The fhuACDB operon, present in a number of Enterobacteriaceae, encodes components essential for the uptake of ferric hydroxamate type siderophores. FhuA acts not only as a transporter for physiologically important chelated ferric iron but also as a receptor for various bacteriophages, toxins, and antibiotics, which are pathogenic to bacterial cells.
KEYWORDS: FhuA, sequence diversity, gene evolution, E. coli, Salmonella
ABSTRACT
The fhuACDB operon, present in a number of Enterobacteriaceae, encodes components essential for the uptake of ferric hydroxamate type siderophores. FhuA acts not only as a transporter for physiologically important chelated ferric iron but also as a receptor for various bacteriophages, toxins, and antibiotics, which are pathogenic to bacterial cells. In this research, fhuA gene distribution and sequence diversity were investigated in Enterobacteriaceae, especially Salmonella and Escherichia. Comparative sequence analysis resulted in a fhuA phylogenetic tree that did not match the expected phylogeny of species or trees of the fhuCDB genes. The fhuA sequences showed a unique mosaic clustering pattern. On the other hand, the gene sequences showed high conservation for strains from the same serovar or serotype. In total, six clusters were identified from FhuA proteins in Salmonella and Escherichia, among which typical peptide fragment variations could be defined. Six fragmental insertions/deletions and two substitution fragments were discovered, for which the combination of polymorphism patterns could well classify the different clusters. Structural modeling demonstrated that all the six featured insertions/deletions and one substitution fragment are located at the apexes of the long loops present as part of the FhuA external pocket. These frequently mutated regions are likely under high selection pressure, with bacterial strains balancing escape from phage infection or toxin/antibiotics attack via fhuA gene mutations while maintaining the siderophore uptake activity essential for bacterial survival. The unusual fhuA clustering suggests that high-frequency exchange of fhuA genes has occurred between enterobacterial strains after distinctive species were established.
INTRODUCTION
FhuA (ferric hydroxamate uptake protein A) is a multifunctional protein with a total length of 747 residues in Escherichia coli strain MG1655, including an N-terminal 33-amino-acid signal peptide and a 714-amino-acid mature protein sequence (1). It represents a transmembrane receptor, residing in the outer membrane (OM), which can mediate the uptake of ferric iron (bound to the siderophore ferrichrome) into the periplasm in a TonB-dependent manner (2–4). Iron is essential for bacterial survival and virulence, but it is mainly present in the nonsoluble ferric form, and the bioavailable concentration in the environment is often extremely low. To increase bioavailability, bacteria secrete siderophore molecules that can chelate ferric iron with high affinity (5, 6). The iron-loaded siderophores can be recognized by bacterial membrane receptors (e.g., FhuA) and transported into the periplasm. Further translocation of siderophores across the inner membrane is mediated by ABC transporters. In the case of ferrichrome, the transport into the cytoplasm depends on the binding protein FhuD, as well as the integral membrane protein FhuB and the ATPase FhuC (reference 7 and references therein). Besides the major physiological activity, FhuA is also usurped pathologically as the target for bacteriophages (T1, T5, ϕ80, and UC-1) and bacterial toxins (colicin M and microcin 25) (8). Some antibiotics also target FhuA as a receptor, e.g., albomycin and rifamycin CGP 4832 (9, 10).
Functional FhuA, embedded in the outer membrane, is a monomeric β-barrel protein, with its C terminus forming 22 antiparallel β-strands and the N terminus folded inside the β-barrel from the periplasmic side to form a cork domain (3, 11). The cork domain separates a pair of pockets, the larger one open to the external medium and the smaller one facing the periplasm (3). The N-terminal portion of FhuA that contains the TonB-interacting region (i.e., the TonB box) lies in the periplasm (4, 12, 13). The β-strands are connected by 11 long loops at the outer surface (L1 to L11) and 10 short turns within the periplasmic side (T1 to T10) (3, 11). The exact residues or motifs in the external pocket of FhuA that specifically interact with ferrichrome remain elusive, although conserved residues in loops L3 and L11 were found to be important for binding (3, 14). Ferrichrome binding induces structural changes that go through the entire FhuA molecule up to the N terminus, further signaling TonB binding and activating the TonB-ExbB-ExbD transporting system to internalize ferrichrome molecules (3, 15, 16). Other FhuA-binding molecules/ligands are also transported via the TonB-dependent pathway, except for phage T5 (15, 17). The binding sites and specificity determinants for these components are frequently located within the external loops of FhuA. For example, loops L4, L5, and L8 are involved in binding specificity of phages T1 and ϕ80, L5 and L8 are important for phage T5, and L3, L4, L7, L8, and L11 are involved in the sensitivity to colicin M and the antibiotics albomycin and rifamycin (14, 15). Microcin J25 (MccJ25) binds at a location in FhuA similar to the location at which ferrichrome binds (18).
In E. coli, fhuA is located within the fhuACDB operon (19); the protein products of fhuC, fhuD, and fhuB form a complex, mediating the translocation of ferrichrome from the periplasm into the cytoplasm (20–22). In other species, fhuA and fhuCDB often appear in the same operon but with a different transcription order, or they are separated into two different operons (23–25). Fragmented gene sequences have been reported for fhuA frequently (26, 27). Gene evolutionary studies also suggested positive selection in fhuA rather than fhuB, fhuC, or fhuD (28, 29). Therefore, it has been hypothesized that FhuA has a different evolutionary route with selection pressures that are independent from the FhuCDB cluster. For example, in some Gram-negative bacterial species, FhuD and FhuB display a broader ligand specificity than that of the OM receptor proteins (such as FhuA) in that they accept and transport not only ferrichromes but also other siderophores of the hydroxamate type, e.g., aerobactin and coprogen (20).
Despite the large amount of research already performed on fhuA, the mechanisms of gene evolution and the relationship with protein function remain largely unknown. Nearly all studies have been limited to fhuA genes that are present in one or a few closely related strains. To examine gene evolution in a larger subset of strains, we compared fhuA genes from a comprehensive list of Salmonella and Escherichia serovars and other close species to determine the gene distribution and overall sequence diversity. This analysis identified evolutionary patterns with direct relevance to the structure and potential interactions of FhuA.
RESULTS
Distribution and diversification of fhuA in Salmonella and Escherichia.
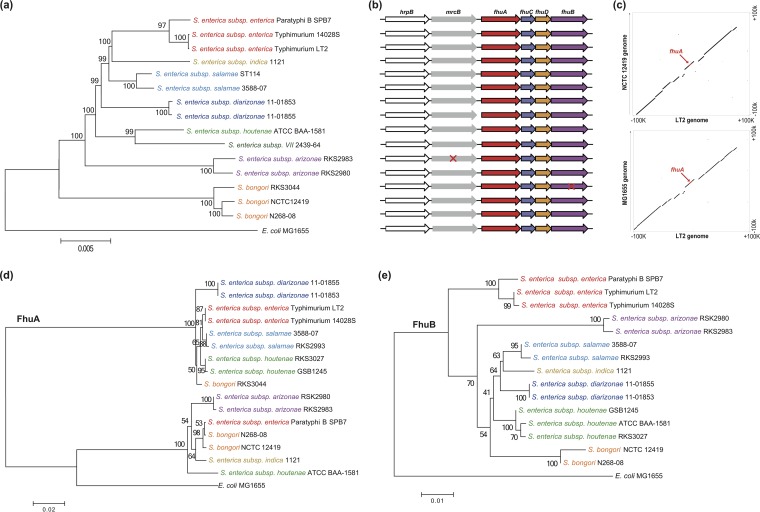
In representative genomes from the two Salmonella species, Salmonella enterica and Salmonella bongori, and six subspecies of S. enterica, including Salmonella enterica subsp. enterica, S. enterica subsp. arizonae, S. enterica subsp. diarizonae, S. enterica subsp. indica, S. enterica subsp. salamae, and S. enterica subsp. houtenae, only one chromosomal copy of fhuA was identified in each strain. The order of fhuA and adjacent genes fhuCDB was conserved among Salmonella phylogenetic clusters (Fig. 1a and b). Expanded analysis of the genomic regions that flanked fhuA showed apparent collinearity between different Salmonella genomes (Fig. 1c, top) and showed high gene synteny and sequence collinearity between Salmonella and E. coli strains (Fig. 1c, bottom). This indicated that the fhuABCD locus existed at least in the most recent common ancestor of the two genera.
FIG 1.
Distribution and phylogenetics of fhuA in Salmonella. (a) Phylogenomic neighbor-joining tree constructed for Salmonella using an Enterobacteriaceae core orthologous gene set. Different Salmonella species or subspecies are shown in different colors. (b) Gene order and distribution of the fhuA locus in Salmonella. The strains were in line with those shown in the tree in panel a. Pseudogenes are indicated with a red “×.” (c) Collinearity of the fhuA locus was compared between S. Typhimurium LT2 and S. bongori NCTC 12419 (top) or E. coli MG1655 (bottom). (d and e) Neighbor-joining trees of FhuA and FhuB proteins in Salmonella, respectively. E. coli MG1655 was used as the outgroup. Percentage scores from 1,000 bootstrapping tests are displayed at the nodes of the trees.
When FhuA proteins were compared between Salmonella strains, the topology of the phylogenetic tree (Fig. 1d) was far different from that of the core genome tree (Fig. 1a). Consistent with previous Salmonella species trees built on 16S rRNA sequences, the phylogenomic tree is predicted to accurately reflect the evolution of Salmonella species or subspecies, with strains clustered together within the same phylogenetic group (Fig. 1a). In the FhuA tree, however, the strains belonging to a unique phylogenetic group diverged, while the ones from different phylogenetic groups clustered (Fig. 1d). For example, S. bongori RKS3044 FhuA clustered away from the other two S. bongori strains, whereas S. enterica subsp. indica 1121 FhuA clustered with S. bongori N268-08 and NCTC12419 (Fig. 1d). The FhuA clustering pattern was independent of the methods used for tree reconstruction or sequence nature (see Fig. S1a and b in the supplemental material). In contrast, trees based on the fhuCDB genes (Fig. 1e; see also Fig. S1c and d) or on the hrpB gene adjacent to fhuA in the genome (Fig. S1e) were similar to the phylogenomics tree based on the Enterobacteriaceae core ortholog gene set (Fig. 1a).
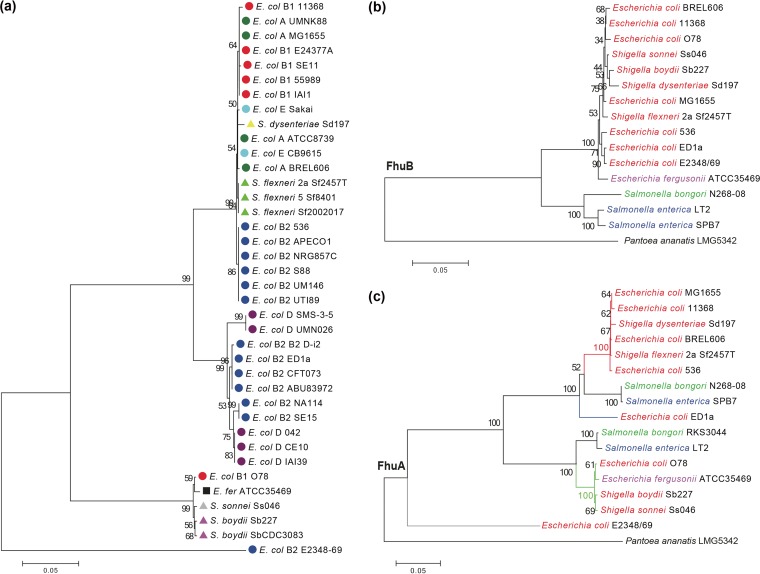
FhuA variance within a phylogenetic group was also confirmed by the tree reconstructed for the Escherichia genus, which included Escherichia fergusonii as an outgroup (Fig. 2a; see also Fig. S2). FhuA clustered into four clearly discernible groups, totally independent of phylogenetic clustering results of Escherichia/Shigella strains based on core gene data sets or oligonucleotide composition (Fig. 2a; see also Fig. S2) (30, 31). In contrast, a FhuB protein tree did not form robust clusters due to the ultraconservation of corresponding sequences (Fig. S3). When proteins from representative Salmonella and Escherichia strains were analyzed together, the FhuB tree was consistent with the established evolutionary patterns of Escherichia and Salmonella species (Fig. 2b), while the FhuA tree was clearly different (Fig. 2c).
FIG 2.
Phylogenetics of FhuA in Escherichia and Salmonella. (a) Neighbor-joining tree of FhuA constructed from E. coli or Shigella strain sequences. Different subgroups of E. coli/Shigella strains are shown in different colors or shapes. Neighbor-joining trees for FhuB (b) and FhuA (c) were constructed from E. coli/Shigella and Salmonella strain sequences using Pantoea ananatis LMG5342 as the outgroup. Percentage scores from 1,000 bootstrapping tests are displayed at the nodes of the trees. E. col, E. coli; E. fer, E. fergusonii.
Within-serovar/serotype conservation of fhuA in Salmonella and E. coli.
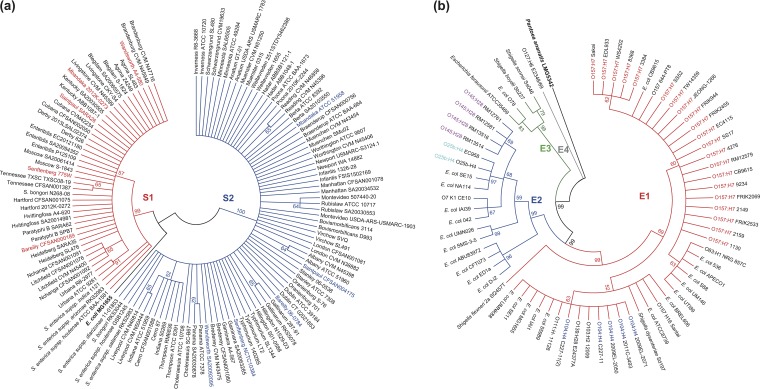
We obtained genome sequences for 66 Salmonella serovars, with draft genomes for two or more strains (Table S1) (http://www.ncbi.nlm.nih.gov/genome). Two or three representative genomes were selected randomly from each serovar, followed by screening and clustering analysis of the fhuA gene. Truncated alleles of fhuA were detected in all strains of serovars Typhi and Paratyphi A and from at least one strain for serovars Mississippi, Give, Javiana, Pullorum, and Johannesburg (Table S1). An unsupervised clustering analysis revealed two robust FhuA clusters (S1 and S2) (Fig. 3a). For 54 of 59 serovars, the FhuA proteins of strains from the same serovar clustered together, with the exceptions of serovars Bareilly, Mbandaka, Saintpaul, Senftenberg, and Wandsworth (Fig. 3a). The conservation within serovars was not the result of selecting a limited number of strains from each serovar, since inclusion of all sequenced genomes of serovars Typhimurium (41 strains), Enteritidis (58 strains), Heidelberg (23 strains), Newport (19 strains), and Anatum (13 strains) led to similar S1 and S2 clustering (Fig. S4a and Table S1). Only serovar Typhimurium FORC 015 and Newport CDC 2010K-2159 strains did not match the observed pattern (Fig. S4a and Table S1). We also included FhuA proteins from 112 Salmonella serovars that had only one genome-sequenced strain (Table S1), but no new pattern was disclosed (Fig. S4b and Table S1).
FIG 3.
Distribution of FhuA clusters among serotypes of Salmonella and E. coli. (a) FhuA clusters identified from a Salmonella-based sequence alignment. Strains belonging to the same serovar but with different clusters of FhuA are shown in red or blue. (b) FhuA clusters identified from an E. coli-based sequence alignment. For both panels, clustering was based on neighbor-joining trees of full-length FhuA protein sequences. Percentage scores from 1,000 bootstrapping tests are displayed at the nodes of the trees.
For E. coli and Shigella, inclusion of FhuA from more strains led to more definitive identification of the four major clusters, E1, E2, E3, and E4 (Fig. 3b). The FhuA proteins of E. coli serotypes with multiple strains, including O157:H7 (19), O145:H28 (4), O104:H4 (4), and O25b:H4 (2), also exhibited conservation within the serotype (Fig. 3b; see also Table S1).
Categorization of FhuA proteins with specific sequence features.
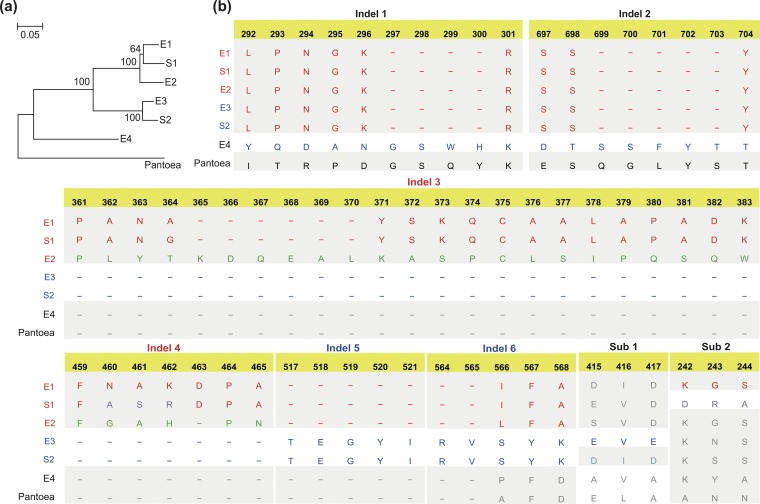
To identify potential sequence signatures within Salmonella and E. coli and between each species, FhuA protein sequences from Salmonella (clusters S1 and S2) and Escherichia (clusters E1 to E4) were compared. The FhuA alleles clustered into three large groups: (i) E1, S1, and E2; (ii) E3 and S2; and (iii) E4 only (Fig. 4a). The FhuA sequence alignment identified insertions or deletions that were unique to each major group, subgroup (i.e., E1/S1 or E2), or individual cluster (Fig. 4b; see also Table S2). Group E4 proteins had unique insertions in regions termed indel 1 and indel 2 (Fig. 4b). There were also fragment insertions in the same regions of FhuA from Pantoea, which served as the outgroup for the phylogenetic tree (Fig. 4b). The E1/S1/E2 FhuA cluster all had peptides inserted in two regions termed indel 3 and indel 4, with more sequence homology between E1 and S1 than E2 (Fig. 4b). For the E3/S2 group, unique peptide insertions were detected at indel 5 and indel 6 (Fig. 4b). The FhuA proteins of E1 and S1 (or E3 and S2) had high similarity, and no unique insertions/deletions could be detected between them. Individual amino acid differences were sufficient to distinguish each individual group, however, since the insertions/deletions were removed before generating the clustering tree (Fig. 4a). For example, distinct amino acid sequences were detected at regions termed Sub1 between groups E3 and S2 and Sub2 and indel 4 between E1 and S1 (Fig. 4b; see also Table S2).
FIG 4.
Typical mutation patterns of FhuA clusters in Salmonella and Escherichia. (a) A diagram tree showing the clusters of FhuA determined for Salmonella and Escherichia. (b) List of indels and substitutions and corresponding sequences that are able to distinguish the different FhuA clusters. Indel 1 and indel 2 can distinguish E4 from E1/S1/E2/E3/S2, indels 3 to 6 can distinguish E3/S2 from E1/S1/E2, Sub1 can distinguish E3 from S2, and Sub2 can distinguish S1 from E1.
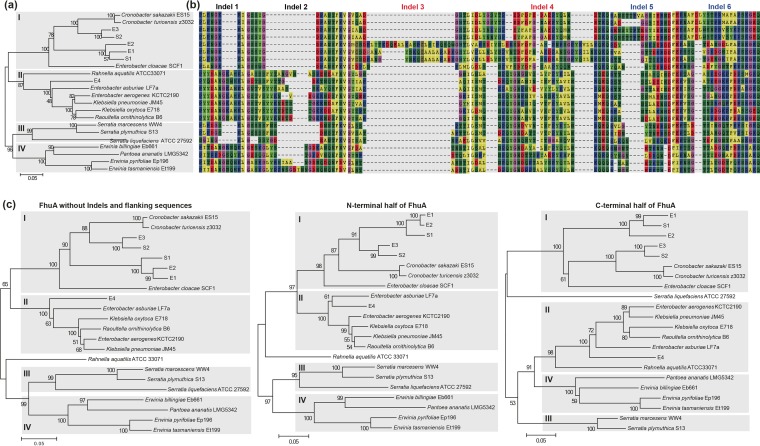
To generate a thorough categorization of FhuA within the Enterobacteriaceae, representative genomes from other species, including Cronobacter, Enterobacter, Erwinia, Klebsiella, Rahnella, Raoultella, and Serratia, were analyzed (Fig. 5a). With the inclusion of these new organisms, the FhuA proteins clustered into four major groups (I to IV), where E1/S1/E2 and E3/S2 belonged to group I, E4 belonged to group II, group III appeared to be unique to Serratia, and Pantoea FhuA belonged to group IV (Fig. 5a). The main variation for insertions/deletions among the four major groups remained in the indel 1 to 6 locations identified previously, indicating that there was easy gain or loss and variation of fragments in these positions (Fig. 5b). The first 50 amino acids in the FhuA N-terminal region, which represents the signal peptide sequences and the unstructured fragment (3, 11), were highly variable. However, the TonB box was conserved (Fig. S5). When FhuA trees were built after removal of all the indels and flanking variable amino acid positions or removal of 200-amino-acid fragments from the N- or C-terminal region, the clustering patterns remained the same as for the full-length FhuA proteins (Fig. 5c). This indicated that the group-specific variations were not constrained to the insertion/deletion regions or to specific fragments but rather that frequent recombination along the entire FhuA length may have occurred.
FIG 5.
Typical mutation patterns of FhuA clusters in Enterobacteriaceae. (a) Neighbor-joining tree of FhuA protein in Enterobacteriaceae species supported grouping FhuA into 4 main clusters (I to IV). (b) The indel patterns with corresponding amino acid sequence of different FhuA clusters. (c) Neighbor-joining tree of FhuA proteins after removing the indels and flanking highly mutated sequences (left) and the N-terminal (middle) and C-terminal (right) FhuA protein sequences. Percentage scores from 1,000 bootstrapping tests are displayed at the nodes of the trees.
Structure variations in the extracellular surface of FhuA proteins.
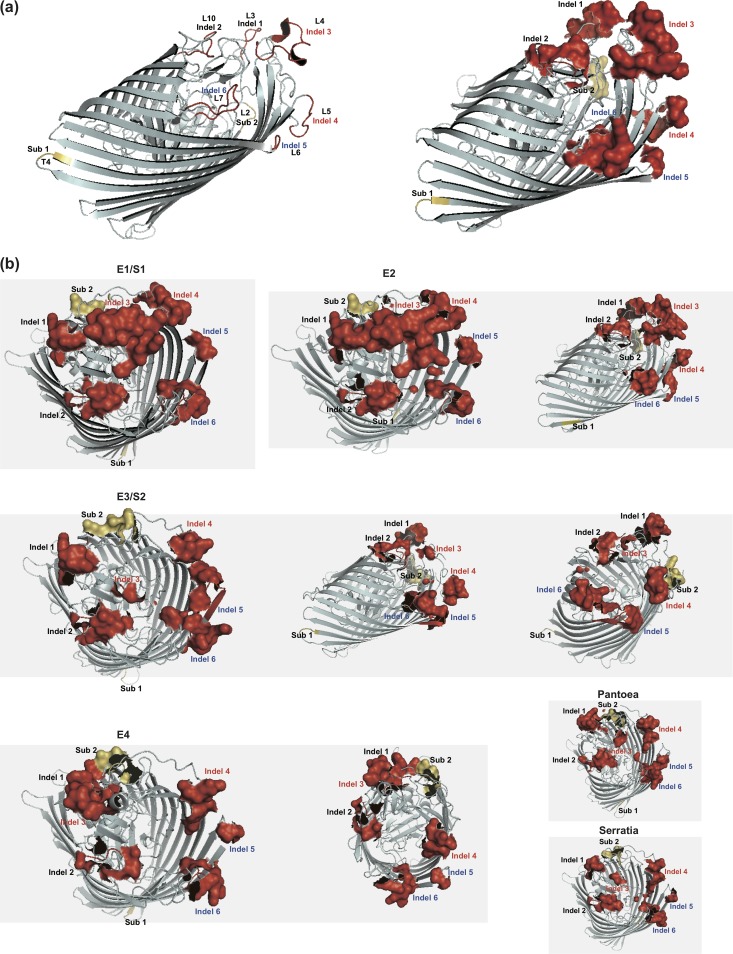
Based on the coordinates of the recently solved structure of FhuA from E. coli K-12 (group E1) (3, 11), we performed homology modeling to compute the structures of FhuA proteins from groups E2, E3, E4, S1, and S2 as well as from Serratia (group III) and Pantoea (group IV) (Fig. 5a). All proteins had similar, predicted cork barrel structures, with cork domains showing high similarity in both sequence and structure (Fig. 6). The six indel regions identified by sequence alignment were located in loops within the external pocket of FhuA that faces the extracellular environment, corresponding to loops L3 (indel 1), L10 (indel 2), L4 (indel 3), L5 (indel 4), L6 (indel 5), and L7 (indel 6) (Fig. 6a; see also Table S2). The Sub2 region corresponded to external surface loop L2, whereas Sub1 was located in the T4 linker between β-strands (Fig. 6a). Few variations were observed in the cork or transmembrane barrels or on the surface facing the periplasm, coinciding with the results of the sequence alignment (Table S2). S1 FhuA proteins had a predicted structure that was highly similar to that of E1, while E2 proteins showed an insertion in L4 (indel 3) compared to E1/S1, causing the approaching of the domain with L3 (indel 1) and the formation of a compact surface structure (Fig. 6b, E2, left and right). In E3 proteins, L4 was shortened extensively, along with L5, but L6 and L7 were both elongated and formed a fused surface (Fig. 6b, E3/S2, left, middle, and right). E4 proteins had a more diverse external surface structure, with insertions in L3 and L10, a deletion in L4, and changes to loops L2, L5, L6, and L7. The loops L2 (Sub2), L3 (indel 1), and L4 (indel 2) became continuous, forming a local surface structure (Fig. 6b, E4, left and right). The Pantoea and Serratia FhuAs also showed a diverse outer surface structure but conservation in other regions (Fig. 6b, Pantoea and Serratia). Taken together, the results of this analysis suggested that the main diversity of FhuA sequences was represented in structural variation in the exposed regions facing the external environment.
FIG 6.
Influence of typical mutations on FhuA structure. (a) Resolved E. coli FhuA structure (PDB accession code 1QFF) with mutation hot spots highlighted. (b) Modeling structure of different FhuA clusters (E1/S1, E2, E3/S2, E4, Pantoea, and Serratia), with mutation hot spots highlighted. The overall protein structure was displayed with PyMOL in cartoon format; the mutation hot spots were shown in either cartoon or surface format.
DISCUSSION
The fhuACDB locus showed high conservation within the genomes of Salmonella and Escherichia. Despite the fragmentation of the fhuA protein-coding frame in some serovars or strains, e.g., Salmonella serovar Typhi, the gene vestige can be well traced like the fhuCDB genes within the same operon. This suggests that fhuA was not acquired by horizontal gene transfer after the divergence of Salmonella and Escherichia. However, the mosaic fhuA gene does not follow the established phylogenetic routes for Salmonella or E. coli, which is in contrast to fhCDB, and tested housekeeping genes, which show a clear separation of species. There have been previous reports that fhuA is subject to positive selection, specifically in uropathogenic E. coli (UPEC) (28) and in additional E. coli strains (29). We detected FhuA conservation within a given serovar/serotype but diversity between serovars or serotypes, which is consistent with the positive-selection hypothesis within a single genus (32). In this hypothesis, the fhuA gene would vary to be better adapted to the specific environment in which strains of the serovar/serotype survive. However, this simple positive-selection model has difficulty in explaining the intergenus/interspecies shared fhuA sequence clusters and mutation patterns disclosed here. Within a single genus or species, the ancient fhuA may have diversified as the strains evolved and adapted to different growth environments. The intergenus/interspecies common sequence clusters and mutation patterns could indicate convergent evolution of fhuA for strains living in similar environments but originating from different genera or species. It suggests for fhuA that the selection pressures based on the specific environment are stronger than taxon conservation.
The intergenus/interspecies common sequence patterns observed for FhuA are more reflected by short-fragment insertions or deletions than by single-nucleotide polymorphisms (29). Common insertions or deletions among widely varied gene sequences from different genera or species are more likely to be the result of recombination. However, in dN/dS ratio (ratio of nonsynonymous to synonymous evolutionary changes)-based selection studies, these inserted/deleted sites are removed before further analysis (33, 34). Previous analyses of fhuA were based on the premise of no recombination happening in the gene (28, 29). Evidence for recombination may also have been missed using the existent algorithms, especially when a low number of strains were analyzed (28, 29). A more simplified explanation for the mosaic fhuA sequences observed is that frequent recombination events have shaped the fhuA sequence patterns. The amino acid substitution patterns within different peptide fragments of FhuA can classify the protein sequences into clusters similar to those grouped by insertion/deletion patterns. Therefore, we hypothesize that recombination has occurred throughout the fhuA gene.
Two distinct clusters for fhuA genes in Salmonella (S1 and S2) and four distinct clusters for fhuA genes in E. coli (E1 to E4) were clearly defined in our study. When the taxa were enlarged to include more enterobacterial species, four fhuA superclusters (I to IV) were detected. From these diverse comparisons, we identified eight clear mutation hotspots in the fhuA protein sequence, corresponding to six insertion/deletion sites and two amino acid substitution fragments that were frequently mutated within all the Enterobacteriaceae strains. The most intriguing aspect of these findings was that each indel region and one of the substitution regions mapped to the apexes of FhuA surface loops that are exposed to the external environment (Fig. 6). In contrast, the TonB box, β-strands, and the cork region showed high sequence conservation. Consequently, the predicted overall structure did not vary greatly between FhuA clusters (Fig. 6). The surface regions, especially the loops, are critical for recognition of substrates and determine the binding specificity (14, 15). It needs to be verified experimentally whether the loop conformation changes in different clusters of FhuA influence the binding patterns to a variety of substrates, including bacteriophages. Selection pressures based on bacteriophage binding have been observed in the past (35, 36); we hypothesize that some of the changes in surface-exposed FhuA loops evolved to provide resistance to key bacteriophages. We recently found that Yersinia leucine-rich repeat genes had frequent mutations that were constrained to maintain protein activity and overall structure (37). Instead of random nucleotide substitutions, the amino acid codons tended to mutate with maximum parsimony (i.e., in the most straightforward and simplest way), resulting in predictable patterns at the peptide level rather than total irregularity (37). For a protein such as FhuA, it is possible that activities essential for organism survival will be conserved and under negative selection, while other activities could be placed under positive selection. Such proteins could adopt comutations at multiple loci to simultaneously change some function, such as alteration of a ligand recognition motif, while still maintaining the essential activity (37). Under these two hypotheses, the mosaic evolution patterns, the mutation loci, and the possible protein activities of FhuA could be well explained.
There are many other questions about fhuA evolution that remain unanswered. For example, the fhuA and fhuCDB genes show the same gene order and were reported to form an operon sharing uniform gene regulation in E. coli (or Salmonella) strains (19, 21). However, in other species, the gene order was often changed and the genes were sometimes divided into multiple operons (23–25). How did the expression regulation of fhuA evolve, and what is its relationship with fhuCDB? We have inspected the sequence patterns of fhuA only within the Enterobacteriaceae, and the distribution and evolution of fhuA beyond the family level are yet largely unknown. Within a bacterial genome, generally very few genes (i.e., ∼30) are thought to be under positive selection pressure (28, 29). Besides FhuA, other β-barrel porins, e.g., OmpF, OmpC, and LamB, and iron acquisition-related proteins, e.g., FepE, EntD, and EntF, were also enriched in the list of proteins undergoing positive selection (28, 29). We anticipate that further analysis of each individual protein and comparisons between them could inspire important insights into the evolution and function of these proteins. Finally, fhuA has been annotated as a “pseudogene” in many strains (26, 27). In the case of Salmonella serovar Typhi, the fhuA pseudogene is the result of a 1- to 2-bp deletion or insertion leading to a frameshift. The loss of a functional fhuA gene might be tolerated in this human-restricted serovar (38) because it has less reliance on long-term survival in the environment (39), where acquisition of ferric iron via ferrichrome would constitute a major fitness advantage. Most frameshift events in fhuA result in a significant polar effect regarding the expression of the downstream fhuCDB genes. Polar fhuA mutants can be tolerated in serovar Typhi but possibly not in other serovars, which would depend for certain periods of their life cycle on the uptake of various hydroxamate siderophores transported by the FhuCDB complex, such as aerobactin, coprogen, and rhodotorulic acid.
In summary, the unusual FhuA mosaic clustering suggests that an exchange of fhuA genes between different enterobacterial strains occurred after distinctive species were established. For the moment, the “driving force” (i.e., selective pressure, phages, and nutritional requirements) and mechanism(s) of genetic exchange (e.g., conjugation, transduction, and mobile elements) remain largely unknown. It is also unclear for which period this genetic exchange was happening and if the possibility of exchange still exists. It appears that there was no significant fhuA gene exchange between Escherichia and Salmonella and between different serovars in the near past, since the FhuA protein shows invariance among strains belonging to the same Salmonella serovar or E. coli serotype. It remains to be determined how the described changes in FhuA can influence or are reflective of the life cycle, environmental niche, or geographical location of a given strain.
MATERIALS AND METHODS
Bacterial strains, genomes, and gene sequences.
The genomes of Salmonella, Escherichia, and other Enterobacteriaceae strains were downloaded from the National Center for Biotechnology Information (NCBI) genome database. For each strain with a publicly available genome, the strain information, including serovar/serotype, taxonomy, and pathogenicity, was manually annotated from the corresponding genome annotation files, original publications, and NCBI taxonomy database. Genome annotation files were the base for retrieving gene and protein sequences. The standalone BLAST suite was downloaded from the NCBI website, installed, and used for screening FhuA, FhuB, FhuC, FhuD, GroEL, and HrpB with E. coli MG1655 counterparts as the query sequences and with an identity cutoff of 70%. The genomic loci of the genes were positioned based on the protein description within the corresponding genome annotation file. For strains without a specific protein captured, the potential genomic locus was reasoned according to its adjacent conserved genes, followed by retrieval, alignment, and comparison of the nucleotide sequences. We have listed all of the fhuA sequence information (e.g., species, serovar/serotype, strain, and protein accession) in Table S3.
Comparative genomics and phylogenetic analysis.
The regions containing fhuA and neighboring genes were depicted according to their order present in their corresponding genome to make synteny comparison among different bacterial strains. The genes of interest and their flanking sequences (100 kb on each side) were also retrieved from the corresponding genomes, and PipMaker was used for collinearity analysis (40). ClustalW was used to align the proteins or genes, with the default parameters (https://www.ebi.ac.uk/). The whole-sequence alignment results with gaps removed were imported to MEGA6.0, where a neigbor-joining method was implemented to calculate the pairwise distances and build trees (41). To construct phylogenetic trees based on specified gene-containing regions, the original multiple whole-gene/protein sequence alignment results (including the gaps) were based, desired fragments were retrieved with in-house scripts written with GO programming language, and neighbor-joining trees were built directly with a similar procedure. For all tree constructions, bootstrapping tests were performed with 1,000 replicates. A node was considered robust only when the bootstrapping score was above 50. Maximum parsimony or maximum likelihood method was also adopted when indicated in context; these were implemented in MEGA6.0 with the default parameter settings.
Bacterial species phylogenomic trees were generated based on a high-confidence core orthologous gene set of Enterobacteriaceae (Z. Chen, Y. Xu, X. Cheng, Y. Hu, M. A. Sun, W. Köster, A. P. White, and Y. Wang, unpublished data). Twenty representative genera of Enterobacteriaceae and 158 strains were selected for analysis, including the Legionella pneumophila Philadelphia 1 strain, which was used as the outgroup. An oligonucleotide composition-based method (6- and 7-mer) was used for the initial round of phylogenomic tree building (42). Based on these rough species clustering results, the Enterobacteriaceae core orthologous gene set was determined with a bottom-up strategy: homologous gene pairs were determined for the stepwise nearest strains or representative strains of the nearest clades (blastp was used with sequence similarity at >60% and length coverage at >70% of the average length of the two aligned proteins). This was repeated until the top of the tree was reached and the Enterobacteriaceae core gene set was identified. The core genes were concatenated according to their mutual order within the corresponding genomes. PipMaker was used to align any pair of concatenated core gene sequences. Since the local collinearity would be broken if a gene was displaced, these genes were removed from the core orthologous gene set. In total, 384 orthologous protein-encoding genes were identified in the Enterobacteriaceae strains, and 246 were also found to be orthologous in Legionella pneumophila Philadelphia 1, which were further used for species phylogenomic analysis (gene lists will be sent by requisition from the authors). The 384 core genes/proteins were retrieved from Salmonella and E. coli strains, and sequences were concatenated according to a predefined order, aligned with ClustalW, and used for tree building with the neighbor-joining method implemented in MEGA6.0, with a procedure similar to that described above for FhuA analysis. Bootstrapping tests were performed with 1,000 replicates. E. coli MG1655 was used as the outgroup for Salmonella trees, and E. fergusonii was used as the outgroup for E. coli trees. Table S4 lists the genome sequence accession numbers of the E. coli and Salmonella strains, while Table S5 gives the locus tags of the 384 core orthologous clusters.
Structure modeling and comparison.
The experimentally resolved structure of E. coli FhuA was used as a reference (PDB accession code 1QFF), and the structures of other FhuA proteins were predicted with PHYRE2 (43). PyMOL was used for structure visualization, analysis, and comparison (http://www.pymol.org).
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by a Natural Science Fund of Shenzhen (JCYJ201607115221141) and a Shenzhen Peacock Start-up Research Fund for Overseas Talents to Y.W. Research in the laboratory of W.K. was supported by the Agriculture Development Fund (ADF) from the Government of Saskatchewan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00573-18.
REFERENCES
- 1.Coulton JW, Mason P, Cameron DR, Carmel G, Jean R, Rode HN. 1986. Protein fusions of beta-galactosidase to the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol 165:181–192. doi: 10.1128/jb.165.1.181-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hantke K, Braun V. 1975. Membrane receptor dependent iron transport in Escherichia coli. FEBS Lett 49:301–305. doi: 10.1016/0014-5793(75)80771-X. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 4.Freed DM, Lukasik SM, Sikora A, Mokdad A, Cafiso DS. 2013. Monomeric TonB and the Ton box are required for the formation of a high-affinity transporter-TonB complex. Biochemistry 52:2638–2648. doi: 10.1021/bi3016108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holden VI, Bachman MA. 2015. Diverging roles of bacterial siderophores during infection. Metallomics 7:986–995. doi: 10.1039/C4MT00333K. [DOI] [PubMed] [Google Scholar]
- 6.Wilson BR, Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. 2016. Siderophores in iron metabolism: from mechanism to therapy potential. Trends Mol Med 22:1077–1090. doi: 10.1016/j.molmed.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köster W. 2005. Cytoplasmic membrane iron permease systems in the bacterial cell envelope. Front Biosci 10:462–477. doi: 10.2741/1542. [DOI] [PubMed] [Google Scholar]
- 8.Braun V. 2009. FhuA (TonA), the career of a protein. J Bacteriol 191:3431–3436. doi: 10.1128/JB.00106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson AD, Braun V, Fiedler HP, Coulton JW, Diederichs K, Welte W. 2000. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci 9:956–963. doi: 10.1110/ps.9.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson AD, Ködding J, Walker G, Bös C, Coulton JW, Diederichs K, Braun V, Welte W. 2001. Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA. Structure 9:707–716. doi: 10.1016/S0969-2126(01)00631-1. [DOI] [PubMed] [Google Scholar]
- 11.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771–778. doi: 10.1016/S0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 12.Schöffler H, Braun V. 1989. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol Gen Genet 217:378–383. [DOI] [PubMed] [Google Scholar]
- 13.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endriss F, Braun V. 2004. Loop deletions indicate regions important for FhuA transport and receptor functions in Escherichia coli. J Bacteriol 186:4818–4823. doi: 10.1128/JB.186.14.4818-4823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moeck GS, Coulton JW. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol 28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 16.Hickman SJ, Cooper REM, Bellucci L, Paci E, Brockwell DJ. 2017. Gating of TonB-dependent transporters by substrate-specific forced remodelling. Nat Commun 8:14804. doi: 10.1038/ncomms14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flayhan A, Wien F, Paternostre M, Boulanger P, Breyton C. 2012. New insights into pb5, the receptor binding protein of bacteriophage T5, and its interaction with its Escherichia coli receptor FhuA. Biochimie 94:1982–1989. doi: 10.1016/j.biochi.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Mathavan I, Zirah S, Mehmood S, Choudhury HG, Goulard C, Li Y, Robinson CV, Rebuffat S, Beis K. 2014. Structural basis for hijacking siderophore receptors by antimicrobial lasso peptides. Nat Chem Biol 10:340–342. doi: 10.1038/nchembio.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fecker L, Braun V. 1983. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J Bacteriol 156:1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köster W. 1991. Iron(III)hydroxamate transport across the cytoplasmic membrane of Escherichia coli. Biol Metals 4:23–32. doi: 10.1007/BF01135553. [DOI] [PubMed] [Google Scholar]
- 21.Mademidis A, Köster W. 1998. Transport activity of FhuA, FhuC, FhuD, and FhuB derivatives in a system free of polar effects, and stoichiometry of components involved in ferrichrome uptake. Mol Gen Genet 258:156–165. [DOI] [PubMed] [Google Scholar]
- 22.Carter DM, Miousse IR, Gagnon JN, Martinez E, Clements A, Lee J, Hancock MA, Gagnon H, Pawelek PD, Coulton JW. 2006. Interactions between TonB from Escherichia coli and the periplasmic protein FhuD. J Biol Chem 281:35413–35424. doi: 10.1074/jbc.M607611200. [DOI] [PubMed] [Google Scholar]
- 23.Mikael LG, Pawelek PD, Labrie J, Sirois M, Coulton JW, Jacques M. 2002. Molecular cloning and characterization of the ferric hydroxamate uptake (fhu) operon in Actinobacillus pleuropneumoniae. Microbiology 148(Part 9):2869–2882. doi: 10.1099/00221287-148-9-2869. [DOI] [PubMed] [Google Scholar]
- 24.del Río ML, Navas J, Martín AJ, Gutiérrez CB, Rodríguez-Barbosa JI, Rodríguez Ferri EF. 2006. Molecular characterization of Haemophilus parasuis ferric hydroxamate uptake (fhu) genes and constitutive expression of the FhuA receptor. Vet Res 37:49–59. doi: 10.1051/vetres:2005039. [DOI] [PubMed] [Google Scholar]
- 25.Abdelhamed H, Lu J, Lawrence ML, Karsi A. 2016. Ferric hydroxamate uptake system contributes to Edwardsiella ictaluri virulence. Microb Pathog 100:195–200. doi: 10.1016/j.micpath.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Stevens JB, Carter RA, Hussain H, Carson KC, Dilworth MJ, Johnston AW. 1999. The fhu genes of Rhizobium leguminosarum, specifying siderophore uptake proteins: fhuDCB are adjacent to a pseudogene version of fhuA. Microbiology 145(Part 3):593–601. [DOI] [PubMed] [Google Scholar]
- 27.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 28.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A 103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. 2007. Genes under positive selection in Escherichia coli. Genome Res 17:1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5(1):e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sims GE, Kim SH. 2011. Whole-genome phylogeny of Escherichia coli/Shigella group by feature frequency profiles (FFPs). Proc Natl Acad Sci U S A 108:8329–8334. doi: 10.1073/pnas.1105168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell-Olds T, Willis JH, Goldstein DB. 2007. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet 8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- 33.Hughes AL, Nei M. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE, Ivers LC, Boncy J, Harris JB, Camilli A. 2014. Evolutionary consequences of intra-patient phage predation on microbial populations. Elife 3:e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Houte S, Buckling A, Westra ER. 2016. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol Mol Biol Rev 80:745–763. doi: 10.1128/MMBR.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, Huang H, Hui X, Cheng X, White AP, Zhao Z, Wang Y. 2016. Distribution and evolution of Yersinia leucine-rich repeat proteins. Infect Immun 84:2243–2254. doi: 10.1128/IAI.00324-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiyoshi H, Tiffany CR, Bronner DN, Bäumler AJ. 2018. Typhoidal Salmonella serovars: ecological opportunity and the evolution of a new pathovar. FEMS Microbiol Rev 42:527–541. [DOI] [PubMed] [Google Scholar]
- 39.MacKenzie KD, Palmer MB, Köster WL, White AP. 2017. Examining the link between biofilm formation and the ability of pathogenic salmonella strains to colonize multiple host species. Front Vet Sci 4:138. doi: 10.3389/fvets.2017.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. 2000. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo G, Hao B. 2015. CVTree3 web server for whole-genome-based and alignment-free prokaryotic phylogeny and taxonomy. Genomics Proteomics Bioinformatics 13:321–331. doi: 10.1016/j.gpb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.