Tissues and organs provide the structural and biochemical landscapes upon which microbial pathogens and commensals function to regulate health and disease. While flat two-dimensional (2-D) monolayers composed of a single cell type have provided important insight into understanding host-pathogen interactions and infectious disease mechanisms, these reductionist models lack many essential features present in the native host microenvironment that are known to regulate infection, including three-dimensional (3-D) architecture, multicellular complexity, commensal microbiota, gas exchange and nutrient gradients, and physiologically relevant biomechanical forces (e.g., fluid shear, stretch, compression).
KEYWORDS: 3-D, 3D, RWV, rotating wall vessel, gut-on-a-chip, host-microbe interaction, host-pathogen interactions, mechanotransduction, organ-on-a-chip, organoid
ABSTRACT
Tissues and organs provide the structural and biochemical landscapes upon which microbial pathogens and commensals function to regulate health and disease. While flat two-dimensional (2-D) monolayers composed of a single cell type have provided important insight into understanding host-pathogen interactions and infectious disease mechanisms, these reductionist models lack many essential features present in the native host microenvironment that are known to regulate infection, including three-dimensional (3-D) architecture, multicellular complexity, commensal microbiota, gas exchange and nutrient gradients, and physiologically relevant biomechanical forces (e.g., fluid shear, stretch, compression). A major challenge in tissue engineering for infectious disease research is recreating this dynamic 3-D microenvironment (biological, chemical, and physical/mechanical) to more accurately model the initiation and progression of host-pathogen interactions in the laboratory. Here we review selected 3-D models of human intestinal mucosa, which represent a major portal of entry for infectious pathogens and an important niche for commensal microbiota. We highlight seminal studies that have used these models to interrogate host-pathogen interactions and infectious disease mechanisms, and we present this literature in the appropriate historical context. Models discussed include 3-D organotypic cultures engineered in the rotating wall vessel (RWV) bioreactor, extracellular matrix (ECM)-embedded/organoid models, and organ-on-a-chip (OAC) models. Collectively, these technologies provide a more physiologically relevant and predictive framework for investigating infectious disease mechanisms and antimicrobial therapies at the intersection of the host, microbe, and their local microenvironments.
INTRODUCTION
Mucosal surfaces lining the gastrointestinal, respiratory and urogenital tracts continuously interface with the external environment and serve as a barrier against pathogens, commensals, chemicals, drugs, and toxins. These tissues possess a complex architecture with multiple cell types organized into three-dimensional (3-D) structures that facilitate tissue-specific functions. The biological, chemical, and biomechanical characteristics that define microenvironmental niches along these surfaces provide the structure and context in which infection takes place. Pathogens have adapted to detect specific host structures, polarity, and changes in local environmental stimuli (pH, temperature, oxygen, nutrients, hormones, physical forces, etc.) to know where and when to activate specific virulence programs during different infection stages (1–7). A major challenge in tissue engineering for infectious disease research is recreating in vivo spatiotemporal properties of dynamic 3-D microenvironments to more accurately model host-pathogen interactions in the laboratory.
Historically, infectious disease has been commonly studied in vitro by assessing the interaction of a single microbe with a single host cell type, with the latter grown as flat 2-D monolayers. This reductionist approach has enabled important discoveries and advanced our understanding of mechanisms that underlie infection and disease. However, the study of disease in isolation or out of context can change the native behavior of both host and microbe, thus creating a barrier for researchers to correlate in vitro and in vivo responses. In this data-rich period where multiple -omics technologies are being synergistically applied for unparalleled insight into host-pathogen interactions, it is critical to consider the context in which these investigations are performed. Reconstructing host microenvironments is key, including 3-D tissue architecture, multicellular complexity, microbiota composition/localization, oxygen tension, transport processes, and biomechanical forces (e.g., fluid shear, stretch, compression) (1, 8–11). Within this context, in vitro models are positioned along a continuum between 2-D and 3-D, with flat monolayers of a single cell type representing the most basic system and more complex models located further down the spectrum that recreate multiple aspects of the native tissue microenvironment (Fig. 1). Since tissues and organs function in a 3-D context, consideration of proper structure is essential for development of models that better mimic in vivo responses. Since no current in vitro model fully accomplishes this task, multidisciplinary teams of biologists, engineers, physicists, mathematicians, and clinicians are creatively working together to develop next-generation 3-D models with enhanced predictive capabilities to open new avenues for clinical translation.
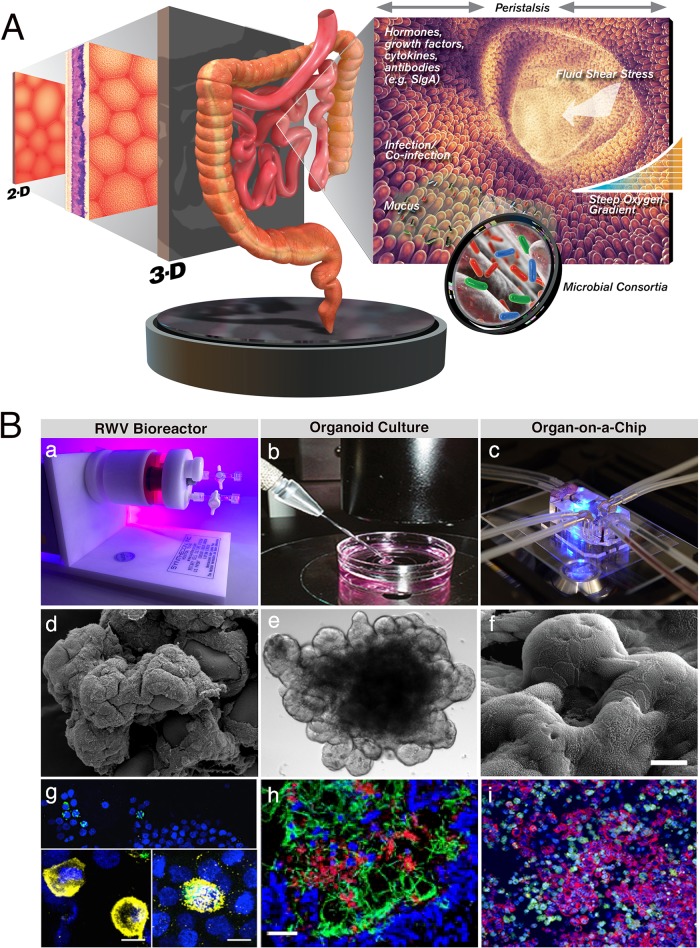
FIG 1.
Recreating the complex intestinal microenvironment to study host-pathogen interactions. (A) In vitro model advancement from 2-D to 3-D by incorporation of physiological factors to better mimic the in vivo environment. (Intestinal lumen, cell, intestine, and intestinal microbe images are republished from references 398 to 401, respectively, with permission of the publisher.) (B) Three-dimensional approaches routinely used to develop advanced intestinal models: (a) RWV bioreactor, (b) (republished from reference 307 with permission of the publisher), and (c) OAC (republished from reference 344 with permission of the publisher). (d) Scanning electron micrograph (SEM) showing an RWV colon model. (Republished from PLoS One [152].) (e) Light micrograph of an enteroid model. (Republished from Physiological Reports [240].) (f) SEM of a gut-on-a-chip model (republished from reference 341 with permission of the publisher). (g) Oxygen-dependent host cell colocalization of S. Typhimurium in an RWV 3-D coculture model of intestinal epithelium and macrophages. Following aerobic culture of bacteria, no macrophages were found, but following microaerobic culture, macrophages were present and either were empty (left inset) or contained internalized bacteria (right inset). Macrophages (CD45; yellow), Salmonella (green; white when overlaid with CD45), and nuclei (4′,6-diamidino-2-phenylindole [DAPI]; blue) are visible. Scale bar = 10 μm. (Republished from npj Microgravity [171].) (h) iHIOs injected with E. coli O157:H7. Nuclei (blue), neutrophils (CD11b; red), and E. coli (green) are visible. Scale bar = 100 μm. (Republished from PLoS One [260].) (i) CVB-infected gut-on-a-chip. CVB (green), F-actin (red), and nuclei (blue) are visible. (Republished from PLoS One [343].)
Present-day 3-D culture techniques result from a series of progressive advances in tissue engineering over the past century to better mimic the native structure and microenvironment of normal and diseased tissues (reviewed in reference 12). Indeed, long ago the cancer research community recognized that appropriate modeling of the 3-D microenvironment is important for mimicking disease, leading to development and application of 3-D organoid models developed within or on top of extracellular matrix (ECM) (12–16). The bidirectional exchange of biological and physical signals between cells and their microenvironment regulates cell structure/function and is largely manifested by tensile connections between ECM, cell surface receptors (e.g., integrins), and the cytoskeleton to transduce signals to and from the nucleus (17–31). This structural network is also engaged by certain invasive pathogens (e.g., Salmonella, Shigella, Listeria, rotavirus, and influenza virus) that hijack and remodel host cell architecture to facilitate their internalization, intracellular trafficking, and/or dissemination (9, 32–34). Similarly, we and others have demonstrated that bacteria also respond to biomechanical forces like fluid shear, which can regulate virulence, gene expression, and/or stress responses (1–5, 35–47). Indeed, the discovery of biomechanical forces as environmental regulators of microbial pathogenesis was made by our team almost 2 decades ago with the finding that fluid shear forces globally reprogram Salmonella gene expression, stress responses, and virulence (35). Fluid shear also plays a central role in regulating a number of host responses, including differentiation (48–50).
Although 3-D models have long been applied for cancer research (12–16), they remained largely unincorporated by the infectious disease community until the late 1990s and early 2000s. As expected for many new ideas in an established field, the use of 3-D models to study host-pathogen interactions was initially met with skepticism. The first reports of 3-D models to study viral infections were by Long et al. in 1998 (rhinovirus) and bacterial infections by Nickerson et al. in 2001 (Salmonella enterica serovar Typhimurium) (11, 51). Recently, infectious disease researchers have broadly embraced 3-D models for studying pathogenesis mechanisms, for biomarker discovery, and for drug candidate screening. In this review, we highlight key microenvironmental factors to consider when selecting in vitro 3-D intestinal models to study host-pathogen interactions. We focus on three key technologies for model development, (i) the rotating wall vessel (RWV) bioreactor, (ii) ECM-embedded/organoid models, and (iii) gut-on-a-chip models, and propose a vision for future model advancements. We also provide proper historical context for use of 3-D cell cultures in studying host-pathogen interactions, which is finally gaining a critical mass of scientists who understand and appreciate the value of studying disease in the proper context of tissue form and function.
MICROENVIRONMENTAL CUES IN HOST-MICROBE INTERACTIONS
Mucosal tissue function and homeostasis are meticulously controlled by complex bidirectional interactions between cells and their microenvironment (15, 20, 25, 27–29, 52–55). The microenvironment includes 3-D tissue architecture, multiple cell types, ECM, innate immunity mediators, indigenous microbiota, and physical forces. These factors are regulatory signals for mucosal pathogens and may be beneficial or detrimental for infection (1–5, 8, 35–45, 47, 56–64). Below we address key cellular, biochemical, and biophysical cues that dictate infection outcome and are important considerations when modeling host-enteric pathogen interactions.
Cellular factors.
Intestinal mucosal epithelium contains an array of specialized epithelial and immune cells that work in synergy to protect against infection by (i) serving as a barrier against luminal toxins, commensals, and pathogens, (ii) sampling microbial antigens, and (iii) recruiting innate and adaptive immune effectors (65). The intestine contains multiple epithelial cell types, including enterocytes (absorptive functions), enteroendocrine cells (hormone secretion), Paneth cells (antimicrobial production), goblet cells (mucin production), M cells (luminal antigen sampling/induction of mucosal immunity), Tuft cells (Th2 immunity), and cup cells (unknown function) (66, 67). The intestine also contains immune cells for innate and adaptive responses to pathogen attack, including macrophages, dendritic cells, and T and B cells, including those organized in lymphoid structures termed Peyer's patches, sites of induction of mucosal immunity. As the body's largest immune organ, the composition, organization, and function of the intestine vary by region and consist of integrated cross-communication networks of different cell types and effectors critical for protection against pathogens (described in references 59, 65, and 68 to 72).
Epithelial cell polarity establishes barrier function, regulates uptake/transport of nutrients, and maintains epithelial architecture (65, 73–75). In the intestine, apical surfaces face the lumen and regions between villi/folds, lateral surfaces face adjacent cells, and basal surfaces face the basement membrane and lamina propria. Along Peyer's patches and isolated lymphoid follicles, the basal side of the follicle-associated epithelium overlies a subepithelial dome region containing a mixture of immune cells (76). The distinct biochemical composition (e.g., protein and lipid) of apical and basolateral surfaces facilitates their specific functions (75). Given that many pathogens have evolved to recognize surface-specific molecules for attachment and/or to disrupt barrier integrity to enable their uptake and dissemination (6, 74, 77–79), appropriately modeling polarity in vitro is critical, as pathogens infect host cells differently depending on whether they are polarized or nonpolarized (80–83). Maintaining barrier integrity requires proper expression and localization of tight and adherens junctions. Adherens junctions are mediated by E-cadherin and catenin interactions, while tight junctions are composed of transmembrane proteins (e.g., claudins and occludins) and cytoplasmic plaque proteins (e.g., zonula occludens). While generally protective, junctional complexes are also exploited by pathogens to facilitate invasion (74) and some enteric viruses utilize receptors localized to these junctions (77, 78).
Another major cellular component encountered by enteric pathogens is the diverse microbial community—termed microbiota (referring to microorganisms) or microbiome (referring to microbial genomes). The intestinal tract contains prokaryotes, viruses, archaea, and eukaryotes, some of which protect the host against pathogen colonization by a variety of mechanisms, including epithelial cell turnover, mucin synthesis, and triggering bacterial sensors on host cells (84–86). Reciprocal interactions between host and microbiota contribute to tissue function and homeostasis and determine microbiota composition, thereby playing an important role in infection and disease (87). For example, members of the intestinal microbiota regulate production of antimicrobial peptides by Paneth cells (88) and shape immune responses by regulating numbers, subsets, and/or functions of T, B, and myeloid cells (65). Microbiota-induced changes in immunity also determine intestinal microbiota composition (85, 89).
The intestinal microbiota is comprised of ∼1014 bacteria (>1,000 species), with Firmicutes and Bacteroidetes most abundant (90–93). Interpersonal variation in intestinal microbiome occurs, with each individual carrying a subset of the total known microbiome (94). Temporal and spatial variation occurs throughout the intestinal tract (95, 96). Increasing data suggest a relationship between an imbalanced intestinal microbiome and various diseases, including obesity, inflammatory bowel disorders, and cancer (97). The importance of the gut microbiota to health is highlighted by successful clinical application of fecal microbiota transplants from healthy individuals to patients with recurrent, antibiotic-resistant Clostridioides difficile infections (98–100).
Biochemical cues.
Mucosal tissues contain an array of small molecules, including innate defense mediators that target pathogens and regulate downstream host defenses. Intestinal mucus harbors compounds from the innate and adaptive systems that protect against microbial insult, including digestive enzymes (e.g., lysozyme), lactoferrin, antimicrobial peptides, complement, and antibodies (e.g., secretory immunoglobulin A [sIgA]) (65). In addition, cells of the innate defense system respond to pathogen-associated molecular patterns (PAMPs) using pathogen recognition receptors (PRRs). Depending on the pathogen, PRR-mediated signal transduction results in different cellular outcomes (e.g., cell proliferation, apoptosis, antimicrobial peptide production, autophagy, and cytokine secretion). Cytokine production leads to recruitment of innate and adaptive immune effectors to the infection site, representing a bridge between these two arms of immunity (65, 101).
Mucins are complex mixtures of high-molecular-weight, glycosylated macromolecules that bind and remove pathogens and their products (7, 101). Enteric pathogens sense and respond to cues within mucus and overcome this barrier to reach underlying epithelium (7). Normal intestinal mucus consists of two layers: an outer layer colonized by microbes and a sterile inner layer (102–104). The composition and thickness of these mucin layers vary throughout intestinal regions to accommodate their different functions and microbial burdens. Within the small intestine, the inner and outer mucosal layers are thinner to facilitate nutrient absorption, with thicker regions found toward the ileum, where microbial burden is heavier (7). In the colon, both layers are thicker to accommodate the burden of several trillion commensals (7). The presence of sIgA and other mucin antimicrobials also serves to reduce bacterial colonization (105).
The ECM is another key contributor to tissue homeostasis. The ECM has historically been neglected as a signaling entity, but seminal discoveries have revealed the central role of ECM in regulating tissue architecture/function (20, 53). The ECM is a three-dimensional noncellular scaffold comprised of proteins (e.g., collagens, elastins, laminins, and fibronectins), proteoglycans, and water. Two main types of ECM include (i) interstitial connective tissue matrix, which serves as a cellular scaffold, and (ii) basement membrane matrix, which separates epithelium from interstitium (106, 107). ECM components also serve as ligands for cell receptors like integrins, which transduce physical forces into biological responses (mechanotransduction). Additionally, immune responses are mediated through interactions with the ECM (107, 108). Furthermore, the ECM controls availability/release of growth factors and other signaling molecules (hormones and cytokines) (107). The complexity, composition, and structure of ECM are highly dynamic and specific (as are the biochemical gradients it contains) and depend on tissue type, developmental stage, and health/disease state (107).
Biophysical forces.
The role of physical forces in cell and tissue development/function is as important as those of genes and biochemical signals (28, 109). Physical forces regulate cell proliferation, differentiation, and homeostasis (110, 111). Forces experienced by intestinal cells include fluid shear, pressure (112), and contractile peristalsis of muscles (113). Hydrodynamic calculations suggest that fluid shear forces on the exposed epithelial brush border microvilli are ∼200 times greater than those between microvilli (<0.01 dynes/cm2) (114).
The cytoskeleton and its linkage with the ECM play an essential role in enabling cells to sense and respond to biophysical forces. While the governing role of the ECM as a dynamic signaling entity that regulates tissue form/function is now appreciated, it was initially considered a purely static scaffold. However, tissue-specific architecture and function are regulated by the biophysical properties of the ECM (20, 115, 116), which exerts physical influences transduced by cell surface receptors through the cytoskeleton to the nucleus to ultimately alter cellular and molecular properties. These structural networks are critical for regulating cell shape/architecture and have been modeled using the principle of tensegrity, which refers to structures that are stabilized under continuous tension by balancing opposing tension and compression forces (27–29, 31). The integration of biophysical forces across cells and tissues using this structural network regulates a wide range of biological processes (e.g., cell proliferation, apoptosis, differentiation, adhesion, migration, gene expression, and architecture) (8, 20, 21, 23, 25, 27, 29–31, 55, 117). Accordingly, ECM composition and stiffness are critical regulators of cellular responses (118, 119). These properties are continuously remodeled through the process of “dynamic reciprocity” (17, 20, 53, 117), theorized by Bissell in 1982 to explain how signaling between the ECM and nucleus regulates tissue function. This laid the foundation for modern 3-D cell culture approaches used today (20, 107, 120). Not surprisingly, pathogen-ECM interactions play an important role in mediating infection (121–126). In addition to impacting the host, physical forces also globally alter bacterial gene expression, stress responses, and virulence in unexpected ways to contribute to infection (5, 36–40, 47, 62–64).
MODELING THE MICROENVIRONMENT: 3-D MODELS FOR INFECTIOUS DISEASE
Several cell culture systems exist for the development and application of 3-D models of human tissues for infectious disease research, including the RWV bioreactor, ECM-embedded scaffolds (e.g., ECM extracts, purified ECM, or synthetic/semisynthetic hydrogels), and organ-on-a-chip (OAC) models. The choice of system to use depends on several factors, including the experimental question being addressed, technical complexity, and cost and expertise for model development. Different cell types in the native tissue (including immune cells) can be cocultured in these models to further enhance physiological relevance. Additionally, a single epithelial cell type can spontaneously differentiate into multiple epithelial cell types normally found in the parental tissue and undergo self-assembly into tissue-like structures using all of these 3-D technologies. To date, most in vitro infection studies have been performed using cell lines; however, there is a push to develop models using either primary or stem cells to better mimic the native tissue. To explore the integration of different environmental signals in regulating infection, a hierarchical series of increasingly complex 3-D model systems comprised of different cells types can be developed and applied in parallel under differing experimental conditions (e.g., different oxygen tensions and physical forces).
RWV-derived 3-D models.
The RWV bioreactor is an optimized form of suspension culture that facilitates formation of self-organizing 3-D tissue-like aggregates by allowing cells the spatial freedom to colocalize and self-assemble based on natural affinities within a low-fluid-shear environment (Fig. 1B, panel a) (8, 127). Fluid shear influences cell proliferation, differentiation, morphology, and function (30, 114, 128–140). Models developed within the dynamic RWV environment experience excellent mass transfer of nutrients/wastes and exhibit enhanced structure, differentiation, function, and multicellular complexity relative to 2-D monolayers (11, 80, 141–154). Along these lines, observations from the 1970s showed that flotation of collagen gels led to a more permissive environment for cellular differentiation (12, 155, 156). Moreover, the low-fluid-shear environment in the RWV is also physiologically relevant to that encountered by pathogens in low-shear regions of the infected host, including the intestine (38, 114, 129–131). Accordingly, the RWV is also used to culture pathogens to study the role of fluid shear and mechanotransduction in regulating microbial pathogenesis and host-pathogen/commensal interactions (1, 35–41, 45–47, 62, 64, 145, 157–167).
The RWV is among the most extensively used approaches to develop 3-D models to study host-pathogen interactions. It was the first technology used to develop 3-D models for infection studies with bacterial (Salmonella) and viral (rhinovirus) pathogens (11, 51). A range of RWV-derived 3-D models have been developed using cell lines, stem cells, and/or primary cells, including small and large intestine (11, 80, 141, 143, 145, 146, 152, 168–177), lung (144, 147, 178–182), liver (148, 153, 174, 183, 184), bladder (8, 185–187), reproductive tissue (149–151, 188–190), heart (191–193), prostate (142, 186, 194), pancreas (195, 196), nervous tissue (182, 197–199), blood-brain barrier (200), skin (201), eye (202), bone, joint, or disc (203–207), and tonsil (208), among others. These studies demonstrated that RWV-derived models exhibit enhanced in vivo-like characteristics, including spontaneous differentiation into multiple cell types that self-organize into 3-D structures (Fig. 1B, panel d), polarization, appropriate expression/localization of adherens/tight junctional complexes, metabolic product secretion, gene expression, cytokine production, responses to antimicrobials and microbial products, support of commensals, and/or susceptibility to infection (8, 11, 80, 141–153, 168–194, 197–208). In addition, RWV models have been advanced to incorporate immune cells to study their role in host-microbe interactions (171, 175, 177, 180).
Models are typically initiated by harvesting monolayers, combining cells with porous ECM-coated microcarrier scaffolds, and loaded into the RWV. Scaffold and ECM porosity allows the basal side of cells to experience autocrine/paracrine communications, aiding cellular differentiation/responses in a manner reflecting in vivo tissues. This differs from monolayers where cells proliferate on impermeable surfaces, thus hindering proper communications across apical and basolateral surfaces. Additionally, models may be developed scaffold free or using nonmicrocarrier scaffolds (e.g., decellularized tissues) for transplantation (179, 181, 209). Once developed, distribution of 3-D models into multiwell plates lends to their experimental tractability for infection assays, as their structural/functional integrity remains intact following seeding. Alternatively, pathogens or compounds can be directly added to the RWV to study interactions under physiological fluid shear. One key advantage of RWV culture is the production of large numbers of cells (∼107 to 108 per culture). Below we discuss RWV-derived 3-D models of human intestinal mucosa.
RWV-derived intestinal models.
We began using the RWV to engineer 3-D models of human intestine for infection studies in the late 1990s after realizing that available models for studying bacterial pathogenesis lacked multiple aspects of the in vivo microenvironment (11). RWV-derived 3-D models have enabled the study of host-microbe interactions relevant to different regions of the intestinal tract, including the small intestine (11, 169) and colon (80, 143, 145, 146, 152, 171, 172, 175, 177). Imaging of these models revealed enhanced 3-D architecture relative to monolayers, including the presence of extensive 3-D folds and microvilli, that more closely resembled what is observed in vivo (Fig. 1B, panel d). These 3-D models are essentially “inside-out” such that the apical/luminal side faces the media and the basal side faces the scaffold, allowing for straightforward introduction of pathogens, toxins, and antimicrobials at the apical surface, as commonly occurs in vivo. Collectively, these models have shown physiologically relevant expression and localization of key cellular components, including junctional proteins (e.g., ZO-1, occludin, symplekin, E-cadherin, β-catenin, and desmosomes), secretion of basal lamina components (e.g., collagen types II, III, and IV, laminin, vimentin, and fibronectin), brush border formation with villin, and/or mucus secretion (11, 80, 143, 145, 146, 152, 169, 171, 172, 175, 177). Spontaneous cellular differentiation into multiple lineages found in the intestinal epithelium is also observed, including enterocytes, M cells, goblet cells, and/or Paneth cells (enteroendocrine cells were not evaluated) (11, 80, 146, 152, 171, 175). The presence of multiple cell types within a model (e.g., epithelial and immune cells) enables study of their combined effects on infection, and in particular, pathogen colocalization patterns with different cell types. An example is described below in which an advanced 3-D RWV coculture model that combined human colonic epithelium with phagocytic macrophages was used to study infection by different Salmonella pathovars (171). Primary human lymphocytes have also been incorporated in a 3-D coculture model of intestinal epithelium to study Salmonella infection (175).
RWV-derived intestinal models have contributed to the study of a variety of pathogens, such as S. Typhimurium (including multidrug resistant ST313), Salmonella enterica serovar Typhi, enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), Cryptosporidium parvum, and human enteroviruses, including coxsackievirus B (CVB) and poliovirus (11, 80, 143, 145, 146, 152, 171, 172, 175). Studies with S. Typhimurium using 3-D models of small and large intestine displayed marked differences from monolayers in colonization, tissue morphology, apoptosis, and prostaglandin and cytokine expression (11, 80, 152). The responses of these 3-D intestinal models to S. Typhimurium challenge were highly predictive of in vivo responses in humans/animals (11, 80, 152), including rapid repair of the small intestine (initial site of Salmonella pathogenesis) and significant damage to the colon (primary site of pathogenesis) (210). These models were also the first in vitro systems to challenge the widely accepted paradigm established using monolayers that the Salmonella pathogenicity island 1 (SPI-1) type 3 secretion system (T3SS) is required for invasion of intestinal epithelium (80, 152). Historically, studies with monolayers contradicted in vivo observations wherein successful animal infections were possible with T3SS SPI-1 mutants (211, 212), and clinical isolates of Salmonella lacking SPI-1 function were isolated from foodborne disease outbreaks in patients experiencing gastroenteritis (213). Using a 3-D intestinal model comprised solely of epithelial cells, Radtke et al. demonstrated that SPI-1 mutants and a Salmonella mutant lacking all known T3SSs (SPI-1, SPI-2, and the flagellar system) still exhibited high levels of invasion relative to the wild type (although approximately 0.5 to 1 log lower) (152). As expected, in monolayers these mutants exhibited little to no invasion (<10 CFU), a finding which does not reflect in vivo observations (152). Thus, for the first time, an in vitro intestinal epithelial model was able to parallel in vivo results by supporting Salmonella invasion independently of SPI-1. These findings demonstrate the enhanced capability of RWV models to predict in vivo-like pathogenic mechanisms.
Host-pathogen-commensal and host-commensal interactions have also been investigated using RWV 3-D intestinal models (172, 177). Commensal microbes naturally enhance intestinal mucosal barrier function against pathogen colonization through complex mechanisms not yet fully characterized (214). Naturally occurring probiotic strains of bacteria are being exploited as a strategy against pathogens to combat ongoing problems of antibiotic resistance. Treatment of a 3-D intestinal model with Lactobacillus reuteri or its antimicrobial metabolite, reuterin, before or after challenge with S. Typhimurium reduced adhesion, invasion, and intracellular survival of this pathogen compared to findings for untreated cells (172). This was the first study to report the effect of reuterin on the enteric infection process for any mammalian cell type. A 3-D intestinal coculture model containing immune cells was used to profile responses to both free secretory IgA (sIgA) and sIgA complexed with a commensal strain of E. coli (177). Application of free sIgA to the model induced upregulation of MUC2, interleukin 8 (IL-8), and polymeric immunoglobulin receptor (pIgR) secretion. When sIgA was complexed with E. coli and applied to the model, these responses were downregulated relative to models treated with free sIgA.
Barrila et al. reported advancement of a 3-D HT-29 colon model to include phagocytic macrophages, thereby improving its physiological relevance to study aspects of the innate immune response to infection (171). Characterization of this coculture model revealed macrophages integrated between and underneath epithelial cells while preserving epithelial tight junctions and the presence of multiple epithelial cell types, including enterocytes, M cells, and goblet cells (171). Macrophage phagocytosis was confirmed by evaluating their ability to engulf inert, bacterium-sized beads. The contribution of macrophages to Salmonella infection was assessed using S. enterica pathovars with differing host tropisms and disease phenotypes, including the well-studied sequence type 19 (ST19) S. Typhimurium strain SL1344, which causes disease in a wide range of hosts, the multidrug-resistant ST313 S. Typhimurium strain D23580, and the human-specific S. Typhi strain Ty2. Although classified as S. Typhimurium, ST313 strains display genome degradation similar to that of human-adapted S. Typhi and are associated with devastating epidemics of blood-borne infections in sub-Saharan Africa (215). Bacteria were cultured aerobically or microaerobically prior to infection to simulate oxygen environments encountered before and during intestinal infection. Colonization of all strains was reduced in the coculture model containing macrophages relative to the epithelial model, indicating antimicrobial function of macrophages. Although ST313 strains are considered highly invasive due to the systemic infection they cause, D23580 was not highly invasive in the 3-D models but instead exhibited enhanced survival/replication, thus providing clues as to what drives this organism's pathogenicity. Pathovar- and oxygen-specific differences in host cell colocalization patterns were also observed (Fig. 1B, panel g), indicating the ability of these advanced models to distinguish between closely related Salmonella serovars, thus providing a unique advantage over models composed of a single cell type (171).
RWV-derived intestinal models are also valuable for investigating host-pathogen interactions for which conventional cultivation strategies are unable to adequately model in vivo complexity. Recently, a 3-D colonic model was applied to study human CVB (146), a pathogen for which in vitro and in vivo models may not fully model the enteral infection route in humans (146, 216–219). Comparisons between polarized 2-D and 3-D cells revealed that the 3-D model displayed an enhanced number of viral particles secreted into the media at early stages of the viral life cycle, which did not coincide with increased host cell destruction relative to monolayers (146). These data suggest that 3-D models exhibit an enhancement in nonlytic release of viral particles, which might result from morphological changes (e.g., enhanced brush border formation) in 3-D cells. Similarly, another 3-D colonic model was used to study Cryptosporidium parvum, a parasite for which there is a lack of physiologically relevant in vitro and in vivo models (143). Following C. parvum infection, morphological changes were observed that were consistent with those from colonic biopsy specimens of infected patients (143). These studies further emphasize the critical importance of model complexity and physiological relevance as determinants in enabling host-pathogen interactions.
In summary, 3-D RWV intestinal models are powerful tractable research tools that advance the study of host-pathogen interactions. These models can be modularly altered to incorporate different cell types (including patient-derived cells), ECM, commensal microbiota, physical forces, etc., akin to in vivo scenarios, increasing their relevance. Their tissue-like architecture, differentiation and polarization, enhanced expression/localization of junctional proteins, and mucin production are necessary components of an effective barrier to invading pathogens.
Limitations and future directions of RWV-derived 3-D models.
Although many key structural/functional characteristics of parental tissues have been successfully recapitulated using RWV models, several limitations remain. The full extent of 3-D architecture, multicellular complexity, and array of physical forces of in vivo tissues has not yet been attained. Ongoing studies are further enhancing these features, plus incorporating patient-specific immune cells and fecal microbiota, and achieving vascularization and innervation. Models can be costly due to high medium consumption required for culturing large numbers of cells; however, researchers can scale down. Although bead porosity facilitates apical/basal cytokine secretion and there is excellent access to the apical side of the models, there is currently an inability to sample the basal side. This also prevents measurement of transepithelial electrical resistance (TEER), which measures electrical resistance across a monolayer as a proxy for assessing barrier integrity (220). The technique involves using two electrodes, one in contact with cells on a semipermeable membrane (e.g., apical side) and the other in a different chamber containing culture medium (e.g., basal side). With most RWV models grown on tiny (∼175 μm) microcarrier beads, these measurements are not currently possible with off-the-shelf technology. This challenge will likely be surmounted with custom electrode design to accommodate current RWV models or the use of alternative scaffolds. Currently, immunofluorescence imaging of cytoskeletal and tight junctional markers represents an alternative method to evaluate model integrity. As these models grow in size and complexity, introduction of vasculature and nerve cells will be important. Finally, current models are not easily amenable to chronic infection due to lack of perfusion once removed from the RWV; however, inclusion of automated waste removal and nutrient delivery during infection will facilitate this approach.
3-D organoid models.
The term organoid (“organ-like”) has been used to describe a variety of 3-D models that resemble in vivo tissues. Historically, this included models engineered with different technologies using cell lines, stem cells, primary cells, or tissue explants either embedded in or cultured on top of ECM scaffolds that allow cells to self-assemble into 3-D structures (8, 12, 143, 145, 146, 169, 171, 221–229). Advances in stem cell biology led to a recent terminology shift to more specifically define organoids as 3-D models derived from stem cells, progenitor cells, or primary explants (222, 230–238). Here we focus on 3-D models cultured within a 3-D ECM that fit this definition. It is important to emphasize that current models are based on decades of work by pioneering cell biologists that laid the foundation for the current organoid field (reviewed in reference 12), representing an advancement and merging of old and new technologies to enable novel discoveries (12, 228, 239). Models cultivated using thick ECMs have deep roots in tissue engineering and cancer biology, in which they were applied to develop advanced models enabling the study of a variety of biological mechanisms, particularly with regard to understanding the interrelationship between tissue structure and function (12). This effort resulted in a critical mass of scientists who now recognize the importance of 3-D models for infection and are bringing elegant advances to the field but may not be fully aware of their historical context.
A range of different organoid models have been established, including small and large intestine (229, 230, 232, 234, 240–268), lung (269–274), stomach (275–282), breast (55, 283, 284), brain (285–287), liver (222, 288, 289), pancreas (222, 290, 291), gallbladder (292), eye (293), kidney (294), prostate (222, 295, 296) and reproductive tract (297, 298), among others. Relative to monolayers, these models more closely mimic endogenous tissues, including organization and spontaneous differentiation of multiple cell types into physiologically relevant 3-D structures (Fig. 1B, panel e), expression and localization of tight junctions, mucus production, polarity, gene expression, cell viability and proliferation, cytokine production, responses to antimicrobials, support of commensals, and susceptibility to infection (12, 55, 222, 226, 228–235, 237, 238, 240–266, 269–319).
To develop 3-D organoid models, stem cells or tissue explants containing stem cells are used. Biopsy specimens may be treated with a dissociation agent and/or mechanically disrupted prior to embedding into ECM. Stem cells isolated from biopsy specimens can be predifferentiated into progenitor cells and further differentiated into ECM-embedded organoids. Differentiation into committed cell types is enabled by stepwise supplementation and/or removal of signaling factors during culture (249, 251, 252, 254, 264, 275, 278, 303, 320–322). Purified ECM components and mixtures can be used, including Matrigel, a laminin-rich ECM isolated from chondrosarcomas (323, 324). Synthetic hydrogels help circumvent challenges associated with Matrigel, including batch-to-batch variation and potential carcinogenic issues connected with tumor-derived matrices (229).
3-D intestinal organoids.
Sato et al. (249) and Ootani et al. (253) independently reported conditions enabling long-term in vitro culture of mouse intestinal crypts containing Lgr5+ stem cells (as well as purified Lgr5+ stem cells that generate villus/crypt-like structures [249]). These approaches used either Matrigel (249) or collagen (253) in combination with supplementation of Wnt agonist R-spondin 1. Sato et al. also included epidermal growth factor to enable crypt growth and noggin to facilitate passaging (249). These models displayed a polarized, multicellular epithelium (enterocytes, goblet cells, Paneth cells, and enteroendocrine cells) organized into a central lumen lined by villus/crypt-like structures (249, 253). Murine intestinal organoids developed from single Lgr5+ stem cells also developed into these multicellular structures (249). Subsequently, additional factors were included to enable human colonoid culture (264).
The NIH Intestinal Stem Cell Consortium defined a standardized nomenclature to reflect model sources, approaches, and in vitro structures (325). Structures directly isolated include epithelial sheets, crypts, and organoids (crypts and surrounding mesenchymal elements) (325). Various structures produced in vitro from small intestine include enterospheres (rounded epithelial cyst-like structures), enteroids (formation of budding crypts from enterospheres), and induced intestinal organoids (multicellular clusters from induced embryonal or pluripotent stem cells, e.g., induced human intestinal organoids [iHIOs]) (325). Analogous colonic structures include colonospheres, colonoids, and colonic organoids (325). It is common to see terms used interchangeably, and the nomenclature will likely evolve as the field expands.
Model infection can be accomplished by (i) addition of pathogen directly to the media (basal side), (ii) microinjection into the lumen (Fig. 1B, panel b), (iii) shearing of models into fragments followed by pathogen addition, and (iv) complete disruption of 3-D models into flat monolayers followed by pathogen addition (230, 237). Consideration of the normal infection route is critical. Direct addition to the media is easiest; however, for pathogens that infect via the apical/luminal side, this represents a nonphysiological route of infection. Microinjection is technically challenging but preferable for pathogens that normally infect from the lumen. Due to challenges associated with microinjection, there is a growing tendency to mechanically dissociate organoids into smaller pieces or completely dissociate them into monolayers on Transwell inserts or plastic (237, 261, 281, 313, 314). This approach has been successful for a number of studies, including cultivation of norovirus (314), a major advance in the field. Use of Transwell inserts also facilitates TEER analysis and easier cytokine sampling from the apical side of the model.
When dissociating 3-D models prior to infection, it is important to note that this disconnects their form and function, similar to disrupting primary tissue into monolayers, and may render them less predictive for some (not all) phenotypes. In this approach, Transwell inserts are preferable over plastic, as the former display improved physiological relevance over conventional monolayers (326). Additional profiling should confirm the extent to which the dissociated model may have dedifferentiated and additional culture time may be required to reestablish polarity/barrier function. Key findings should be validated using intact organoids and microinjection to avoid artifacts. Additionally, since ECM-pathogen interactions are important for infection dynamics (61), infection surfaces should not contain ECM components not typically found in that location in vivo (e.g., lumen) if the pathogen would not normally encounter it.
A variety of pathogens have been studied using 3-D enteroid/colonoid/organoid models, including Salmonella, C. difficile, EHEC, EPEC, enterotoxigenic E. coli (ETEC), norovirus, rotavirus, enteroviruses, Toxoplasma gondii, and coronaviruses (230, 231, 233–235, 238, 240–245, 257–263, 266, 268, 307–319, 327). The first infection using iHIOs was performed using human rotavirus, which lacks robust in vitro culture systems (315). Both laboratory and clinical rotaviruses replicated in iHIOs and were detected in epithelial and mesenchymal cells (315). Crypt-derived enteroids also supported rotavirus replication and were used to assess antiviral efficacy against patient isolates (244, 266). Ettayebi et al. made a significant advance by the successful in vitro culture of human norovirus (HuNoV), known for its lack of a reproducible culture system (314). The authors initially cultured 3-D intestinal organoids and then dissociated them into monolayers on plastic or Transwell inserts (314). Successful viral replication was observed and only enterocytes were infected with HuNoVs, regardless of the strain or the intestinal region from which the model was derived. Additional viral models, including those using enteroviruses (e.g., CVB, echovirus 11, and enterovirus 71) have identified the cell-type-specific nature of these infections and the virus-specific nature of innate immune signaling in response to infection (327).
Enteroid models were also used to study S. Typhimurium and E. coli. Zhang et al. (240) and Wilson et al. (243) used crypt-derived enteroids to study Salmonella infection. S. Typhimurium successfully colonized the model (240, 243), and infection responses aligned well with in vivo observations, including disruption of tight junctions, inflammatory responses, and decreased stem cell numbers (240). Forbester et al. infected iHIOs with S. Typhimurium and observed physiological transcriptomic and cytokine profiles (257). Injection of E. coli O157:H7 into iHIOs containing neutrophils led to loss of actin, epithelial integrity disruption, induction of inflammatory cytokines, and neutrophil recruitment (Fig. 1B, panel h) (260). In contrast, commensal E. coli was retained within the lumen, with no loss of model integrity. Infection of colonoid-derived Transwell models identified MUC2 and protocadherin-24 as early EHEC infection targets (261). Colonoids were initially cultured in 3-D, followed by dissociation onto Transwells. Model differentiation correlated with expression of differentiation markers, increased TEER, and microvilli (261). EHEC preferentially colonized the differentiated model relative to an undifferentiated control, reducing colonic mucus and inducing damage to microvilli. A similar approach was applied to study EPEC and ETEC infections in coculture models containing macrophages (313). Inclusion of macrophages in the bottom chamber of the enteroid-derived Transwell model enhanced barrier function, increased epithelial height, and altered cytokine responses relative to the control. EPEC increased total macrophage numbers and induced projections that extended into the epithelium, while ETEC induced macrophage extensions across the epithelium to the apical surface. The presence of macrophages in the coculture model enhanced barrier function and correlated with decreased numbers of ETEC organisms relative to the model lacking immune cells.
iHIOs were also used to study C. difficile infection (CDI) (258, 259, 262, 263). CDI patients secrete acidic mucus consisting primarily of MUC1, with decreased MUC2 and altered oligosaccharide composition relative to that in healthy patients (259). Injection of the pathogen alone into iHIOs decreased MUC2, while whole CDI stool supernatant was required to induce patient-like oligosaccharide composition changes (259). iHIOs were also used to investigate nontoxigenic and toxigenic strains of C. difficile and purified toxins TcdA and TcdB (262). Injection of the toxigenic isolate or purified TcdA led to a loss of barrier function, while iHIOs injected with the nontoxigenic strain remained intact. Separately, colonoids helped identify Frizzled proteins as receptors for the TcdB toxin (263).
In summary, 3-D organoid models are advancing mechanistic understanding of host-microbe interactions due to their enhanced 3-D architecture and presence of Lgr5+ stem cells together with multiple cell types and other functional properties. In addition, patient organoid “biobanks” have been established and are facilitating fundamental research and clinical applications (230, 231, 328, 329). One exciting example of the applicability of these models is the use of patient-derived organoids to predict drug responses for cystic fibrosis treatment (222, 231, 250, 307, 329, 330).
Limitations and future directions of 3-D organoids.
As for other models, organoids have limitations that researchers are working to overcome. Variability and quality control challenges between experimental preparations includes (i) heterogeneity in size, shape, and viability of organoids within a culture and across different samples, (ii) batch-to-batch variability in Matrigel or other ECM, and (iii) batch-to-batch variability in growth factor sources. Organoid infection presents challenges as described above. Medium cost is high if scaling up due to reliance on specific growth factors. Incorporation of the full array of cell types found in vivo, including the diverse collection of immune cells and microbiota, has not been attained. Organoid models also lack spontaneous M cell formation (251, 331). Pretreatment of in vitro models with RANKL, exposure to lymphocytes, or infection with pathogens like S. Typhimurium can induce M cell formation (331–333). Although the mechanism by which M cells spontaneously differentiate in RWV models (11, 152, 171, 175) is unknown, it is possible that the low-fluid-shear suspension culture environment is important, since flotation of ECM scaffolds was more permissive for differentiation than surface-attached ECM (12, 155, 156). Since organoid models are typically ECM embedded, another limitation is that the application of the range of biomechanical forces found in vivo is limited; however, an iHIO model containing functional neurons that enabled peristalsis-like contractions was reported (256). Combinations of technologies, including organoid-derived 3-D models developed using the RWV bioreactor (202) and organ-on-a-chip (OAC) (334), are further expanding these capabilities. TEER measurements are also not currently possible with intact organoid models due to their size and structure and because they are ECM embedded. Some studies have dissociated organoids into 2-D on Transwells to facilitate these measurements, although there can be disadvantages to using this approach, as discussed.
Organ-on-a-chip models.
Advanced microfluidic and microfabrication technologies are being broadly applied to develop organ-on-a-chip models that mimic key aspects of in vivo microenvironments. Rather than focusing on recreating the 3-D structure of the entire tissue, this technology aims to recreate a microscale model of the local 3-D architecture and spatial distribution of dynamic tissue interfaces to mimic tissue- and organ-level functions (335). These devices are designed with micrometer-sized fluidic channels separated by thin, flexible porous membranes that enable development of different tissues in adjacent chambers while retaining their ability to interact (Fig. 1B, panel c) (335–339). These features allow flexibility to model active processes within a tissue, such as vascular-like perfusion. One exciting functional feature engineered into the design of many of these devices is the capability to apply dynamic forces across the tissue to model fluid shear and peristalsis (334, 340–343).
OAC models vary in complexity, ranging from simple systems containing a single perfused chamber and cell type to more advanced chips that contain several microchannels, membranes, and assorted cell types, thereby allowing the reconstruction of multiple tissue interfaces (335). Microengineering techniques for these devices have been extensively reviewed (335, 338, 344–349). Chips are commonly made of a silicone polymer called polydimethylsiloxane (PDMS), which is compatible with many cell types and has several advantages, including optical transparency for easy imaging, low cost, flexibility, and high gas permeability (335, 339, 344, 350). PDMS does carry some disadvantages (discussed below), so other options are being explored (350, 351). Depending on experimental requirements, chip design and approaches for tissue development can be altered. Porous membranes can be coated with a variety of matrices/scaffolds (335, 339, 344, 345, 352). Moreover, 3-D bioprinting techniques are facilitating complex spatial patterning of cells and scaffolds (352). Although traditional electrodes used for TEER measurements do not accommodate the small culture area of most OAC models (220), recent studies have integrated custom electrodes (353).
A variety of OAC platforms have been derived from cell lines, stem cells, and/or primary cells, including small and large intestine (334, 340–342, 353–356), lung (357–361), liver (362–369), kidney (370–372), heart (373–377), cornea (378), skin (379), nervous tissue (380–383), bone (384, 385), reproductive tract (386), and blood/endothelium and blood-brain barrier (387–393), among others. Once developed, these models typically retain their structural and functional integrity for several weeks (model specific), further lending to their experimental tractability. Similar to the other 3-D models discussed, OAC models exhibit in vivo-like characteristics, including spontaneous differentiation into multiple cell types, polarity/barrier function, formation of local 3-D structures (Fig. 1B, panel f), responses to biophysical forces, cytokine production, gene expression, mucus production, responses to nanoparticles and drugs, support of commensals, responses to microbial components (e.g., LPS), and/or susceptibility to microbial infections (334, 335, 339–342, 354–377, 379–394). The application of physical forces across several of these models alters physiological responses, including changes in expression/localization of tight junctions, barrier integrity/function, polarity and differentiation, cell viability, size, morphology, ECM production, integrin expression, enzyme activity, cytokine responses, chemical/gas exchange gradients, molecular transport, drug responses, bacterial colonization, virion-related cytopathic effects, and/or formation of 3-D structures (e.g., villi) (334, 340–343, 345, 358, 359, 361, 371, 372, 376, 377, 384, 387, 388, 395). Importantly, several models have been advanced to incorporate immune cells (342, 359, 396). Below we discuss examples of gut-on-a-chip models that have been applied to study pathogens or commensals.
Gut-on-a-chip models.
The Ingber laboratory developed a series of “mechanically active” gut-on-a-chip models and applied them to study host-microbe interactions (340, 342, 343). They initially constructed a PDMS chip containing two microfluidic channels separated by a flexible, porous ECM-coated membrane (340). Colonic cells were seeded in the upper channel under low-fluid-shear stress (0.006 to 0.06 dyne/cm2), and medium also flowed in the bottom chamber. The chip was engineered with dual vacuum chambers on either side of the main microchamber to enable application of a physiological cyclic strain across the membrane to mimic intestinal peristalsis. This led to a highly polarized columnar epithelium and spontaneous formation of 3-D villus-like folds with basal proliferative cells in the crypt region. Model characterization revealed well-formed tight junctions, mucus production, and multiple intestinal epithelial cell types (absorptive, goblet, enteroendocrine, and Paneth cells) (340, 341). The ability of this model to support commensal colonization was assessed using Lactobacillus rhamnosus LGG. Colonization of LGG improved barrier function and was supported for greater than a week without impacting model integrity, consistent with previous in vivo observations for probiotics. The model was also applied to study host-virus interactions using CVB (Fig. 1B, panel i) (343). Exposure of CVB to the apical surface led to successful viral replication, induction of cytopathic effects (CPE), and polarized (apical) release of proinflammatory cytokines. Infection of the basal side led to decreased viral titers and lower CPE, with apical secretion of proinflammatory cytokines.
The above-described gut-on-a-chip model was further advanced to include immune cells (peripheral blood mononuclear cells [PBMCs]) and/or endothelial cells (vascular or lymphatic) (342). This combination of models enabled exploration of the interplay between these factors (and others) in bacterial overgrowth and inflammation in the onset of intestinal injury. Synergistic effects between PBMCs and either nonpathogenic E. coli, pathogenic enteroinvasive E. coli (EIEC), or purified lipopolysaccharide (LPS) led to altered barrier function and changes in villus architecture. Similarly, the presence of both PBMCs and LPS led to polarized secretion of basal proinflammatory cytokines, which stimulates recruitment of additional immune cells in an in vivo scenario. Exposure of the PBMC-containing model to a therapeutic formulation of probiotic bacteria increased barrier function. The formulation reduced EIEC-induced intestinal damage in the model lacking PBMCs but in the presence of immune cells only delayed injury onset. Cessation of cyclic stretching led to enhanced bacterial overgrowth, even under constant medium flow.
Limitations and future directions of OAC models.
While there are many advantages to OAC models, there are limitations. Many of these models have multiple cell types which exhibit enhanced 3-D architecture; however, the vast array of native heterogeneous cell types found in vivo still need to be incorporated and different laboratories are optimizing ECM composition and structure. Along these lines, to our knowledge, no one has yet reported the presence of M cells in gut-on-a-chip models. There is also a strong push for physically linked multiorgan models, or “humans-on-chips” (338, 397). Another limitation is the PDMS material commonly used for chip construction, which can absorb small hydrophobic molecules and interfere with drug screening and cell signaling analysis (338, 350, 351). There are also risks of uncross-linked PDMS leaching into the culture if the curing process is incomplete, causing cell damage (350, 351). While the small number of cells required can be considered advantageous, in some cases, larger numbers of cells (106, 107) may be required depending on the experiment. Infection studies typically involve many permutations, and it is not uncommon to use several multiwell plates within a single experiment. For example, during colonization assays, samples are harvested at different times and plated for viable bacteria, while others are processed for downstream analyses. Thus, it will be beneficial to incorporate multiple 3-D model systems into infectious disease research depending on the experimental question being addressed, as no single model system is sufficient to address all infectious disease experimental scenarios.
CONCLUSIONS
Over the past 2 decades, a multidisciplinary consortium of researchers has been creative in developing 3-D intestinal models of increasing complexity that better mimic the biological, chemical, and physical microenvironments of the endogenous tissue for studying host-microbe interactions. These models have been developed using a variety of approaches and are being applied to understand the dynamic relationship between the host, pathogens, and commensals that dictate infection outcome and for development of new treatment/prevention strategies. Collectively, these models have ushered in a new era for infectious disease research by offering predictive in vitro translational platforms. Indeed, the establishment of 3-D intestinal models and their application as human surrogates for infectious disease research have provided specific examples of how the study of microbial pathogenesis can be advanced by using appropriate, biologically meaningful models.
We are still in the infancy of learning how to build more realistic 3-D tissue models, and there remain an endless number of questions and hypotheses to test about how infection actually happens in the body. Continued model advancement to better recapitulate the in vivo tissue microenvironment coupled with the application of multiple 3-D model systems will lead to increased translation of research discoveries to practical and significant outcomes. Such advances will be pivotal for the success of personalized medicine approaches using patient-specific normal and diseased cells and of incorporation of the full repertoire of immune cells to predict clinical correlates of protection for vaccine development.
Toward this goal, we must deeply comprehend 3-D tissue/organ structure and function, the associated microenvironment, and the microorganisms to be studied. It is likewise important that we are aware of and acknowledge the rich history and work of researchers who have long applied 3-D tissue modeling to study host-pathogen interactions. Accordingly, we should revisit past research in the field to help us understand and guide our direction. While it remains a daunting task to gain a complete understanding of infectious disease, the alignment of multidisciplinary research teams dedicated to the establishment of 3-D models that reconstruct the architecture and function of the in vivo organ and their application for host-pathogen interaction studies make this an exciting time to be a scientist!
ACKNOWLEDGMENTS
We thank Michael Northrop for his illustration in Fig. 1. We apologize to authors whose work we were unable to cite due to length limitations given the extensive literature available for each model system.
This work was funded by NASA grants NNX13AM01G and NNX15AL06G (C.A.N., J.B., and C.M.O.) and NIH R01-AI081759 (C.B.C. and C.A.N.).
Biographies

Jennifer Barrila is an Assistant Research Professor at the Biodesign Institute, Arizona State University (ASU). She received her B.S. in biochemistry in 2002 from Syracuse University. In 2008 she received her Ph.D. in biology from Johns Hopkins University, where she structurally characterized the severe acute respiratory syndrome (SARS) coronavirus main protease to facilitate structure-based drug design. Her postdoctoral work at the Biodesign Institute used innovative culture systems to investigate how biomechanical forces regulate host and pathogen responses during infection. In 2010 she was promoted to Assistant Research Scientist and in 2013 to Assistant Research Professor. Her current research involves the development and application of multicellular 3-D models of human intestine to investigate the role of cellular biomechanics in host-pathogen-microbiota interactions. Her biomedical research has flown on several NASA spaceflight missions to the International Space Station to study the influence of biophysical forces on infection. In 2014 she received the Thora W. Halstead Young Investigator's Award.

Aurélie Crabbé obtained her Ph.D. in Bioscience Engineering at the Vrije Universiteit Brussel, Belgium. During her Ph.D. studies she received a fellowship of the Belgian American Educational Foundation to perform research in the laboratory of Prof. Cheryl Nickerson at the Biodesign Institute at Arizona State University. She then received a postdoctoral position in the Nickerson lab, where she developed and used physiologically relevant models of the lung mucosa to explore host-pathogen interactions. She is currently a team leader in the Laboratory of Pharmaceutical Microbiology at Ghent University under the tutelage of Prof. Tom Coenye, through an Odysseus fellowship of the Research Foundation Flanders. Her research focuses on understanding how the host, microbiome, and their interactions influence antimicrobial agent efficacy and inflammation in chronic lung infections. To this end, in vivo-like models of lung epithelium, the microbiome, or both are used to facilitate translation of in vitro results to novel therapeutic approaches.

Jiseon Yang is an Alfred P. Sloan postdoctoral fellow (NASA joint program) at the Biodesign Institute, ASU. She received her M.S. in microbiology studying Salmonella pathogenesis from Pusan National University (South Korea). She subsequently worked with Dr. Roy Curtiss III and Dr. Josephine Clark-Curtiss (ASU) on developing a 2nd-generation lysis system in recombinant attenuated Salmonella vaccines to deliver Mycobacterium tuberculosis antigens. She received her Ph.D. in microbiology under Dr. Cheryl Nickerson (ASU), in whose laboratory she characterized virulence, stress, and molecular genetic responses of invasive, multidrug-resistant nontyphoidal Salmonella to physiological fluid shear. She was co-first author on the first report of an RWV-derived 3-D intestinal coculture model (epithelial cells and macrophages) to study Salmonella pathogenesis. She currently studies how microbes inhabiting built environments influence human health and systems integrity using the International Space Station as a microbial observatory to reveal new insight into how interspecies interactions within microbial populations can adapt/evolve over time.

Karla Franco received her bachelor's degree in general science from Pontificia Universidad Catolica de Puerto Rico. She became interested in microbial pathogenesis during a year-long NIH training fellowship (ASU PREP). She enrolled in the microbiology Ph.D. program at Arizona State University, where she received a 2-year fellowship from the NIH IMSD program. As a member of the Nickerson laboratory, she has spent the last 3 years investigating the role of mechanotransduction in regulating the phenotypic and molecular genetic responses of Salmonella Typhimurium.

Seth D. Nydam received his doctorate in veterinary medicine from Cornell University and his Ph.D. from Washington State University. His graduate studies in Dr. Douglas Call's laboratory centered on the type 3 secretion system of Vibrio parahaemolyticus, after which he joined Dr. Cheryl Nickerson at Arizona State University for postdoctoral training to explore 3-D cell culture and its interactions with resident microflora and Salmonella pathogenesis. He is currently the clinical veterinarian at Arizona State University's Department of Animal Care and Technologies, where he leads the clinical team and provides research support and oversight. His continuing interests include microbial pathogenesis, animal models in infectious disease research, and teaching.

Rebecca J. Forsyth received her B.S. in microbiology from Arizona State University in 2008. She first joined the Nickerson laboratory as an undergraduate student, was subsequently hired as an Assistant Research Technologist, and was later promoted to the positions of Associate Research Specialist and Senior Research Specialist. She used a variety of 3-D models, microbes, and model host organisms in her infectious disease research. She was passionate about using “outside-of-the-box” approaches to solve important biomedical health issues, including the use of the spaceflight platform and the RWV bioreactor to study microbial physiology and host-pathogen interactions.

Richard R. Davis is a Senior Research Specialist in the Biodesign Center for Immunotherapy, Vaccines, and Virotherapy at Arizona State University. He earned his B.A. in anthropology in 2001 and a B.S. in microbiology in 2007 from Arizona State University. He joined the Nickerson laboratory as an undergraduate student, was subsequently hired as an Assistant Research Technician, and was later promoted to the positions of Research Specialist and Senior Research Specialist. His research over the past 11 years has focused on using both the microgravity platform (including six spaceflight experiments) and the RWV bioreactor to study the effect of physical forces on microbial pathogenesis and host-pathogen interactions.

Sandhya Gangaraju received her master's degree in biochemistry in 2003 from the University of Ottawa, Canada. In 2003, she joined the National Research Council, Canada, as a research technical officer and viral facility manager in the department of neurogenesis and brain repair. She implemented lentiviral technology to deliver neurotropic factors to neural cells and developed Transwell assays to study neutrophil transmigration through endothelial cells. In 2014, she joined the Biodesign Institute, Arizona State University, as a principal research specialist and compliance officer for the Center for Biosignatures Discovery Automation, where she managed the cell culture facility, led experimental design for students and junior staff members, and optimized working protocols for microfluidic devices for studying three-dimensional tissue environments. Most recently, she joined the Nickerson laboratory as a principal research specialist; at this laboratory, she uses the RWV and the spaceflight platform to study microbial pathogenesis and for 3-D tissue engineering.

C. Mark Ott received his B.S. in chemical engineering from the University of Texas at Austin in 1982, his M.B.A. from Louisiana State University in 1989, and his Ph.D. in microbiology from Louisiana State University in 1998. He has published extensively in the areas of microbial ecology in spacecraft, human and microbial responses to spaceflight, and the development of advanced tissue culture models to investigate infectious disease. For the past 20 years, Dr. Ott has served as a technical lead in the Johnson Space Center Microbiology Laboratory, which is responsible for mitigating infectious disease risk during human spaceflight. His responsibilities include the assessment of microbial risk and development of spaceflight requirements based on vehicle and mission architecture as well as crewmember, food, and environmental monitoring.

Carolyn B. Coyne completed her Ph.D. at the University of North Carolina at Chapel Hill, where she studied the human respiratory epithelium. She then carried out her postdoctoral fellowship at the Children's Hospital of Philadelphia (CHOP), Philadelphia, PA, and the University of Pennsylvania, Philadelphia, PA, where her research focused on identifying the mechanisms by which enteroviruses invade the gastrointestinal epithelium and blood-brain barrier endothelium. She joined the University of Pittsburgh, Pittsburgh, PA, as a faculty member in 2007; at this institution, her work continued to focus on defining the mechanisms by which viruses breach cellular barriers. Dr. Coyne's laboratory also studies how the human placenta restricts viral infections. Her research interests also include the development of primary and cell line-based models of cellular barriers, focusing on both the gastrointestinal tract and placenta. Her research interests include enteroviruses and flaviviruses, with a particular emphasis on the strategies by which these viruses bypass cellular barriers.

Mina J. Bissell is a Distinguished Scientist, the highest rank bestowed at Lawrence Berkeley National Laboratory, and serves as Senior Advisor to the Laboratory Director on Biology. She is also Faculty of four graduate groups in UC Berkeley: Comparative Biochemistry, Endocrinology, Molecular Toxicology, and Bioengineering (UCSF/UCB joint). Having challenged several established paradigms, Bissell is a pioneer in breast cancer research, and her body of work has provided much impetus for the current recognition of the significant role that extracellular matrix signaling and the microenvironment play in gene expression regulation in both normal and malignant cells. Her laboratory developed novel 3-D assays and techniques that demonstrate her signature phrase: after conception, “phenotype is dominant over genotype.” Bissell has received numerous honors and awards and is an elected fellow of most U.S. honorary scientific academies. She has published over 400 publications and continues to engage in full-time research, among other scientific activities.

Cheryl A. Nickerson is a Professor in the School of Life Sciences at the Biodesign Institute, Arizona State University. She received her Ph.D. in microbiology from Louisiana State University. Her postdoctoral training in Salmonella pathogenesis was done with Dr. Roy Curtiss III at Washington University in St. Louis, MO. She initiated her ongoing studies into the connection between cellular biomechanics/mechanotransduction and host-pathogen systems biology after joining the faculty at the Tulane University School of Medicine in 1998. Her development of innovative model pathogenesis systems includes 3-D organotypic tissue culture models to study host-pathogen interactions and approaches that characterize pathogen responses to physiological fluid shear forces encountered in the infected host and in the microgravity environment of spaceflight. She received the Presidential Early Career Award for Scientists and Engineers, NASA's Exceptional Scientific Achievement Medal, is an American Society for Microbiology Distinguished Lecturer, and was selected as a NASA astronaut candidate finalist.
REFERENCES
- 1.Nickerson C, Ott CM, Wilson JW, Pierson DL. 2004. Microbial responses to microgravity and other low shear environment. Microbiol Mol Biol Rev 68:345–361. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persat A. 2017. Bacterial mechanotransduction. Curr Opin Microbiol 36:1–6. doi: 10.1016/j.mib.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, Wingreen NS, Bassler BL, Gitai Z, Stone HA. 2015. The mechanical world of bacteria. Cell 161:988–997. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109:913–923. doi: 10.1016/S0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 5.Alsharif G, Ahmad S, Islam MS, Shah R, Busby SJ, Krachler AM. 2015. Host attachment and fluid shear are integrated into a mechanical signal regulating virulence in Escherichia coli O157:H7. Proc Natl Acad Sci U S A 112:5503–5508. doi: 10.1073/pnas.1422986112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang FC, Frawley ER, Tapscott T, Vazquez-Torres A. 2016. Bacterial stress responses during host infection. Cell Host Microbe 20:133–143. doi: 10.1016/j.chom.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuckin MA, Linden SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 8.Barrila J, Radtke AL, Crabbe A, Sarker SF, Herbst-Kralovetz MM, Ott CM, Nickerson CA. 2010. Organotypic 3D cell culture models: using the rotating wall vessel to study host-pathogen interactions. Nat Rev Microbiol 8:791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- 9.Ibarra JA, Steele-Mortimer O. 2009. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol 11:1579–1586. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costello CM, Sorna RM, Goh YL, Cengic I, Jain NK, March JC. 2014. 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol Pharm 11:2030–2039. doi: 10.1021/mp5001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nickerson CA, Goodwin TJ, Terlonge J, Ott CM, Buchanan KL, Uicker WC, Emami K, LeBlanc CL, Ramamurthy R, Clarke MS, Vanderburg CR, Hammond T, Pierson DL. 2001. Three-dimensional tissue assemblies: novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun 69:7106–7120. doi: 10.1128/IAI.69.11.7106-7120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simian M, Bissell MJ. 2017. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol 216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bissell MJ. 1981. The differentiated state of normal and malignant cells or how to define a “normal” cell in culture. Int Rev Cytol 70:27–100. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz RI, Bissell MJ. 1977. Dependence of the differentiated state on the cellular environment: modulation of collagen synthesis in tendon cells. Proc Natl Acad Sci U S A 74:4453–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith LG, Swartz MA. 2006. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol 7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 16.Yamada KM, Cukierman E. 2007. Modeling tissue morphogenesis and cancer in 3D. Cell 130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Bissell MJ, Aggeler J. 1987. Dynamic reciprocity: how do extracellular matrix and hormones direct gene expression? Prog Clin Biol Res 249:251–262. [PubMed] [Google Scholar]
- 18.Jorgens DM, Inman JL, Wojcik M, Robertson C, Palsdottir H, Tsai WT, Huang H, Bruni-Cardoso A, Lopez CS, Bissell MJ, Xu K, Auer M. 2017. Deep nuclear invaginations are linked to cytoskeletal filaments—integrated bioimaging of epithelial cells in 3D culture. J Cell Sci 130:177–189. doi: 10.1242/jcs.190967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maniotis AJ, Chen CS, Ingber DE. 1997. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A 94:849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissell MJ, Hall HG, Parry G. 1982. How does the extracellular matrix direct gene expression? J Theor Biol 99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 21.Boudreau N, Myers C, Bissell MJ. 1995. From laminin to lamin: regulation of tissue-specific gene expression by the ECM. Trends Cell Biol 5:1–4. doi: 10.1016/S0962-8924(00)88924-2. [DOI] [PubMed] [Google Scholar]
- 22.Tapley EC, Starr DA. 2013. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr Opin Cell Biol 25:57–62. doi: 10.1016/j.ceb.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu R, Spencer VA, Groesser DL, Bissell MJ. 2010. Laminin regulates PI3K basal localization and activation to sustain STAT5 activation. Cell Cycle 9:4315–4322. doi: 10.4161/cc.9.21.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DH, Wirtz D. 2015. Cytoskeletal tension induces the polarized architecture of the nucleus. Biomaterials 48:161–172. doi: 10.1016/j.biomaterials.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagios C, Lochter A, Bissell MJ. 1998. Tissue architecture: the ultimate regulator of epithelial function? Philos Trans R Soc Lond B Biol Sci 353:857–870. doi: 10.1098/rstb.1998.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle AD, Yamada KM. 2016. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp Cell Res 343:60–66. doi: 10.1016/j.yexcr.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingber DE. 2003. Tensegrity I. Cell structure and hierarchical systems biology. J Cell Sci 116:1157–1173. doi: 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- 28.Ingber DE. 2003. Mechanobiology and diseases of mechanotransduction. Ann Med 35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 29.Ingber DE. 2003. Tensegrity II. How structural networks influence cellular information processing networks. J Cell Sci 116:1397–1408. doi: 10.1242/jcs.00360. [DOI] [PubMed] [Google Scholar]
- 30.Ingber DE. 2008. Tensegrity-based mechanosensing from macro to micro. Prog Biophys Mol Biol 97:163–179. doi: 10.1016/j.pbiomolbio.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingber DE. 2006. Cellular mechanotransduction: putting all the pieces together again. FASEB J 20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 32.Gruenheid S, Finlay BB. 2003. Microbial pathogenesis and cytoskeletal function. Nature 422:775–781. doi: 10.1038/nature01603. [DOI] [PubMed] [Google Scholar]
- 33.Taylor MP, Koyuncu OO, Enquist LW. 2011. Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol 9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delorme-Axford E, Coyne CB. 2011. The actin cytoskeleton as a barrier to virus infection of polarized epithelial cells. Viruses 3:2462. doi: 10.3390/v3122462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickerson CA, Ott CM, Mister SJ, Morrow BJ, Burns-Keliher L, Pierson DL. 2000. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect Immun 68:3147–3152. doi: 10.1128/IAI.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson JW, Ott CM, Ramamurthy R, Porwollik S, McClelland M, Pierson DL, Nickerson CA. 2002. Low-shear modeled microgravity alters the Salmonella enterica serovar Typhimurium stress response in an RpoS-independent manner. Appl Environ Microbiol 68:5408–5416. doi: 10.1128/AEM.68.11.5408-5416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson JW, Ramamurthy R, Porwollik S, McClelland M, Hammond T, Allen P, Ott CM, Pierson DL, Nickerson CA. 2002. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc Natl Acad Sci U S A 99:13807–13812. doi: 10.1073/pnas.212387899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nauman EA, Ott CM, Sander E, Tucker DL, Pierson D, Wilson JW, Nickerson CA. 2007. Novel quantitative biosystem for modeling physiological fluid shear stress on cells. Appl Environ Microbiol 73:699–705. doi: 10.1128/AEM.02428-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crabbé A, De Boever P, Van Houdt R, Moors H, Mergeay M, Cornelis P. 2008. Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01. Environ Microbiol 10:2098–2110. doi: 10.1111/j.1462-2920.2008.01631.x. [DOI] [PubMed] [Google Scholar]
- 40.Crabbé A, Pycke B, Van Houdt R, Monsieurs P, Nickerson C, Leys N, Cornelis P. 2010. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ Microbiol 12:1545–1564. doi: 10.1111/j.1462-2920.2010.02184.x. [DOI] [PubMed] [Google Scholar]
- 41.Pacello F, Rotilio G, Battistoni A. 2012. Low-shear modeled microgravity enhances Salmonella enterica resistance to hydrogen peroxide through a mechanism involving KatG and KatN. Open Microbiol J 6:53–64. doi: 10.2174/1874285801206010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aprikian P, Interlandi G, Kidd BA, Le Trong I, Tchesnokova V, Yakovenko O, Whitfield MJ, Bullitt E, Stenkamp RE, Thomas WE, Sokurenko EV. 2011. The bacterial fimbrial tip acts as a mechanical force sensor. PLoS Biol 9:e1000617. doi: 10.1371/journal.pbio.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isberg RR, Barnes P. 2002. Dancing with the host; flow-dependent bacterial adhesion. Cell 110:1–4. doi: 10.1016/S0092-8674(02)00821-8. [DOI] [PubMed] [Google Scholar]
- 44.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Barrila J, Roland KL, Ott CM, Nickerson CA. 2016. Physiological fluid shear alters the virulence potential of invasive multidrug-resistant non-typhoidal Salmonella Typhimurium D23580. NPJ Microgravity 2:16021. doi: 10.1038/npjmgrav.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dingemans J, Monsieurs P, Yu SH, Crabbe A, Forstner KU, Malfroot A, Cornelis P, Van Houdt R. 2016. Effect of shear stress on Pseudomonas aeruginosa isolated from the cystic fibrosis lung. mBio 7:e00813-. doi: 10.1128/mBio.00813-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castro SL, Nelman-Gonzalez M, Nickerson CA, Ott CM. 2011. Induction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl Environ Microbiol 77:6368–6378. doi: 10.1128/AEM.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarbell JM, Weinbaum S, Kamm RD. 2005. Cellular fluid mechanics and mechanotransduction. Ann Biomed Eng 33:1719–1723. doi: 10.1007/s10439-005-8775-z. [DOI] [PubMed] [Google Scholar]
- 49.Kaul H, Ventikos Y. 2015. Dynamic reciprocity revisited. J Theor Biol 370:205–208. doi: 10.1016/j.jtbi.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 50.Rutkowski JM, Swartz MA. 2007. A driving force for change: interstitial flow as a morphoregulator. Trends Cell Biol 17:44–50. doi: 10.1016/j.tcb.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Long JP, Pierson SS, Hughes JH. 1998. Rhinovirus replication in HeLa cells cultured under conditions of simulated microgravity. Aviat Space Environ Med 69:851–856. [PubMed] [Google Scholar]
- 52.Bissell MJ, Rizki A, Mian IS. 2003. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol 15:753–762. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Correia AL, Bissell MJ. 2012. The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist Updat 15:39–49. doi: 10.1016/j.drup.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson CM, Bissell MJ. 2006. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol 22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmeichel KL, Bissell MJ. 2003. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci 116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bals R, Hiemstra PS. 2004. Innate immunity in the lung: how epithelial cells fight against respiratory pathogens. Eur Respir J 23:327–333. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- 57.Crabbé A, Ledesma MA, Nickerson CA. 2014. Mimicking the host and its microenvironment in vitro for studying mucosal infections by Pseudomonas aeruginosa. Pathog Dis 71:1–19. doi: 10.1111/2049-632X.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duerkop BA, Vaishnava S, Hooper LV. 2009. Immune responses to the microbiota at the intestinal mucosal surface. Immunity 31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Kraehenbuhl J-P, Corbett M. 2004. Keeping the gut microflora at bay. Science 303:1624–1625. doi: 10.1126/science.1096222. [DOI] [PubMed] [Google Scholar]
- 60.Lopetuso LR, Scaldaferri F, Franceschi F, Gasbarrini A. 2014. The gastrointestinal microbiome—functional interference between stomach and intestine. Best Pract Res Clin Gastroenterol 28:995–1002. doi: 10.1016/j.bpg.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Singh B, Fleury C, Jalalvand F, Riesbeck K. 2012. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev 36:1122–1180. doi: 10.1111/j.1574-6976.2012.00340.x. [DOI] [PubMed] [Google Scholar]
- 62.Allen CA, Niesel DW, Torres AG. 2008. The effects of low-shear stress on adherent-invasive Escherichia coli. Environ Microbiol 10:1512–1525. doi: 10.1111/j.1462-2920.2008.01567.x. [DOI] [PubMed] [Google Scholar]
- 63.Fonseca AP, Sousa JC. 2007. Effect of shear stress on growth, adhesion and biofilm formation of Pseudomonas aeruginosa with antibiotic-induced morphological changes. Int J Antimicrob Agents 30:236–241. doi: 10.1016/j.ijantimicag.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 64.Lynch SV, Mukundakrishnan K, Benoit MR, Ayyaswamy PS, Matin A. 2006. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl Environ Microbiol 72:7701–7710. doi: 10.1128/AEM.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mestecky J, Strober W, Russell MW, Kelsall BL, Cheroutre H, Lambrecht BN. 2015. Mucosal immunology. Elsevier, San Diego, CA. [Google Scholar]
- 66.Gerbe F, Legraverend C, Jay P. 2012. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci 69:2907–2917. doi: 10.1007/s00018-012-0984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 68.Mowat AM, Agace WW. 2014. Regional specialization within the intestinal immune system. Nat Rev Immunol 14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 69.Cerutti A. 2008. The regulation of IgA class switching. Nat Rev Immunol 8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cerutti A, Rescigno M. 2008. The biology of intestinal immunoglobulin A responses. Immunity 28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holmgren J, Czerkinsky C. 2005. Mucosal immunity and vaccines. Nat Med 11:S45–S53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 72.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. 2008. The immune geography of IgA induction and function. Mucosal Immunol 1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 73.Engel J, Eran Y. 2011. Subversion of mucosal barrier polarity by Pseudomonas aeruginosa. Front Microbiol 2:114. doi: 10.3389/fmicb.2011.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zihni C, Balda MS, Matter K. 2014. Signalling at tight junctions during epithelial differentiation and microbial pathogenesis. J Cell Sci 127:3401–3413. doi: 10.1242/jcs.145029. [DOI] [PubMed] [Google Scholar]
- 75.Schneeberger K, Roth S, Nieuwenhuis EES, Middendorp S. 2018. Intestinal epithelial cell polarity defects in disease: lessons from microvillus inclusion disease. Dis Model Mech 11:dmm031088. doi: 10.1242/dmm.031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jung C, Hugot JP, Barreau F. 2010. Peyer's patches: the immune sensors of the intestine. Int J Inflam 2010:823710. doi: 10.4061/2010/823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen CJ, Shieh JTC, Pickles RJ, Okegawa T, Hsieh J-T, Bergelson JM. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci U S A 98:15191–15196. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451. doi: 10.1016/S0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 79.Bonazzi M, Cossart P. 2011. Impenetrable barriers or entry portals? The role of cell-cell adhesion during infection. J Cell Biol 195:349–358. doi: 10.1083/jcb.201106011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Höner zu Bentrup K, Ramamurthy R, Ott CM, Emami K, Nelman-Gonzalez M, Wilson JW, Richter EG, Goodwin TJ, Alexander JS, Pierson DL, Pellis N, Buchanan KL, Nickerson CA. 2006. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes Infect 8:1813–1825. doi: 10.1016/j.micinf.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 81.Fleiszig SM, Evans DJ, Do N, Vallas V, Shin S, Mostov KE. 1997. Epithelial cell polarity affects susceptibility to Pseudomonas aeruginosa invasion and cytotoxicity. Infect Immun 65:2861–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hurley BP, McCormick BA. 2003. Translating tissue culture results into animal models: the case of Salmonella typhimurium. Trends Microbiol 11:562–569. doi: 10.1016/j.tim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 83.Law RJ, Gur-Arie L, Rosenshine I, Finlay BB. 2013. In vitro and in vivo model systems for studying enteropathogenic Escherichia coli infections. Cold Spring Harb Perspect Med 3:a009977. doi: 10.1101/cshperspect.a009977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ashida H, Ogawa M, Kim M, Mimuro H, Sasakawa C. 2011. Bacteria and host interactions in the gut epithelial barrier. Nat Chem Biol 8:36–45. doi: 10.1038/nchembio.741. [DOI] [PubMed] [Google Scholar]
- 85.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krishnan S, Alden N, Lee K. 2015. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr Opin Biotechnol 36:137–145. doi: 10.1016/j.copbio.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chu H, Mazmanian SK. 2013. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 14:668–675. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maynard CL, Elson CO, Hatton RD, Weaver CT. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, MetaHIT Consortium, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lavelle A, Lennon G, O'Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O'Donoghue D, Hyland J, Ross RP, Coffey JC, Sheahan K, Cotter PD, Shanahan F, Winter DC, O'Connell PR. 2015. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 64:1553–1561. doi: 10.1136/gutjnl-2014-307873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A. 2016. The gut microbiota and host health: a new clinical frontier. Gut 65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lynch SV, Pedersen O. 2016. The human intestinal microbiome in health and disease. N Engl J Med 375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 99.Kassam Z, Lee CH, Yuan Y, Hunt RH. 2013. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol 108:500–508. doi: 10.1038/ajg.2013.59. [DOI] [PubMed] [Google Scholar]
- 100.Gianotti RJ, Moss AC. 2017. Fecal microbiota transplantation: from Clostridium difficile to inflammatory bowel disease. Gastroenterol Hepatol (N Y) 13:209–213. [PMC free article] [PubMed] [Google Scholar]
- 101.Parker D, Prince A. 2011. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol 45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johansson ME, Ambort D, Pelaseyed T, Schutte A, Gustafsson JK, Ermund A, Subramani DB, Holmen-Larsson JM, Thomsson KA, Bergstrom JH, van der Post S, Rodriguez-Pineiro AM, Sjovall H, Backstrom M, Hansson GC. 2011. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci 68:3635–3641. doi: 10.1007/s00018-011-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Johansson ME, Larsson JM, Hansson GC. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 108(Suppl 1):S4659–S4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, Stolp B, Stein JV, Stecher B, Sauer U, McCoy KD, Macpherson AJ. 2015. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun 6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rogier EW, Frantz AL, Bruno MEC, Kaetzel CS. 2014. Secretory IgA is concentrated in the outer layer of colonic mucus along with gut bacteria. Pathogens 3:390–403. doi: 10.3390/pathogens3020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Frantz C, Stewart KM, Weaver VM. 2010. The extracellular matrix at a glance. J Cell Sci 123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bonnans C, Chou J, Werb Z. 2014. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ray SJ, Franki SN, Pierce RH, Dimitrova S, Koteliansky V, Sprague AG, Doherty PC, de Fougerolles AR, Topham DJ. 2004. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity 20:167–179. doi: 10.1016/S1074-7613(04)00021-4. [DOI] [PubMed] [Google Scholar]
- 109.Mammoto T, Mammoto A, Ingber DE. 2013. Mechanobiology and developmental control. Annu Rev Cell Dev Biol 29:27–61. doi: 10.1146/annurev-cellbio-101512-122340. [DOI] [PubMed] [Google Scholar]
- 110.Button B, Boucher RC, University of North Carolina Virtual Lung Group. 2008. Role of mechanical stress in regulating airway surface hydration and mucus clearance rates. Respir Physiol Neurobiol 163:189–201. doi: 10.1016/j.resp.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gayer CP, Basson MD. 2009. The effects of mechanical forces on intestinal physiology and pathology. Cell Signal 21:1237–1244. doi: 10.1016/j.cellsig.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scott SM, Knowles CH, Wang D, Yazaki E, Picon L, Wingate DL, Lindberg G. 2006. The nocturnal jejunal migrating motor complex: defining normal ranges by study of 51 healthy adult volunteers and meta-analysis. Neurogastroenterol Motil 18:927–935. doi: 10.1111/j.1365-2982.2006.00824.x. [DOI] [PubMed] [Google Scholar]
- 113.Otterson MF, Sarr MG. 1993. Normal physiology of small intestinal motility. Surg Clin North Am 73:1173–1192. doi: 10.1016/S0039-6109(16)46186-4. [DOI] [PubMed] [Google Scholar]
- 114.Guo P, Weinstein AM, Weinbaum S. 2000. A hydrodynamic mechanosensory hypothesis for brush border microvilli. Am J Physiol Renal Physiol 279:F698–F712. doi: 10.1152/ajprenal.2000.279.4.F698. [DOI] [PubMed] [Google Scholar]
- 115.Ashida N, Takechi H, Kita T, Arai H. 2003. Vortex-mediated mechanical stress induces integrin-dependent cell adhesion mediated by inositol 1,4,5-trisphosphate-sensitive Ca2+ release in THP-1 cells. J Biol Chem 278:9327–9331. doi: 10.1074/jbc.M212316200. [DOI] [PubMed] [Google Scholar]
- 116.Kim M, Javed NH, Yu JG, Christofi F, Cooke HJ. 2001. Mechanical stimulation activates Galphaq signaling pathways and 5-hydroxytryptamine release from human carcinoid BON cells. J Clin Invest 108:1051–1059. doi: 10.1172/JCI12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Roskelley CD, Bissell MJ. 1995. Dynamic reciprocity revisited: a continuous, bidirectional flow of information between cells and the extracellular matrix regulates mammary epithelial cell function. Biochem Cell Biol 73:391–397. doi: 10.1139/o95-046. [DOI] [PubMed] [Google Scholar]
- 118.Zhang J, Owen CR, Sanders MA, Turner JR, Basson MD. 2006. The motogenic effects of cyclic mechanical strain on intestinal epithelial monolayer wound closure are matrix dependent. Gastroenterology 131:1179–1189. doi: 10.1053/j.gastro.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 119.Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Baumler AJ. 2003. Molecular pathogenesis of Salmonella enterica serotype typhimurium-induced diarrhea. Infect Immun 71:1–12. doi: 10.1128/IAI.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu P, Takai K, Weaver VM, Werb Z. 2011. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 3:a005058. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Abbott A. 2003. Cell culture: biology's new dimension. Nature 424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 122.de Bentzmann S, Plotkowski C, Puchelle E. 1996. Receptors in the Pseudomonas aeruginosa adherence to injured and repairing airway epithelium. Am J Respir Crit Care Med 154:S155–S162. doi: 10.1164/ajrccm/154.4_Pt_2.S155. [DOI] [PubMed] [Google Scholar]
- 123.de Bentzmann S, Roger P, Dupuit F, Bajolet-Laudinat O, Fuchey C, Plotkowski MC, Puchelle E. 1996. Asialo GM1 is a receptor for Pseudomonas aeruginosa adherence to regenerating respiratory epithelial cells. Infect Immun 64:1582–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ljungh A, Moran AP, Wadstrom T. 1996. Interactions of bacterial adhesins with extracellular matrix and plasma proteins: pathogenic implications and therapeutic possibilities. FEMS Immunol Med Microbiol 16:117–126. doi: 10.1111/j.1574-695X.1996.tb00128.x. [DOI] [PubMed] [Google Scholar]
- 125.Ljungh A, Wadstrom T. 1996. Interactions of bacterial adhesins with the extracellular matrix. Adv Exp Med Biol 408:129–140. doi: 10.1007/978-1-4613-0415-9_15. [DOI] [PubMed] [Google Scholar]
- 126.Plotkowski MC, Tournier JM, Puchelle E. 1996. Pseudomonas aeruginosa strains possess specific adhesins for laminin. Infect Immun 64:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wolf DA, Schwarz RP. 1991. Analysis of gravity-induced particle motion and fluid perfusion flow in the NASA-designed rotating zero-head space tissue culture vessel. NASA technical paper 3143; NASA Johnson Space Center, Houston, TX. [Google Scholar]
- 128.Basson MD. 2003. Paradigms for mechanical signal transduction in the intestinal epithelium. Digestion 68:217–225. doi: 10.1159/000076385. [DOI] [PubMed] [Google Scholar]
- 129.Beeson JG, Rogerson SJ, Cooke BM, Reeder JC, Chai W, Lawson AM, Molyneux ME, Brown GV. 2000. Adhesion of Plasmodium falciparum-infected erythrocytes to hyaluronic acid in placental malaria. Nat Med 6:86–90. doi: 10.1038/71582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cai Z, Xin J, Pollock DM, Pollock JS. 2000. Shear stress-mediated NO production in inner medullary collecting duct cells. Am J Physiol Renal Physiol 279:F270–F274. doi: 10.1152/ajprenal.2000.279.2.F270. [DOI] [PubMed] [Google Scholar]
- 131.Creasy RK, Reznik R. 1984. Maternal-fetal medicine: principles and practice. WB Saunders Company, Philadelphia, PA. [Google Scholar]
- 132.Stock UA, Vacanti JP. 2001. Cardiovascular physiology during fetal development and implications for tissue engineering. Tissue Eng 7:1–7. doi: 10.1089/107632701300003241. [DOI] [PubMed] [Google Scholar]
- 133.Vetsch JR, Betts DC, Muller R, Hofmann S. 2017. Flow velocity-driven differentiation of human mesenchymal stromal cells in silk fibroin scaffolds: a combined experimental and computational approach. PLoS One 12:e0180781. doi: 10.1371/journal.pone.0180781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu YS, Lee OK. 2014. In search of the pivot point of mechanotransduction: mechanosensing of stem cells. Cell Transplant 23:1–11. doi: 10.3727/096368912X659925. [DOI] [PubMed] [Google Scholar]
- 135.Geuss LR, Suggs LJ. 2013. Making cardiomyocytes: how mechanical stimulation can influence differentiation of pluripotent stem cells. Biotechnol Prog 29:1089–1096. doi: 10.1002/btpr.1794. [DOI] [PubMed] [Google Scholar]
- 136.Chen G, Lv Y, Guo P, Lin C, Zhang X, Yang L, Xu Z. 2013. Matrix mechanics and fluid shear stress control stem cells fate in three dimensional microenvironment. Curr Stem Cell Res Ther 8:313–323. doi: 10.2174/1574888X11308040007. [DOI] [PubMed] [Google Scholar]
- 137.Adamo L, Garcia-Cardena G. 2011. Directed stem cell differentiation by fluid mechanical forces. Antioxid Redox Signal 15:1463–1473. doi: 10.1089/ars.2011.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ando J, Yamamoto K. 2009. Vascular mechanobiology: endothelial cell responses to fluid shear stress. Circ J 73:1983–1992. doi: 10.1253/circj.CJ-09-0583. [DOI] [PubMed] [Google Scholar]
- 139.Patwari P, Lee RT. 2008. Mechanical control of tissue morphogenesis. Circ Res 103:234–243. doi: 10.1161/CIRCRESAHA.108.175331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Miura S, Sato K, Kato-Negishi M, Teshima T, Takeuchi S. 2015. Fluid shear triggers microvilli formation via mechanosensitive activation of TRPV6. Nat Commun 6:8871. doi: 10.1038/ncomms9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jessup JM, Frantz M, Sonmez-Alpan E, Locker J, Skena K, Waller H, Battle P, Nachman A, Weber ME, Thomas DA, Curbeam RL Jr, Baker TL, Goodwin TJ. 2000. Microgravity culture reduces apoptosis and increases the differentiation of a human colorectal carcinoma cell line. In Vitro Cell Dev Biol Anim 36:367–373. [DOI] [PubMed] [Google Scholar]
- 142.Zhau HE, Goodwin TJ, Chang SM, Baker TL, Chung LW. 1997. Establishment of a three-dimensional human prostate organoid coculture under microgravity-simulated conditions: evaluation of androgen-induced growth and PSA expression. In Vitro Cell Dev Biol Anim 33:375–380. [DOI] [PubMed] [Google Scholar]
- 143.Alcantara Warren C, Destura RV, Sevilleja JE, Barroso LF, Carvalho H, Barrett LJ, O'Brien AD, Guerrant RL. 2008. Detection of epithelial-cell injury, and quantification of infection, in the HCT-8 organoid model of cryptosporidiosis. J Infect Dis 198:143–149. doi: 10.1086/588819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carterson AJ, Honer zu Bentrup K, Ott CM, Clarke MS, Pierson DL, Vanderburg CR, Buchanan KL, Nickerson CA, Schurr MJ. 2005. A549 lung epithelial cells grown as three-dimensional aggregates: alternative tissue culture model for Pseudomonas aeruginosa pathogenesis. Infect Immun 73:1129–1140. doi: 10.1128/IAI.73.2.1129-1140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Carvalho HM, Teel LD, Goping G, O'Brien AD. 2005. A three-dimensional tissue culture model for the study of attach and efface lesion formation by enteropathogenic and enterohaemorrhagic Escherichia coli. Cell Microbiol 7:1771–1781. doi: 10.1111/j.1462-5822.2004.00594.x. [DOI] [PubMed] [Google Scholar]
- 146.Drummond CG, Nickerson CA, Coyne CB. 2016. A three-dimensional cell culture model to study enterovirus infection of polarized intestinal epithelial cells. mSphere 1:e00030-. doi: 10.1128/mSphere.00030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.David J, Sayer NM, Sarkar-Tyson M. 2014. The use of a three-dimensional cell culture model to investigate host-pathogen interactions of Francisella tularensis in human lung epithelial cells. Microbes Infect 16:735–745. doi: 10.1016/j.micinf.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 148.Chang TT, Hughes-Fulford M. 2014. Molecular mechanisms underlying the enhanced functions of three-dimensional hepatocyte aggregates. Biomaterials 35:2162–2171. doi: 10.1016/j.biomaterials.2013.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hjelm BE, Berta AN, Nickerson CA, Arntzen CJ, Herbst-Kralovetz MM. 2010. Development and characterization of a three-dimensional organotypic human vaginal epithelial cell model. Biol Reprod 82:617–627. doi: 10.1095/biolreprod.109.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.LaMarca HL, Ott CM, Honer Zu Bentrup K, Leblanc CL, Pierson DL, Nelson AB, Scandurro AB, Whitley GS, Nickerson CA, Morris CA. 2005. Three-dimensional growth of extravillous cytotrophoblasts promotes differentiation and invasion. Placenta 26:709–720. doi: 10.1016/j.placenta.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 151.McConkey CA, Delorme-Axford E, Nickerson CA, Kim KS, Sadovsky Y, Boyle JP, Coyne CB. 2016. A three-dimensional culture system recapitulates placental syncytiotrophoblast development and microbial resistance. Sci Adv 2:e1501462. doi: 10.1126/sciadv.1501462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Radtke AL, Wilson JW, Sarker S, Nickerson CA. 2010. Analysis of interactions of Salmonella type three secretion mutants with 3-D intestinal epithelial cells. PLoS One 5:e15750. doi: 10.1371/journal.pone.0015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sainz B Jr, TenCate V, Uprichard SL. 2009. Three-dimensional Huh7 cell culture system for the study of hepatitis C virus infection. Virol J 6:103. doi: 10.1186/1743-422X-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Duray PH, Hatfill SJ, Pellis NR. 1997. Tissue culture in microgravity. Sci Med 4:46–55. [PubMed] [Google Scholar]
- 155.Michalopoulos G, Pitot HC. 1975. Primary culture of parenchymal liver cells on collagen membranes. Morphological and biochemical observations. Exp Cell Res 94:70–78. [DOI] [PubMed] [Google Scholar]
- 156.Emerman JT, Pitelka DR. 1977. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro 13:316–328. [DOI] [PubMed] [Google Scholar]
- 157.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, LeBlanc CL, Honer zu Bentrup K, Hammond T, Pierson DL. 2003. Low-shear modeled microgravity: a global environmental regulatory signal affecting bacterial gene expression, physiology, and pathogenesis. J Microbiol Methods 54:1–11. doi: 10.1016/S0167-7012(03)00018-6. [DOI] [PubMed] [Google Scholar]
- 158.Soni A, O'Sullivan L, Quick LN, Ott CM, Nickerson CA, Wilson JW. 2014. Conservation of the low-shear modeled microgravity response in Enterobacteriaceae and analysis of the trp genes in this response. Open Microbiol J 8:51–58. doi: 10.2174/1874285801408010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Grant KC, Khodadad CLM, Foster JS. 2013. Role of Hfq in an animal-microbe symbiosis under simulated microgravity conditions. Int J Astrobiol 13:53–61. doi: 10.1017/S1473550413000359. [DOI] [Google Scholar]
- 160.Altenburg SD, Nielsen-Preiss SM, Hyman LE. 2008. Increased filamentous growth of Candida albicans in simulated microgravity. Genomics Proteomics Bioinformatics 6:42–50. doi: 10.1016/S1672-0229(08)60019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Baker PW, Meyer ML, Leff LG. 2004. Escherichia coli growth under modeled reduced gravity. Microgravity Sci Technol 15:39–44. doi: 10.1007/BF02870967. [DOI] [PubMed] [Google Scholar]
- 162.Demain AL, Fang A. 2001. Secondary metabolism in simulated microgravity. Chem Rec 1:333–346. doi: 10.1002/tcr.1018. [DOI] [PubMed] [Google Scholar]
- 163.Fang A, Pierson DL, Koenig DW, Mishra SK, Demain AL. 1997. Effect of simulated microgravity and shear stress on microcin B17 production by Escherichia coli and on its excretion into the medium. Appl Environ Microbiol 63:4090–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lynch SV, Brodie EL, Matin A. 2004. Role and regulation of sigma S in general resistance conferred by low-shear simulated microgravity in Escherichia coli. J Bacteriol 186:8207–8212. doi: 10.1128/JB.186.24.8207-8212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tucker DL, Ott CM, Huff S, Fofanov Y, Pierson DL, Willson RC, Fox GE. 2007. Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment. BMC Microbiol 7:15. doi: 10.1186/1471-2180-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Abshire CF, Prasai K, Soto I, Shi R, Concha M, Baddoo M, Flemington EK, Ennis DG, Scott RS, Harrison L. 2016. Exposure of Mycobacterium marinum to low-shear modeled microgravity: effect on growth, the transcriptome and survival under stress. NPJ Microgravity 2:16038. doi: 10.1038/npjmgrav.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Orsini SS, Lewis AM, Rice KC. 2017. Investigation of simulated microgravity effects on Streptococcus mutans physiology and global gene expression. NPJ Microgravity 3:4. doi: 10.1038/s41526-016-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Goodwin TJ, Schroeder WF, Wolf DA, Moyer MP. 1993. Rotating-wall vessel coculture of small intestine as a prelude to tissue modeling: aspects of simulated microgravity. Proc Soc Exp Biol Med 202:181–192. [DOI] [PubMed] [Google Scholar]
- 169.Straub TM, Honer zu Bentrup K, Orosz-Coghlan P, Dohnalkova A, Mayer BK, Bartholomew RA, Valdez CO, Bruckner-Lea CJ, Gerba CP, Abbaszadegan M, Nickerson CA. 2007. In vitro cell culture infectivity assay for human noroviruses. Emerg Infect Dis 13:396–403. doi: 10.3201/eid1303.060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Skardal A, Sarker SF, Crabbé A, Nickerson CA, Prestwich GD. 2010. The generation of 3-D tissue models based on hyaluronan hydrogel-coated microcarriers within a rotating wall vessel bioreactor. Biomaterials 31:8426–8435. doi: 10.1016/j.biomaterials.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 171.Barrila J, Yang J, Crabbe A, Sarker SF, Liu Y, Ott CM, Nelman-Gonzalez MA, Clemett SJ, Nydam SD, Forsyth RJ, Davis RR, Crucian BE, Quiriarte H, Roland KL, Brenneman K, Sams C, Loscher C, Nickerson CA. 2017. Three-dimensional organotypic co-culture model of intestinal epithelial cells and macrophages to study Salmonella enterica colonization patterns. NPJ Microgravity 3:10. doi: 10.1038/s41526-017-0011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.De Weirdt R, Crabbe A, Roos S, Vollenweider S, Lacroix C, van Pijkeren JP, Britton RA, Sarker S, Van de Wiele T, Nickerson CA. 2012. Glycerol supplementation enhances L. reuteri's protective effect against S. Typhimurium colonization in a 3-D model of colonic epithelium. PLoS One 7:e37116. doi: 10.1371/journal.pone.0037116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Goodwin TJ, Jessup JM, Wolf DA. 1992. Morphologic differentiation of colon carcinoma cell lines HT-29 and HT-29KM in rotating-wall vessels. In Vitro Cell Dev Biol 28A:47–60. [DOI] [PubMed] [Google Scholar]
- 174.Devarasetty M, Wang E, Soker S, Skardal A. 2017. Mesenchymal stem cells support growth and organization of host-liver colorectal-tumor organoids and possibly resistance to chemotherapy. Biofabrication 9:021002. doi: 10.1088/1758-5090/aa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Salerno-Goncalves R, Fasano A, Sztein MB. 2011. Engineering of a multicellular organotypic model of the human intestinal mucosa. Gastroenterology 141:e18–e20. doi: 10.1053/j.gastro.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Salerno-Goncalves R, Fasano A, Sztein MB. 2016. Development of a multicellular three-dimensional organotypic model of the human intestinal mucosa grown under microgravity. J Vis Exp 2016(113):e54148. doi: 10.3791/54148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Salerno-Goncalves R, Safavie F, Fasano A, Sztein MB. 2016. Free and complexed-secretory immunoglobulin A triggers distinct intestinal epithelial cell responses. Clin Exp Immunol 185:338–347. doi: 10.1111/cei.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Crabbé A, Liu Y, Matthijs N, Rigole P, De La Fuente-Nunez C, Davis R, Ledesma MA, Sarker S, Van Houdt R, Hancock RE, Coenye T, Nickerson CA. 2017. Antimicrobial efficacy against Pseudomonas aeruginosa biofilm formation in a three-dimensional lung epithelial model and the influence of fetal bovine serum. Sci Rep 7:43321. doi: 10.1038/srep43321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Crabbé A, Liu Y, Sarker SF, Bonenfant NR, Barrila J, Borg ZD, Lee JJ, Weiss DJ, Nickerson CA. 2015. Recellularization of decellularized lung scaffolds is enhanced by dynamic suspension culture. PLoS One 10:e0126846. doi: 10.1371/journal.pone.0126846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Crabbé A, Sarker SF, Van Houdt R, Ott CM, Leys N, Cornelis P, Nickerson CA. 2011. Alveolar epithelium protects macrophages from quorum sensing-induced cytotoxicity in a three-dimensional co-culture model. Cell Microbiol 13:469–481. doi: 10.1111/j.1462-5822.2010.01548.x. [DOI] [PubMed] [Google Scholar]
- 181.Cortiella J, Niles J, Cantu A, Brettler A, Pham A, Vargas G, Winston S, Wang J, Walls S, Nichols JE. 2010. Influence of acellular natural lung matrix on murine embryonic stem cell differentiation and tissue formation. Tissue Eng Part A 16:2565–2580. doi: 10.1089/ten.tea.2009.0730. [DOI] [PubMed] [Google Scholar]
- 182.Goodwin TJ, McCarthy M, Cohrs RJ, Kaufer BB. 2015. 3D tissue-like assemblies: a novel approach to investigate virus-cell interactions. Methods 90:76–84. doi: 10.1016/j.ymeth.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Skardal A, Devarasetty M, Rodman C, Atala A, Soker S. 2015. Liver-tumor hybrid organoids for modeling tumor growth and drug response in vitro. Ann Biomed Eng 43:2361–2373. doi: 10.1007/s10439-015-1298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Khaoustov VI, Darlington GJ, Soriano HE, Krishnan B, Risin D, Pellis NR, Yoffe B. 1999. Induction of three-dimensional assembly of human liver cells by simulated microgravity. In Vitro Cell Dev Biol Anim 35:501–509. [DOI] [PubMed] [Google Scholar]
- 185.Prewett TL, Goodwin TJ, Spaulding GF. 1993. Three-dimensional modeling of T-24 human bladder carcinoma cell line: a new simulated microgravity culture vessel. J Tissue Cult Methods 15:29–36. [Google Scholar]
- 186.Ingram M, Techy GB, Saroufeem R, Yazan O, Narayan KS, Goodwin TJ, Spaulding GF. 1997. Three-dimensional growth patterns of various human tumor cell lines in simulated microgravity of a NASA bioreactor. In Vitro Cell Dev Biol Anim 33:459–466. [DOI] [PubMed] [Google Scholar]
- 187.Smith YC, Grande KK, Rasmussen SB, O'Brien AD. 2006. Novel three-dimensional organoid model for evaluation of the interaction of uropathogenic Escherichia coli with terminally differentiated human urothelial cells. Infect Immun 74:750–757. doi: 10.1128/IAI.74.1.750-757.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Łaniewski P, Gomez A, Hire G, So M, Herbst-Kralovetz MM. 2017. Human three-dimensional endometrial epithelial cell model to study host interactions with vaginal bacteria and Neisseria gonorrhoeae. Infect Immun 85:e01049-. doi: 10.1128/IAI.01049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.McGowin CL, Radtke AL, Abraham K, Martin DH, Herbst-Kralovetz M. 2013. Mycoplasma genitalium infection activates cellular host defense and inflammation pathways in a 3-dimensional human endocervical epithelial cell model. J Infect Dis 207:1857–1868. doi: 10.1093/infdis/jit101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Doerflinger SY, Throop AL, Herbst-Kralovetz MM. 2014. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J Infect Dis 209:1989–1999. doi: 10.1093/infdis/jiu004. [DOI] [PubMed] [Google Scholar]
- 191.Bursac N, Loo Y, Leong K, Tung L. 2007. Novel anisotropic engineered cardiac tissues: studies of electrical propagation. Biochem Biophys Res Commun 361:847–853. doi: 10.1016/j.bbrc.2007.07.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rungarunlert S, Klincumhom N, Bock I, Nemes C, Techakumphu M, Pirity MK, Dinnyes A. 2011. Enhanced cardiac differentiation of mouse embryonic stem cells by use of the slow-turning, lateral vessel (STLV) bioreactor. Biotechnol Lett 33:1565–1573. doi: 10.1007/s10529-011-0614-8. [DOI] [PubMed] [Google Scholar]
- 193.Papadaki M, Bursac N, Langer R, Merok J, Vunjak-Novakovic G, Freed LE. 2001. Tissue engineering of functional cardiac muscle: molecular, structural, and electrophysiological studies. Am J Physiol Heart Circ Physiol 280:H168–H178. doi: 10.1152/ajpheart.2001.280.1.H168. [DOI] [PubMed] [Google Scholar]
- 194.Margolis L, Hatfill S, Chuaqui R, Vocke C, Emmert-Buck M, Linehan WM, Duray PH. 1999. Long term organ culture of human prostate tissue in a NASA-designed rotating wall bioreactor. J Urol 161:290–297. [PubMed] [Google Scholar]
- 195.Rose MI, Brown DC, Pellis NR, Crisera CA, Colen KL, Longaker MT, Gittes GK. 1999. Effects of microgravity on the embryonic pancreas. In Vitro Cell Dev Biol Anim 35:560–563. [DOI] [PubMed] [Google Scholar]
- 196.Murray HE, Paget MB, Downing R. 2005. Preservation of glucose responsiveness in human islets maintained in a rotational cell culture system. Mol Cell Endocrinol 238:39–49. doi: 10.1016/j.mce.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 197.Lelkes PI, Galvan DL, Hayman GT, Goodwin TJ, Chatman DY, Cherian S, Garcia RM, Unsworth BR. 1998. Simulated microgravity conditions enhance differentiation of cultured PC12 cells towards the neuroendocrine phenotype. In Vitro Cell Dev Biol Anim 34:316–325. [DOI] [PubMed] [Google Scholar]
- 198.Myers TA, Nickerson CA, Kaushal D, Ott CM, Höner zu Bentrup K, Ramamurthy R, Nelman-Gonzalez M, Pierson DL, Philipp MT. 2008. Closing the phenotypic gap between transformed neuronal cell lines in culture and untransformed neurons. J Neurosci Methods 174:31–41. doi: 10.1016/j.jneumeth.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Valmikinathan CM, Hoffman J, Yu X. 2011. Impact of scaffold micro and macro architecture on Schwann cell proliferation under dynamic conditions in a rotating wall vessel bioreactor. Mater Sci Eng C Mater Biol Appl 31:22–29. doi: 10.1016/j.msec.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bramley JC, Drummond CG, Lennemann NJ, Good CA, Kim KS, Coyne CB. 2017. A three-dimensional cell culture system to model RNA virus infections at the blood-brain barrier. mSphere 2:e00206-. doi: 10.1128/mSphere.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lei XH, Ning LN, Cao YJ, Liu S, Zhang SB, Qiu ZF, Hu HM, Zhang HS, Liu S, Duan EK. 2011. NASA-approved rotary bioreactor enhances proliferation of human epidermal stem cells and supports formation of 3D epidermis-like structure. PLoS One 6:e26603. doi: 10.1371/journal.pone.0026603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.DiStefano T, Chen HY, Panebianco C, Kaya KD, Brooks MJ, Gieser L, Morgan NY, Pohida T, Swaroop A. 2018. Accelerated and improved differentiation of retinal organoids from pluripotent stem cells in rotating-wall vessel bioreactors. Stem Cell Rep 10:300–313. doi: 10.1016/j.stemcr.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Botchwey EA, Pollack SR, Levine EM, Laurencin CT. 2001. Bone tissue engineering in a rotating bioreactor using a microcarrier matrix system. J Biomed Mater Res 55:242–253. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Detamore MS, Athanasiou KA. 2005. Use of a rotating bioreactor toward tissue engineering the temporomandibular joint disc. Tissue Eng 11:1188–1197. doi: 10.1089/ten.2005.11.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yang X, Wang D, Hao J, Gong M, Arlet V, Balian G, Shen FH, Li XJ. 2011. Enhancement of matrix production and cell proliferation in human annulus cells under bioreactor culture. Tissue Eng Part A 17:1595–1603. doi: 10.1089/ten.tea.2010.0449. [DOI] [PubMed] [Google Scholar]
- 206.Rauh J, Milan F, Gunther KP, Stiehler M. 2011. Bioreactor systems for bone tissue engineering. Tissue Eng Part B Rev 17:263–280. doi: 10.1089/ten.teb.2010.0612. [DOI] [PubMed] [Google Scholar]
- 207.Ulbrich C, Wehland M, Pietsch J, Aleshcheva G, Wise P, van Loon J, Magnusson N, Infanger M, Grosse J, Eilles C, Sundaresan A, Grimm D. 2014. The impact of simulated and real microgravity on bone cells and mesenchymal stem cells. Biomed Res Int 2014:928507. doi: 10.1155/2014/928507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Duray PH, Yin SR, Ito Y, Bezrukov L, Cox C, Cho MS, Fitzgerald W, Dorward D, Zimmerberg J, Margolis L. 2005. Invasion of human tissue ex vivo by Borrelia burgdorferi. J Infect Dis 191:1747–1754. doi: 10.1086/429632. [DOI] [PubMed] [Google Scholar]
- 209.Duke J, Daane E, Arizpe J, Montufar-Solis D. 1996. Chondrogenesis in aggregates of embryonic limb cells grown in a rotating wall vessel. Adv Space Res 17:289–293. [DOI] [PubMed] [Google Scholar]
- 210.Darwin KH, Miller VL. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev 12:405–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. 2005. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun 73:7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt WD. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 213.Hu Q, Coburn B, Deng W, Li Y, Shi X, Lan Q, Wang B, Coombes BK, Finlay BB. 2008. Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J Clin Microbiol 46:1330–1336. doi: 10.1128/JCM.01255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V, Marsh K, Achtman M, Molyneux ME, Cormican M, Parkhill J, MacLennan CA, Heyderman RS, Dougan G. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res 19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shieh JTC, Bergelson JM. 2002. Interaction with decay-accelerating factor facilitates coxsackievirus B infection of polarized epithelial cells. J Virol 76:9474–9480. doi: 10.1128/JVI.76.18.9474-9480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Patel KP, Coyne CB, Bergelson JM. 2009. Dynamin- and lipid raft-dependent entry of decay-accelerating factor (DAF)-binding and non-DAF-binding coxsackieviruses into nonpolarized cells. J Virol 83:11064–11077. doi: 10.1128/JVI.01016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Coyne CB, Bergelson JM. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 219.Pan J, Zhang L, Odenwald MA, Shen L, Turner JR, Bergelson JM. 2015. Expression of human decay-accelerating factor on intestinal epithelium of transgenic mice does not facilitate infection by the enteral route. J Virol 89:4311–4318. doi: 10.1128/JVI.03468-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ. 2015. TEER measurement techniques for in vitro barrier model systems. J Lab Autom 20:107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nickerson CA, Ott CM. 2004. A new dimension in modeling infectious disease. ASM News 70:169–175. [Google Scholar]
- 222.Fatehullah A, Tan SH, Barker N. 2016. Organoids as an in vitro model of human development and disease. Nat Cell Biol 18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 223.Flora AD, Teel LD, Smith MA, Sinclair JF, Melton-Celsa AR, O'Brien AD. 2013. Ricin crosses polarized human intestinal cells and intestines of ricin-gavaged mice without evident damage and then disseminates to mouse kidneys. PLoS One 8:e69706. doi: 10.1371/journal.pone.0069706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bergmann S, Steinert M. 2015. From single cells to engineered and explanted tissues: new perspectives in bacterial infection biology. Int Rev Cell Mol Biol 319:1–44. doi: 10.1016/bs.ircmb.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 225.Astashkina A, Grainger DW. 2014. Critical analysis of 3-D organoid in vitro cell culture models for high-throughput drug candidate toxicity assessments. Adv Drug Deliv Rev 69–70:1–18. [DOI] [PubMed] [Google Scholar]
- 226.Shamir ER, Ewald AJ. 2014. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol 15:647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bhat R, Bissell MJ. 2014. Of plasticity and specificity: dialectics of the microenvironment and macroenvironment and the organ phenotype. Wiley Interdiscip Rev Dev Biol 3:147–163. doi: 10.1002/wdev.130. [DOI] [PubMed] [Google Scholar]
- 228.Bissell MJ. 2017. Goodbye flat biology—time for the 3rd and the 4th dimensions. J Cell Sci 130:3–5. doi: 10.1242/jcs.200550. [DOI] [PubMed] [Google Scholar]
- 229.Cruz-Acuña R, Quiros M, Farkas AE, Dedhia PH, Huang S, Siuda D, Garcia-Hernandez V, Miller AJ, Spence JR, Nusrat A, Garcia AJ. 2017. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol 19:1326–1335. doi: 10.1038/ncb3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bartfeld S. 2016. Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev Biol 420:262–270. doi: 10.1016/j.ydbio.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 231.Bartfeld S, Clevers H. 2017. Stem cell-derived organoids and their application for medical research and patient treatment. J Mol Med 95:729–738. doi: 10.1007/s00109-017-1531-7. [DOI] [PubMed] [Google Scholar]
- 232.Finkbeiner SR, Spence JR. 2013. A gutsy task: generating intestinal tissue from human pluripotent stem cells. Dig Dis Sci 58:1176–1184. doi: 10.1007/s10620-013-2620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hill DR, Spence JR. 2017. Gastrointestinal organoids: understanding the molecular basis of the host-microbe interface. Cell Mol Gastroenterol Hepatol 3:138–149. doi: 10.1016/j.jcmgh.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.In JG, Foulke-Abel J, Estes MK, Zachos NC, Kovbasnjuk O, Donowitz M. 2016. Human mini-guts: new insights into intestinal physiology and host-pathogen interactions. Nat Rev Gastroenterol Hepatol 13:633–642. doi: 10.1038/nrgastro.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, de Jonge HR, Estes MK, Donowitz M. 2016. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J Biol Chem 291:3759–3766. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Merker SR, Weitz J, Stange DE. 2016. Gastrointestinal organoids: how they gut it out. Dev Biol 420:239–250. doi: 10.1016/j.ydbio.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 237.Dutta D, Clevers H. 2017. Organoid culture systems to study host-pathogen interactions. Curr Opin Immunol 48:15–22. doi: 10.1016/j.coi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. 2016. Organoid models of human gastrointestinal development and disease. Gastroenterology 150:1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Anonymous. 2018. Method of the year 2017: organoids. Nat Methods 15:1. [Google Scholar]
- 240.Zhang YG, Wu S, Xia Y, Sun J. 2014. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep 2:e12147. doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bhushal S, Wolfsmuller M, Selvakumar TA, Kemper L, Wirth D, Hornef MW, Hauser H, Koster M. 2017. Cell polarization and epigenetic status shape the heterogeneous response to type iii interferons in intestinal epithelial cells. Front Immunol 8:671. doi: 10.3389/fimmu.2017.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Davies JM, Santaolalla R, von Furstenberg RJ, Henning SJ, Abreu MT. 2015. The viral mimetic polyinosinic:polycytidylic acid alters the growth characteristics of small intestinal and colonic crypt cultures. PLoS One 10:e0138531. doi: 10.1371/journal.pone.0138531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wilson SS, Tocchi A, Holly MK, Parks WC, Smith JG. 2014. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol 8:352–361. doi: 10.1038/mi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yin Y, Bijvelds M, Dang W, Xu L, van der Eijk AA, Knipping K, Tuysuz N, Dekkers JF, Wang Y, de Jonge J, Sprengers D, van der Laan LJW, Beekman JM, ten Berge D, Metselaar HJ, de Jonge H, Koopmans MPG, Peppelenbosch MP, Pan Q. 2015. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res 123:120–131. doi: 10.1016/j.antiviral.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 245.Aladegbami B, Barron L, Bao J, Colasanti J, Erwin CR, Warner BW, Guo J. 2017. Epithelial cell specific Raptor is required for initiation of type 2 mucosal immunity in small intestine. Sci Rep 7:5580. doi: 10.1038/s41598-017-06070-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Cao L, Kuratnik A, Xu W, Gibson JD, Kolling F IV, Falcone ER, Ammar M, Van Heyst MD, Wright DL, Nelson CE, Giardina C. 2015. Development of intestinal organoids as tissue surrogates: cell composition and the epigenetic control of differentiation. Mol Carcinog 54:189–202. doi: 10.1002/mc.22089. [DOI] [PubMed] [Google Scholar]
- 247.Fischer JC, Bscheider M, Eisenkolb G, Lin CC, Wintges A, Otten V, Lindemans CA, Heidegger S, Rudelius M, Monette S, Porosnicu Rodriguez KA, Calafiore M, Liebermann S, Liu C, Lienenklaus S, Weiss S, Kalinke U, Ruland J, Peschel C, Shono Y, Docampo M, Velardi E, Jenq RR, Hanash AM, Dudakov JA, Haas T, van den Brink MRM, Poeck H. 2017. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci Transl Med 9:eaag2513. doi: 10.1126/scitranslmed.aag2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Foulke-Abel J, In J, Yin J, Zachos NC, Kovbasnjuk O, Estes MK, de Jonge H, Donowitz M. 2016. Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology 150:638–649.e8. doi: 10.1053/j.gastro.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 250.Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NWM, Bijvelds MJC, Scholte BJ, Nieuwenhuis EES, van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM. 2013. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 19:939. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 251.de Lau W, Kujala P, Schneeberger K, Middendorp S, Li VS, Barker N, Martens A, Hofhuis F, DeKoter RP, Peters PJ, Nieuwenhuis E, Clevers H. 2012. Peyer's patch M cells derived from Lgr5+ stem cells require SpiB and are induced by RankL in cultured “miniguts.” Mol Cell Biol 32:3639–3647. doi: 10.1128/MCB.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.McCracken KW, Howell JC, Wells JM, Spence JR. 2011. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc 6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. 2009. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med 15:701. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM. 2011. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rodríguez-Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras-Raves M, Sacchetti A, Hornsveld M, Oost KC, Snippert HJ, Verhoeven-Duif N, Fodde R, Burgering BMT. 2017. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543:424. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- 256.Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, Chang C-F, Schiesser J, Aubert P, Stanley EG, Elefanty AG, Miyaoka Y, Mandegar MA, Conklin BR, Neunlist M, Brugmann SA, Helmrath MA, Wells JM. 2017. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 23:49–59. doi: 10.1038/nm.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Forbester JL, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, Mukhopadhyay S, Dougan G. 2015. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun 83:2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Engevik MA, Engevik KA, Yacyshyn MB, Wang J, Hassett DJ, Darien B, Yacyshyn BR, Worrell RT. 2015. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol 308:G497–G509. doi: 10.1152/ajpgi.00090.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Engevik MA, Yacyshyn MB, Engevik KA, Wang J, Darien B, Hassett DJ, Yacyshyn BR, Worrell RT. 2015. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 308:G510–G524. doi: 10.1152/ajpgi.00091.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Karve SS, Pradhan S, Ward DV, Weiss AA. 2017. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS One 12:e0178966. doi: 10.1371/journal.pone.0178966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.In J, Foulke-Abel J, Zachos NC, Hansen A-M, Kaper JB, Bernstein HD, Halushka M, Blutt S, Estes MK, Donowitz M, Kovbasnjuk O. 2016. Enterohemorrhagic Escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol 2:48–62.e3. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, Spence JR. 2015. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun 83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tao L, Zhang J, Meraner P, Tovaglieri A, Wu X, Gerhard R, Zhang X, Stallcup WB, Miao J, He X, Hurdle JG, Breault DT, Brass AL, Dong M. 2016. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538:350–355. doi: 10.1038/nature19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. 2011. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 265.Cristobal A, van den Toorn HWP, van de Wetering M, Clevers H, Heck AJR, Mohammed S. 2017. Personalized proteome profiles of healthy and tumor human colon organoids reveal both individual diversity and basic features of colorectal cancer. Cell Rep 18:263–274. doi: 10.1016/j.celrep.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 266.Yin Y, Wang Y, Dang W, Xu L, Su J, Zhou X, Wang W, Felczak K, van der Laan LJ, Pankiewicz KW, van der Eijk AA, Bijvelds M, Sprengers D, de Jonge H, Koopmans MP, Metselaar HJ, Peppelenbosch MP, Pan Q. 2016. Mycophenolic acid potently inhibits rotavirus infection with a high barrier to resistance development. Antiviral Res 133:41–49. doi: 10.1016/j.antiviral.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 267.Senger S, Ingano L, Freire R, Anselmo A, Zhu W, Sadreyev R, Walker WA, Fasano A. 2018. Human fetal-derived enterospheres provide insights on intestinal development and a novel model to study necrotizing enterocolitis (NEC). Cell Mol Gastroenterol Hepatol 5:549–568. doi: 10.1016/j.jcmgh.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nickerson KP, Senger S, Zhang Y, Lima R, Patel S, Ingano L, Flavahan WA, Kumar DKV, Fraser CM, Faherty CS, Sztein MB, Fiorentino M, Fasano A. 2018. Salmonella Typhi colonization provokes extensive transcriptional changes aimed at evading host mucosal immune defense during early infection of human intestinal tissue. EBioMedicine 31:92–109. doi: 10.1016/j.ebiom.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Chen YW, Huang SX, de Carvalho A, Ho SH, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova JL, Bhattacharya J, Liang AF, Palermo LM, Porotto M, Moscona A, Snoeck HW. 2017. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19:542–549. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, Spence JR. 2015. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Firth AL, Dargitz CT, Qualls SJ, Menon T, Wright R, Singer O, Gage FH, Khanna A, Verma IM. 2014. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc Natl Acad Sci U S A 111:E1723–E1730. doi: 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hegab AE, Arai D, Gao J, Kuroda A, Yasuda H, Ishii M, Naoki K, Soejima K, Betsuyaku T. 2015. Mimicking the niche of lung epithelial stem cells and characterization of several effectors of their in vitro behavior. Stem Cell Res 15:109–121. doi: 10.1016/j.scr.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 273.Nikolić MZ, Rawlins EL. 2017. Lung organoids and their use to study cell-cell interaction. Curr Pathobiol Rep 5:223–231. doi: 10.1007/s40139-017-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Shen Y, Chen L, Wang M, Lin D, Liang Z, Song P, Yuan Q, Tang H, Li W, Duan K, Liu B, Zhao G, Wang Y. 2017. Flagellar hooks and hook protein FlgE participate in host microbe interactions at immunological level. Sci Rep 7:1433. doi: 10.1038/s41598-017-01619-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, Clevers H. 2015. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148:126–136.e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Mahe MM, Aihara E, Schumacher MA, Zavros Y, Montrose MH, Helmrath MA, Sato T, Shroyer NF. 2013. Establishment of gastrointestinal epithelial organoids. Curr Protoc Mouse Biol 3:217–240. doi: 10.1002/9780470942390.mo130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Bertaux-Skeirik N, Feng R, Schumacher MA, Li J, Mahe MM, Engevik AC, Javier JE, Peek RM Jr, Ottemann K, Orian-Rousseau V, Boivin GP, Helmrath MA, Zavros Y. 2015. CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS Pathog 11:e1004663. doi: 10.1371/journal.ppat.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM. 2014. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, Passarelli B, Camorlinga M, Bouley DM, Alvarez G, Nusse R, Torres J, Amieva MR. 2015. Helicobacter pylori activates and expands Lgr5+ stem cells through direct colonization of the gastric glands. Gastroenterology 148:1392–1404.e21. doi: 10.1053/j.gastro.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 280.McCracken KW, Aihara E, Martin B, Crawford CM, Broda T, Treguier J, Zhang X, Shannon JM, Montrose MH, Wells JM. 2017. Wnt/beta-catenin promotes gastric fundus specification in mice and humans. Nature 541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Schlaermann P, Toelle B, Berger H, Schmidt SC, Glanemann M, Ordemann J, Bartfeld S, Mollenkopf HJ, Meyer TF. 2016. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Burkitt MD, Duckworth CA, Williams JM, Pritchard DM. 2017. Helicobacter pylori-induced gastric pathology: insights from in vivo and ex vivo models. Dis Model Mech 10:89–104. doi: 10.1242/dmm.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Li ML, Aggeler J, Farson DA, Hatier C, Hassell J, Bissell MJ. 1987. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc Natl Acad Sci U S A 84:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Vidi PA, Bissell MJ, Lelievre SA. 2013. Three-dimensional culture of human breast epithelial cells: the how and the why. Methods Mol Biol 945:193–219. doi: 10.1007/978-1-62703-125-7_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 287.Qian X, Nguyen Ha N, Song Mingxi M, Hadiono C, Ogden Sarah C, Hammack C, Yao B, Hamersky Gregory R, Jacob F, Zhong C, Yoon K-J, Jeang W, Lin L, Li Y, Thakor J, Berg Daniel A, Zhang C, Kang E, Chickering M, Nauen D, Ho C-Y, Wen Z, Christian Kimberly M, Shi P-Y, Maher Brady J, Wu H, Jin P, Tang H, Song H, Ming G-l. 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Huch M, Dorrell C, Boj SF, van Es JH, Li VSW, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, Vries RG, Grompe M, Clevers H. 2013. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang R-R, Ueno Y, Zheng Y-W, Koike N, Aoyama S, Adachi Y, Taniguchi H. 2013. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 290.Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJM, van de Wetering M, Sojoodi M, Li VSW, Schuijers J, Gracanin A, Ringnalda F, Begthel H, Hamer K, Mulder J, van Es JH, de Koning E, Vries RGJ, Heimberg H, Clevers H. 2013. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Greggio C, De Franceschi F, Figueiredo-Larsen M, Gobaa S, Ranga A, Semb H, Lutolf M, Grapin-Botton A. 2013. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development 140:4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song JY, Neefjes-Borst EA, te Riele H, Holden DW, Nath G, Neefjes J. 2015. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 17:763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 293.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. 2011. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 294.Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, Nishinakamura R. 2014. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 295.Karthaus Wouter R, Iaquinta Phillip J, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling Catherine M, Gao D, Begthel H, Sachs N, Vries Robert GJ, Cuppen E, Chen Y, Sawyers Charles L, Clevers Hans C. 2014. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, Badani KK, McKiernan JM, Benson MC, Hibshoosh H, Shen MM. 2014. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol 16:951–961. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zhu Y, Yang Y, Guo J, Dai Y, Ye L, Qiu J, Zeng Z, Wu X, Xing Y, Long X, Wu X, Ye L, Wang S, Li H. 2017. Ex vivo 2D and 3D HSV-2 infection model using human normal vaginal epithelial cells. Oncotarget 8:15267–15282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B, Berger H, Mollenkopf HJ, Mangler M, Sehouli J, Fotopoulou C, Meyer TF. 2015. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun 6:8989. doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ghaedi M, Calle EA, Mendez JJ, Gard AL, Balestrini J, Booth A, Bove PF, Gui L, White ES, Niklason LE. 2013. Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J Clin Invest 123:4950–4962. doi: 10.1172/JCI68793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Finkbeiner SR, Freeman JJ, Wieck MM, El-Nachef W, Altheim CH, Tsai YH, Huang S, Dyal R, White ES, Grikscheit TC, Teitelbaum DH, Spence JR. 2015. Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids. Biol Open 4:1462–1472. doi: 10.1242/bio.013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, Morty RE, Vadasz I, Mayer K, Gattenloehner S, Fink L, Matrosovich M, Li X, Seeger W, Lohmeyer J, Bellusci S, Herold S. 2016. Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b-driven epithelial repair. PLoS Pathog 12:e1005544. doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Al Shammari B, Shiomi T, Tezera L, Bielecka MK, Workman V, Sathyamoorthy T, Mauri F, Jayasinghe SN, Robertson BD, D'Armiento J, Friedland JS, Elkington PT. 2015. The extracellular matrix regulates granuloma necrosis in tuberculosis. J Infect Dis 212:463–473. doi: 10.1093/infdis/jiv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Clevers H. 2016. Modeling development and disease with organoids. Cell 165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 304.Lancaster MA, Knoblich JA. 2014. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 305.Bissell MJ, Labarge MA. 2005. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell 7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Chingwaru W, Glashoff RH, Vidmar J, Kapewangolo P, Sampson SL. 2016. Mammalian cell cultures as models for Mycobacterium tuberculosis-human immunodeficiency virus (HIV) interaction studies: a review. Asian Pac J Trop Med 9:832–838. doi: 10.1016/j.apjtm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 307.Dutta D, Heo I, Clevers H. 2017. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med 23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 308.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A. 2017. A single-cell survey of the small intestinal epithelium. Nature 551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Ettayebi K, Blutt SE, Hyser JM, Zeng XL, Crawford SE, Broughman JR, Estes MK, Donowitz M. 2014. Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Exp Biol Med 239:1124–1134. doi: 10.1177/1535370214529398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G. 2014. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio 5:e01438-. doi: 10.1128/mBio.01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Naito T, Mulet C, De Castro C, Molinaro A, Saffarian A, Nigro G, Bérard M, Clerc M, Pedersen AB, Sansonetti PJ, Pédron T. 2017. Lipopolysaccharide from crypt-specific core microbiota modulates the colonic epithelial proliferation-to-differentiation balance. mBio 8:e01680-. doi: 10.1128/mBio.01680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ. 2014. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 15:792–798. doi: 10.1016/j.chom.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 313.Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF, Zachos NC. 2017. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci Rep 7:45270. doi: 10.1038/srep45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. 2012. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3:e00159-. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Huang L, Hou Q, Ye L, Yang Q, Yu Q. 2017. Crosstalk between H9N2 avian influenza virus and crypt-derived intestinal organoids. Vet Res 48:71. doi: 10.1186/s13567-017-0478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Zhang D, Tan M, Zhong W, Xia M, Huang P, Jiang X. 2017. Human intestinal organoids express histo-blood group antigens, bind norovirus VLPs, and support limited norovirus replication. Sci Rep 7:12621. doi: 10.1038/s41598-017-12736-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zhou J, Li C, Zhao G, Chu H, Wang D, Yan HH-N, Poon VK-M, Wen L, Wong BH-Y, Zhao X, Chiu MC, Yang D, Wang Y, Au-Yeung RKH, Chan IH-Y, Sun S, Chan JF-W, To KK-W, Memish ZA, Corman VM, Drosten C, Hung IF-N, Zhou Y, Leung SY, Yuen K-Y. 2017. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Science Advances 3:eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Burger E, Araujo A, Lopez-Yglesias A, Rajala MW, Geng L, Levine B, Hooper LV, Burstein E, Yarovinsky F. 2018. Loss of Paneth cell autophagy causes acute susceptibility to Toxoplasma gondii-mediated inflammation. Cell Host Microbe 23:177–190.e4. doi: 10.1016/j.chom.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 321.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, Clevers H, Batlle E. 2011. Isolation and in vitro expansion of human colonic stem cells. Nat Med 17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 322.Hannan NR, Fordham RP, Syed YA, Moignard V, Berry A, Bautista R, Hanley NA, Jensen KB, Vallier L. 2013. Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Rep 1:293–306. doi: 10.1016/j.stemcr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Kleinman HK, Martin GR. 2005. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol 15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 324.Orkin RW, Gehron P, McGoodwin EB, Martin GR, Valentine T, Swarm R. 1977. A murine tumor producing a matrix of basement membrane. J Exp Med 145:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Stelzner M, Helmrath M, Dunn JC, Henning SJ, Houchen CW, Kuo C, Lynch J, Li L, Magness ST, Martin MG, Wong MH, Yu J, Consortium NIHISC. 2012. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol 302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Nossol C, Diesing AK, Walk N, Faber-Zuschratter H, Hartig R, Post A, Kluess J, Rothkotter HJ, Kahlert S. 2011. Air-liquid interface cultures enhance the oxygen supply and trigger the structural and functional differentiation of intestinal porcine epithelial cells (IPEC). Histochem Cell Biol 136:103–115. doi: 10.1007/s00418-011-0826-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Drummond CG, Bolock AM, Ma C, Luke CJ, Good M, Coyne CB. 2017. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc Natl Acad Sci U S A 114:1672–1677. doi: 10.1073/pnas.1617363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kingwell K. 2016. 3D cell technologies head to the R&D assembly line. Nat Rev Drug Discov 16:6–7. doi: 10.1038/nrd.2016.282. [DOI] [PubMed] [Google Scholar]
- 330.Noordhoek J, Gulmans V, van der Ent K, Beekman JM. 2016. Intestinal organoids and personalized medicine in cystic fibrosis: a successful patient-oriented research collaboration. Curr Opin Pulm Med 22:610–616. doi: 10.1097/MCP.0000000000000315. [DOI] [PubMed] [Google Scholar]
- 331.Ohno H, Kanaya T, Williams IR. 2012. M cell differentiation: distinct lineage or phenotypic transition? Salmonella provides answers. Cell Host Microbe 12:607–609. doi: 10.1016/j.chom.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 332.Tahoun A, Mahajan S, Paxton E, Malterer G, Donaldson DS, Wang D, Tan A, Gillespie TL, O'Shea M, Roe AJ, Shaw DJ, Gally DL, Lengeling A, Mabbott NA, Haas J, Mahajan A. 2012. Salmonella transforms follicle-associated epithelial cells into M cells to promote intestinal invasion. Cell Host Microbe 12:645–656. doi: 10.1016/j.chom.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 333.Kernéis S, Bogdanova A, Kraehenbuhl J-P, Pringault E. 1997. Conversion by Peyer's patch lymphocytes of human enterocytes into M cells that transport bacteria. Science 277:949–952. doi: 10.1126/science.277.5328.949. [DOI] [PubMed] [Google Scholar]
- 334.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Li H, Breault DT, Ingber DE. 2018. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep 8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Bhatia SN, Ingber DE. 2014. Microfluidic organs-on-chips. Nat Biotechnol 32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 336.Benam KH, Dauth S, Hassell B, Herland A, Jain A, Jang KJ, Karalis K, Kim HJ, MacQueen L, Mahmoodian R, Musah S, Torisawa YS, van der Meer AD, Villenave R, Yadid M, Parker KK, Ingber DE. 2015. Engineered in vitro disease models. Annu Rev Pathol 10:195–262. doi: 10.1146/annurev-pathol-012414-040418. [DOI] [PubMed] [Google Scholar]
- 337.Selimovic S, Dokmeci MR, Khademhosseini A. 2013. Organs-on-a-chip for drug discovery. Curr Opin Pharmacol 13:829–833. doi: 10.1016/j.coph.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 338.Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. 2012. Microengineered physiological biomimicry: organs-on-chips. Lab Chip 12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 339.Esch EW, Bahinski A, Huh D. 2015. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 14:248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Kim HJ, Huh D, Hamilton G, Ingber DE. 2012. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 341.Kim HJ, Ingber DE. 2013. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb) 5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 342.Kim HJ, Li H, Collins JJ, Ingber DE. 2016. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A 113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Villenave R, Wales SQ, Hamkins-Indik T, Papafragkou E, Weaver JC, Ferrante TC, Bahinski A, Elkins CA, Kulka M, Ingber DE. 2017. Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS One 12:e0169412. doi: 10.1371/journal.pone.0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Huh D, Hamilton GA, Ingber DE. 2011. From 3D cell culture to organs-on-chips. Trends Cell Biol 21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Huh D, Kim HJ, Fraser JP, Shea DE, Khan M, Bahinski A, Hamilton GA, Ingber DE. 2013. Microfabrication of human organs-on-chips. Nat Protoc 8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- 346.Folch A, Toner M. 2000. Microengineering of cellular interactions. Annu Rev Biomed Eng 2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 347.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. 2006. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A 103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. 2001. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng 3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 349.Whitesides GM. 2006. The origins and the future of microfluidics. Nature 442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 350.Halldorsson S, Lucumi E, Gomez-Sjoberg R, Fleming RM. 2015. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron 63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 351.Sackmann EK, Fulton AL, Beebe DJ. 2014. The present and future role of microfluidics in biomedical research. Nature 507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 352.Yi H-G, Lee H, Cho D-W. 2017. 3D printing of organs-on-chips. Bioengineering 4:10. doi: 10.3390/bioengineering4010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Henry OYF, Villenave R, Cronce MJ, Leineweber WD, Benz MA, Ingber DE. 2017. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 17:2264–2271. doi: 10.1039/C7LC00155J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Kim J, Hegde M, Jayaraman A. 2010. Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions. Lab Chip 10:43–50. doi: 10.1039/B911367C. [DOI] [PubMed] [Google Scholar]
- 355.Esch MB, Sung JH, Yang J, Yu C, Yu J, March JC, Shuler ML. 2012. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic ‘body-on-a-chip’ devices. Biomed Microdevices 14:895–906. doi: 10.1007/s10544-012-9669-0. [DOI] [PubMed] [Google Scholar]
- 356.Mahler GJ, Esch MB, Glahn RP, Shuler ML. 2009. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol Bioeng 104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- 357.Huh D, Fujioka H, Tung YC, Futai N, Paine R III, Grotberg JB, Takayama S. 2007. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc Natl Acad Sci U S A 104:18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. 2012. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med 4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. 2010. Reconstituting organ-level lung functions on a chip. Science 328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Huh DD. 2015. A human breathing lung-on-a-chip. Ann Am Thorac Soc 12(Suppl 1):S42–S44. doi: 10.1513/AnnalsATS.201410-442MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Tavana H, Zamankhan P, Christensen PJ, Grotberg JB, Takayama S. 2011. Epithelium damage and protection during reopening of occluded airways in a physiologic microfluidic pulmonary airway model. Biomed Microdevices 13:731–742. doi: 10.1007/s10544-011-9543-5. [DOI] [PubMed] [Google Scholar]
- 362.Kane BJ, Zinner MJ, Yarmush ML, Toner M. 2006. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem 78:4291–4298. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- 363.Lee PJ, Hung PJ, Lee LP. 2007. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol Bioeng 97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 364.Chao P, Maguire T, Novik E, Cheng KC, Yarmush ML. 2009. Evaluation of a microfluidic based cell culture platform with primary human hepatocytes for the prediction of hepatic clearance in human. Biochem Pharmacol 78:625–632. doi: 10.1016/j.bcp.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Novik E, Maguire TJ, Chao P, Cheng KC, Yarmush ML. 2010. A microfluidic hepatic coculture platform for cell-based drug metabolism studies. Biochem Pharmacol 79:1036–1044. doi: 10.1016/j.bcp.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Legendre A, Baudoin R, Alberto G, Paullier P, Naudot M, Bricks T, Brocheton J, Jacques S, Cotton J, Leclerc E. 2013. Metabolic characterization of primary rat hepatocytes cultivated in parallel microfluidic biochips. J Pharm Sci 102:3264–3276. doi: 10.1002/jps.23466. [DOI] [PubMed] [Google Scholar]
- 367.Cheng S, Prot JM, Leclerc E, Bois FY. 2012. Zonation related function and ubiquitination regulation in human hepatocellular carcinoma cells in dynamic vs. static culture conditions. BMC Genomics 13:54. doi: 10.1186/1471-2164-13-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Sivaraman A, Leach JK, Townsend S, Iida T, Hogan BJ, Stolz DB, Fry R, Samson LD, Tannenbaum SR, Griffith LG. 2005. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr Drug Metab 6:569–591. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- 369.Toh YC, Lim TC, Tai D, Xiao G, van Noort D, Yu H. 2009. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip 9:2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 370.Baudoin R, Griscom L, Monge M, Legallais C, Leclerc E. 2007. Development of a renal microchip for in vitro distal tubule models. Biotechnol Prog 23:1245–1253. doi: 10.1021/bp0603513. [DOI] [PubMed] [Google Scholar]
- 371.Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY, Ingber DE. 2013. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol (Camb) 5:1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 372.Jang KJ, Suh KY. 2010. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 10:36–42. doi: 10.1039/B907515A. [DOI] [PubMed] [Google Scholar]
- 373.Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. 2013. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Khanal G, Chung K, Solis-Wever X, Johnson B, Pappas D. 2011. Ischemia/reperfusion injury of primary porcine cardiomyocytes in a low-shear microfluidic culture and analysis device. Analyst 136:3519–3526. doi: 10.1039/c0an00845a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Grosberg A, Alford PW, McCain ML, Parker KK. 2011. Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip 11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Cheng W, Klauke N, Sedgwick H, Smith GL, Cooper JM. 2006. Metabolic monitoring of the electrically stimulated single heart cell within a microfluidic platform. Lab Chip 6:1424–1431. doi: 10.1039/b608202e. [DOI] [PubMed] [Google Scholar]
- 377.Giridharan GA, Nguyen M-D, Estrada R, Parichehreh V, Hamid T, Ismahil MA, Prabhu SD, Sethu P. 2010. Microfluidic cardiac cell culture model (μCCCM). Anal Chem 82:7581–7587. doi: 10.1021/ac1012893. [DOI] [PubMed] [Google Scholar]
- 378.Puleo CM, McIntosh Ambrose W, Takezawa T, Elisseeff J, Wang TH. 2009. Integration and application of vitrified collagen in multilayered microfluidic devices for corneal microtissue culture. Lab Chip 9:3221–3227. doi: 10.1039/b908332d. [DOI] [PubMed] [Google Scholar]
- 379.O'Neill AT, Monteiro-Riviere NA, Walker GM. 2008. Characterization of microfluidic human epidermal keratinocyte culture. Cytotechnology 56:197–207. doi: 10.1007/s10616-008-9149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Xiao RR, Zeng WJ, Li YT, Zou W, Wang L, Pei XF, Xie M, Huang WH. 2013. Simultaneous generation of gradients with gradually changed slope in a microfluidic device for quantifying axon response. Anal Chem 85:7842–7850. doi: 10.1021/ac4022055. [DOI] [PubMed] [Google Scholar]
- 381.Tsantoulas C, Farmer C, Machado P, Baba K, McMahon SB, Raouf R. 2013. Probing functional properties of nociceptive axons using a microfluidic culture system. PLoS One 8:e80722. doi: 10.1371/journal.pone.0080722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Shi M, Majumdar D, Gao Y, Brewer BM, Goodwin CR, McLean JA, Li D, Webb DJ. 2013. Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab Chip 13:3008–3021. doi: 10.1039/c3lc50249j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Shayan G, Shuler ML, Lee KH. 2011. The effect of astrocytes on the induction of barrier properties in aortic endothelial cells. Biotechnol Prog 27:1137–1145. doi: 10.1002/btpr.620. [DOI] [PubMed] [Google Scholar]
- 384.Park SH, Sim WY, Min BH, Yang SS, Khademhosseini A, Kaplan DL. 2012. Chip-based comparison of the osteogenesis of human bone marrow- and adipose tissue-derived mesenchymal stem cells under mechanical stimulation. PLoS One 7:e46689. doi: 10.1371/journal.pone.0046689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Zhang Y, Gazit Z, Pelled G, Gazit D, Vunjak-Novakovic G. 2011. Patterning osteogenesis by inducible gene expression in microfluidic culture systems. Integr Biol (Camb) 3:39–47. doi: 10.1039/C0IB00053A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, McKinnon KE, Dokic D, Rashedi AS, Haisenleder DJ, Malpani SS, Arnold-Murray CA, Chen K, Jiang M, Bai L, Nguyen CT, Zhang J, Laronda MM, Hope TJ, Maniar KP, Pavone ME, Avram MJ, Sefton EC, Getsios S, Burdette JE, Kim JJ, Borenstein JT, Woodruff TK. 2017. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun 8:14584. doi: 10.1038/ncomms14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Griep LM, Wolbers F, de Wagenaar B, ter Braak PM, Weksler BB, Romero IA, Couraud PO, Vermes I, van der Meer AD, van den Berg A. 2013. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed Microdevices 15:145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- 388.Booth R, Kim H. 2012. Characterization of a microfluidic in vitro model of the blood-brain barrier (muBBB). Lab Chip 12:1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 389.van der Helm MW, van der Meer AD, Eijkel JC, van den Berg A, Segerink LI. 2016. Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 4:e1142493. doi: 10.1080/21688370.2016.1142493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Liu MC, Shih HC, Wu JG, Weng TW, Wu CY, Lu JC, Tung YC. 2013. Electrofluidic pressure sensor embedded microfluidic device: a study of endothelial cells under hydrostatic pressure and shear stress combinations. Lab Chip 13:1743–1753. doi: 10.1039/c3lc41414k. [DOI] [PubMed] [Google Scholar]
- 391.Hasenberg T, Muhleder S, Dotzler A, Bauer S, Labuda K, Holnthoner W, Redl H, Lauster R, Marx U. 2015. Emulating human microcapillaries in a multi-organ-chip platform. J Biotechnol 216:1–10. doi: 10.1016/j.jbiotec.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 392.Shin M, Matsuda K, Ishii O, Terai H, Kaazempur-Mofrad M, Borenstein J, Detmar M, Vacanti JP. 2004. Endothelialized networks with a vascular geometry in microfabricated poly(dimethyl siloxane). Biomed Microdevices 6:269–278. doi: 10.1023/B:BMMD.0000048559.29932.27. [DOI] [PubMed] [Google Scholar]
- 393.van der Meer AD, Orlova VV, ten Dijke P, van den Berg A, Mummery CL. 2013. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab Chip 13:3562–3568. doi: 10.1039/c3lc50435b. [DOI] [PubMed] [Google Scholar]
- 394.Wikswo JP. 2014. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med 239:1061–1072. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Stucki AO, Stucki JD, Hall SR, Felder M, Mermoud Y, Schmid RA, Geiser T, Guenat OT. 2015. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip 15:1302–1310. doi: 10.1039/C4LC01252F. [DOI] [PubMed] [Google Scholar]
- 396.Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee HH, Alves SE, Salmon M, Ferrante TC, Weaver JC, Bahinski A, Hamilton GA, Ingber DE. 2016. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods 13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 397.Abaci HE, Shuler ML. 2015. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr Biol (Camb) 7:383–391. doi: 10.1039/C4IB00292J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.nobeastsofierce (Contributor). Date unknown Stock image of intestinal lining and villi [digital image]. Retrieved from https://www.123rf.com/ (123RF Image ID 55676294). Accessed 8 February 2017.
- 399.Ivan Moshe (Contributor). Date unknown Stock image of cells [digital image]. Retrieved from https://www.shutterstock.com/ (Shutterstock Image ID 174738014). Accessed 23 January 2018.
- 400.sciencepics (Contributor). Date unknown Stock image of small and large intestine [digital image]. Retrieved from https://www.shutterstock.com/ (Shutterstock Image ID 541068199) Accessed 23 January 2018.
- 401.Kateryna Kon (Contributor). Date unknown Stock image of human intestine with intestinal bacteria [digital image]. Retrieved from https://www.shutterstock.com/ (Shutterstock Image ID 400088722). Accessed 23 January 2018.