Mutations in σE-regulated lipoproteins have previously been shown to impact bacterial viability under conditions of stress and during in vivo infection. YraP is conserved across a number of Gram-negative pathogens, including Neisseria meningitidis, where the homolog is a component of the Bexsero meningococcal group B vaccine.
KEYWORDS: Salmonella, YraP, virulence
ABSTRACT
Mutations in σE-regulated lipoproteins have previously been shown to impact bacterial viability under conditions of stress and during in vivo infection. YraP is conserved across a number of Gram-negative pathogens, including Neisseria meningitidis, where the homolog is a component of the Bexsero meningococcal group B vaccine. Investigations using laboratory-adapted Escherichia coli K-12 have shown that yraP mutants have elevated sensitivity to a range of compounds, including detergents and normally ineffective antibiotics. In this study, we investigate the role of the outer membrane lipoprotein YraP in the pathogenesis of Salmonella enterica serovar Typhimurium. We show that mutations in S. Typhimurium yraP result in a defective outer membrane barrier with elevated sensitivity to a range of compounds. This defect is associated with attenuated virulence in an oral infection model and during the early stages of systemic infection. We show that this attenuation is not a result of defects in lipopolysaccharide and O-antigen synthesis, changes in outer membrane protein levels, or the ability to adhere to and invade eukaryotic cell lines in vitro.
INTRODUCTION
The outer membrane (OM) forms the interface between a Gram-negative bacterium and its environment. The OM is composed of lipopolysaccharide (LPS), phospholipids, integral outer membrane proteins (OMPs), and peripheral lipoproteins. These components act in concert to create a semipermeable barrier that excludes noxious substances, while allowing nutrients to enter the cell. Each component of the OM is synthesized in the cytoplasm and then trafficked and incorporated into the OM by dedicated proteinaceous nanomachines. Specifically, LPS is trafficked to the OM by the Lpt pathway, OMPs are inserted into the OM by the BAM system, and the OM lipoproteins are trafficked by the Lol system (1–3). Reflecting the importance of the OM, many of the genes encoding components of the OM and their assembly machines are essential for viability. However, numerous studies have demonstrated that many of the OM components that are not essential for growth in vitro are critical for survival of the bacterium in its natural environment; examples include the periplasmic chaperone SurA and the BAM complex component BamB (4–6). Loss of these components often compromise the barrier function of the OM rendering the bacterium susceptible to environmental stresses such as temperature, host defense peptides, or phage infection.
To maintain the barrier function of the OM, bacteria must be able to sense and respond to stresses that perturb it. There are several recognized sensory pathways for detecting and responding to envelope stress: Cpx, Rcs, Psp, and the extracytoplasmic stress response (7–11). The extracytoplasmic stress response is controlled by RpoE or σE, which is essential in Escherichia coli (12). Under nonstressed conditions, σE is sequestered at the cytoplasmic face of the inner membrane by RseA (11, 13). Cell envelope stress (e.g., the accumulation of OMPs, LPS, or high temperatures) induces a catalytic cascade resulting in the release of σE through the degradation of RseA by DegS and RseP (11, 13, 14). σE alters the expression of many genes, including those encoding periplasmic chaperones or proteases and genes required for OM biosynthesis such as fkpA, surA, lpxP, and bamB (15–18). The resulting stress responses rescue the integrity of the cell envelope, ensuring survival of the cell in an unfavorable environment.
Recent studies have described several σE-regulated genes of unknown function in E. coli, including yraP, which was predicted to encode an OM lipoprotein (15, 18, 19). Previous investigations noted that disruption of yraP in E. coli and the yraP homolog (GNA2091) in Neisseria meningitidis severely perturbed the OM barrier function (19–21). Surprisingly, while GNA2091 forms part of the recently licensed N. meningitidis 4CMenB vaccine (22, 23), the contribution of yraP to bacterial virulence has not been investigated.
In Salmonella enterica, σE is not essential even though it functions in a similar manner to E. coli σE and regulates many of the same genes found in E. coli (17, 18). However, S. enterica and several other Gram-negative species harboring null mutations in rpoE are avirulent in animal models of infection (24, 25). Several studies have demonstrated that mutations in genes regulated by σE attenuate S. Typhimurium virulence to various degrees (26–28). For example, disruption of genes encoding the OM lipoproteins BamB, BamE, and Lpp cause significant attenuation in a murine model of infection (26, 29–33). Here, we demonstrate that yraP is widely conserved in Gram-negative bacteria and that loss of yraP compromises the integrity of the S. enterica OM. Furthermore, an S. enterica yraP-null mutant is attenuated in oral and systemic murine models of infection.
RESULTS
YraP is conserved across Gram-negative bacteria.
To determine the prevalence of yraP across Gram-negative bacterial species, the predicted amino acid sequence of the complete E. coli protein was used to interrogate all sequenced bacterial genomes available in the NCBI database using BLAST. Homologues of yraP were identified across the alpha-, beta-, and gammaproteobacteria (see Fig. S1A in the supplemental material). The 573-bp yraP gene (STM3267) of S. enterica serovar Typhimurium, which is 82% identical to the E. coli gene, is located downstream of genes yraM, yraN, and yraO (Fig. S1B). The gene is present across all Salmonella spp. and is highly conserved, with >98% nucleotide identity. yraP is not predicted to be in an operon, having two independent σE promoter sites located 60 (yraPp1) and 337 (yraPp2) bp upstream of the start codon (34). The conserved RpoE binding motifs are homologous to those previously reported for the σE-regulated E. coli yraP (Fig. S1D) (19). RpoE proteins from both species possess greater than 99% sequence identity (Fig. S1E). The SalCom database (Fig. S1C) indicates that transcription from yraPp2 is maximal in growth conditions associated with the intracellular lifestyle of S. enterica (35). These data indicate that YraP is σE regulated, that it is maximally expressed under conditions of OM stress, and that it may play a role in virulence.
YraP is an outer membrane lipoprotein.
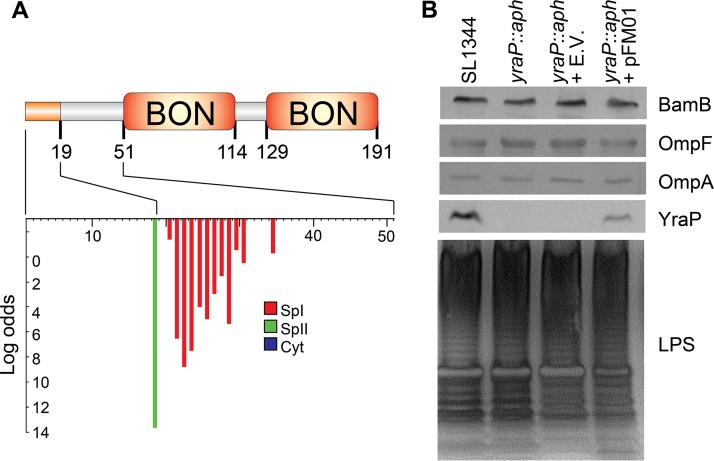
Bioinformatic analyses predict the yraP gene encodes a protein (YraP) with three distinct domains: a Sec-dependent signal sequence and two bacterial and OsmY nodulation (BON) (Pfam PF04972) domains (Fig. 1A). The signal sequence is predicted to be cleaved by signal peptidase II (LspA) after amino acid residue 18, producing a mature protein of approximately 18 kDa with an N-terminal acylated cysteine (Fig. 1A). These predictions are consistent with a protein trafficked to and anchored in the inner leaflet of the OM by the Lol system (3). The BON domains, which are located between residues 46 to 115 and residues 124 to 191, have no proven function but are proposed to interact with phospholipid (36).
FIG 1.
Bioinformatic and experimental analysis of yraP and the impact of isogenic S. Typhimurium yraP mutant. (A) Bioinformatic analysis of YraP, including the N-terminal signal peptidase II cleavage site, after position 18, acylated cysteine residue at position 19, and two bacterial and OsmY nodulation domains (Pfam PF04972). (B) Western immunoblot analysis of OM fractions for S. Typhimurium parental strain SL1344, mutant SL1344 yraP::aph, vector-only SL1344 yraP::aph pQE60NdeI, and complement SL1344 yraP::aph pFM01 using anti-YraP, -BamB, -OmpA, or -OmpF specific primary antibodies and anti-rabbit or anti-mouse total IgG antibody conjugated to alkaline phosphatase for the detection with NBP-BCIP. LPS preparations from S. Typhimurium strains (described above), prepared as described previously (64), were electrophoresed using SDS-PAGE and stained with an Invitrogen silver staining kit. The Western immunoblot and SDS-PAGE gels shown are representative of results obtained from individual experiments conducted on three separate days.
To determine whether YraP from Salmonella localized to the OM, we investigated the cellular localization of the mature lipoprotein. OM fractions from S. Typhimurium SL1344, the isogenic yraP mutant, and the complemented mutant (SL1344 yraP::aph pFM01) were separated by SDS-PAGE and YraP was detected by Western immunoblotting with a monospecific polyclonal antibody generated toward E. coli YraP. A band of ∼18 kDa was detectable in the OM fraction of the parental strain (SL1344) and the complement (SL1344 yraP::aph pFM01) but absent in the mutant (SL1344 yraP::aph) and empty vector control (Fig. 1B). Notably, the protein was detectable in fractions containing known OMPs (Fig. 1B). These results are consistent with the predictions of YraP as an OM lipoprotein.
Loss of YraP does not perturb the biogenesis of OM components.
Previously, Bos et al. reported that the YraP homolog from Neisseria meningitidis, GNA2091, was required for the correct assembly of a subset of OMPs (21). Therefore, we investigated whether the loss of YraP altered the levels of OMPs or OM lipoproteins. The OM fractions were probed using antibodies specific to OmpA/F or the BAM complex lipoprotein BamB. However, no obvious defect in the levels of any of these proteins was observed (Fig. 1B).
Since the yraP mutation had no significant effect on the levels of OMPs or BamB, we examined whether it was required for normal LPS biosynthesis. LPS fractions from the S. enterica SL1344 parental strain, the isogenic yraP mutant and the complemented strain were analyzed by SDS-PAGE and subsequent silver staining. No detectable differences in either the chain length or quantity of the LPS and O antigen were observed (Fig. 1B). These data suggest that loss of YraP does not have a global effect on OMP, LPS, or lipoprotein production.
YraP is required for the integrity of the S. Typhimurium OM.
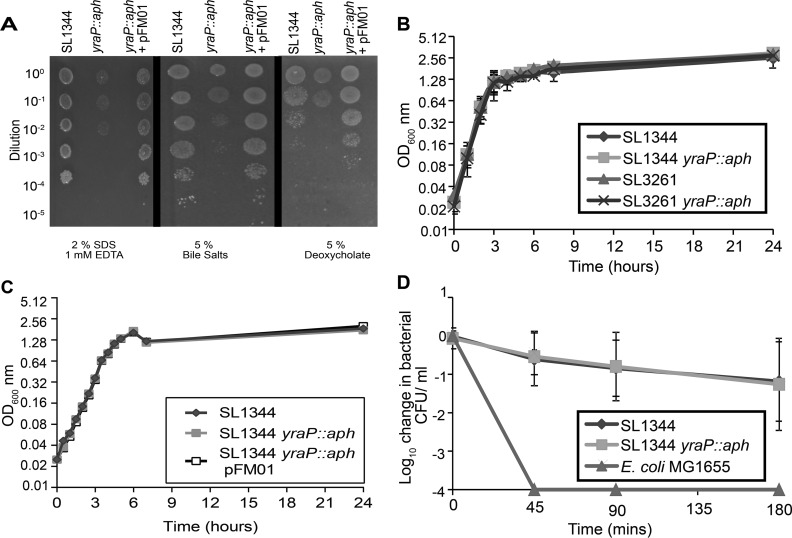
Having established that YraP was an OM localized lipoprotein, we sought to determine whether loss of YraP had an impact on OM integrity. We investigated the susceptibility of S. Typhimurium SL1344 and the isogenic yraP mutant to SDS-EDTA, bile salts, and deoxycholate. Previous investigations had demonstrated that OM homeostasis and active efflux are required for resistance to these compounds (37, 38). Compared to the parental strain, the yraP mutant showed heightened sensitivity to a range of compounds, a phenotype readily complemented by supplying the gene in trans, an observation that is consistent with the mutant having a perturbed OM (Fig. 2A). To ensure that loss of yraP did not induce a growth defect that might be responsible for these observations, we compared the growth rates of the parental strains with their mutant derivatives under standard laboratory conditions in lysogeny broth (LB) or M9 minimal medium supplemented with Casamino Acids and glucose. No differences in growth rate could be discerned for either S. enterica SL1344 or SL3261 derivatives (Fig. 2B and C), indicating there was a defect in OM integrity.
FIG 2.
Impact of yraP disruption on in vitro sensitivity. (A) Growth of S. Typhimurium SL1344 and the yraP mutant in the presence of 2% sodium dodecyl sulfate, 1 mM EDTA, 5% deoxycholate, and 5% bile salts. Stationary-phase bacterial cultures were adjusted to an OD600 of 1 (109 CFU/ml) and serially diluted (10-fold), with 3 μl spotted onto LB agar containing the above-described compounds, followed by incubation at 37°C for 18 h prior to the inspection of growth. The images shown are representative of results obtained from individual experiments conducted on three separate days. (B) Growth kinetics for S. Typhimurium, parental strains SL1344 and SL3261, and the respective yraP mutants in LB at 37°C with continuous agitation, measured as the OD600. Each value represents the mean of triplicate experiments, with error bars denoting standard deviations. (C) Growth kinetics for S. Typhimurium strain SL1344, the isogenic yraP mutant, and the complemented mutant in M9 minimal medium supplemented with 0.1% Casamino Acids and 0.4% glucose. Samples were grown at 37°C with continuous agitation and were measured as the OD600. Data points represent the means of three biological repeats, with error bars representing standard deviations. (D) Bacterial survival of S. Typhimurium parental strain SL1344 and the respective yraP mutant, compared to serum-sensitive E. coli strain MG1655, in the presence of healthy human acellular serum. The results presented are the means of three independent experiments, with error bars denoting standard deviations.
Since there was a perturbation of OM integrity in the absence of yraP, we investigated whether this translated into heightened susceptibility to complement-mediated killing by human serum from healthy donors. Although previous studies have shown that O-antigen chain length is a major contributor to serum resistance of Salmonella (39), the degree by which a destabilized OM barrier can affect resistance has, to our knowledge, not been investigated. Thus, to address this possibility, we investigated bacterial survival in the presence of healthy human serum in vitro. Sera were obtained from volunteers at University of Birmingham and incubated with 106 CFU/ml of either the parental strain SL1344 or the yraP mutant. As expected, the serum-sensitive E. coli K-12 strain MG1655 was killed rapidly, and no viable cells were detected 45 min postinoculation. While the yraP mutant was sensitive to serum, this was at a level comparable with the parental strain (Fig. 2C). This suggests that the defect in OM homeostasis induced by loss of yraP did not confer susceptibility to serum-mediated killing.
YraP mutants are attenuated in a murine model of infection.
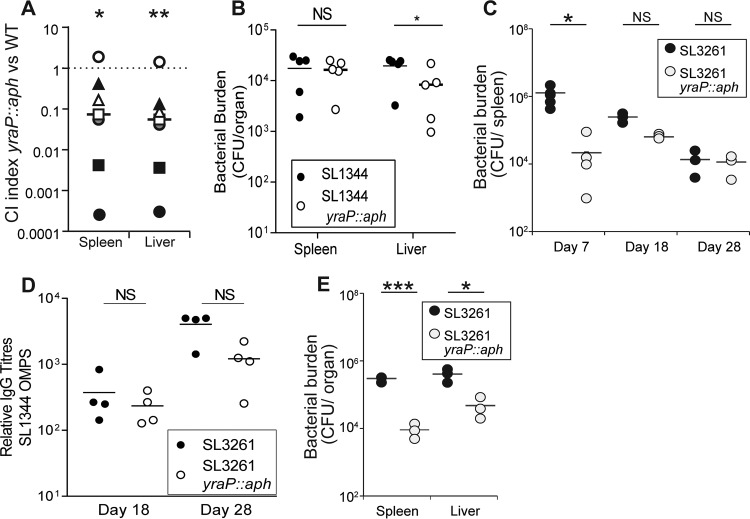
Since OM homeostasis is critical for virulence of numerous bacterial pathogens, we investigated the impact of yraP on virulence after oral challenge with Salmonella. To do this, we examined the capacity of the two bacterial strains to survive within the same C57BL/6 mouse, generating a competitive index as a comparative measure of in vivo fitness for a bacterial mutant. The competitive index was calculated as the ratio of the CFU per gram of each organ of S. Typhimurium parental strain to the CFU per gram of organ of the isogenic yraP mutant. At 4 days after infection by oral gavage with 109 CFU of a 1:1 mix of both strains, the bacterial numbers were enumerated from the spleens and livers of mice. Analysis of the competitive indices from oral infections indicated that in 6 of 7 mice the yraP mutant was significantly outcompeted by the parental strain in both the spleen and the liver (Fig. 3A). The difference was sufficiently marked that in most mice 10-fold more SL1344 CFU were recovered than for the yraP mutant.
FIG 3.
Impact of yraP disruption on in vivo virulence. (A) Competitive indexes of the S. Typhimurium SL1344 parental strain versus the isogenic yraP mutant at 4 days postinfection were determined from the spleens and livers of C57BL/6 mice. Statistical significance was determined using a Mann-Whitney t test (*, P < 0.05; **, P < 0.01). (B) Bacterial burdens per organ of CD1 mice infected intraperitoneally with 3 × 103 CFU of S. Typhimurium SL1344 and the isogenic yraP mutant. Results were obtained at 3 days postinfection. Statistical significance was calculated using a Mann-Whitney test (*, P < 0.05; NS, not significant). (C) Bacterial burdens per spleen at various time points after intraperitoneal infection of C57BL/6 mice with 5 × 105 CFU of attenuated S. Typhimurium SL3261 and the respective yraP mutant. Bars represent the average burdens calculated from four mice. Statistical significance was calculated using a Mann-Whitney test (*, P < 0.05). (D) Relative total IgG titers to purified OMPs in mouse sera compared to control (noninfected-mouse sera) from days 18 and 28 postinfection with the attenuated S. Typhimurium SL3261 parental stain or isogenic yraP mutant. Bars represent the average titers calculated from four mice per group. (E) Bacterial burdens per organ of RagB6 mice 24 h after intraperitoneal infection with 5 × 105 CFU of attenuated S. Typhimurium strain SL3261 or isogenic yraP mutant. Bars represent the average burdens calculated from four mice. *, P < 0.05; ***, P < 0.001.
In addition to assessing virulence via the oral route, we examined infection in a systemic model using the virulent S. Typhimurium strain SL1344 in CD1 mice. Mice were infected via the intraperitoneal route with either parental S. Typhimurium SL1344 or an isogenic yraP mutant, and the bacterial burdens were enumerated from the spleens and livers at day 3 postinfection. The yraP mutant demonstrated a significant attenuation in the liver compared to the parental strain (Fig. 3B). To further explore the impact of yraP on longer-term systemic virulence and bacterial clearance over time, we elected to utilize a resolving model of infection in which intrinsically susceptible C57BL/6 mice can be infected with attenuated bacteria lacking the aroA gene (S. Typhimurium SL3261) for a month or longer. Mice were infected via the intraperitoneal route with S. Typhimurium SL3261 and an isogenic yraP mutant. The numbers of bacteria recovered in the liver and spleen were enumerated at days 7, 18, and 28 postinfection. The bacterial numbers at day 7 were markedly lower in the mutant than in the SL3261 parental strain (Fig. 3C); ∼60-fold fewer bacteria were recovered in the spleen at this time (P < 0.05). Nevertheless, after this time, when T helper 1 cells are necessary for bacterial clearance (40, 41), this was not further enhanced (Fig. 3C). Both strains induced similar levels of IgG to OMPs at days 18 and 28 postinfection (Fig. 3D).
We hypothesized that the failure to observe a difference in bacterial numbers at the later time points might be due to the accumulation of suppressor mutations in the yraP-deficient strains. Such secondary mutations would restore the barrier function of the OM. To investigate this possibility, we tested the SDS-EDTA sensitivity of 16 ex vivo yraP isolates (four isolates per mouse) obtained from spleens and livers harvested from mice at 18 days postinfection. All isolates retained sensitivity to SDS-EDTA, suggesting no second-site mutations occurred that could restore the barrier function of the OM (Fig. S2). However, to ensure no other compensatory mutations occurred, which could explain the ability of the mutant to proliferate at the later time points, each isolate was subjected to whole-genome sequencing using Illumina-based sequencing. Comparison of these sequences to the genome of the parent S. Typhimurium revealed that no additional mutations were present in the mutant bacteria after in vivo growth.
That differences in bacterial numbers were predominantly observed at early infection time points suggested that the yraP mutant was more susceptible to innate mechanisms of immunity. To investigate this possibility, we infected Rag1-deficient mice, which lack T and B cells, with attenuated S. Typhimurium SL3261 or the yraP mutant and examined the bacterial numbers at 24 h postinfection. The yraP mutants again showed significantly reduced bacterial burdens in both the liver and the spleen (Fig. 3E, P < 0.05). This indicates that the yraP mutation results in initial colonization defects, potentially due to increased sensitivity to innate immune responses.
Loss of YraP does not impact adhesion, invasion, or intracellular survival.
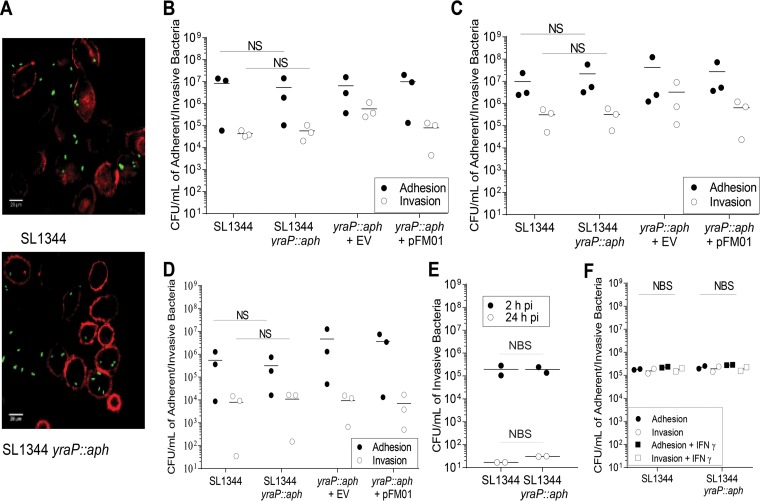
Transcriptomic data from the Hinton laboratory (35, 42) indicated that under the 22 conditions tested, yraP was upregulated under conditions that favored the induction of a type III secretion system located on the SPI-2 pathogenicity island (43). This secretion system is required for Salmonella to survive intracellularly within the Salmonella-containing vacuole. Furthermore, Srikumar et al. (42) demonstrated that yraP was most significantly upregulated within the macrophage. This suggested that the defect in in vivo survival of S. Typhimurium yraP mutants might be due to a deficiency in the ability of the bacteria to either adhere or invade and survive intracellularly. To test this hypothesis, we examined the impact of yraP disruption on S. Typhimurium SL1344 adhesion to and invasion of the murine macrophage cell line J774 and the semiadherent epithelial cell line Caco2. First, cells were infected with S. Typhimurium SL1344 and SL1344 yraP::aph constitutively expressing green fluorescent protein (GFP) for 2 h, before staining with ActinRed (Thermo Fisher Scientific). Adhered and intracellular bacteria were then visualized with a Nikon AR1 inverted confocal microscope using 540/565-nm and 485/510-nm excitation and emission for ActinRed and GFP, respectively. Microscopy indicated that both strains were capable of adhesion and invasion of these cell types in vitro (Fig. 4A). For definitive quantification, these assays were repeated, lysing the cells and enumerating the recovered bacteria for the S. Typhimurium SL1344 and SL1344 yraP::aph. The yraP mutant adhered to and invaded both cell types at levels comparable to the parental strain (Fig. 4B and C). Since J774 and Caco2 cells are immortalized cell lines, we next sought to investigate whether the yraP mutation affected the ability of Salmonella to adhere to or invade primary bone marrow-derived macrophages (BMDMs). Similar to the observations for the immortalized cell lines, disruption of yraP had no impact on either adhesion or invasion (Fig. 4D). Mild increases in bacterial burdens observed in the yraP mutants containing the vector only were not statistically significant. The ability of Salmonella to survive for extended periods inside macrophages is a key virulence phenotype critical to the success of this pathogen. To investigate whether yraP contributed to survival in macrophages, we infected J774 macrophages with the yraP mutant or the isogenic parental strain for 2 or 24 h. Figure 4E shows that yraP does not contribute to the ability of Salmonella to persist in macrophages. However, the experiments described above reflect an environment in which other host immune factors are missing. Therefore, we sought to investigate whether activation of macrophages using gamma interferon (IFN-γ) treatment would impact the adhesion and invasion of Salmonella. IFN-γ activation of J774 macrophages did not affect the adhesion or invasion of either the yraP mutant or the parental strain (Fig. 4F).
FIG 4.
Adherence and invasion of the S. Typhimurium SL1344 and isogenic yraP mutant to various cell types. (A) Images of J774 murine macrophages infected with the S. Typhimurium SL1344 parental strain and the respective yraP mutant. Samples expressing GFP for 2 h were fixed and stained with ActinRed before visualization with a Nikon AR1 inverted confocal microscope using 540/565-nm and 485/510-nm excitation and emission for ActinRed and GFP, respectively. (B to D) Adherence and invasion of S. Typhimurium SL1344 and yraP mutants to the murine macrophage cell line J774 (B), the human intestinal cell line CaCo2 (C), and bone marrow-derived macrophages from C57BL/6 mice (D). Cells were infected at an MOI of 1:100 and incubated for 2 h before lysing and enumeration of the bacterial CFU (adhesion) or treatment with gentamicin for a further 2 h (invasion). Individual data points represent the means of three technical replicates from a single experiment; the bars represent the means of the three independent experiments. (E) The survival of S. Typhimurium SL1344 and isogenic yraP mutant in the murine macrophage cell line, J774, during a time course of 2 and 24 h postinfection (pi). Individual data points represent the means of three technical replicates from two independent experiments. (F) Adherence and invasion of S. Typhimurium SL1344 and yraP mutant to the murine macrophage cell line, J774, in the presence or absence of IFN-γ. Individual data points represent the means of five technical replicates from two biological repeats. NBS, not biologically significant.
DISCUSSION
During this study, we sought to investigate the functional significance of YraP, a σE-regulated lipoprotein from S. enterica serovar Typhimurium. YraP is an OM-localized lipoprotein, as predicted and confirmed by bioinformatic and experimental analysis, respectively. Despite the significant phylogenetic distribution of yraP, the gene is largely uncharacterized. Previously, Bos et al. reported that the N. meningitidis homolog GNA2091 is an uncharacterized component of the BAM complex, responsible for the biogenesis of a subset of trimeric OMPs. However, these researchers were unable to demonstrate a direct interaction between GNA2091 and any BAM component (21). Consistent with previous reports, the yraP mutants display a perturbed OM (20), with heightened sensitivity to a number of anionic detergents. The results presented as part of this study are consistent with the idea that yraP mutants may alter the composition of the OM through a currently unknown mechanism, resulting in increased sensitivity to anionic compounds. Although we have not examined an exhaustive list of potential candidates that may be affected by yraP disruption, we have revealed that the levels of LPS, O antigen, and major OMPs were comparable in our strains, suggesting changes in other components, such as phospholipids. Unfortunately, the mechanism by which yraP alters the composition or integrity of the membrane is not understood and is outside the scope of this investigation.
The differences in bacterial burdens observed after oral or systemic infection strongly suggest that there is an increased susceptibility of the yraP mutant to innate immune defenses. This is not due to an increased sensitivity to complement, since bacteria did not show enhanced susceptibility in serum bactericidal activity (SBA) and murine complement is not efficient at promoting cell-free bacterial killing (44). Moreover, our in vitro adhesion/invasion studies suggested that there was no defect in bacterial binding to or entering host cells. We initially hypothesized that reduced in vivo survival of the yraP mutant was due to increased killing by cellular mechanisms, such as reactive oxygen or nitrogen intermediates from cells such as macrophages. However, this appears not to be the case since the yraP mutant and parental strain survived equally well over an extended period inside macrophages and in IFN-γ-activated macrophages. These results suggest that the disruption of yraP impacts other functions, such as resistance to an as-yet-unknown antimicrobial substance. Interestingly, after the early, profound decrease in bacterial numbers, the level of infection remained relatively constant in the weeks that followed. In the attenuated model of infection, bacterial burdens at day 28 were similar in the yraP mutant- and SL3261 parental strain-infected mice. One potential reason for this persistence in bacterial numbers could be increased mutations leading to compensation for the loss of yraP. Yet, assessment of bacteria by ex vivo functional studies and genome sequencing of recovered bacteria showed that this was not the case. Alternatively, this could be because the bacteria are located in a niche in which they can persist that differs from that for SL3261 or because the lower numbers of yraP mutant bacteria that survive the first few days of infection do not induce as great a Th1 response as SL3261 to promote bacterial clearance.
The results presented as part of this study highlight the need for further analysis of the OM composition of yraP mutants. The increased susceptibility to both anionic compounds and innate immune responses suggests that this lipoprotein may be a potential target for small molecules that render bacteria more susceptible to antibiotics. Furthermore, by enhancing our understanding of how this lipoprotein contributes to vaccine efficacy against N. meningitidis, we will enhance our understanding of the function of YraP and improve the design of future vaccines against a range of Gram-negative pathogens.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this study are detailed in Table 1. Bacteria were routinely cultured in lysogeny broth (LB) or M9 minimal medium (45) with shaking at 37°C. Where appropriate, LB was supplemented with 1.5% agar bacteriological no. 1, 50 μg/ml kanamycin sulfate (Sigma), 100 μg/ml carbenicillin (Melford), or 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Bioline), and M9 minimal medium was supplemented with 0.1% Casamino Acids (BD) and 0.4% glucose (Sigma). Sensitivity to various compounds was determined by plating serial dilutions of cultures adjusted to 109 CFU/ml onto LB agar supplemented with 2% (wt/vol) sodium dodecyl sulfate (SDS) and 1 mM EDTA, 5% (wt/vol) bile salts, or 5% (wt/vol) deoxycholate. The semiadherent murine macrophage-like cell line J774 and the colorectal carcinoma cell line Caco-2 were cultured in Dulbecco modified Eagle medium and Eagle minimum essential medium, as previously described (46). Bone marrow-derived macrophages (BMDMs) were isolated from uninfected C57BL/6 mice and differentiated in complete RPMI 1640 supplemented with 15% L929 conditioned medium (L-cell) (47) for 7 days prior to use. Briefly BMDMs were harvested from the tibias and femurs in complete RPMI prior to treatment with red blood cell lysing buffer (Sigma) and seeded into nonadherent 10-cm petri dishes at a density of 106. The following day, medium was supplemented by the addition of an equal volume of complete RPMI containing 15% L-cell medium. At day 4 postharvest, 50% of the medium was replaced with fresh medium. Adherent cells were collected at day 7 and seeded at the required density 24 h prior to use.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Comment | Source or reference |
|---|---|---|
| Strains | ||
| SL1344 | S. enterica serovar Typhimurium parental strain | 65 |
| SL1344 yraP::aph | yraP gene (formally STM3267) disrupted by a kanamycin resistance cassette | This study |
| SL3261 | Attenuated S. enterica serovar Typhimurium SL1344 ΔaroA | 66 |
| SL3261 yraP::aph | yraP gene (formally STM3267) disrupted by a kanamycin resistance cassette | This study |
| Plasmids | ||
| pQE60NdeI | IPTG-inducible expression vector with added NdeI restriction site in the multiple-cloning site and the previously existing NdeI site removed | 46 |
| pFM01 | pQE60NdeI with yraP cloned in between NdeI and HindIII | This study |
| pFM02 | pET16b with yraP cloned in between NdeI and XhoI | This study |
| pJB39 | Derived from pDOC-C, constitutively expressing GFP | 67 |
DNA manipulation and analyses.
The primers used in this study are listed in Table S1 in the supplemental material. Plasmid DNA, genomic DNA, and PCR products were purified using relevant kits from Qiagen according to the manufacturer's instructions. All restriction enzymes (New England BioLabs) and T4 DNA ligases (Promega) were used in accordance with the manufacturers' specifications. PCR amplifications were performed using Phusion high-fidelity DNA polymerase (New England BioLabs) or 1× ReddyMix PCR master mix (Thermo Scientific). Plasmid sequencing and whole-genome sequencing were done at the University of Birmingham's Functional Genomics and Proteomics Laboratory and MicrobesNG, respectively. PSI-BLAST was used to identify orthologs of YraP. Protein sequences were aligned using ClustalX (48), and phylogenetic distances were estimated using the neighbor-joining method of Saitou and Nei (49). Phylogenetic trees were drawn using Phylodraw 0.8 (50), incorporating bootstrap values obtained from 1,000 replicates. Pfam (51) and LipoP (52) were used to determine the presence of conserved domains within the predicted amino acid sequence.
To generate pFM01, yraP was amplified from S. enterica SL1344, using primers F1 and F2; the products were digested with NdeI and HindIII and subsequently ligated into the modified pQE60NdeI vector (46). To generate pFM02, yraP was amplified from E. coli BW25113, using the primers F6 and F7; the products were digested with NdeI and XhoI and ligated into pET16b. The yraP mutant strains were generated using the one-step gene inactivation method described for E. coli and S. Typhimurium (53, 54). Briefly, the kanamycin resistance cassette was amplified using the primers F3 and F4; the resulting DNA was transformed into S. enterica SL1344 carrying the pKD46 plasmid. Disruption of yraP was confirmed by PCR by using the primers K1 and K2 in combination with the primers F5 (upstream check primer) and F6 (downstream check primer) and subsequent whole-genome DNA sequencing. Using P22 phage, the resulting mutation was transduced into the respective parental strains to generate S. Typhimurium SL1344 yraP::aph and SL3261 yraP::aph.
YraP antibody production.
Antibodies against E. coli YraP were raised in New Zealand White rabbits using His-tagged protein purified from cytosolic inclusion bodies. E. coli BL21(DE3) pFM02 was grown to an optical density at 600 nm (OD600) of 0.6, induced with 1 mM IPTG, and cultured at 18°C for approximately 20 h. Cultures were harvested by centrifugation at 9,000 × g and resuspended in 50 mM sodium phosphate, 500 mM NaCl, 50 mM imidazole, and 0.5 mM TCEP (C4706; Sigma) supplemented with protease inhibitor cocktail (Roche) and lysed using an Emulsiflex (Avestin). Insoluble material was isolated by ultracentrifugation at 75,000 × g for 45 min, washed three times in resuspension buffer supplemented with 0.5% (vol/vol) Triton X-100, and finally resuspended in 50 mM sodium phosphate, 500 mM NaCl, 50 mM imidazole, and 8 M urea. The protein was purified with a Ni-NTA column (Bioline) and eluted in 50 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, and 8 M urea. The resulting eluent was examined by SDS-PAGE, and the YraP protein was gel extracted for immunization of the rabbits.
Preparation and analysis of cellular fractions.
Cellular fractions were prepared as described previously (55). Cellular fractions were electrophoresed on SDS–15% PAGE gels and were either stained with Coomassie blue or transferred to a polyvinylidene difluoride membrane for Western blotting, as previously described (56). Western blots were probed with a 1:1,000, 1:5,000, 1:5,000, or 1:10,000 dilution of the anti-YraP, anti-OmpA (57), anti-BamB, or anti-OmpF antibody, respectively. Secondary anti-rabbit or anti-mouse (1:30,000) of IgG alkaline phosphatase was used before detection with the addition of the substrate, NBP-BCIP (nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolylphosphate; Sigma).
LPS was isolated as previously described (58). In brief, cultures were grown overnight at 37°C. The equivalent of 1 ml of culture at OD600 of 1.0 was pelleted and then resuspended in 100 μl of lysis buffer (1 M Tris [pH 6.8], 2% SDS, and 4% 2-mercaptoethanol). The suspension was then boiled for 10 min and pelleted, and the supernatant was transferred to a fresh tube. Proteinase K (5 μl at 5 mg/ml) was added to each sample before incubation at 60°C for 1 h. Finally, the LPS preparations were heated at 98°C for 10 min and stored at −20°C. LPS was visualized on a 4 to 20% Bis-Tris SDS-PAGE gel and stained using a SilverQuest kit (Invitrogen) in accordance with manufacturer's instructions.
Adherence and invasion/intracellular survival assays.
Cell culture assays were performed as previously described (59). Briefly, bacterial cultures were grown to an OD600 of 1.0, washed three times in 1× phosphate-buffered saline (PBS; Invitrogen), and resuspended in serum-free culture medium (108 CFU/ml). Plates (24-well) were seeded with 105 J774 cells, 106 Caco-2 cells, or 2 × 105 BMDM cells per well. For adherence and invasion assays, cells were infected with an MOI of 1:100, followed by incubation at 37°C in 5% CO2 for 2 h. J774 cells were activated by treatment with 100 U/ml recombinant IFN-γ (R&D Systems) for 20 h before infection with an MOI of 1:10. To calculate the number of adherent bacteria, cells were washed three times with 1× Dulbecco PBS (DPBS) to remove the nonadherent bacteria. Intact cells were then disrupted with 1× DPBS containing 1% (vol/vol) Triton X-100 solution. Serial dilutions of the lysates were then plated onto LB agar supplemented with appropriate antibiotics and CFU were enumerated the following day. Intracellular/invasive bacteria were determined by washing infected cells several times with 1× DPBS and treating them with 100 μg/ml gentamicin for 2 h at 37°C and 5% CO2. The cells were then lysed and plated, and the CFU were enumerated, as described above. The results are expressed as the means of three technical replicates, with three individual experiments conducted on separate days. To determine intracellular bacterial survival over time, plates were seeded as described above and infected with an MOI of 1:10. Samples were incubated at 37°C in 5% CO2 for 30 min, prior to washing three times with DPBS. Extracellular bacteria were eliminated by treatment with 100 μg/ml gentamicin for 1.5 h at 37°C and 5% CO2. Cells were then washed several times with DPBS, lysed, and plated, and the CFU were enumerated, or the medium was replaced with one containing 10 μg/ml gentamicin for a further 22 h, prior to lysis and CFU enumeration as described above.
In vivo assessment of virulence and immune response.
Animal experiments were performed with full ethical approval under Home Office license 30/2850 and P4B8D3A46. Virulence was assessed using C57BL/6, CD1, or RagB6 mice, housed at the Biomedical Services Unit, University of Birmingham. Competitive indices were determined using an oral infection model. Briefly, mice were dosed with 109 CFU in total, composed of a 1:1 mix of the parental S. Typhimurium strain SL1344 and the isogenic yraP mutant, by oral gavage and sacrificed at 4 days postinfection. The numbers of yraP mutant bacteria were calculated by plating homogenized tissue samples on LB agar containing kanamycin and counting the number of CFU. The numbers of wild-type bacteria were determined by plating tissue samples on LB agar and subtracting the number of yraP mutant bacteria present on the kanamycin plates. To assess systemic virulence, mice were infected intraperitoneally with either 5 × 105 CFU (SL3261) or 3 × 103 CFU (SL1344) and sacrificed at the indicated times postinfection, as previously described (60). The blood, spleens, livers, Peyer's patches, and mesenteric lymph nodes were harvested, and bacterial burdens were measured by homogenizing the tissues and plating on LB agar as described previously (61). Each data point represents a single tissue or mouse, with means and standard deviations represented by the horizontal and vertical lines, respectively.
Relative antibody titers against purified OMPs present in harvested serum were determined by enzyme-linked immunosorbent assay (ELISA), as previously described (61). Briefly, MaxiSorp 96-well plates (Nunc) were coated with 5 μg/ml of purified OMPs (purified as described in reference 62) in carbonate coating buffer. After washing, the plates were blocked for 1 h at 37°C with 1% bovine serum albumin in 1× PBS. Subsequently, serum samples were added in blocking buffer and serially diluted, and incubation was continued for 1 h at 37°C. Secondary goat anti-mouse IgG conjugated to alkaline phosphatase (Southern Biotech) was diluted in blocking buffer and added for 1 h at 37°C. The signal was detected using SigmaFAST p-nitrophenyl phosphate system, and the plates were read at 405 nm. Each data point represents the sera from an individual mouse, with means and standard deviations represented by the horizontal and vertical lines, respectively.
Serum bactericidal assay.
Serum bactericidal assays were performed as described previously (63). Briefly, serum-mediated killing of bacterial strains was investigated using healthy human serum. Log-phase cultures were prepared in 1× PBS, and 10 μl of each bacterial sample was added to 90 μl of freshly thawed, undiluted serum to give a final bacterial concentration of 106 CFU/ml. The mixtures were incubated at 37°C with gentle rocking (20 rpm), and 10-μl samples were withdrawn at 45, 90, and 180 min. Samples were serially diluted, and viable bacterial counts were determined. The results are means of three individual experiments conducted in triplicate on three separate days.
Statistical analysis.
Statistical significance was determined using a Mann-Whitney t test. P values of ≤0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeff Cole and Xenia Kostoulias for critical reading of the manuscript. We thank MicrobesNG for support in genome sequencing. This study was supported by BBSRC and MRC grants to I.R.H.
F.C.M. and T.J.W. performed the majority of the experiments (F.C.M., in vitro and in vivo assays; T.J.W., in vivo assays and phylogenetic analysis) and assisted in manuscript preparation. A.E.R., A.E.S., Y.R.S., J.R., G.L.I., and J.M. performed ELISAs and assisted with in vitro assays and in vivo infections. J.A.B. performed sensitivity testing. D.J.P.S. constructed the mutant, and T.J.K. and R.M. purified YraP for antibody production. All work was performed in the laboratories of and under the supervision of I.R.H., M.O., and A.F.C. The authors confirm no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00829-17.
REFERENCES
- 1.Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- 2.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zückert WR. 2014. Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim Biophys Acta 1843:1509–1516. doi: 10.1016/j.bbamcr.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ranallo RT, Kaminski RW, George T, Kordis AA, Chen Q, Szabo K, Venkatesan MM. 2010. Virulence, inflammatory potential, and adaptive immunity induced by Shigella flexneri msbB mutants. Infect Immun 78:400–412. doi: 10.1128/IAI.00533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somerville JE Jr, Cassiano L, Darveau RP. 1999. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect Immun 67:6583–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nevola JJ, Stocker BA, Laux DC, Cohen PS. 1985. Colonization of the mouse intestine by an avirulent Salmonella Typhimurium strain and its lipopolysaccharide-defective mutants. Infect Immun 50:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duguay AR, Silhavy TJ. 2004. Quality control in the bacterial periplasm. Biochim Biophys Acta 1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 9.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet 5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alba BM, Gross CA. 2004. Regulation of the Escherichia coli σE-dependent envelope stress response. Mol Microbiol 52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 12.De Las Peñas A, Connolly L, Gross CA. 1997. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol 179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ades SE. 2008. Regulation by destruction: design of the σE envelope stress response. Curr Opin Microbiol 11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. 2013. Dual molecular signals mediate the bacterial response to outer membrane stress. Science 340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skovierova H, Rowley G, Rezuchova B, Homerova D, Lewis C, Roberts M, Kormanec J. 2006. Identification of the σE regulon of Salmonella enterica serovar Typhimurium. Microbiology 152:1347–1359. doi: 10.1099/mic.0.28744-0. [DOI] [PubMed] [Google Scholar]
- 16.Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA. 2009. Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica. J Bacteriol 191:7279–7287. doi: 10.1128/JB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redford P, Welch RA. 2006. Role of sigma E-regulated genes in Escherichia coli uropathogenesis. Infect Immun 74:4030–4038. doi: 10.1128/IAI.01984-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dartigalongue C, Missiakas D, Raina S. 2001. Characterization of the Escherichia coli σE regulon. J Biol Chem 276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- 19.Onufryk C, Crouch M-L, Fang FC, Gross CA. 2005. Characterization of six lipoproteins in the σE regulon. J Bacteriol 187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bos MP, Grijpstra J, Tommassen-van Boxtel R, Tommassen J. 2014. Involvement of Neisseria meningitidis lipoprotein GNA2091 in the assembly of a subset of outer membrane proteins. J Biol Chem 289:15602–15610. doi: 10.1074/jbc.M113.539510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esposito S, Tagliabue C, Bosis S. 2015. Meningococcal B vaccination (4CMenB) in infants and toddlers. J Immunol Res 2015:402381. doi: 10.1155/2015/402381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernikos G, Medini D. 2014. Bexsero® chronicle. Pathogens Global Health 108:305–316. doi: 10.1179/2047773214Y.0000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M-C, Kuo K-T, Chien H-F, Tsai Y-L, Liaw S-J. 2015. New aspects of RpoE in uropathogenic Proteus mirabilis. Infect Immun 83:966–977. doi: 10.1128/IAI.02232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rattanama P, Thompson JR, Kongkerd N, Srinitiwarawong K, Vuddhakul V, Mekalanos JJ. 2012. Sigma E regulators control hemolytic activity and virulence in a shrimp pathogenic Vibrio harveyi. PLoS One 7:e32523. doi: 10.1371/journal.pone.0032523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. 1999. The alternative sigma factor, σE, is critically important for the virulence of Salmonella Typhimurium. Infect Immun 67:1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sydenham M, Douce G, Bowe F, Ahmed S, Chatfield S, Dougan G. 2000. Salmonella enterica serovar Typhimurium surA mutants are attenuated and effective live oral vaccines. Infect Immun 68:1109–1115. doi: 10.1128/IAI.68.3.1109-1115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horne SM, Kottom TJ, Nolan LK, Young KD. 1997. Decreased intracellular survival of an fkpA mutant of Salmonella Typhimurium Copenhagen. Infect Immun 65:806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fardini Y, Chettab K, Grépinet O, Rochereau S, Trotereau J, Harvey P, Amy M, Bottreau E, Bumstead N, Barrow PA, Virlogeux-Payant I. 2007. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica serovar Enteritidis. Infect Immun 75:358–370. doi: 10.1128/IAI.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. 2002. The alternative sigma factor σE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol 43:771–782. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- 31.Sha J, Fadl AA, Klimpel GR, Niesel DW, Popov VL, Chopra AK. 2004. The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect Immun 72:3987–4003. doi: 10.1128/IAI.72.7.3987-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis C, Skovierova H, Rowley G, Rezuchova B, Homerova D, Stevenson A, Sherry A, Kormanec J, Roberts M. 2008. Small outer membrane lipoprotein, SmpA, is regulated by σE and has a role in cell envelope integrity and virulence of Salmonella enterica serovar Typhimurium. Microbiology 154:979–988. doi: 10.1099/mic.0.2007/011999-0. [DOI] [PubMed] [Google Scholar]
- 33.Nickerson CA, Curtiss R. 1997. Role of sigma factor RpoS in initial stages of Salmonella Typhimurium infection. Infect Immun 65:1814–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gama-Castro S, Salgado H, Santos-Zavaleta A, Ledezma-Tejeida D, Muñiz-Rascado L, García-Sotelo JS, Alquicira-Hernández K, Martínez-Flores I, Pannier L, Castro-Mondragón JA, Medina-Rivera A, Solano-Lira H, Bonavides-Martínez C, Pérez-Rueda E, Alquicira-Hernández S, Porrón-Sotelo L, López-Fuentes A, Hernández-Koutoucheva A, Moral-Chávez VD, Rinaldi F, Collado-Vides J. 2016. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 44:D133–D143. doi: 10.1093/nar/gkv1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran Sathesh K, Hammarlöf Disa L, Canals R, Grissom Joe E, Conway T, Hokamp K, Hinton Jay CD. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Yeats C, Bateman A. 2003. The BON domain: a putative membrane-binding domain. Trends Biochem Sci 28:352–355. doi: 10.1016/S0968-0004(03)00115-4. [DOI] [PubMed] [Google Scholar]
- 37.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prouty AM, Van Velkinburgh JC, Gunn JS. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J Bacteriol 184:1270–1276. doi: 10.1128/JB.184.5.1270-1276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray GL, Attridge SR, Morona R. 2005. Inducible serum resistance in Salmonella Typhimurium is dependent on wzzfepE-regulated very long O antigen chains. Microbes Infect 7:1296–1304. doi: 10.1016/j.micinf.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. 2005. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol 175:4603. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 41.Bobat S, Flores-Langarica A, Hitchcock J, Marshall JL, Kingsley RA, Goodall M, Gil-Cruz C, Serre K, Leyton DL, Letran SE, Gaspal F, Chester R, Chamberlain JL, Dougan G, López-Macías C, Henderson IR, Alexander J, MacLennan ICM, Cunningham AF. 2011. Soluble flagellin, FliC, induces an Ag-specific Th2 response, yet promotes T-bet-regulated Th1 clearance of Salmonella Typhimurium infection. Eur J Immunol 41:1606–1618. doi: 10.1002/eji.201041089. [DOI] [PubMed] [Google Scholar]
- 42.Srikumar S, Kröger C, Hébrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron ADS, Hokamp K, Hinton JCD. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desvaux M, Hébraud M, Henderson IR, Pallen MJ. 2006. Type III secretion: what's in a name? Trends Microbiol 14:157–160. doi: 10.1016/j.tim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Siggins MK, Cunningham AF, Marshall JL, Chamberlain JL, Henderson IR, MacLennan CA. 2011. Absent bactericidal activity of mouse serum against invasive African nontyphoidal Salmonella results from impaired complement function but not a lack of antibody. J Immunol 186:2365–2371. doi: 10.4049/jimmunol.1000284. [DOI] [PubMed] [Google Scholar]
- 45.Eaves DJ, Ricci V, Piddock LJ. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob Agents Chemother 48:1145–1150. doi: 10.1128/AAC.48.4.1145-1150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raghunathan D, Wells TJ, Morris FC, Shaw RK, Bobat S, Peters SE, Paterson GK, Tveen Jensen K, Leyton DL, Blair JM, Browning DF, Pravin J, Flores-Langarica A, Hitchcock JR, Moraes CT, Piazza RM, Maskell DJ, Webber MA, May RC, Maclennan CA, Piddock LJ, Cunningham AF, Henderson IR. 2011. SadA, a trimeric autotransporter from Salmonella enterica serovar Typhimurium, can promote biofilm formation and provides limited protection against infection. Infect Immun 79:4342–4352. doi: 10.1128/IAI.05592-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinar A, Dowling JK, Bitto NJ, Robertson AAB, Latz E, Stewart CR, Drummond GR, Cooper MA, McAuley JL, Tate MD, Mansell A. 2017. PB1-F2 peptide derived from avian influenza A virus H7N9 induces inflammation via activation of the NLRP3 inflammasome. J Biol Chem 292:826–836. doi: 10.1074/jbc.M116.756379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 50.Choi J-H, Jung H-Y, Kim H-S, Cho H-G. 2000. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics 16:1056–1058. doi: 10.1093/bioinformatics/16.11.1056. [DOI] [PubMed] [Google Scholar]
- 51.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Juncker AS, Willenbrock H, von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP, Piddock LJV. 2004. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother 48:4012–4015. doi: 10.1128/AAC.48.10.4012-4015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parham NJ, Srinivasan U, Desvaux M, Foxman B, Marrs CF, Henderson IR. 2004. PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol Lett 230:73–83. doi: 10.1016/S0378-1097(03)00862-0. [DOI] [PubMed] [Google Scholar]
- 56.Wells TJ, McNeilly TN, Totsika M, Mahajan A, Gally DL, Schembri MA. 2009. The Escherichia coli O157:H7 EhaB autotransporter protein binds to laminin and collagen I and induces a serum IgA response in O157:H7 challenged cattle. Environ Microbiol 11:1803–1814. doi: 10.1111/j.1462-2920.2009.01905.x. [DOI] [PubMed] [Google Scholar]
- 57.Marshall JL, Flores-Langarica A, Kingsley RA, Hitchcock JR, Ross EA, Lopez-Macias C, Lakey J, Martin LB, Toellner K-M, MacLennan CA, MacLennan IC, Henderson IR, Dougan G, Cunningham AF. 2012. The capsular polysaccharide Vi from Salmonella Typhi is a B1b antigen. J Immunol 189:5527–5532. doi: 10.4049/jimmunol.1103166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Browning DF, Whitworth DE, Hodgson DA. 2003. Light-induced carotenogenesis in Myxococcus xanthus: functional characterization of the ECF sigma factor CarQ and anti-sigma factor CarR. Mol Microbiol 48:237–251. doi: 10.1046/j.1365-2958.2003.03431.x. [DOI] [PubMed] [Google Scholar]
- 59.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol 10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 60.Bobat S, Darby M, Mrdjen D, Cook C, Logan E, Auret J, Jones E, Schnoeller C, Flores-Langarica A, Ross EA, Vira A, López-Macías C, Henderson IR, Alexander J, Brombacher F, Horsnell WG, Cunningham AF. 2014. Natural and vaccine-mediated immunity to Salmonella Typhimurium is impaired by the helminth Nippostrongylus brasiliensis. PLoS Negl Trop Dis 8:e3341. doi: 10.1371/journal.pntd.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cunningham AF, Gaspal F, Serre K, Mohr E, Henderson IR, Scott-Tucker A, Kenny SM, Khan M, Toellner K-M, Lane PJL, MacLennan ICM. 2007. Salmonella induces a switched antibody response without germinal centers that impedes the extracellular spread of infection. J Immunol 178:6200–6207. doi: 10.4049/jimmunol.178.10.6200. [DOI] [PubMed] [Google Scholar]
- 62.Gil-Cruz C, Bobat S, Marshall JL, Kingsley RA, Ross EA, Henderson IR, Leyton DL, Coughlan RE, Khan M, Jensen KT, Buckley CD, Dougan G, MacLennan ICM, López-Macías C, Cunningham AF. 2009. The porin OmpD from nontyphoidal Salmonella is a key target for a protective B1b cell antibody response. Proc Natl Acad Sci U S A 106:9803–9808. doi: 10.1073/pnas.0812431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wells TJ, Whitters D, Sevastsyanovich YR, Heath JN, Pravin J, Goodall M, Browning DF, O'Shea MK, Cranston A, De Soyza A, Cunningham AF, MacLennan CA, Henderson IR, Stockley RA. 2014. Increased severity of respiratory infections associated with elevated anti-LPS IgG2 which inhibits serum bactericidal killing. J Exp Med 211:1893–1904. doi: 10.1084/jem.20132444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Browning D, Wells T, Franca F, Morris F, Sevastsyanovich Y, Bryant J, Johnson M, Lund P, Cunningham A, Hobman J, May R, Webber M, Henderson I. 2013. Laboratory adapted Escherichia coli K-12 becomes a pathogen of Caenorhabditis elegans upon restoration of O antigen biosynthesis. Mol Microbiol 87:939–950. doi: 10.1111/mmi.12144. [DOI] [PubMed] [Google Scholar]
- 65.Wray C, Sojka WJ. 1978. Experimental Salmonella Typhimurium infection in calves. Res Vet Sci 25:139–143. [PubMed] [Google Scholar]
- 66.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella Typhimurium are nonvirulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 67.Bryant JA, Sellars LE, Busby SJW, Lee DJ. 2014. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res 42:11383–11392. doi: 10.1093/nar/gku828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.