Enterotoxigenic Escherichia coli (ETEC), a heterogeneous diarrheal pathovar defined by production of heat-labile (LT) and/or heat-stable (ST) toxins, causes substantial morbidity among young children in the developing world. Studies demonstrating a major burden of ST-producing ETEC have focused interest on ST toxoids for ETEC vaccines.
KEYWORDS: diarrhea, enterotoxigenic, heat-stable toxin, pathogenesis, toxoids, vaccines
ABSTRACT
Enterotoxigenic Escherichia coli (ETEC), a heterogeneous diarrheal pathovar defined by production of heat-labile (LT) and/or heat-stable (ST) toxins, causes substantial morbidity among young children in the developing world. Studies demonstrating a major burden of ST-producing ETEC have focused interest on ST toxoids for ETEC vaccines. We examined fundamental aspects of ST biology using ETEC strain H10407, which carries estH and estP genes encoding STh and STp, respectively, in addition to eltAB genes responsible for LT. Here, we found that deletion of estH significantly diminished cyclic GMP (cGMP) activation in target epithelia, while deletion of estP had a surprisingly modest impact, and a dual estH estP mutant was not appreciably different from the estH mutant. However, we noted that either STh or STp recombinant peptides stimulated cGMP production and that the loss of estP was compensated by enhanced estH transcription. We also found that the TolC efflux protein was essential for toxin secretion and delivery, providing a potential avenue for efflux inhibitors in treatment of acute diarrheal illness. In addition, we demonstrated that the EtpA adhesin is required for optimal delivery of ST and that antibodies against either the adhesin or STh significantly impaired toxin delivery and cGMP activation in target T84 cells. Finally, we used FLAG epitope fusions to demonstrate that the STh propeptide sequence is secreted by ETEC, potentially providing additional epitopes for antibody neutralization. These studies collectively extend our understanding of ETEC pathogenesis and potentially inform additional avenues to mitigate disease by these common diarrheal pathogens.
INTRODUCTION
Diarrheal illnesses in low-income countries continue to cause substantial morbidity and remain one of the leading causes of death in young children under the age of 5 years in developing countries. Among the bacterial causes of diarrheal illness, enterotoxigenic Escherichia coli strains (ETEC) are commonly linked to more-severe forms of illness in young children (1). These organisms are perennially the most common cause of diarrhea in those who travel to areas of endemicity where sanitation is poor (2, 3); however, they have been identified repeatedly as the etiology of diarrheal outbreaks and sporadic cases of illness in industrialized countries, including the United States (4–8).
Acute clinical presentations of ETEC infection may range from mild self-limited illness to severe choleralike diarrhea (9–11). In addition, ETEC and other diarrheal pathogens have been linked to pernicious sequelae, including malnutrition, growth stunting, and impaired cognitive development (12). Presently, there are no vaccines to protect against these common infections. ETEC strains are a genetically (13) and serotypically (14) diverse pathovar of E. coli defined by the production of heat-labile (LT) and/or heat-stable (ST) enterotoxins that activate production of host cyclic nucleotides to alter intestinal salt and water transport that culminate in net fluid losses and secretory diarrhea.
Heat-stable toxins are synthesized as 72-amino-acid proteins consisting of a signal peptide, a propeptide, and a carboxy-terminal region of 18 to 19 amino acids, which forms the mature active enterotoxin (15). Two enterotoxins cause disease in humans: STp (ST1a), 18 amino acids, and STh (ST1b), 19 amino acids. Both of the mature toxins contain a total of six cysteine residues that form three intramolecular disulfide bonds (16). Their overall structure is shared with two similar mammalian peptides, guanylin and uroguanylin. Each of the bacterial and mammalian peptides binds to guanylate cyclase C (17, 18), leading to increases in intracellular cyclic GMP (cGMP) (19). Increases in cGMP result in activation of protein kinases that phosphorylate and activate the cystic fibrosis transmembrane regulatory (CFTR) channel and inhibit sodium-hydrogen ion exchange via sodium/hydrogen exchanger 3 (NHE3) (20). These effects lead to a net loss of salt and water into the intestinal lumen with ensuing watery diarrhea.
Bacteria producing any of the toxins LT, STh, or STp have been linked to diarrheal illness in humans (21–24), and recent studies suggest that ST-producing ETEC is commonly represented among the pathogens that cause severe diarrheal illness among young children in low-income countries, leading to substantial interest in the development of a vaccine that incorporates ST toxoids (25).
Enterotoxigenic E. coli strain H10407, originally isolated from a case of severe choleralike diarrheal illness in Bangladesh, is to date the most extensively characterized isolate of this pathovar. Interestingly, this isolate encodes all three canonical enterotoxins associated with ETEC diarrheal illness in humans (26, 27), with the gene for STh on the largest (94,797-bp) virulence plasmid p948 (NCBI GenBank accession number NC_017724.1) and the genes for both LT and STp on a 66,681-bp plasmid, p666 (NCBI GenBank accession number NC_017722.1) (28). In addition, H10407 encodes a copy of the ST-like EAST1 peptide (29) on the large p948 plasmid (30). H10407 is frequently used as the challenge strain in controlled human infection models to test candidate vaccines. Therefore, we set out to examine the relative contribution of STh and STp to the accumulation of cGMP in host epithelia by H10407 and the ability of anti-ST and antiadhesin antibodies to mitigate effective toxin delivery by the bacteria.
RESULTS
Contributions of STh, STp, and EAST1 to activation of cGMP in target epithelial cells.
Understanding the individual contributions of ST and ST-like molecules of ETEC is relevant to development and testing of toxin neutralization strategies. H10407 encodes three peptides with the potential to activate cGMP in target intestinal epithelial cells: STh, STp, and EAST1, a heat-stable toxin originally identified in enteroaggregative E. coli (29). STh is encoded by the estH gene on the large, 94,797-bp plasmid p948 (NCBI GenBank accession NC_017724.1). The astA gene, which encodes the EAST1 peptide (30), is embedded within an IS1414 insertion sequence (31) and is also located on the p948 plasmid immediately downstream from the etpBAC adhesin locus (28, 32). STp is encoded by the estP gene on a second, 66-kb virulence plasmid (NCBI GenBank accession number FN649417.1) in H10407.
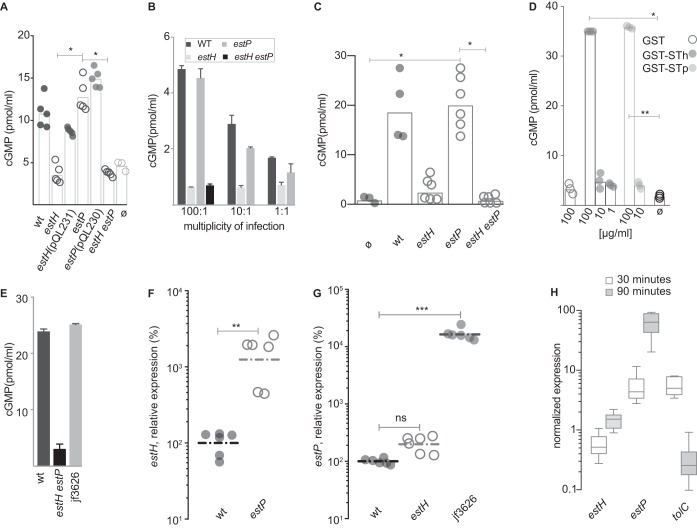
We found that deletion of estH resulted in appreciable decreases in cGMP production at or near background levels of cGMP production by uninfected cells and that complementation of estH in trans restored activation of this cyclic nucleotide (Fig. 1A). In contrast, the estP mutant was not appreciably different from the wild-type strain, and the introduction of the estP mutation to the estH strain did not yield further measurable decreases in cGMP production by target epithelial cells. Similar results were obtained at various multiplicities of infection (MOIs) (Fig. 1B), suggesting that this was not related to a spurious inoculum effect. In addition, infection of small intestinal enteroids demonstrated that while deletion of estH alone resulted in a substantial decrease in the production of cGMP in target epithelia to background levels, deletion of estP had no demonstrable effect on cyclic nucleotide production (Fig. 1C). However, we found that in distinct contrast to an A14Q mutant STh peptide (25), the target T84 cells did respond to glutathione S transferase (GST) fusions to either STh or STp (Fig. 1D) and that estP complementation of the estH estP dual deletion mutant in trans was sufficient to completely restore activation of cGMP to wild-type levels (Fig. 1E), clearly suggesting that enterocytes have the capacity to respond to either peptide. These results suggested that deletion of estP results in a compensatory increase in estH transcription and/or secretion of the encoded peptide. Interestingly, we found that relative to what was seen in the wild-type strain, estH transcription was significantly increased in the estP mutant (Fig. 1F), while transcription of estP in the estH mutant was not substantially increased (Fig. 1G). Collectively, these results suggest that both ST peptides likely act in conjunction with heat-labile toxin to cause the diarrheal illness associated with the H10407 strain, but the molecular details governing transcriptional regulation of toxin production have not been completely elucidated.
FIG 1.
Relative contributions of STh and STp to cGMP production in target epithelia. (A) cGMP production in T84 target epithelial cells following infection with wild-type estH, the complemented estH mutant estH(pQL231), the estP mutant, the complemented estP mutant estP(pQL230), and the estH estP dual deletion mutant. ø, uninfected cells. Data represent the combined results of two experimental replicates with each data point representing a technical replicate. (B) cGMP production in T84 cells at various multiplicities of infection with H10407 (wt), the estH or estP mutant, or the estH estP dual deletion mutant (n = 3 technical replicates; shown are mean values ± standard errors of the means [SEM]). (C) cGMP production in small intestinal enteroids following infection with the wild type (wt) , the estH or estP mutant, or the estH estP dual deletion mutant. (D) cGMP production in T84 cells following addition of purified GST, GST-STh, or GST-STp fusions. Numbers on the x axis represent final concentrations of protein in micrograms per milliliter. ø, untreated cells. Data include three technical replicates where geometric means are represented as bars. In panels A, C, and D, bars represent geometric means, with statistical comparisons by analysis of variance (ANOVA) (Kruskal-Wallis); *, P < 0.05; **, P < 0.01. (E) Complementation of the estH estP mutant with estP in trans (jf3626) restores wild-type levels of toxin activity following infection of T84 cells (bars represent results for 3 technical replicates ± SEM). (F) Transcription of estH gene in wild-type and estP mutant bacteria. Individual symbols represent 6 technical replicates combined from two independent experiments. Horizonal lines represent geometric means of expression relative to the wild type (H10407); **, P = 0.002 by Mann-Whitney (two-tailed) nonparametric comparison. (G) Transcription of estP gene in estH mutant bacteria relative to the wild type. The estP-complemented estH estP mutant, jf3626, is shown as a positive transcription control (***, P < 0.0002 by ANOVA, Kruskal-Wallis testing). (H) Transcription of genes encoding ST toxins STh (estH), STp (estP), and TolC. Data are normalized relative to the arcA housekeeping gene and represent the ratio of transcripts attached to planktonic bacteria at 30 and 90 min after infection of monolayers. Whisker plots represent the range of data obtained over six replicates from two independent experiments. Horizontal lines represent mean values. Data for each group represent mean of 3 replicates ± SEM.
Because we previously demonstrated that a number of virulence factors are transcriptionally modulated by ETEC pathogen-host cell interaction (33) we also examined transcription of both ST toxin genes and tolC, which encodes the putative export channel for ST. Interestingly, we found that in contrast to estH, estP transcription was significantly influenced by bacterial cell contact, with transcription of the gene encoding STp enhanced by bacterial adhesion, as was the tolC gene (Fig. 1H). These data suggest that similar to LT, the secretion and delivery of one or both of the STh and STp heat-stable toxins are likely highly orchestrated by contact with the intestinal epithelia.
In addition to STh and STp, earlier studies demonstrated that ETEC strains, including H10407 (31), also encode EAST, a heat-stable toxin originally identified in enteroaggregative E. coli (29, 31). Although the structures of the mature EAST and STh peptides are similar, there is only 37% identity (7/19 amino acids) between the two molecules (29), a finding that could impact the rational design of toxoids if EAST contributes to ETEC diarrheal illness. However, induction of cGMP in monolayers infected with a strain carrying an isogenic mutation of the astA gene, encoding EAST on the large p948 virulence plasmid of H10407 (3.8 ± 0.42 pmol/ml), was not significantly different from that of the wild-type parent (4.0 ± 0.35 pmol/ml), while deletion of both estP and estH resulted in complete abrogation of cGMP production (0.52 ± 0.06 pmol/ml), findings that suggest that EAST may not contribute significantly to diarrheal illness caused by this particular ETEC isolate.
TolC is required for effective ETEC secretion of STh and STp.
The precise mechanism for secretion of heat-stable toxins from ETEC strains associated with disease in humans is presently uncertain. Prior studies of heat-stable toxin investigated the secretion of STb (ST-II) (34) or STp (STIa) (35) from laboratory strains of E. coli containing recombinant expression plasmids. While both studies suggested that the outer membrane protein TolC is involved in secreting these toxins from the recombinant E. coli background, there are conflicting data regarding the involvement of the STb (STII) toxin in human disease (36, 37), and unlike the STh and STp toxins, STb does not bind to guanylate cyclase C. Similarly, to our knowledge, there has been no verification of the role of tolC in mediating the secretion of either of the ST1 toxins (STh and STp) from strains of ETEC isolated from humans. Therefore, to verify the importance of TolC in secretion of both STh and STp from ETEC strains that cause human illness, we constructed an isogenic tolC mutant in the ETEC H10407 strain and tested the ability of the mutant bacteria to deliver ST to target epithelial cells.
We found that mutants lacking tolC were devoid of detectable toxin in culture supernatants (Fig. 2A) and were markedly deficient in their ability to deliver ST toxins to epithelial monolayers, as we observed only background levels of cGMP production following infection with the tolC mutant strain, and complementation with tolC in trans restored the ability of the bacteria to provoke a cGMP response in targeted cells (Fig. 2B).
FIG 2.
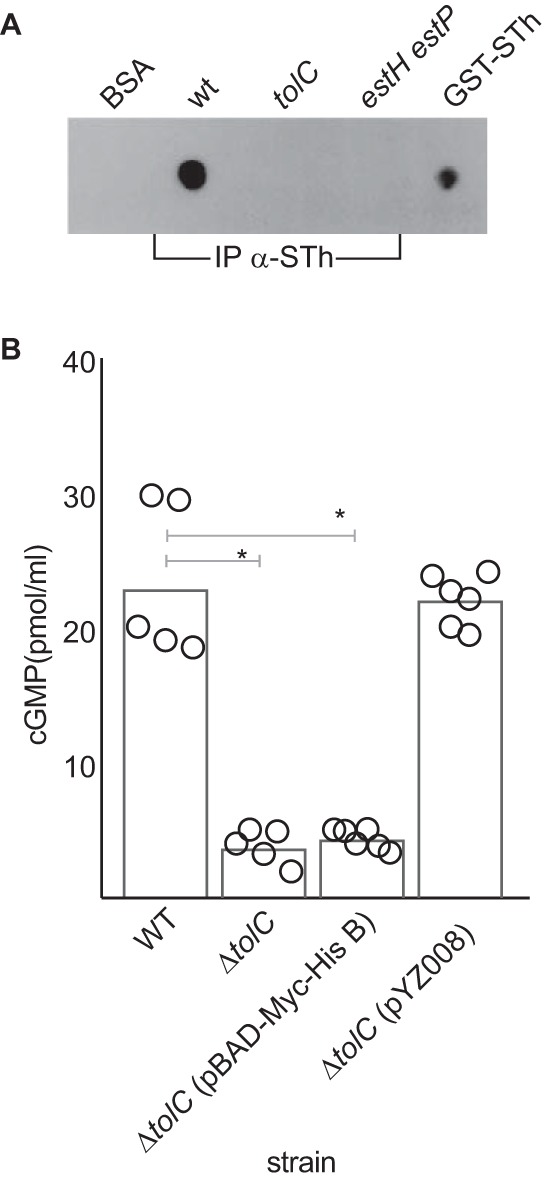
Role of tolC in ST secretion and delivery to target epithelial cells. (A) Immunoblot detection of ST toxins in culture supernatants of the wild-type H10407 strain, tolC mutant, or estH estP dual deletion mutant following immunoprecipitation of culture supernatants with affinity-purified anti-STh antibodies (IP α-STh). BSA and GST-STh fusion are shown as negative and positive controls, respectively. (B) cGMP production by T84 target epithelial cells following infection with wild-type (wt) ETEC strain H10407 (STh, STp), the tolC mutant, the vector-complemented mutant (pBAD-Myc-HisB) or the tolC-complemented mutant (pZY008). Data shown represent the combined results of two experimental replicates. Individual data points represent technical replicates, and bars indicate the geometric mean values, obtained in the presence (+) of phosphodiesterase inhibitors (PDE-I). Statistical comparisons by ANOVA (Kruskal-Wallis): *, P < 0.05.
Optimal delivery of ST requires the EtpA adhesin.
We have previously demonstrated that intimate interaction of ETEC with intestinal epithelial cells is essential for efficient delivery of heat-labile toxin to intestinal epithelial cells (38). Moreover, delivery of LT requires the concerted action of several ETEC adhesins with different receptor specificities (39, 40, 52) including EtpA, a plasmid-encoded secreted adhesin (32) that serves as a molecular bridge between bacteria and intestinal epithelial cells (42). EtpA is expressed by a diverse population of ETEC isolates (43), is recognized during the course of infection (44), and is a protective antigen in preclinical models of ETEC infection (40, 45, 46).
Because we are exploring the role of EtpA as a protective immunogen (47) and ST-producing ETEC strains have predominated in recent epidemiologic studies (1, 48), we set out to examine the dependence of ST delivery on EtpA-mediated bacterial adhesion by comparing cGMP activation of target intestinal epithelial cells infected with wild-type ETEC to that in cells infected with an isogenic etpA mutant. These studies demonstrated that cGMP activation in target epithelia by wild-type bacteria was significantly accelerated relative to that seen with the etpA mutant (Fig. 3), suggesting that efficient delivery of these small peptides also requires effective bacterium-host interactions.
FIG 3.

The EtpA adhesin is required for optimal delivery of heat-stable toxins. Shown are comparisons of cGMP activation in target T84 intestinal epithelial cells following infection with wild-type ETEC H10407 or the etpA mutant (n = 4 replicates), or the estH estP dual deletion mutant and uninfected monolayers (n = 3). Dashed lines in each group connect geometric mean values obtained at 30 and 60 min following the addition of bacteria. *, P < 0.05; obtained by Mann-Whitney (two-tailed) comparisons.
Similarly, we found that antibodies directed against either the EtpA adhesin molecule or STh significantly inhibited the delivery of heat-stable toxins to target cells (Fig. 4). Although we were not able to demonstrate that the combination of these antibodies was synergistic, these data provide additional support to the concept that EtpA could be useful as a target to engender coverage against a wide variety of ETEC isolates that produce heat-stable and/or heat-labile enterotoxins (43, 49).
FIG 4.
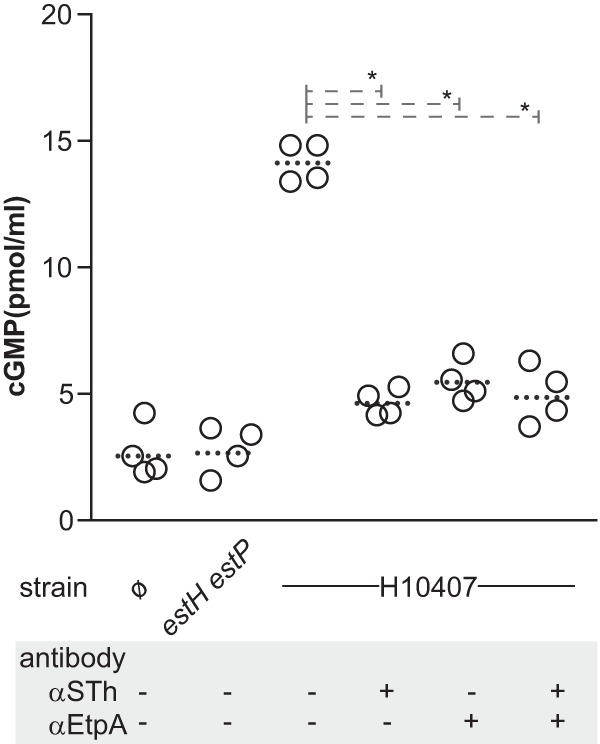
Antitoxin and antiadhesin antibodies inhibit heat-stable toxin delivery. Shown are monolayers infected with the dual heat-stable toxin mutant (estH estP), wild-type H10407, or uninfected control monolayers (ø). Dotted horizontal lines for each group represent geometric means. *, P = 0.028 by Mann Whitney two-tailed nonparametric comparisons. Both anti-STh and anti-EtpA antibodies were affinity purified and cross adsorbed and used at final dilution of 1:25.
Delivery and secretion of epitope-tagged STh.
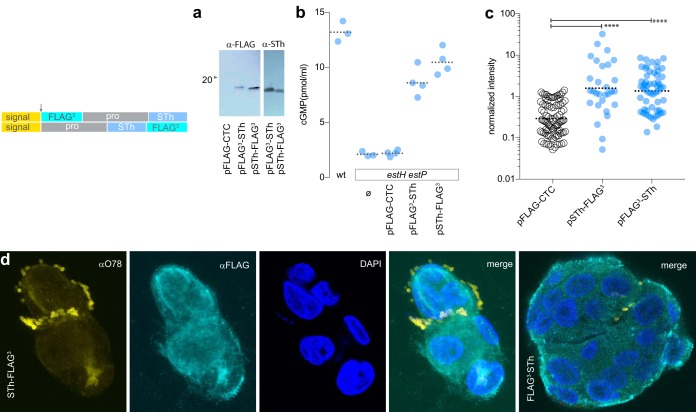
Understanding the nature of ST secretion and its delivery to epithelial receptors could be relevant to an informed development of ST toxoid molecules. STh is synthesized as a 72-amino-acid molecule that includes a 19-amino-acid signal peptide, followed by a 34-amino-acid propeptide and a mature ST molecule of 19 amino acids. Although most attempts to develop ST toxoids have targeted the mature peptides (16, 25), there are earlier but conflicting data regarding the precise form of ST that is secreted into the extracellular milieu (15, 50), with some data suggesting that the propeptide may be exported with subsequent processing to the mature peptide outside the bacteria (51). To investigate the potential secretion of the propeptide, we engineered 3×FLAG-epitope fusions to the amino-terminal end of the propeptide region (FLAG3-pro-STh) of STh and compared the export and delivery of the resulting peptides to those of carboxy-terminal fusions to the mature peptide (pro-STh-FLAG3) (Fig. 5).
FIG 5.
Secretion and delivery of FLAG-epitope-tagged STh. The schematic (top left) depicts the location of the region encoding the 3×FLAG peptide relative to the estH gene. The arrow shows the location of the predicted signal peptide cleavage site which is retained in both constructs. (a) Anti-FLAG and anti-STh immunoblots of FLAG3-STh and STh-FLAG3 peptides recovered by anti-FLAG immunoprecipitation from culture supernatants of the ETEC estH estP mutant bearing pFLAG3-STh and pSTh-FLAG3. (b) Complementation of the estP estH mutant strain with pSTh-FLAG3 or pFLAG3-STh restores toxicity upon infection of T84 cells. (c) Quantitation of FLAG3-tagged ST delivered to epithelial cells by estH estP bacteria complemented with pSTh-FLAG3 or pFLAG3-STh. Values represent fluorescence intensity per field. ****, P < 0.0001 by ANOVA (Kruskal-Wallis test). (d) Confocal immunofluorescence images of bacteria expressing pST-FLAG3 (anti-O78, yellow) attached to T84 cells (nuclei, blue) and the distribution of FLAG epitope-tagged toxin (cyan).
We were able to recover either the amino-terminal or carboxy-terminal FLAG-tagged molecules from culture supernatants of estH estP mutant ETEC transformed with the pSTh-FLAG3 or pFLAG3-STh plasmids, respectively (Fig. 5a). Both molecules appeared to yield functional ST mature peptides, as either plasmid was sufficient to complement the ability of the estH estP mutant to activate cGMP in target epithelial cells (Fig. 5b). Likewise, we were able to demonstrate binding of FLAG-epitope-tagged molecules to target epithelial cells following infection with either of the complemented strains (Fig. 5c and d). Collectively, these data appear to reaffirm earlier observations of STh (51), suggesting that both the mature form (19 amino acids) of the peptide and the extended pro-STh (53 amino acids) may be found outside the bacteria.
DISCUSSION
Because of the significant global burden of diarrheal illness caused by ETEC, these pathogens have been a target for vaccine development since they were first identified as a cause of severe diarrheal illness more than 4 decades ago. Currently, there is no vaccine available that affords broad-based protection against ETEC, in part due to the substantial antigenic diversity within the pathovar and limited mechanistic insight into immunologic correlates of the protection that appears to follow early infections among young children in areas of endemicity. Moreover, many features of the molecular pathogenesis of these common pathogens have not been explored in sufficient detail to inform vaccine development.
Because ST-producing ETEC isolates comprise a large proportion of strains associated with symptomatic diarrheal illness (1), we investigated the molecular contributions of the known heat-stable toxins, their proposed secretion apparatus, and bacterial adhesion to toxin delivery. The present studies suggest that efficient delivery of ST toxins is a complex process that requires the ability to effectively engage host cells and produce at least transient adhesion afforded by EtpA and other adhesins (52). Moreover, as antibodies directed at either the EtpA adhesin or ST impaired toxin delivery, our studies provide further support for development of a vaccine platform that combines ST toxoid (25) and antiadhesin approaches. However, further studies will be needed to define an optimal vaccination approach capable of generating sufficient antibodies to neutralize binding of the small ST peptides to cognate receptors in the small intestine.
Our data provide an important validation of the potential biologic importance of both STh and STp toxins. While recent large-scale epidemiologic data have highlighted the importance of ETEC strains producing STh as a cause of moderate to severe diarrhea in young children (1), subsequent reevaluation of these data has suggested that STp-producing strains also contribute significantly to the overall burden of illness associated with ETEC (48). Indeed, earlier clinical studies demonstrated that while there were regional differences in the distribution of STp and STh among ETEC isolates, both peptides were commonly associated with diarrheal illness, and the clinical presentations attributed to ETEC producing either ST peptide were indistinguishable (24). In keeping with these observations, we found no difference in the ability of STh or STp peptides to elicit cGMP production (19) in target epithelia. Although deletion of the gene encoding STh resulted in lower cGMP production in infected monolayers than when the estP gene was deleted from the ETEC H10407 strain, we discovered a compensatory increase in transcription of estH in the estP mutant. Collectively, therefore, our results suggest that both peptides likely contribute to diarrheal illness caused by H10407. Indeed, deletion of the tolC gene, which encodes the membrane transporter for both peptides, resulted in complete abrogation of cGMP activation in target epithelial cells, similar to deletion of both the estH and estP toxin genes from H10407.
Earlier studies also noted that many ETEC strains, including H10407, bear copies of an insertion sequence (31) that encompasses the astA gene encoding EAST1 (30), a cGMP-activating peptide structurally similar to ST1 that was originally identified as a heat-stable enterotoxin in enteroaggregative E. coli (29). However, we saw similar increases in cGMP production by target epithelial cells following infection with the astA mutant and wild-type ETEC. We cannot rule out the possibility of EAST1 expression from additional copies of astA not residing on the large, p948 virulence plasmid or that EAST1 is not optimally expressed in vitro. However, the complete absence of a cGMP response in cells infected with the STh/STp deletion mutant might alternatively suggest that EAST1 does not contribute to ETEC virulence and that further toxoid vaccine development can simply focus on engendering neutralizing antibody responses to the established ST and LT enterotoxins.
The reaffirmation of TolC as a key mechanism for export of ST1 toxins could be relevant to the management of diarrheal illness. The rapid emergence of multidrug resistance in the Enterobacteriaceae that is in part dependent on drug efflux through TolC has stimulated interest in efflux inhibitors to enhance the potency of available antimicrobial agents (53–55). Theoretically, these inhibitors could offer novel therapeutic agents for treatment of ETEC.
Our studies also revisit the concept that the larger propeptide form of STh may be exported by the bacteria, and the data presented here are consistent with prior observations suggesting that some processing of the propeptide occurs outside the bacteria (15, 51). Further study will be needed, however, to determine whether the propeptide sequence contributes to immunologic recognition of ST following infection and whether these larger molecules might be exploited in the development of improved toxoids to neutralize ST.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used are listed in Table 1. For general purposes, ETEC bacteria were grown in lysogeny broth at 37°C overnight from frozen glycerol stocks. To optimize synthesis and secretion of ST toxin, bacteria were grown in Casamino Acids-yeast extract-sucrose medium (CAYE-ST) (2% Casamino Acids, 0.6% yeast extract, 43 mM NaCl, 38 mM K2HPO4, 203 mM MgSO4, 25 mM MnCl2, 18 mM FeCl3, 2% sucrose) at 37°C overnight from frozen glycerol stocks.
TABLE 1.
Bacterial strains and plasmids used in this studyd
| Strain or plasmid | Description | Source or reference(s) |
|---|---|---|
| Strains | ||
| H10407 | Wild-type ETEC strain O78:H11; CFA/1; LT, STh, STp; EtpA | 28, 32, 64 |
| jf2656 | H10407 derivative with isogenic deletion of estP | This study |
| jf2649 | H10407 derivative with isogenic deletion of estH | This study |
| jf2847 | H10407 derivative with isogenic deletion of estP and estH | This study |
| jf3038 | Deletion of astA on the large p948 virulence plasmid of H10407; Kmr | This study |
| jf3076 | jf2656 complemented with pQL230 | This study |
| jf3081 | jf2649 complemented with pQL231 | This study |
| JW5503-1 | E. coli K-12 in-frame ΔtolC732::Kmr | 58 |
| jf4652 | H010407 derivative with tolC::Kmr mutation | This study |
| jf4709 | jf4652 with pBAD/mycHisB | This study |
| jf4712 | jf4652 complemented with pYZ008 | This study |
| jf4644 | jf2847 with pFLAG-CTC, Cmr Kmr Ampr | This study |
| jf4648 | jf2847 with pFLAG-STh, Cmr Kmr Ampr | This study |
| jf4651 | jf2847 with pSTh-FLAG, Cmr Kmr Ampr | This study |
| jf3626 | jf2847 with pQL239, Cmr Kmr Ampr | This study |
| AAEC191A | Afimbriate E. coli K-12 derivative | 61 |
| Plasmids | ||
| pBAD/mycHisA | pBR322ori; PBAD; Ampr; araC; C-terminal myc epitope tag; arabinose-inducible expression plasmid | Invitrogen |
| pBAD/mycHisB | pBR322ori; PBAD; Ampr; araC; C-terminal myc epitope tag; arabinose-inducible expression plasmid | Invitrogen |
| pKD13 | oriRSKγ Tn5 neomycin phosphotransferase (Kmr), FRT, β-lactamase (Ampr) | 56 |
| pKD46 | Lambda red recombinase helper plasmid | 56 |
| pQL239 | estP cloned into XhoI site of pBAD/myc-HisA in frame with myc-His tags | This study |
| pACYC184 | p15Aori, Cmr Tcr | 65 |
| pSMD001 | estH cloned into EcoRV site of pACYC184 | This study |
| pGPS4 | oriRSKγ Cmr Tcr | NEB |
| pETDuet-1 | pBR322ori; lacI; Ampr; IPTG-inducible expression plasmid | Novagen |
| pQL230 | estP cloned into BglII and XhoI sites of pETDeut-1 | This study |
| pQL231 | estH cloned into NcoI and EcoRI sites of pETDeut-1 | This study |
| pQL238 | estP cloned into BglII and XhoI sites and estH cloned into NcoI and EcoRI sites of pETDeut-1 | This study |
| pGEX-4T1 | lacIq, Ampr; expression plasmid for N-terminal GST fusions | GE Healthcare Life Sciences |
| pCW002a | 5,047-bp GST-STh expression plasmid, Ampr | This study |
| pCW003b | estP cloned into BamHI and EcoRI sites of pGEX | This study |
| pYZ011c | 5,047-bp GST-mSTh expression plasmid for mSTh (A14Q), Ampr | This study, 25 |
| pYZ008 | H10407 tolC cloned into pBAD/mycHisB | This study |
| pFLAG-CTC | 5.3-kb plasmid, expressing C-terminal FLAG fusion proteins, Ampr | Sigma |
| pFLAG3-STh | 5,609-bp Flag-pro-STh expression plasmid, Ampr | This study |
| pSTh-FLAG3 | 5,612-bp pro-STh-Flag expression plasmid, Ampr | This study |
Addgene accession number 90225 (https://www.addgene.org/90225/).
Addgene accession number 90226 (https://www.addgene.org/90226/).
Addgene accession number 110570 (https://www.addgene.org/110570/).
Abbreviations: Kmr, kanamycin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; NEB, New England Biolabs; FRT, FLP recombination target.
Construction of mutants lacking production and secretion of heat-stable toxins.
Lambda red recombinase-mediated recombination (56) was used to disrupt genes encoding heat-stable toxins in H10407. In brief, we used the primers jf070212.1 and jf070212.2 to amplify the kanamycin (Km) cassette from pKD13 with a 36-bp overhang sequence from the estH gene and primers jf070212.3 and jf070212.4 to amplify the kanamycin cassette from pKD13 with a 36-bp overhang sequence from the estP gene. The PCR product was electroporated into competent H10407 containing helper plasmid pKD46 and selected on LB plates with 50 μg/ml kanamycin. Kanamycin-resistant (Kmr) colonies were screened by PCR with primers jf071612.1 and jf071612.2 for the estH locus and with primers jf071612.3 and k2 for estP. The deletion mutants were further confirmed by toxin multiplex PCR as previously described (57).
To construct complemented strains, we amplified the estP gene with primers jf030316.3 and jf060614.1 for cloning into the XhoI site of pBAD/mycHisA, yielding pQL239; estH was amplified with primers jf072414.5 and jf072414.6 and cloned into the EcoRV site of pACYC184 to produce pSMD001. We also cloned estH and estP genes individually and together in pETDuet-1 at NcoI and EcoRI sites and at BglII and XhoI sites with primer pairs jf060614.2/jf060614.3 and jf060614.4/jf060614.5, respectively. This resulted in plasmids pQL230 (estP), pQL231(estH), and pQL238 (estP estH) (Table 1). After confirmation by sequencing, the respective plasmids were transformed into the deletion strains, and complementation was confirmed by PCR.
To generate an ETEC tolC deletion mutant, primers jf110716.44/45 were first used to amplify a ΔtolC732::Kmr fragment from JQ55031 (58). Next, the regions flanking tolC in the H10407 genome were amplified with primer pairs jf110716.38/jf110716.43 and jf110716.46/jf110716.39 to amplify 1,027 bp of upstream sequence and 985 bp of downstream sequence, respectively. The three resulting fragments were then fused by PCR with primer pairs jf110716.38/jf110716.39, using high-fidelity polymerase Phusion (Thermo Fisher Scientific), with denaturing for 2 min at 98°C, followed by 30 cycles of 10 s at 98°C, 30 s at 65°C, and 1.5 min at 72°C and a final extension for 10 min at 72°C. The resulting 3,325-bp amplicon was then introduced into H10407(pKD46) as described above. Kanamycin-resistant, ampicillin-sensitive colonies were then screened by colony PCR using primer pair jf112016.50/51 flanking the entire amplicon for a 4,269-bp product and primers jf101716.21/22 specific to tolC gene (603-bp product). Primers k2/jf112016.51 (2,024-bp product) were then used to confirm the tolC gene deletion and the Kmr cassette integration in the H10407 genome. To complement the tolC mutant, the tolC gene was amplified from H10407 genomic DNA using primers jf120716.59/jf120716.60, and the plasmid vector backbone was amplified from plasmid pBAD/myc-His B using primers jf120716.52/jf120716.53. The recombinant pYZ008 complementation plasmid was assembled using an adaptation of circular polymerase extension cloning (CPEC) (59) (30 s of denaturation at 98°C, followed by 20 cycles of 10 s at 98°C, 30 s at 55°C, and 1.5 min at 72°C and a final extension for 10 min at 72°C). Following sequence verification, pYZ008 or the pBAD/myc-His B vector control plasmids were then used to transform the ΔtolC mutant. Complementation was confirmed by PCR with primers jf101716.21/22.
FLAG-STh fusions.
FLAG epitope fusions were constructed to introduce the 3×FLAG sequence between the signal peptide of estH and the beginning of the propeptide-encoding region (on plasmid pFLAG3-STh) or at the 3′ end of estH (on pSTh-FLAG3). The 3×FLAG fragment was first constructed by annealing complementary synthetic oligonucleotides jf092616.9 encoding positive-strand bases 1 to 43 and jf092616.10 representing negative-strand bases 66 to 19 of a 66-bp sequence encoding the 3×FLAG peptide (DYKDHDGDYKDHDIDYKDDDDK).
To generate plasmid pFLAG3-STh, in which the 3×FLAG-encoding sequence was inserted between the STh signal peptide and the STh propeptide, primers jf101916.33/jf092616.4 were first used to amplify the nucleotides (1 to 63) of estH encoding the signal peptide. Next, primers jf092616.1/jf092616.2 were used to amplify the 3×FLAG fragment from the above-described synthetic oligonucleotide 3×FLAG template flanked by nucleotide extensions representing nucleotides 44 to 60 and 64 to 99 of estH, while primers jf092616.3/jf101916.34 were used to amplify the 3′ end of estH from nucleotide 64 to the native stop codon. The three fragments were fused in a single PCR using primers jf101916.33/jf101916.34. Next, the vector backbone was amplified using primers jf101916.31/jf101916.32 from pFLAG-CTC (Sigma), followed by the final assembly of pFLAG3-STh by CPEC. Similarly, to make plasmid pSTh-FLAG3, primers jf092616.5/jf101916.35 were used to amplify 3×FLAG with a 5′ nucleotide extension representing nucleotides 197 to 216 of estH, while primers jf101916.33/jf092616.8 were used to amplify the estH gene with a 5′ nucleotide extension corresponding to pFLAG-CTC. The resulting amplicons were then fused in a single PCR using primers jf101916.33/jf101916.35 and assembled with the pFLAG-CTC backbone by CPEC.
Cloning, expression, and purification of recombinant ST proteins peptides.
To construct a GST-STh or mutant GST-mSTh (A14Q) expression plasmid, synthetic oligonucleotides that encompassed the region of the estH gene corresponding to the mature peptide minus the native start codon, preceded by an in-frame flexible linker sequence (60), were synthesized (IDT, Coralville, IA). The forward sequence (jf042715.1 for STh and jf042517.1 for mSTh), preceded by a BamHI overhang sequence (GATCC), and the reverse sequence (jf042715.2 for STh and jf042517.2 for mSTh), preceded by an EcoRI overhang sequence (AATTC), were mixed in a 1:1 molar ratio, heated to 95°C for 5 min, and cooled to room temperature. The annealed double-stranded 91-bp DNA fragments were then cloned directly into the pGEX-4T1 vector digested with BamHI and EcoRI, yielding plasmids pCW002 and pYZ011, respectively. A similar strategy was to construct a GST-STp expression plasmid using forward oligonucleotide jf031915.5 and the reverse sequence jf031915.6, resulting in plasmid pCW003. E. coli TOP10(pCW002), TOP10(pCW003), and TOP10(pYZ011) were then used to express recombinant GST-STh, GST-STp, and GST-mSTh, and the resulting fusion proteins were purified by affinity chromatography. In brief, the bacterial cultures were grown in Luria broth at 37°C to an optical density at 600 nm (OD600) between 0.6 and 0.7 and induced with 1 mM (final concentration) IPTG (isopropyl-β-d-thiogalactopyranoside) for 1 to 3 h. The cell pellets were resuspended in 30 ml cold phosphate-buffered saline (PBS) containing 5 mM dithiothreitol (DTT), 1 protease inhibitor tablet (Roche), and 0.1 mg/ml lysozyme. Following sonication, supernatants were clarified by centrifugation at 12,000 rpm for 20 min at 4°C, followed by passage through a 0.45-μm filter. Filtered supernatants were then loaded onto columns (glutathione Sepharose High Performance; GE Healthcare) preequilibrated with PBS, pH 7.3 (140 mM NaCl, 2.7 mM KCl,10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]). After washing with 20 column volumes of PBS containing 1 mM DTT, GST fusion proteins were eluted in fresh buffer containing 100 mM Tris-HCl (pH 8.0) and 10 mM reduced glutathione and dialyzed against PBS.
To liberate native STh or mutant STh (mSTh) from its GST fusion partner, GST-fusion protein was dialyzed in 50 mM NH4HCO3 (pH 8.5) containing 1 unit of thrombin (Sigma) per mg GST-fusion protein at 4°C overnight, centrifuged at 4,000 × g for 20 min with a 10-kDa-molecular-weight cutoff (MWCO) centrifugal filter to collect the filtrate, and dried by vacuum centrifugation. Peptide concentrations were then determined by measurement of the molar extinction coefficient at 280 nm (Take3; BioTek).
Transcriptional analysis of estH, estP, and tolC genes.
To assess transcription of toxin genes in mutant strains relative to wild-type H10407, the bacteria were grown overnight in 2-ml cultures of lysogeny broth at 37°C and 225 rpm and then diluted 1:100 into fresh medium and grown for an additional 3 h. To assess modulation of transcription upon cell contact, confluent T84 cells were inflected with early-log-phase H10407 bacteria (∼109 CFU) for 30 or 90 min. Total RNA was isolated from adherent and planktonic (nonadherent) bacterial fractions using an RNAspin Mini isolation kit (25050070; GE Life Sciences) and treated with DNase I (18068015; Thermo Fisher). PCR for the arcA housekeeping gene was used to confirm the removal of DNA. Total RNA was reverse transcribed (SuperScript VILO cDNA synthesis kit, 11754250; Thermo Fisher). RNA transcripts were quantified by real-time (RT) PCR (Fast SYBR green master mix, number 4385612 from Thermo Fisher; ViiA 7 real-time PCR system from Applied Biosystems). Primers specific to the arcA, estH, estP, and tolC genes are listed in Table 2. All transcripts were normalized to arcA and are presented as a ratio of transcripts in adherent bacteria relative to transcripts in planktonic bacteria.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′)a | Description |
|---|---|---|
| jf070212.1 | ATGAAAAAATCAATATTATTTATTTTTCTTTCTGTAGTGTAGGCTGGAGCTGCTTCG | Forward primer for amplifying estH flanking from pKD13 |
| jf070212.2 | TTAATAGCACCCGGTACAAGCAGGATTACAACACAAATTCCGGGGATCCGTCGACC | Reverse primer for amplifying estH flanking from pKD13 |
| jf070212.3 | ATGAAAAAGCTAATGTTGGCAATTTTTATTTCTGTAGTGTAGGCTGGAGCTGCTTC | Forward primer for amplifying estP flanking from pKD13 |
| jf070212.4 | TTAATAACATCCAGCACAGGCAGGATTACAACAAAGATTCCGGGGATCCGTCGACC | Reverse primer for amplifying estP flanking from pKD13 |
| jf071612.1 | GGCGCACACAAATATAAAG | 368-bp amplicon in H10407-estH downstream primer |
| jf071612.2 | AGCGGAGAGTATAGTATGA | estH upstream |
| jf071612.3 | AAAACCAGATAGCCAGAC | 168 bp upstream from estP on H10407 plasmid p666 |
| k2 | CGG TGC CCT GAA TGA ACT GC | Primer binding Kmr cassette for confirming estP and tolC deletion |
| jf072312.1 | ATGAAAAAATCAATATTATTTATTTTTCTTTCTGTACCCTGTTATCCCTAGATTT | Forward primer for amplifying estH from pGPS4 |
| jf072312.2 | TTAATAGCACCCGGTACAAGCAGGATTACAACACAATAACGGTCCTAAGGTAGC | Reverse primer for amplifying estH from pGPS4 |
| jf072412.5 | GGCCTCTTGCGGGATATCTAAATGAAAAAATCAATATTA | pACYC184_ECORV_TAA_STh forward primer for in-fusion cloning |
| jf072412.6 | GTCGGAATGGACGATATCTTAATAGCACCCGGTACA | pACYC184_ECORV_TAA_STh reverse primer for in-fusion cloning |
| jf060614.2 | AGGAGATATACCATGGATGAAAAAATCAATATTA | Forward primer for STh in NcoI and EcoRI sites on pETDuet-1 vector |
| jf060614.3 | CGCCGAGCTCGAATTCTTAATAGCACCCGGTACA | Reverse primer for STh in NcoI and EcoRI sites on pETDuet-1 vector |
| jf060614.4 | TACATATGGCAGATCTATGAAAAAGCTAATGTTG | Forward primer for STp in BglII and XhoI sites on pETDuet-1 vector |
| jf060614.5 | CTTTACCAGACTCGAGTTAATAACATCCAGCACA | Reverse primer for STp in BglII and XhoI sites on pETDuet-1 vector |
| jf042715.1 | GATCCGATCCCCGGGTACCGAGCTCGAATAGTAGCAATTACTGCTGTGAATTGTGTTGTAATCCTGCTTGTACCGGGTGCTATTGAG | 5′-Mature estH gene with BamHI, linker, gene, stop codon |
| jf042715.2 | AATTCTCAATAGCACCCGGTACAAGCAGGATTACAACACAATTCACAGCAGTAATTGCTACTATTCGAGCTCGGTACCCGGGGATCG | 3′-Mature estH gene with linker, gene, stop codon, EcoRI |
| jf042517.1 | GATCCGATCCCCGGGTACCGAGCTCGAATAGTAGCAATTACTGCTGTGAATTGTGTTGTAATCCTcaaTGTACCGGGTGCTATTGAG | 5′-Mutant estH gene with BamHI, linker, gene, stop codon |
| jf042517.2 | AATTCTCAATAGCACCCGGTACAttgAGGATTACAACACAATTCACAGCAGTAATTGCTACTATTCGAGCTCGGTACCCGGGGATCG | 3′-Mutant estH gene with linker, gene, stop codon, EcoRI |
| jf031915.5 | GATCCGATCCCCGGGTACCGAGCTCGAACACATTTTACTGCTGTGAACTTTGTTGTAATCCTGCCTGTGCTGGATGTTATTGAG | 5′-Mature estP gene with BamHI, linker, gene |
| jf031915.6 | AATTCTCAATAACATCCAGCACAGGCAGGATTACAACAAAGTTCACAGCAGTAAAATGTGTTCGAGCTCGGTACCCGGGGATCG | 3′-Mature estP gene with linker, gene, EcoRI |
| jf021915.1 | CGCTTACAGACAAGCTGTG | Reverse sequencing primer that binds 70 bp downstream of the pGEX cloning site |
| jf021915.2 | CCAGCAAGTATATAGCATGG | Forward sequencing primer that binds 117 bp upstream of pGEX cloning site |
| jf110716.38 | CGGGCGCAGTCTGTTCTATTG | Forward primer for amplifying 1,027-bp left-flank segment of H10407 tolC |
| jf110716.43 | TCATTTGCATTCCTTGTGGTGAAGCAGTATTTAGCGC | Reverse primer for amplifying 1,027-bp left-flank segment of H10407 tolC |
| jf110716.44 | AAAGGGTTATGTGTAGGCTGGAGCTGCTTCG | Reverse primer for amplifying Kmr cassette from strain JQ5503-1 |
| jf110716.45 | CTTCACCACAAGGAATGCAAATGATTCCGGGGATCC | Forward primer for amplifying Kmr cassette from strain JQ5503-1 |
| jf110716.39 | CTTTTCAACCTGGGCGAGGG | Reverse primer for amplifying 985-bp right-flank segment of H10407 tolC |
| jf110716.46 | CCTACACATAACCCTTTCCGTAACTGATGACGACGACGGGGCTTCGG | Forward primer for amplifying 985-bp right-flank segment of H10407 tolC |
| jf101716.21 | CGATCGTGATGCTGCCTTTG | 603-bp amplicon in H10407-tolC forward primer |
| jf101716.22 | AGCGACAGGTTGCGTTTTTC | 603-bp amplicon in H10407-tolC downstream primer |
| jf112016.50 | ATTTGCCATTGCTCACCAATAAAC | Forward primer binding 2 kb upstream of H10407 tolC |
| jf112016.51 | CTTGCAGACTGTTAAACTGGTCG | Reverse primer binding 2 kb downstream of H10407 tolC |
| jf120716.52 | GAACAAAAACTCATCTCAGAAGAGGATCTGAATAGCG | Forward primer to amplify 4,036 bp of pBAD/myc-His B |
| jf120716.53 | GGTTAATTCCTCCTGTTAGCCCAAAAAACGG | Reverse primer to amplify 4,036 bp of pBAD/myc-His B |
| jf120716.59 | GGCTAACAGGAGGAATTAACCATGCAAATGAAGAAATTGCTCCCCATTCT | Forward primer H10407 tolC gene |
| jf120716.60 | CTCTTCTGAGATGAGTTTTTGTTCTCAGTTACGGAAAGGGTTATGACCGTTACT | Reverse primer H10407 tolC gene |
| jf092616.1 | TTTCACCTTTCGCTCAGGATTACAAAGACCAC | Forward bp 44–60 of estH-5′3×FLAG |
| jf092616.2 | GAAGACCCTGCTGGTTTAGCCTTGTCATCGTC | Reverse bp 83–64 of estH-3′3×FLAG |
| jf092616.3 | GCTAAACCAGCAGGGTCTTCAAAAGAAAAAATTACA | Forward primer bp 64–99 of estH |
| jf092616.4 | ATCCTGAGCGAAAGGTGAAAAAGATAATACAGAAAGA | Reverse primer bp 63–27 of estH |
| jf092616.5 | CTGCTTGTACCGGGTGCTATGATTACAAAGAC | Forward primer bp 197–216 of estH-5′ 3×FLAG |
| jf092616.8 | ATAGCACCCGGTACAAGCAGGATTACA | Reverse primer representing bp 216–190 of estH |
| jf092616.9 | GATTACAAAGACCACGATGGTGACTATAAAGACCATGATATCG | Base pairs 1–43 and 66 through 19 of the 3×FLAG-encoding sequence |
| jf092616.10 | CTTGTCATCGTCGTCTTTATAATCGATATCATGGTCTTTATAGTCACC | |
| jf101916.31 | CAGATCTGGTACCCGGGAATTCT | For amplifying pFLAG-CTC vector backbone |
| jf101916.32 | TGAAGATCGATCTCTCGATCGAGTGA | |
| jf101916.33 | ATTCCCGGGTACCAGATCTGATGAAAAAATCAATATTATTTATTTTTCTTTCTGTATT | Forward primer beginning with pflag-CTC- sequence followed by estH sequence bp 1–38 |
| jf101916.34 | GATCGAGAGATCGATCTTCATTAATAGCACCCGGTACAAGCAGG | Reverse primer beginning with pflag-CTC sequence followed by 3′estH native reverse sequence (bp 219–196) |
| jf101916.35 | GATCGAGAGATCGATCTTCATTACTTGTCATCGTCGTCTTTATAATCGATATCATG | Reverse primer beginning with pflag-CTC-sequence followed by bases 69–34 of the 3×FLAG sequence |
| jf092313.5 | TCTTTCCCCTCTTTTAGTCAG | estP RT-PCR forward primer |
| jf092313.6 | ACAGGCAGGATTACAACAAAG | estP RT-PCR reverse primer |
| jf092313.7 | TACAAGCAGGATTACAACAC | estH RT-PCR forward primer |
| jf092313.8 | AGTGGTCCTGAAAGCATG | estH RT-PCR reverse primer |
| jf092210.1 | ATCAATCTGCCGGGTAAGAACGGT | arcA RT-PCR forward primer |
| jf092210.2 | TCCAGATCACCGCAGAAGCGATAA | arcA RT-PCR reverse primer |
| jf030316.3 | TGGATGCGAGCTCGAGCATGAAAAAGCTAATGTTG | estP forward primer for pBAD/mycHisA-STp cloning |
| jf060614.1 | AGCTGCAGATCTCGAGTTAATAACATCCAGCACA | estP reverse primer for pBAD/mycHisA-STp cloning |
See “Description” column for explanation of underlined bases.
Production and purification of anti-ST polyclonal antibody.
To generate rabbit polyclonal antibody that recognizes STh, New Zealand White rabbits were immunized (Rockland) with recombinant GST-STh. We used lyophilized E. coli AAEC191A (61) and an immobilized E. coli lysate column (44938; Pierce) to adsorb E. coli-reactive antibodies, followed by protein G column purification (HiTrap, GE17-0404-01). IgG GST-STh antibodies were affinity purified using GST-STh immobilized on nitrocellulose as previously described, and anti-GST-reactive antibodies were removed by cross-adsorption against GST coupled to glutathione agarose resin (G-250-100; Gold Biotechnology, St. Louis, MO).
Anti-STp antibody was purified from rabbit antisera (provided by Weiping Zhang, Kansas State University) by cross-adsorption against an immobilized E. coli lysate column, which was followed by affinity purification against recombinant GST-STp immobilized on nitrocellulose membranes for affinity purification.
Immunoprecipitation and detection of heat-stable toxins in culture supernatants.
Purified anti-STh polyclonal IgG was immobilized with AminoLink Plus coupling resin (Pierce Direct IP kit, 26148; Thermo Scientific). Clarified culture supernatants of overnight cultures of ETEC were mixed with protease inhibitor cocktail (88666; Thermo Fisher) and filtered through a 0.45-μm filter. Sixty milliliters of supernatant was then filtered through a 10-kDa-molecular-weight-cutoff membrane (Amicon). Filtrates were then dried by vacuum centrifugation and dialyzed in PBS against a 1-kDa-cutoff membrane (Float-A-Lyzer G2, MWCO 0.5 to 1 kDa; Spectrum Labs). Immunoprecipitations were conducted by incubation of filtrates with anti-STh-immobilized resin for 2 h at room temperature followed by elution with 4.5% acetic acid. Eluates were dried by vacuum centrifugation and reconstituted in PBS. Immunoprecipitated samples or purified protein/peptide as controls were applied directly to 0.22-μm-pore-size polyvinylidene difluoride (PVDF) membranes (Bio-Rad) and detected by anti-STh and/or anti-STp primary antibody and anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody using Clarity Western ECL substrate (Bio-Rad).
FLAG-STh fusion peptides were prepared from strain jf2847 carrying pFLAG3-STh, pSTh-FLAG3, or the blank vector pFLAG-CTC. Briefly, following overnight growth at 37°C in lysogeny broth containing ampicillin (100 μg/ml), cultures were diluted 1:100 in fresh medium, grown to an OD600 of ∼0.2, and then induced with 1 mM IPTG for 7 h. Forty milliliters of clarified supernatant was mixed with 200 μl of anti-FLAG (M2) affinity gel (A2220; Sigma), and then the mixture was incubated overnight at 4°C with agitation. After washing with Tris-buffered saline (TBS) buffer, bound proteins were eluted with 0.1 M glycine–HCl, pH 3.5, separated by SDS-PAGE, transferred to nitrocellulose for subsequent immunoblotting, and detected by anti-FLAG antibody (F1804; Sigma).
Confocal immunofluorescence imaging and quantification.
To examine the delivery of FLAG-tagged STh toxin to intestinal epithelial cells, estP estH mutant strains with or without plasmids encoding FLAG-tagged STh were grown overnight and diluted 1:50 in CAYE-ST medium, grown to an OD600 of ∼0.2, and then induced with 1 mM IPTG for 2 h. The bacteria were added to T84 cells at a multiplicity of infection (MOI) of ∼1:50, maintaining the induction with 1 mM IPTG. After infection with the bacteria, T84 cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min at room temperature, permeabilized with 0.1% Triton X-100 for 5 min, and blocked with 1.5% bovine serum albumin (BSA)–PBS at 37°C for 1 h. Anti-O78 rabbit polyclonal antiserum (Penn State University) diluted 1:300 in PBS with 0.02% Tween 20 (PBST) and 1.5% BSA was used to identify H10407 and monoclonal anti-FLAG M2 antibody (number F1804; Sigma) diluted 1:500 in PBST with 1.5% BSA to detect FLAG-tagged STh. Following incubation overnight at 4°C, slides were washed 3 times with PBS and incubated for 1 h at 37°C with goat-anti-rabbit IgG (H&L) Alexa Fluor 488 conjugate (A11070; Thermo Fisher) and goat-anti-mouse IgG (H&L) conjugated to Alexa Fluor 594 (A11032; Thermo Fisher) at a dilution of 1:500 in PBST containing 1.5% BSA. After washing, DAPI (4′,6-diamidino-2-phenylindole) was added at 1:6,000. Confocal microscopy images were acquired using a Nikon C2+ confocal microscope system. To quantitate binding of FLAG-epitope-tagged STh molecules, fluorescence detection was normalized to the DAPI signal using NIS-Elements DUO software (v4.4).
In vitro assessment of toxin delivery.
Cultures of bacteria were grown overnight in lysogeny broth from frozen glycerol stocks in CAYE-ST medium. Phosphodiesterase inhibitors (PDE-I) vardenafil hydrochloride trihydrate (number Y0001647), rolipram (number R6520), and cilostazol (number C0737) (all from Sigma-Aldrich) were each added to target intestinal epithelial cells at a final concentration of 16.7 or 50 μM and incubated with cells for 1.5 h (T84 cells) or 2 h (enteroids). Bacteria or toxin was added to T84 monolayers seeded into 96-well plates, and the treatment was continued for the indicated duration. After washing in PBS, cyclic GMP (cGMP) levels were determined by enzyme immunoassay (EIA; K020-H1; Arbor Assays) using the acetylated protocol as directed by the manufacturer.
To examine the capacity of antibodies to neutralize ST delivery, antibody against STh and/or EtpA was added directly to T84 cell monolayers at the indicated dilution at the time of infection. After 1.5 h, cells were washed by PBS and lysed, and cGMP assays were performed as described above.
Growth and differentiation of T84 cells and small intestinal enteroids.
T84 intestinal epithelial cells (ATCC CCL-248) were cultured in 96-well plates for 24 to 48 h at 37°C, 5% CO2 incubator, to >90% confluence in a 1:1 mixture of Dulbecco's modified Eagle medium (DMEM)/F12 medium supplemented with l-glutamine (2.5 mM) and fetal bovine serum (FBS; 5% [vol/vol]). Enteroids were grown as previously described in detail (62, 63). Briefly, cells obtained from the Washington University Digestive Diseases Research Core Center BioSpecimens Core were propagated in Matrigel (BD Biosciences, San Jose, CA) in 24-well plates at 37°C with a 1:1 mixture of L-WRN (Wnt3a, R-spondin, and Noggin) conditioned medium (CM) and primary culture medium (Advanced DEM/F12; Invitrogen) supplemented with 20% FBS, 2 mM l-glutamine, 100 units/ml penicillin, 0.1 mg/ml streptomycin, 10 μM Y-27632 (ROCK inhibitor; Tocris Bioscience, R and D Systems, Minneapolis, MN), and 10 μM SB 431542 (TGFBR1 inhibitor; Tocris Bioscience, R and D Systems). Cells were then added to Transwell filters (Corning) and incubated in differentiation medium (1:20 mixture of L-WRN CM and primary culture medium) lacking SB 431542 and were grown 2 to 3 days until confluence. Prior to addition of bacteria, cells were switched to differentiation medium lacking antibiotics.
Accession number(s).
Plasmids pCW002, pCW003, and pYZ011 have been deposited in Addgene (https://www.addgene.org/) under accession numbers 90225, 90226, and 110570, respectively.
ACKNOWLEDGMENTS
These studies were supported in part by funds from the National Institute for Allergy and Infectious Diseases (NIAID) grants R01AI89894 and R01AI126887 and the Department of Veterans Affairs (5I01BX001469-05); C.W. was supported in part by a Washington University/HHMI Summer Undergraduate Research Fellowship (HHMI SURF).
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.Hameed JM, McCaffrey RL, McCoy A, Brannock T, Martin GJ, Scouten WT, Brooks K, Putnam SD, Riddle MS. 2016. Incidence, etiology and risk factors for travelers' diarrhea during a hospital ship-based military humanitarian mission: Continuing Promise 2011. PLoS One 11:e0154830. doi: 10.1371/journal.pone.0154830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah N, DuPont HL, Ramsey DJ. 2009. Global etiology of travelers' diarrhea: systematic review from 1973 to the present. Am J Trop Med Hyg 80:609–614. [PubMed] [Google Scholar]
- 4.Medus C, Besser JM, Juni BA, Koziol B, Lappi V, Smith KE, Hedberg CW. 2016. Long-term sentinel surveillance for enterotoxigenic Escherichia coli and non-O157 Shiga toxin-producing E. coli in Minnesota. Open Forum Infect Dis 3:ofw003. doi: 10.1093/ofid/ofw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beatty ME, Adcock PM, Smith SW, Quinlan K, Kamimoto LA, Rowe SY, Scott K, Conover C, Varchmin T, Bopp CA, Greene KD, Bibb B, Slutsker L, Mintz ED. 2006. Epidemic diarrhea due to enterotoxigenic Escherichia coli. Clin Infect Dis 42:329–334. doi: 10.1086/499246. [DOI] [PubMed] [Google Scholar]
- 6.Devasia RA, Jones TF, Ward J, Stafford L, Hardin H, Bopp C, Beatty M, Mintz E, Schaffner W. 2006. Endemically acquired foodborne outbreak of enterotoxin-producing Escherichia coli serotype O169:H41. Am J Med 119:168.e7–168.e10. doi: 10.1016/j.amjmed.2005.07.063. [DOI] [PubMed] [Google Scholar]
- 7.Jain S, Chen L, Dechet A, Hertz AT, Brus DL, Hanley K, Wilson B, Frank J, Greene KD, Parsons M, Bopp CA, Todd R, Hoekstra M, Mintz ED, Ram PK. 2008. An outbreak of enterotoxigenic Escherichia coli associated with sushi restaurants in Nevada, 2004. Clin Infect Dis 47:1–7. doi: 10.1086/588666. [DOI] [PubMed] [Google Scholar]
- 8.Beatty ME, Bopp CA, Wells JG, Greene KD, Puhr ND, Mintz ED. 2004. Enterotoxin-producing Escherichia coli O169:H41, United States. Emerg Infect Dis 10:518–521. doi: 10.3201/eid1003.030268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sack RB, Gorbach SL, Banwell JG, Jacobs B, Chatterjee BD, Mitra RC. 1971. Enterotoxigenic Escherichia coli isolated from patients with severe cholera-like disease. J Infect Dis 123:378–385. doi: 10.1093/infdis/123.4.378. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter CC, Barua D, Wallace CK, Sack RB, Mitra PP, Werner AS, Duffy TP, Oleinick A, Khanra SR, Lewis GW. 1965. Clinical and physiological observations during an epidemic outbreak of non-vibrio cholera-like disease in Calcutta. Bull World Health Organ 33:665–671. [PMC free article] [PubMed] [Google Scholar]
- 11.Vicente AC, Teixeira LF, Iniguez-Rojas L, Luna MG, Silva L, Andrade JR, Guth BE. 2005. Outbreaks of cholera-like diarrhoea caused by enterotoxigenic Escherichia coli in the Brazilian Amazon Rainforest. Trans R Soc Trop Med Hyg 99:669–674. doi: 10.1016/j.trstmh.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Guerrant RL, DeBoer MD, Moore SR, Scharf RJ, Lima AA. 2013. The impoverished gut—a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 10:220–229. doi: 10.1038/nrgastro.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahl JW, Steinsland H, Redman JC, Angiuoli SV, Nataro JP, Sommerfelt H, Rasko DA. 2011. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect Immun 79:950–960. doi: 10.1128/IAI.00932-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf MK. 1997. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin Microbiol Rev 10:569–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasheed JK, Guzman-Verduzco LM, Kupersztoch YM. 1990. Two precursors of the heat-stable enterotoxin of Escherichia coli: evidence of extracellular processing. Mol Microbiol 4:265–273. doi: 10.1111/j.1365-2958.1990.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 16.Taxt A, Aasland R, Sommerfelt H, Nataro J, Puntervoll P. 2010. Heat-stable enterotoxin of enterotoxigenic Escherichia coli as a vaccine target. Infect Immun 78:1824–1831. doi: 10.1128/IAI.01397-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz S, Green CK, Yuen PS, Garbers DL. 1990. Guanylyl cyclase is a heat-stable enterotoxin receptor. Cell 63:941–948. doi: 10.1016/0092-8674(90)90497-3. [DOI] [PubMed] [Google Scholar]
- 18.Giannella RA, Mann EA. 2003. E. coli heat-stable enterotoxin and guanylyl cyclase C: new functions and unsuspected actions. Trans Am Clin Climatol Assoc 114:67–85. [PMC free article] [PubMed] [Google Scholar]
- 19.Field M, Graf LH Jr, Laird WJ, Smith PL. 1978. Heat-stable enterotoxin of Escherichia coli: in vitro effects on guanylate cyclase activity, cyclic GMP concentration, and ion transport in small intestine. Proc Natl Acad Sci U S A 75:2800–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zachos NC, Tse M, Donowitz M. 2005. Molecular physiology of intestinal Na+/H+ exchange. Annu Rev Physiol 67:411–443. doi: 10.1146/annurev.physiol.67.031103.153004. [DOI] [PubMed] [Google Scholar]
- 21.Sack DA, Merson MH, Wells JG, Sack RB, Morris GK. 1975. Diarrhoea associated with heat-stable enterotoxin-producing strains of Escherichia coli. Lancet ii:239–241. [DOI] [PubMed] [Google Scholar]
- 22.Ryder RW, Wachsmuth IK, Buxton AE, Evans DG, DuPont HL, Mason E, Barrett FF. 1976. Infantile diarrhea produced by heat-stable enterotoxigenic Escherichia coli. N Engl J Med 295:849–853. doi: 10.1056/NEJM197610142951601. [DOI] [PubMed] [Google Scholar]
- 23.Levine MM, Caplan ES, Waterman D, Cash RA, Hornick RB, Snyder MJ. 1977. Diarrhea caused by Escherichia coli that produce only heat-stable enterotoxin. Infect Immun 17:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolin I, Wiklund G, Qadri F, Torres O, Bourgeois AL, Savarino S, Svennerholm AM. 2006. Enterotoxigenic Escherichia coli with STh and STp genotypes is associated with diarrhea both in children in areas of endemicity and in travelers. J Clin Microbiol 44:3872–3877. doi: 10.1128/JCM.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taxt AM, Diaz Y, Aasland R, Clements JD, Nataro JP, Sommerfelt H, Puntervoll P. 2016. Towards rational design of a toxoid vaccine against the heat-stable toxin of Escherichia coli. Infect Immun 84:1239–1249. doi: 10.1128/IAI.01225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moseley SL, Samadpour-Motalebi M, Falkow S. 1983. Plasmid association and nucleotide sequence relationships of two genes encoding heat-stable enterotoxin production in Escherichia coli H-10407. J Bacteriol 156:441–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto T, Yokota T. 1983. Plasmids of enterotoxigenic Escherichia coli H10407: evidence for two heat-stable enterotoxin genes and a conjugal transfer system. J Bacteriol 153:1352–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SG, Henderson IR. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol 192:5822–5831. doi: 10.1128/JB.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savarino SJ, Fasano A, Watson J, Martin BM, Levine MM, Guandalini S, Guerry P. 1993. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci U S A 90:3093–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto T, Echeverria P. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect Immun 64:1441–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McVeigh A, Fasano A, Scott DA, Jelacic S, Moseley SL, Robertson DC, Savarino SJ. 2000. IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect Immun 68:5710–5715. doi: 10.1128/IAI.68.10.5710-5715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleckenstein JM, Roy K, Fischer JF, Burkitt M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect Immun 74:2245–2258. doi: 10.1128/IAI.74.4.2245-2258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kansal R, Rasko DA, Sahl JW, Munson GP, Roy K, Luo Q, Sheikh A, Kuhne KJ, Fleckenstein JM. 2013. Transcriptional modulation of enterotoxigenic Escherichia coli virulence genes in response to epithelial cell interactions. Infect Immun 81:259–270. doi: 10.1128/IAI.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foreman DT, Martinez Y, Coombs G, Torres A, Kupersztoch YM. 1995. TolC and DsbA are needed for the secretion of STB, a heat-stable enterotoxin of Escherichia coli. Mol Microbiol 18:237–245. doi: 10.1111/j.1365-2958.1995.mmi_18020237.x. [DOI] [PubMed] [Google Scholar]
- 35.Yamanaka H, Nomura T, Fujii Y, Okamoto K. 1998. Need for TolC, an Escherichia coli outer membrane protein, in the secretion of heat-stable enterotoxin I across the outer membrane. Microb Pathog 25:111–120. doi: 10.1006/mpat.1998.0211. [DOI] [PubMed] [Google Scholar]
- 36.Weikel CS, Tiemens KM, Moseley SL, Huq IM, Guerrant RL. 1986. Species specificity and lack of production of STb enterotoxin by Escherichia coli strains isolated from humans with diarrheal illness. Infect Immun 52:323–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lortie LA, Dubreuil JD, Harel J. 1991. Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. J Clin Microbiol 29:656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorsey FC, Fischer JF, Fleckenstein JM. 2006. Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell Microbiol 8:1516–1527. doi: 10.1111/j.1462-5822.2006.00736.x. [DOI] [PubMed] [Google Scholar]
- 39.Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J Biol Chem 286:29771–29779. doi: 10.1074/jbc.M111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy K, Hamilton DJ, Fleckenstein JM. 2012. Cooperative role of antibodies against heat-labile toxin and the EtpA adhesin in preventing toxin delivery and intestinal colonization by enterotoxigenic Escherichia coli. Clin Vaccine Immunol 19:1603–1608. doi: 10.1128/CVI.00351-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Roy K, Hilliard GM, Hamilton DJ, Luo J, Ostmann MM, Fleckenstein JM. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594–598. doi: 10.1038/nature07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo Q, Qadri F, Kansal R, Rasko DA, Sheikh A, Fleckenstein JM. 2015. Conservation and immunogenicity of novel antigens in diverse isolates of enterotoxigenic Escherichia coli. PLoS Negl Trop Dis 9:e0003446. doi: 10.1371/journal.pntd.0003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakraborty S, Randall A, Vickers TJ, Molina D, Harro CD, DeNearing B, Brubaker J, Sack DA, Bourgeois AL, Felgner PL, Liang X, Mani S, Wenzel H, Townsend RR, Gilmore PE, Darsley MJ, Rasko DA, Fleckenstein JM. 2018. Human experimental challenge with enterotoxigenic Escherichia coli elicits immune responses to canonical and novel antigens relevant to vaccine development. J Infect Dis doi: 10.1093/infdis/jiy312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy K, Hamilton D, Allen KP, Randolph MP, Fleckenstein JM. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect Immun 76:2106–2112. doi: 10.1128/IAI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy K, Hamilton D, Ostmann MM, Fleckenstein JM. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601–4608. doi: 10.1016/j.vaccine.2009.05.076. [DOI] [PubMed] [Google Scholar]
- 47.Fleckenstein J, Sheikh A, Qadri F. 2014. Novel antigens for enterotoxigenic Escherichia coli vaccines. Expert Rev Vaccines 13:631–639. doi: 10.1586/14760584.2014.905745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, Operario DJ, Uddin J, Ahmed S, Alonso PL, Antonio M, Becker SM, Blackwelder WC, Breiman RF, Faruque AS, Fields B, Gratz J, Haque R, Hossain A, Hossain MJ, Jarju S, Qamar F, Iqbal NT, Kwambana B, Mandomando I, McMurry TL, Ochieng C, Ochieng JB, Ochieng M, Onyango C, Panchalingam S, Kalam A, Aziz F, Qureshi S, Ramamurthy T, Roberts JH, Saha D, Sow SO, Stroup SE, Sur D, Tamboura B, Taniuchi M, Tennant SM, Toema D, Wu Y, Zaidi A, Nataro JP, Kotloff KL, Levine MM, Houpt ER. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Canto F, Valenzuela P, Cantero L, Bronstein J, Blanco JE, Blanco J, Prado V, Levine M, Nataro J, Sommerfelt H, Vidal R. 2011. Distribution of classical and nonclassical virulence genes in enterotoxigenic Escherichia coli isolates from Chilean children and tRNA gene screening for putative insertion sites for genomic islands. J Clin Microbiol 49:3198–3203. doi: 10.1128/JCM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamoto K, Takahara M. 1990. Synthesis of Escherichia coli heat-stable enterotoxin STp as a pre-pro form and role of the pro sequence in secretion. J Bacteriol 172:5260–5265. doi: 10.1128/jb.172.9.5260-5265.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Gao Z, Guzman-Verduzco LM, Tachias K, Kupersztoch YM. 1992. Secretion of the STA3 heat-stable enterotoxin of Escherichia coli: extracellular delivery of Pro-STA is accomplished by either Pro or STA. Mol Microbiol 6:3521–3529. doi: 10.1111/j.1365-2958.1992.tb01787.x. [DOI] [PubMed] [Google Scholar]
- 52.Sheikh A, Rashu R, Begum YA, Kuhlman FM, Ciorba MA, Hultgren SJ, Qadri F, Fleckenstein JM. 2017. Highly conserved type 1 pili promote enterotoxigenic E. coli pathogen-host interactions. PLoS Negl Trop Dis 11:e0005586. doi: 10.1371/journal.pntd.0005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bohnert JA, Schuster S, Kern WV, Karcz T, Olejarz A, Kaczor A, Handzlik J, Kiec-Kononowicz K. 2016. Novel piperazine arylideneimidazolones inhibit the AcrAB-TolC pump in Escherichia coli and simultaneously act as fluorescent membrane probes in a combined real-time influx and efflux assay. Antimicrob Agents Chemother 60:1974–1983. doi: 10.1128/AAC.01995-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahamoud A, Chevalier J, Alibert-Franco S, Kern WV, Pages JM. 2007. Antibiotic efflux pumps in Gram-negative bacteria: the inhibitor response strategy. J Antimicrob Chemother 59:1223–1229. doi: 10.1093/jac/dkl493. [DOI] [PubMed] [Google Scholar]
- 55.Koronakis V, Eswaran J, Hughes C. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem 73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 56.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodas C, Iniguez V, Qadri F, Wiklund G, Svennerholm AM, Sjoling A. 2009. Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. J Clin Microbiol 47:1218–1220. doi: 10.1128/JCM.00316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quan J, Tian J. 2009. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One 4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clements JD. 1990. Construction of a nontoxic fusion peptide for immunization against Escherichia coli strains that produce heat-labile and heat-stable enterotoxins. Infect Immun 58:1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blomfield IC, McClain MS, Eisenstein BI. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol Microbiol 5:1439–1445. doi: 10.1111/j.1365-2958.1991.tb00790.x. [DOI] [PubMed] [Google Scholar]
- 62.Kumar P, Kuhlmann FM, Chakroborty S, Bourgeois AL, Foulke-Abel J, Tumala B, Vickers TJ, Sack DA, DeNearing B, Harro CD, Wright WS, Gildersleeve JC, Ciorba MA, Santhanam S, Porter CK, Gutierrez RL, Prouty MG, Riddle MS, Polino A, Sheikh A, Donowitz M, Fleckenstein JM. 2018. Enterotoxigenic Escherichia coli blood group A interactions intensify diarrheal severity. J Clin Invest 128:3298–3311. doi: 10.1172/JCI97659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. 2015. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans DJ Jr, Evans DG. 1973. Three characteristics associated with enterotoxigenic Escherichia coli isolated from man. Infect Immun 8:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose RE. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res 16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]