |
| SP0899b | Hypothetical protein |
| SP1032a,b,c | Iron compound ABC transporter |
| SP1069d | Hypothetical protein |
| SP1154-2a | IgA1-specific metallopeptidase |
| SP1386a,c | Spermidine/putrescine ABC transporter |
| SP1479b | Peptidoglycan N-acetylglucosamine deacetylase A |
| SP1500b,c | Amino acid ABC transporter substrate-binding protein |
| SP1683a,b,c | Carbohydrate ABC transporter substrate-binding protein |
Among the top 50% in the human IL-17A screen.
Among the top 50% in the murine WCV screen.
Among the top 50% in the murine exposed screen.
Not in the top 50% of any screen.
SP0648 consisted of three separate constructs, all of which were in the top 50% of all screens.
Protection against colonization is IL-17A dependent.
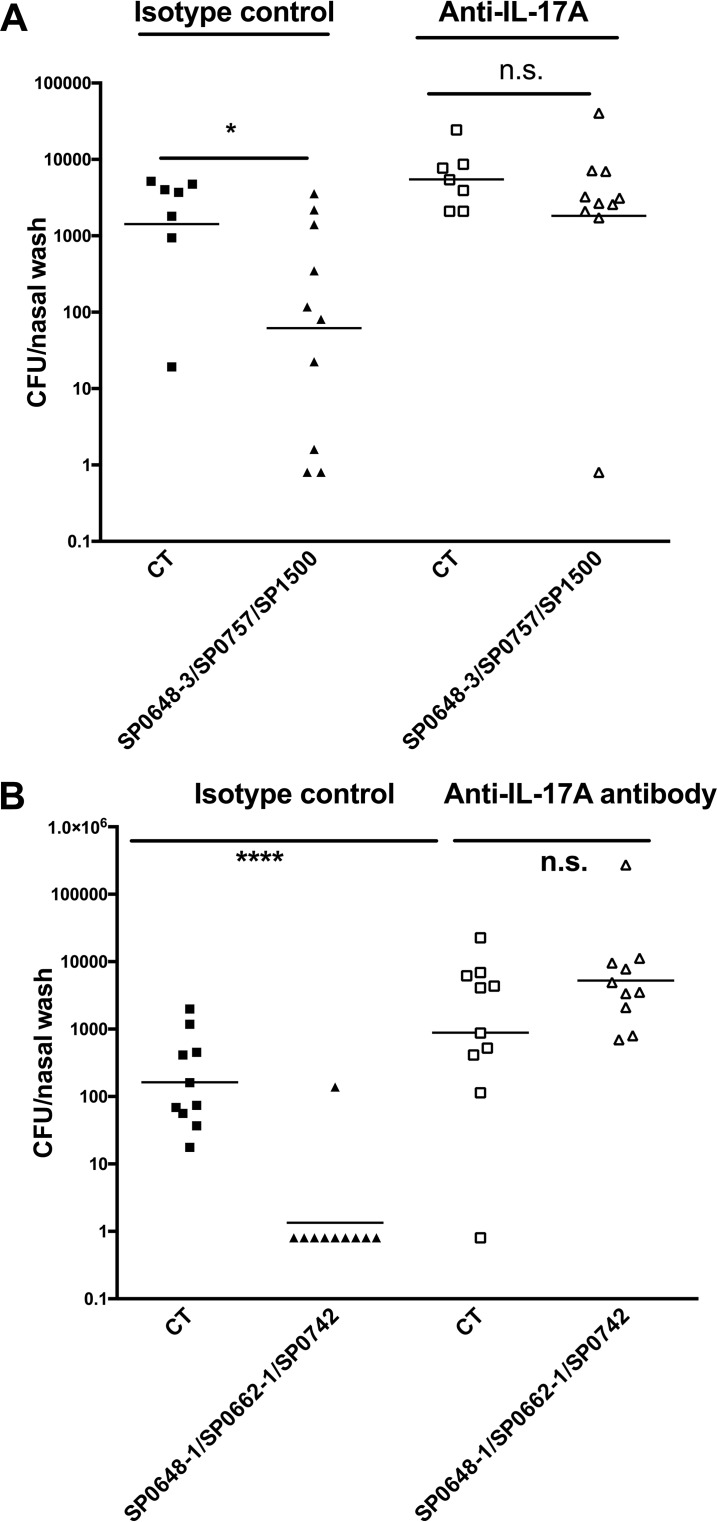
We have shown previously that protection against colonization is dependent on CD4+ Th17 cells, whereas antibodies directed against antigens contained in the whole-cell vaccine were not protective in the colonization model (3, 13, 28). We then tested whether the protection afforded by these antigens is dependent on IL-17A production. To facilitate the experiment, we chose combinations of the three antigens that induced the greatest responses either in murine screens (SP0648-3, SP0757, and SP1500) or in the human IL-17A screen (SP0648-1, SP0662-1, and SP0742) and tested the dependence of protection on IL-17A. Mice were immunized intranasally and were given either an anti-IL17A antibody or an isotype control antibody both 1 day before and 3 days after challenge inoculation with S. pneumoniae. As shown in Fig. 5, immunization with either antigen combination protected mice from colonization, while protection was abolished in immunized mice treated with an antibody directed against IL-17A, confirming the role of this cytokine in this model.
FIG 5.
The protection against nasal colonization conferred by proteins is IL-17A dependent. Mice were immunized intranasally with top-ranked antigens identified via the mouse screen (SP0648-3, SP0757, and SP1500) (A) or the human screen (SP0648-1, SP0662-1, and SP0742) (B). Mice were immunized with 10 μg of each protein and 1 μg of cholera toxin twice (1 week apart) and were challenged 4 weeks after the last immunization with a serotype 6B clinical strain. Mice received either anti-IL-17A or an isotype control antibody 1 day prior to and 3 days after the infection. Statistical analysis was performed with an unpaired nonparametric Mann-Whitney test. n.s, not significant; *, P < 0.05; ****, P < 0.0001.
Ranking of antigens and correlation between screens.
Proteins were ranked in each screen as explained in Materials and Methods. We compared the ranking results of SPWCV-immunized mice, pneumococcus-exposed mice, and human samples with respect to IL-17A and IL-22. There was a weak correlation between the results from immune cells in the two different mouse models (see Fig. S1A in the supplemental material), while there was a strong positive correlation between the IL-17A and IL-22 screens using human cells (Fig. 6). No statistically significant correlation was found between the IL-17A screen results for human cells and the results for cells from SPWCV-immunized mice (Fig. S1B) or from mice previously exposed to pneumococcus (Fig. S1C), nor was any correlation found between IL-22 production by human cells and that by murine cells in either mouse screen (data not shown).
FIG 6.
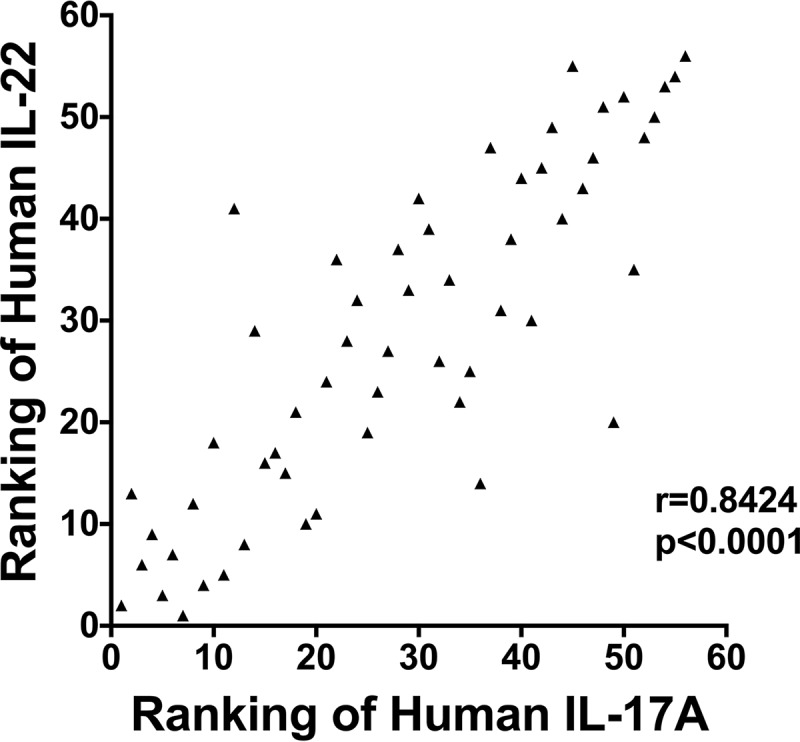
Correlation between human IL-17A and IL-22 screens. The ranking of each antigen was analyzed for correlation between the two screens by use of the nonparametric Spearman method.
Validation of the screening method.
We analyzed how well each screen predicted protection against colonization. Overall, the rate of protection among the proteins in the library is quite high (18/56 [32.1%]) (Fig. 4) in comparison with that for regular screening with the whole genome (21), which probably reflects the way the library was designed. Antigens were categorized as belonging either to the top or the bottom 50%, based on their rankings in each screen. We then compared the “hit rate” (i.e., the rate of identification of an antigen that is protective in the colonization model) in the top 50% of responders with that in the bottom 50% of responders in the two murine screens and the one human IL-17A screen. While the hit rates were higher in the top 50% of responders, none of these differences was significant (Table 2). In contrast, when we identified antigens that were highly ranked either in two screens or among all three screens, the hit rates were significantly higher: for example, for antigens that were in the top 50% rank in all three screens, the hit rate was 61.5%, significantly higher than the 11.1% hit rate for antigens that fell into the bottom 50% in all three screens (P, 0.0306 by Fisher's exact test), and about twice the hit rate for the whole library, which trended toward significance (P, 0.06 by Fisher's exact test). When the same analysis was done using results from two murine screens and the human IL-22 screen, a similar result was obtained. The hit rate in the top 50% rank in all three screens was 58.5%, in contrast to 8.3% in the bottom half of all three screens (P = 0.009).
TABLE 2.
Hit rates for the top 50% of responders in different screensa
| Screen(s) | Hit rateb in: |
|
|---|---|---|
| Top 50% | Bottom 50% | |
| All three | 8/13 (61.5) | 1/9 (11.1) |
| Human–WCV | 8/14 (57.1) | 1/13 (7.7) |
| Human–exposed | 10/21 (47.6) | 4/18 (22.2) |
| WCV–exposed | 9/16 (56.3) | 3/17 (17.6) |
| Human | 12/27 (44.4) | 6/27 (22.2) |
| WCV | 12/28 (42.9) | 6/28 (21.4) |
| Exposed | 11/28 (39.3) | 7/28 (25) |
Shown are data for the top 50% of responders from ranked screen results for immunized mice (WCV), exposed mice (exposed), and human IL-17A (human). Each screen was ranked; then the top 50% or bottom 50% of proteins in the rankings were selected, and the overlap between these antigens is shown here.
Expressed as the number of hits/total number of proteins (percentage).
DISCUSSION
Using a conserved pneumococcal surface protein library, we stimulated human and murine immune cells in order to identify antigens that elicit IL-17A and IL-22 responses, and we tested these antigens to determine which of them confer protection against pneumococcal colonization following intranasal immunization of mice. Overall, we found that antigens that more consistently induced cytokine responses across both murine and human cell screens were more likely to be protective. In addition, we identified many antigens that showed IL-17A-dependent protection against pneumococcal colonization in mice.
One goal of this study was to investigate whether the immune responses to pneumococcus in murine and human cells are correlated and whether murine screens could predict the results of the human screen. The IL-17A and IL-22 responses of human cells were highly correlated to each other, confirming our previous findings that IL-17A and IL-22 responses to WCA in children's PBMCs are highly correlated (6). However, there was no correlation between the rankings of the IL-17A responses to our protein library in murine screens and human screens, suggesting that there are differences in responses that are both host dependent (mouse versus human) and exposure dependent (live versus killed bacteria).
The lack of correlation between exposed mouse and human cells could be explained by many factors. First, there are clear differences between the mouse and human immune systems (30). Second, the local environment for pneumococcal colonization might not be the same in mice and humans, given growing evidence of the influence of the microbiota on immune development (31). Third, most children may have been exposed to pneumococcus many times, after which they may have generated high Th17 responses to nonprotective proteins, whereas the mice were exposed to pneumococcus or pneumococcal antigens only for a short, defined period in our experiment. Fourth, and finally, in addition to intrinsic differences between mouse and human immune cells, we are comparing human adenoidal mucosal cells to murine splenocytes—cells from very different compartments.
Despite these differences, we were able to identify several protective candidates by combining the three screens, suggesting that screening for cytokine production using cells from different sources may be a useful method for identifying cytokine-mediated protection. Indeed, we found that the protective hit rate was highest in the pool of proteins that induced higher responses in all the screens. The 16 protective antigens that we identified have not been reported previously as conferring protection, with the single exception of SP1683, which was reported as a Th17-dependent antigen that protects against colonization while this article was being prepared (32). While the protection against colonization by any single antigen did not approach that of SPWCV in mouse models (Fig. 4), a combination of three antigens significantly improved on the protective efficacy of any individual protein antigen (Fig. 5). This suggests that a candidate protein-based vaccine should likely comprise several antigens in order to maximize protection and coverage.
Another important implication of our work is that immune responses demonstrable in mouse models do not accurately predict those of humans. In light of the recent failure of at least two different protein vaccine formulations (33, 34) to provide protection against colonization, one is left wondering whether excessive reliance on mouse models may be responsible. Clearly, murine models are much more convenient and practical for large-scale screening and identification of potential candidates, as performed here. However, before one performs expensive and time-consuming proof-of-concept studies of impact on colonization in toddler or infant subjects, other approaches that can serve to minimize the risk of the process may be useful. One approach would be to try to establish correlates of protection by performing longitudinal studies of colonization and systemic T cell responses in children. Another approach may be to test promising candidates in intentional challenge studies in humans (8, 35). A potential caveat to this strategy is that intentional challenge studies are performed in adults, whereas the intended target populations of these vaccines are generally toddlers and infants. However, such studies could still be helpful in providing a gating strategy: the impact on carriage density or duration of carriage in a properly powered study of intentionally challenged adult volunteers could be used to decide whether or not to pursue studies of the candidate vaccine in younger subjects.
A potential limitation of our study is that children undergoing adenoidectomy, who are the only practical and ethically acceptable source of pediatric nasopharynx-associated lymphoid tissue (NALT), may be immunologically distinct from a randomly selected sample of healthy children. Nevertheless, previous studies have confirmed very similar patterns of acquisition of serum antibodies with age in both these children and healthy controls (36). Furthermore, the rates of pneumococcal colonization at the time of surgery are similar to those seen in healthy children of comparable age (36). Another potential limitation is that some children may have had limited prior pneumococcal exposure at the time of surgery. However, previous studies have shown that the majority of such children have mucosa-specific immune responses to pneumococcal protein antigens (37).
In summary, we report here the results of screening of human and mouse cells following exposure to pneumococcus by use of a library of conserved pneumococcal proteins. We believe that the use of human and murine cells for this type of screening can inform the selection of potential candidates worthy of further study. This approach could also be applied to the identification of other important mucosal pathogens whose route of entry begins in the nasopharynx.
MATERIALS AND METHODS
Materials.
Cholera toxin (CT) was purchased from List Biological Laboratories. Ni-NTA resin was purchased from Qiagen. CloneEZ PCR cloning kits were obtained from GenScript, Inc. All other reagents were obtained from Sigma or Thermo Fisher Scientific.
Selection of protein candidates by bioinformatic analysis in silico.
We chose 42 S. pneumoniae sequences (including some finished and others in draft form) from the Integrated Microbial Genomes website (http://img.jgi.doe.gov/cgi-bin/w/main.cgi). Beginning with the TIGR4 strain, we identified 335 proteins with a secretion signal peptide and 15 proteins with possible cell wall anchor motifs. The protein library was then narrowed down to 76 proteins based on the following parameters, chosen a priori: (i) conservation across all 42 sequences, defined as >90% identity at the amino acid level (reducing the library to 203 proteins); (ii) exclusion of proteins exhibiting >40% homology with proteins in the human genome (reducing the library to 160); and (iii) exclusion of proteins containing an extracellular domain smaller than 100 amino acids (in order to focus on proteins more likely to be accessible to antibodies in the presence of polysaccharide capsule; reducing the library to 88).
We specifically excluded previously studied antigens (including PsaA [38], SP2018 and SP0148 [21], StkP and PcsB [19], and Pht family proteins [39]) in order to focus on novel antigens. The breakdown of the 76 proteins (see Table S1 in the supplemental material) is as follows: 23 hypothetical proteins, 17 proteins proposed to play roles in substrate binding and transportation, 17 proteins with predicted enzymatic activity, and 19 others with unknown or hypothetical functions. Only extracellular domains without signal peptides or transmembrane regions were cloned.
Construction of the pneumococcal expression library.
The extracellular domains of selected proteins were amplified by PCR using TIGR4 genomic DNA as the template and were then integrated into pET21b expression vectors using the CloneEZ PCR cloning kit. Two large (>250-kDa) proteins (SP0648 and SP1154) proved difficult to purify at full length. We divided each amino acid sequence into three parts based on predictions of their secondary structures by BCL::Jufo (http://meilerlab.org/index.php/servers/show?s_id=5), making truncations in unconserved sequence areas, and purified each fragment separately. The two possible extracellular domains of one protein, SP0662, were both cloned and were designated SP0662-1 and SP0662-2. Thus, the final protein library consists of 81 proteins and peptides. Plasmid inserts were sequenced by Genewiz, Inc., for confirmation.
Protein purification.
E. coli transformants containing the cloned proteins were grown to an optical density at 600 nm (OD600) of 0.6, and protein expression was induced overnight with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C. Cells were spun down, and pellets were resuspended in lysis buffer (20 mM Tris-HCl, 500 mM NaCl [pH 8.0]) and lysed by sonication. The proteins of interest were purified from supernatants over a Ni-NTA column and were eluted in imidazole buffer. Elutions containing each protein were combined and purified over a gel filtration column in 20 mM Tris-HCl, 150 mM NaCl (pH 8.0).
Stimulation of mouse immune cells and human adenoidal cells.
Splenocytes were harvested from mice that had been either immunized intranasally (i.n.) with SPWCV or colonized i.n. with S. pneumoniae strain 0603 (serotype 6B) (29). Stimulations were carried out for 3 days with 5 μg/ml of each protein at 37°C under 5% CO2. The plates were then spun to pellet the cells, after which the supernatants were collected and were assayed for IL-17A using a mouse IL-17A enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems, Inc.).
Human adenoidal mononuclear cells from children undergoing adenoidectomy were separated on Ficoll gradients and were cultured at 1 × 106 cells per well in 48-well plates. Cells were stimulated for 7 days either with 8 μg/ml of recombinant proteins or with SPWCA at a concentration representing 1 × 106 CFU of killed bacteria/ml, and supernatants were assayed for IL-17A and IL-22 concentrations using human IL-17A and IL-22 ELISA kits (eBioscience).
Ranking of antigens.
Cytokine responses to each protein were ranked by averaging the rankings among all responses to all the proteins in the cells from each donor (child or mouse) in each screen, rather than according to actual values, in order to minimize the effects of variation in the responsiveness of the cells of different individuals. In each experiment, the protein with the greatest cytokine response was allocated the highest rank, and each subsequent protein was assigned a chronological rank in the decreasing order of their responses. All proteins that had responses lower than those of unstimulated cells were numbered “1.” The ranking numbers from each experiment were averaged to calculate the final rank of each protein.
Immunization and challenge of mice.
Female C57BL/6J mice (The Jackson Laboratory) were used in all experiments. All animal studies were approved by the animal ethics committee at Boston Children's Hospital. The age at the time of first immunization was 4 to 6 weeks. Intranasal immunization was carried out by instilling either 20 μl of saline with 1 μg of CT, as a control, or CT mixed with 10 μg of antigen as specified, atraumatically, into unanesthetized mice twice, at a 1-week interval. Blood was drawn 3 weeks after the last immunization and was assayed for IL-17A production upon stimulation with 5 μg/ml of the corresponding protein for 6 days. Nasopharyngeal colonization with clinical pneumococcal isolate 0603 (serotype 6B) was carried out as described previously (29).
IL-17A depletion.
An anti-IL-17A monoclonal antibody (clone 17F3) and matching isotype control antibodies were purchased from Bio X Cell. Mice were injected intraperitoneally with a dose of 150 μg per mouse 24 h before and 3 days after infection.
Statistical analysis.
Correlation was analyzed by the nonparametric Spearman method. Nasopharyngeal colonization densities were compared by the Mann-Whitney U test. Both analyses were carried out using Prism (version 7.0a; GraphPad Software, Inc.).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R21-AI103480 to Y.-J.L. from the National Institute of Allergy and Infectious Diseases. R.M. gratefully acknowledges support from the Translational Research Program at Boston Children's Hospital.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00490-18.
REFERENCES
- 1.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, Reingold A, Thomas A, Schaffner W, Craig AS, Smith PJ, Beall BW, Whitney CG, Moore MR. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis 201:32–41. doi: 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 2.Balsells E, Guillot L, Nair H, Kyaw MH. 2017. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: a systematic review and meta-analysis. PLoS One 12:e0177113. doi: 10.1371/journal.pone.0177113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu YJ, Gross J, Bogaert D, Finn A, Bagrade L, Zhang Q, Kolls JK, Srivastava A, Lundgren A, Forte S, Thompson CM, Harney KF, Anderson PW, Lipsitch M, Malley R. 2008. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog 4:e1000159. doi: 10.1371/journal.ppat.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malley R, Trzcinski K, Srivastava A, Thompson CM, Anderson PW, Lipsitch M. 2005. CD4+ T cells mediate antibody-independent acquired immunity to pneumococcal colonization. Proc Natl Acad Sci U S A 102:4848–4853. doi: 10.1073/pnas.0501254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, Clarke TB, Weiser JN. 2009. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest 119:1899–1909. doi: 10.1172/JCI36731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundgren A, Bhuiyan TR, Novak D, Kaim J, Reske A, Lu YJ, Qadri F, Malley R. 2012. Characterization of Th17 responses to Streptococcus pneumoniae in humans: comparisons between adults and children in a developed and a developing country. Vaccine 30:3897–3907. doi: 10.1016/j.vaccine.2012.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pido-Lopez J, Kwok WW, Mitchell TJ, Heyderman RS, Williams NA. 2011. Acquisition of pneumococci specific effector and regulatory CD4+ T cells localising within human upper respiratory-tract mucosal lymphoid tissue. PLoS Pathog 7:e1002396. doi: 10.1371/journal.ppat.1002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright AK, Bangert M, Gritzfeld JF, Ferreira DM, Jambo KC, Wright AD, Collins AM, Gordon SB. 2013. Experimental human pneumococcal carriage augments IL-17A-dependent T-cell defence of the lung. PLoS Pathog 9:e1003274. doi: 10.1371/journal.ppat.1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, Fulcher DA, Tangye SG, Cook MC. 2008. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med 205:1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minegishi Y, Saito M, Nagasawa M, Takada H, Hara T, Tsuchiya S, Agematsu K, Yamada M, Kawamura N, Ariga T, Tsuge I, Karasuyama H. 2009. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J Exp Med 206:1291–1301. doi: 10.1084/jem.20082767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vuononvirta J, Peltola V, Ilonen J, Mertsola J, He Q. 2015. The gene polymorphism of IL-17 G-152A is associated with increased colonization of Streptococcus pneumoniae in young Finnish children. Pediatr Infect Dis J 34:928–932. doi: 10.1097/INF.0000000000000691. [DOI] [PubMed] [Google Scholar]
- 12.Hoe E, Boelsen LK, Toh ZQ, Sun GW, Koo GC, Balloch A, Marimla R, Dunne EM, Tikoduadua L, Russell FM, Satzke C, Mulholland EK, Licciardi PV. 2015. Reduced IL-17A secretion is associated with high levels of pneumococcal nasopharyngeal carriage in Fijian children. PLoS One 10:e0129199. doi: 10.1371/journal.pone.0129199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu YJ, Leite L, Goncalves VM, Dias WDO, Liberman C, Fratelli F, Alderson M, Tate A, Maisonneuve JF, Robertson G, Graca R, Sayeed S, Thompson CM, Anderson P, Malley R. 2010. GMP-grade pneumococcal whole-cell vaccine injected subcutaneously protects mice from nasopharyngeal colonization and fatal aspiration-sepsis. Vaccine 28:7468–7475. doi: 10.1016/j.vaccine.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson R, Cohen JM, Reglinski M, Jose RJ, Chan WY, Marshall H, de Vogel C, Gordon S, Goldblatt D, Petersen FC, Baxendale H, Brown JS. 2017. Naturally acquired human immunity to pneumococcus is dependent on antibody to protein antigens. PLoS Pathog 13:e1006137. doi: 10.1371/journal.ppat.1006137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darrieux M, Goulart C, Briles D, Leite LC. 2015. Current status and perspectives on protein-based pneumococcal vaccines. Crit Rev Microbiol 41:190–200. doi: 10.3109/1040841X.2013.813902. [DOI] [PubMed] [Google Scholar]
- 16.Moffitt K, Malley R. 2016. Rationale and prospects for novel pneumococcal vaccines. Hum Vaccin Immunother 12:383–392. doi: 10.1080/21645515.2015.1087625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling E, Feldman G, Portnoi M, Dagan R, Overweg K, Mulholland F, Chalifa-Caspi V, Wells J, Mizrachi-Nebenzahl Y. 2004. Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin Exp Immunol 138:290–298. doi: 10.1111/j.1365-2249.2004.02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wizemann TM, Heinrichs JH, Adamou JE, Erwin AL, Kunsch C, Choi GH, Barash SC, Rosen CA, Masure HR, Tuomanen E, Gayle A, Brewah YA, Walsh W, Barren P, Lathigra R, Hanson M, Langermann S, Johnson S, Koenig S. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect Immun 69:1593–1598. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giefing C, Meinke AL, Hanner M, Henics T, Bui MD, Gelbmann D, Lundberg U, Senn BM, Schunn M, Habel A, Henriques-Normark B, Ortqvist A, Kalin M, von Gabain A, Nagy E. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med 205:117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffitt KL, Malley R, Lu YJ. 2012. Identification of protective pneumococcal TH17 antigens from the soluble fraction of a killed whole cell vaccine. PLoS One 7:e43445. doi: 10.1371/journal.pone.0043445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffitt KL, Gierahn TM, Lu YJ, Gouveia P, Alderson M, Flechtner JB, Higgins DE, Malley R. 2011. TH17-based vaccine design for prevention of Streptococcus pneumoniae colonization. Cell Host Microbe 9:158–165. doi: 10.1016/j.chom.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Gierahn T, Thompson CM, Trzcinski K, Ford CB, Croucher N, Gouveia P, Flechtner JB, Malley R, Lipsitch M. 2012. Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog 8:e1002989. doi: 10.1371/journal.ppat.1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pope C, Oliver EH, Ma J, Langton Hewer C, Mitchell TJ, Finn A. 2015. Genetic conjugation of components in two pneumococcal fusion protein vaccines enhances paediatric mucosal immune responses. Vaccine 33:1711–1718. doi: 10.1016/j.vaccine.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed MS, Derbyshire S, Flanagan B, Loh C, McCormick M, Barocchi M, Masignani V, Finn A, Zhang Q. 2014. Immune responses to pneumococcal pilus RrgA and RrgB antigens and their relationship with pneumococcal carriage in humans. J Infect 68:562–571. doi: 10.1016/j.jinf.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Leong SC, McNamara PS, Mubarak A, Malley R, Finn A. 2011. Characterisation of regulatory T cells in nasal associated lymphoid tissue in children: relationships with pneumococcal colonization. PLoS Pathog 7:e1002175. doi: 10.1371/journal.ppat.1002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray C, Ahmed MS, Mubarak A, Kasbekar AV, Derbyshire S, McCormick MS, Mughal MK, McNamara PS, Mitchell T, Zhang Q. 2014. Activation of memory Th17 cells by domain 4 pneumolysin in human nasopharynx-associated lymphoid tissue and its association with pneumococcal carriage. Mucosal Immunol 7:705–717. doi: 10.1038/mi.2013.89. [DOI] [PubMed] [Google Scholar]
- 27.Oliver E, Pope C, Clarke E, Langton Hewer C, Ogunniyi A, Paton J, Mitchell T, Malley R, Finn A. Th17 responses to pneumococcus in blood and adenoidal cells in children. Clin Exp Immunol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trzcinski K, Thompson CM, Srivastava A, Basset A, Malley R, Lipsitch M. 2008. Protection against nasopharyngeal colonization by Streptococcus pneumoniae is mediated by antigen-specific CD4+ T cells. Infect Immun 76:2678–2684. doi: 10.1128/IAI.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malley R, Lipsitch M, Stack A, Saladino R, Fleisher G, Pelton S, Thompson C, Briles D, Anderson P. 2001. Intranasal immunization with killed unencapsulated whole cells prevents colonization and invasive disease by capsulated pneumococci. Infect Immun 69:4870–4873. doi: 10.1128/IAI.69.8.4870-4873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mestas J, Hughes CC. 2004. Of mice and not men: differences between mouse and human immunology. J Immunol 172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 31.Lee YK, Mazmanian SK. 2010. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moens L, Hermand P, Wellens T, Wuyts G, Derua R, Waelkens E, Ysebaert C, Godfroid F, Bossuyt X. 2018. Identification of SP1683 as a pneumococcal protein that is protective against nasopharyngeal colonization. Hum Vaccin Immunother 14:1234–1242. doi: 10.1080/21645515.2018.1430541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odutola A, Ota MOC, Antonio M, Ogundare EO, Saidu Y, Foster-Nyarko E, Owiafe PK, Ceesay F, Worwui A, Idoko OT, Owolabi O, Bojang A, Jarju S, Drammeh I, Kampmann B, Greenwood BM, Alderson M, Traskine M, Devos N, Schoonbroodt S, Swinnen K, Verlant V, Dobbelaere K, Borys D. 2017. Efficacy of a novel, protein-based pneumococcal vaccine against nasopharyngeal carriage of Streptococcus pneumoniae in infants: a phase 2, randomized, controlled, observer-blind study. Vaccine 35:2531–2542. doi: 10.1016/j.vaccine.2017.03.071. [DOI] [PubMed] [Google Scholar]
- 34.Chang L, Brook WA, Bruyn G, Bologa M, Hopfer R, Kirby D, Sheng X, Neveu D, Menezes J, Ochs M, Visan L, Dacosta X, Yuan T, Hinds J, Jordanov E. 2014. A multi-component pneumococcal protein vaccine is safe and immunogenic in a phase I randomized, placebo-controlled study. Ninth International Symposium on Pneumococci and Pneumococcal Diseases, Hyderabad, India. [Google Scholar]
- 35.Wright AK, Ferreira DM, Gritzfeld JF, Wright AD, Armitage K, Jambo KC, Bate E, El Batrawy S, Collins A, Gordon SB. 2012. Human nasal challenge with Streptococcus pneumoniae is immunising in the absence of carriage. PLoS Pathog 8:e1002622. doi: 10.1371/journal.ppat.1002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Bernatoniene J, Bagrade L, Pollard AJ, Mitchell TJ, Paton JC, Finn A. 2006. Serum and mucosal antibody responses to pneumococcal protein antigens in children: relationships with carriage status. Eur J Immunol 36:46–57. doi: 10.1002/eji.200535101. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q, Bagrade L, Clarke E, Paton JC, Nunez DA, Finn A. 2010. Bacterial lipoproteins differentially regulate human primary and memory CD4+ T and B cell responses to pneumococcal protein antigens through Toll-like receptor 2. J Infect Dis 201:1753–1763. doi: 10.1086/652495. [DOI] [PubMed] [Google Scholar]
- 38.Briles DE, Ades E, Paton JC, Sampson JS, Carlone GM, Huebner RC, Virolainen A, Swiatlo E, Hollingshead SK. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun 68:796–800. doi: 10.1128/IAI.68.2.796-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godfroid F, Hermand P, Verlant V, Denoel P, Poolman JT. 2011. Preclinical evaluation of the Pht proteins as potential cross-protective pneumococcal vaccine antigens. Infect Immun 79:238–245. doi: 10.1128/IAI.00378-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.