Listeria monocytogenes is a facultative intracellular pathogen that infects a wide variety of cells, causing the life-threatening disease listeriosis. L. monocytogenes virulence factors include two surface invasins, InlA and InlB, known to promote bacterial uptake by host cells, and the secreted pore-forming toxin listeriolysin O (LLO), which disrupts the phagosome to allow bacterial proliferation in the cytosol.
KEYWORDS: InlA, InlB, internalin, Listeria monocytogenes, listeriolysin O, listeriosis, host cell invasion, pore-forming toxins
ABSTRACT
Listeria monocytogenes is a facultative intracellular pathogen that infects a wide variety of cells, causing the life-threatening disease listeriosis. L. monocytogenes virulence factors include two surface invasins, InlA and InlB, known to promote bacterial uptake by host cells, and the secreted pore-forming toxin listeriolysin O (LLO), which disrupts the phagosome to allow bacterial proliferation in the cytosol. In addition, plasma membrane perforation by LLO has been shown to facilitate L. monocytogenes internalization into epithelial cells. In this work, we tested the host cell range and importance of LLO-mediated L. monocytogenes internalization relative to the canonical invasins, InlA and InlB. We measured the efficiencies of L. monocytogenes association with and internalization into several human cell types (hepatocytes, cytotrophoblasts, and endothelial cells) using wild-type bacteria and isogenic single, double, and triple deletion mutants for the genes encoding InlA, InlB and LLO. No role for InlB was detected in any tested cells unless the InlB expression level was substantially enhanced, which was achieved by introducing a mutation (prfA*) in the gene encoding the transcription factor PrfA. In contrast, InlA and LLO were the most critical invasion factors, although they act in a different manner and in a cell-type-dependent fashion. As expected, InlA facilitates both bacterial attachment and internalization in cells that express its receptor, E-cadherin. LLO promotes L. monocytogenes internalization into hepatocytes, but not into cytotrophoblasts and endothelial cells. Finally, LLO and InlA cooperate to increase the efficiency of host cell invasion by L. monocytogenes.
INTRODUCTION
Listeria monocytogenes is a Gram-positive, facultative intracellular bacterium responsible for the foodborne disease listeriosis. Listeriosis is a life-threatening condition for elderly and immunocompromised individuals (1). In these populations, the bacterium can propagate from the intestines to the blood and further disseminate, causing septicemia and meningoencephalitis (1–3, 6). During pregnancy, susceptibility to L. monocytogenes infection is drastically increased and the bacterium can cross the placental barrier, leading to spontaneous abortion, preterm labor, stillbirth, and severe infections of the newborn (1a–1c). An important virulence attribute of L. monocytogenes is its ability to infect numerous cell types, from macrophages to normally nonphagocytic cells such as intestinal and placental epithelial cells, endothelial cells, and neurons (1). The wide host cell range of this pathogen is thought to be critical for crossing the tightest barriers of the human host, i.e., the placental and blood-brain barriers.
The expression of major virulence factors that mediate the L. monocytogenes intracellular life cycle is controlled by PrfA (8–10), which activates transcription in response to a variety of environmental signals, including temperature (11) and nutrient availability (12–14). Two of these virulence factors are the surface proteins InlA and InlB, depicted as the major invasins responsible for L. monocytogenes uptake by normally nonphagocytic cells (4, 15, 16). InlA (internalin) is covalently anchored to the peptidoglycan through its C-terminal LPXTG motif (16, 17), whereas InlB is retained noncovalently at the cell surface via electrostatic interaction between three C-terminal glycine and tryptophan (GW) repeat domains and lipoteichoic acids of the bacterial cell wall (18). The adherens junction protein E-cadherin has been identified as the sole InlA receptor (19), and several host surface proteins, c-Met (or HGF receptor) (20), gC1Q receptor (21), and surface glycosaminoglycans (22), have been identified as InlB receptors. The N-terminal leucine-rich repeat (LRR) domain of InlB binds to c-Met, whereas its C-terminal moiety binds to glycosaminoglycans and gC1Q receptor in addition to being the lipoteichoic acid anchor (21, 22). InlA mediates bacterial entry only into cells expressing E-cadherin, whereas InlB is a more versatile invasin, as its receptors are widely expressed. Importantly, InlA and InlB are species specific: humans and gerbils are permissive to both InlA and InlB, while rabbits/guinea pigs and mice are permissive only to InlA and InlB, respectively (24). It has been proposed that InlB acts as a facilitator of the InlA-dependent invasion pathway in enterocytes (25, 26) and that InlA and InlB, but not listeriolysin O (LLO), are the two most important invasion factors for crossing the intestinal barrier (6, 25, 26).
Upon ingestion by host cells, L. monocytogenes is confined within a vacuole or phagosome that is disrupted by the secreted pore-forming toxin LLO and phospholipases to release the bacterium into the cytosol, where it divides and from which it infects other cells by cell-to-cell spreading (27–30). The role of LLO in mediating vacuolar escape is certainly a major role of this toxin, as the absence of LLO leads to a marked deficiency in intracellular replication of phagocytosed bacteria (30). The role of LLO was considered to be specifically restricted to the disruption of the phagosome (31), but additional roles have been attributed to this toxin. In particular, it has been shown that LLO, secreted by extracellular bacteria, perforates the host cell plasma membrane during the early stage of infection; therefore, LLO secretion and membrane perforation precede the formation of the phagosome (32, 35). Perforation of the host cell plasma membrane activates several signaling pathways (28). One outcome of LLO-induced signaling is the internalization of L. monocytogenes into epithelial cell lines (HepG2, HeLa, and Hep2 cells) (33–35) and professional phagocytes (human neutrophils and murine bone marrow-derived macrophages) (36). However, once bacteria are opsonized, the contribution of LLO in bacterial uptake by professional phagocytes becomes negligible. In addition, LLO-mediated plasma membrane perforation by cytosolic bacteria was recently proposed to facilitate cell-to-cell spreading (37).
Because InlA and InlB are described as the most important factors controlling L. monocytogenes uptake by normally nonphagocytic cells, it was necessary to establish whether the role of LLO is significant in comparison to these two canonical invasins. It was also necessary to determine if LLO plays a general role in inducing L. monocytogenes internalization in all cell types. To address these questions, we used human hepatocytes and cytotrophoblasts, because they are known to be infected by L. monocytogenes during listeriosis (1). It is also known that L. monocytogenes can infect endothelial cells in vitro and may infect these cells in vivo to cross the blood-brain and placental-fetal barriers (38–43). As such, endothelial cells were included in this work. Although enterocytes that make up the intestinal barrier are of critical importance for the establishment of listeriosis, previous work has convincingly shown that crossing the intestinal barrier is InlA dependent and LLO independent, so enterocytes were not included (6). To quantify and compare the roles of the three invasins, we used a fluorescence-based microscopy assay that directly measures the efficiency of bacterial association with host cells and the efficiency of their internalization.
RESULTS
LLO, InlA, and InlB expression levels in single and double deletion mutants.
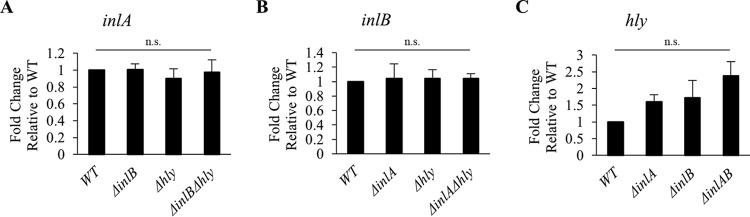
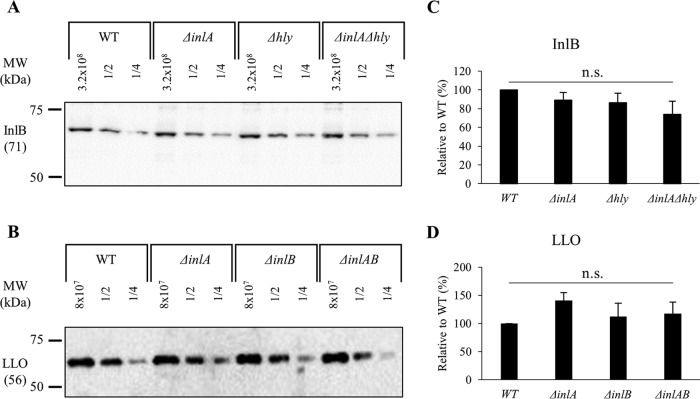
To ensure that deletion of the virulence genes hly, inlA, and inlB, in the single and double deletion L. monocytogenes 10403S mutants, does not affect the expression of the others, the levels of mRNA and proteins of the three invasion factors were measured. Bacteria were grown under the same experimental conditions as for the cell invasion assay and reverse transcription quantitative real-time PCR (RT-qPCR) was used to measure hly, inlA, and inlB mRNA levels. As expected, deletion of one or two virulence genes does not significantly affect the expression of the other genes in comparison to the wild-type (WT) strain (Fig. 1). We then measured the protein expression levels by Western blotting, which required antibodies against LLO, InlB, and InlA. Anti-LLO antibodies are commercially available, but not anti-InlA and anti-InlB. Therefore, we cloned inlA and inlB genes (without the signal peptide-encoding sequence) into an expression vector (pET29b), purified the recombinant proteins, and obtained purified polyclonal rabbit anti-InlB and -InlA. The anti-InlB antibodies could efficiently detect InlB (see Fig. S1 in the supplemental material), but we were not successful with the anti-InlA antibodies. We then measured LLO and InlB protein expression levels by Western blotting. For rigorous evaluation, we analyzed serial dilutions of cell lysates and performed densitometry analyses of the corresponding bands. As expected, single and double deletions of the inlA, inlB, or hly genes do not significantly affect the expression levels of LLO or InlB (Fig. 2).
FIG 1.
inlA, inlB, and hly mRNA quantification. Reverse transcription quantitative real-time PCR (RT-qPCR) was performed to measure inlA, inlB, hly, gap, and rpoB transcripts from L. monocytogenes WT and isogenic deletion mutants. The housekeeping genes, gap and rpoB, were used to normalize the expression of inlA (A), inlB (B), and hly (C). Results are the average fold change in gene expression ± standard error of the mean (SEM) relative to the WT (n ≥ 3). Statistical differences from the WT are indicated (n.s., non-statistically significant).
FIG 2.
InlB and LLO protein levels. (A and B) L. monocytogenes cell lysates, undiluted and at dilutions of 1/2 and 1/4, were subjected to Western blot analysis using anti-InlB and anti-LLO antibodies. (C and D) Densitometry analysis was performed using ImageJ software. Representative Western blots are shown. Results are the mean ± SEM relative to the WT (n ≥ 3). Statistical differences from the WT using data prior to normalization are indicated (n.s., non-statistically significant).
InlA and LLO, but not InlB, control L. monocytogenes uptake by human hepatocytes.
To establish the relative roles of the three virulence factors in L. monocytogenes uptake by human hepatocytes, we used four human hepatocyte cell lines (HepG2, Hep3B, PLC5, and Huh7) to rule out any cell line-specific phenotype and draw conclusions that can generally apply to hepatocytes. Hepatocytes were incubated with L. monocytogenes (WT or Δhly, ΔinlA, ΔinlB, ΔinlAB, ΔinlB Δhly, ΔinlA Δhly, or ΔinlAB Δhly mutants) for 30 min at 37°C and were processed for fluorescence microscopy analysis. Full data sets, including association and internalization efficiencies of the eight bacterial strains into the four cell lines, are presented in Fig. S2. We first focused on analyzing data obtained with the single and triple deletion mutants in comparison to WT L. monocytogenes (Fig. 3). Data show that LLO does not promote L. monocytogenes association with hepatocytes. In one of the hepatocyte cell lines (Hep3B), LLO even significantly decreases bacterial association. In contrast, InlA is the only factor that promotes bacterial association with hepatocytes, in three out of the four cell lines. The decreases in association of the inlA single deletion mutant and the triple deletion mutant were similar in all hepatocyte cell lines, confirming that among the three factors, InlA is the only adhesin. LLO and InlA, but not InlB, promote internalization of L. monocytogenes, although the role of LLO was more prominent in that function than the role of InlA. In one cell line (Hep3B), single deletion mutants had no internalization phenotype, whereas the triple (ΔinlAB Δhly) and double (ΔinlA Δhly) (Fig. S2) deletion mutants displayed a significant decrease in internalization. The latter result shows that LLO and InlA can exert a redundant role in L. monocytogenes internalization. To our surprise, no role for InlB was detected in L. monocytogenes association and internalization into the four hepatocyte cell lines when single, double, and triple deletion mutants were considered (Fig. 3 and Fig. S2). This prompted us to clarify this result.
FIG 3.
Relative roles of LLO, InlA, and InlB in L. monocytogenes invasion of human hepatocytes. HepG2, Hep3B, PLC5, and Huh7 cells were infected with WT, InlA-deficient (ΔinlA), InlB-deficient (ΔinlB), LLO-deficient (Δhly), or InlAB- and LLO-deficient (ΔinlAB Δhly) bacteria (MOI of 20) for 30 min at 37°C. Cells were washed, fixed, and labeled with fluorescent antibodies and DAPI. (A) The bacterial association efficiency was calculated as the total number of bacteria associated per host cell. The average bacterial association values for the WT strain before normalization were as follows: HepG2, 0.14; Hep3B, 3.13; PLC5, 1.34; Huh7, 0.77. (B) The bacterial internalization efficiency was calculated as the percentage of intracellular bacteria. The average percentages of internalization for the WT strain before normalization were as follows: HepG2, 26.45%; Hep3B, 38.77%; PLC5, 18.29%; Huh7, 33.12%. The minimum numbers of host cells counted were as follows: HepG2, 1,000; Hep3B, 150; PLC5, 600; Huh7, 2,000. The average numbers of WT bacteria counted per experiment were as follows: HepG2, 600; Hep3B, 4,000; PLC5, 2,000; Huh7, 3,000 (with a minimum count of 100 bacteria being required for any mutant with reduced association efficiency). Results are expressed as the mean ± SEM relative to the WT (n ≥ 3). Statistical analyses compared each deletion strain to the WT strain and were performed on raw data before normalization (*, P < 0.01, **, P < 0.001).
InlB-mediated L. monocytogenes internalization is dependent on InlB expression level.
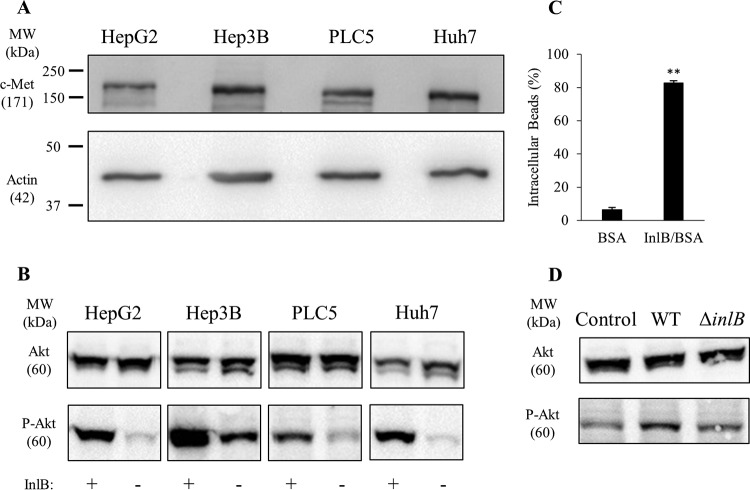
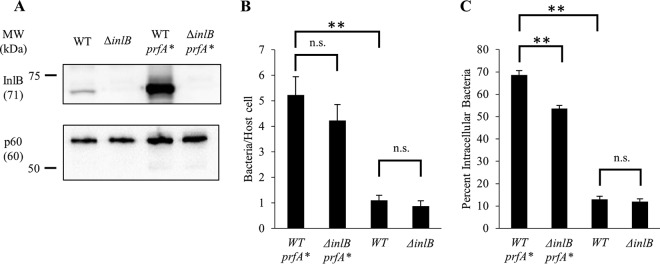
The absence of a role for InlB led us to verify that its receptor, c-Met, was expressed and functional in the hepatocyte cell lines used in these studies. As expected, c-Met was expressed in all tested hepatocyte cell lines (Fig. 4A). Previous studies established that InlB activates c-Met-dependent Akt phosphorylation and F-actin remodeling (44–46). As expected, cell exposure to recombinant InB induced a significant increase in Akt phosphorylation in all cell lines (Fig. 4B). As a second approach, live cell imaging showed that hepatocytes exposed to InlB formed dynamic membrane ruffles, which were not observed in the absence of InlB (see Movies S1 to S4 in the supplemental material). Finally, to evaluate if hepatocytes could undergo InlB-dependent phagocytic uptake, we exposed cells to polystyrene beads (1-μm diameter) that were covalently coated with saturating concentrations of InlB or bovine serum albumin (BSA), used as negative control. As shown in Fig. 4C, 80% of InlB-coated beads were internalized by hepatocytes. We then established if InlB produced by L. monocytogenes could stimulate c-Met. HepG2 cells were incubated with WT and ΔinlB 10403S strains for 30 min at a multiplicity of infection (MOI) of 20, as performed in the invasion assays. As shown in Fig. 4D, WT but not InlB-deficient bacteria induced Akt phosphorylation. Together, these results demonstrate that the hepatocyte cell lines express a functional c-Met and that InlB from 10403S is expressed in a sufficient amount to activate c-Met signaling. However, InlB produced by 10403S failed to induce significant bacterial entry. We then tested the hypothesis that InlB was not produced in sufficient amounts by strain 10403S to promote bacterial uptake. This hypothesis was based on the fact that the bead surface was coated with a saturating amount of recombinant InlB and the fact that laboratory strains used to show a role for InlB in bacterial internalization express high levels of InlB (15, 18, 20, 39, 40, 44, 47–51). Indeed, the commonly studied laboratory strain EGD expresses a constitutively active variant of the transcriptional regulatory factor PrfA, known as a PrfA* (G145S) variant, which is responsible for high production levels of InlB and other PrfA-regulated virulence factors (14, 47, 52–57). To test if an increase in InlB production in the 10403S background would result in InlB-mediated internalization of L. monocytogenes, we generated prfA* and ΔinlB prfA* strains in the 10403S background by phage transduction (47, 57, 58). We compared the production of InlB between 10403S WT and prfA* strains and report a marked increase in InlB production, as expected (47) (Fig. 5A). The replacement of WT prfA with prfA* led to a 5-fold increase in bacterial association (Fig. 5B) and a 7-fold increase in bacterial entry into host cells (Fig. 5C). A comparison of prfA* and ΔinlB prfA* strains showed that, in the prfA* background, InlB plays a significant role in bacterial entry (Fig. 5C), while a comparison of the WT and ΔinlB strains shows no difference in either bacterial association or bacterial entry (Fig. 5B and C). Collectively, these data show that a bacterial strain such as 10304S produces enough InlB to activate c-Met, but this amount is not sufficient to affect L. monocytogenes internalization.
FIG 4.
The InlB/c-Met signaling pathway is functional in hepatocytes. (A) HepG2, Hep3B, PLC5, and Huh7 cell lysates were subjected to Western blot analysis using anti-c-Met and anti-actin (loading control) antibodies. (B) Cells were exposed, or not, to 1.25 nM InlB for 5 min, and cell lysates were subjected to Western blot analysis using anti-Akt and anti-phospho-Akt antibodies. A representative Western blot is presented (n = 3). (C) HepG2 cells were incubated with BSA- or BSA/InlB-coated beads for 30 min at 37°C (MOI of 5). Results are expressed as the average percentage of internalization ± SEM (n = 4; *, P < 0.01; **, P < 0.001). (D) After infection with WT or ΔinlB bacteria (MOI of 20) for 30 min, HepG2 cells were lysed and lysates were subjected to Western blot analysis using anti-Akt and anti-phospho-Akt antibodies. A representative Western blot is shown (n = 3). MW, molecular weight.
FIG 5.
A prfA* mutation in L. monocytogenes strain 10403S leads to increased production of InlB and InlB-dependent hepatocyte invasion. (A) Bacterial lysates (1.6 × 108 cells) were subjected to Western blot analysis using anti-InlB and anti-p60 (loading control) antibodies. A representative Western blot is shown (n = 3). (B and C) PLC5 cells were infected with WT, ΔinlB, WT-prfA*, or ΔinlB-prfA* bacteria (MOI of 5) for 30 min at 37°C. Cells were washed, fixed, and labeled with fluorescent antibodies and DAPI. (B) The bacterial association efficiency was calculated as the total number of bacteria associated per host cell. (C) The bacterial internalization efficiency was calculated as the percentage of intracellular bacteria. (B and C) A minimum of 2,000 bacteria were counted per condition, and a minimum of 500 host cells were counted per condition. Results are expressed as the mean ± SEM (n = 4; *, P < 0.01; **, P < 0.001, n.s., non-statistically significant).
Only InlA, not InlB or LLO, controls L. monocytogenes uptake by human cytotrophoblasts.
We next determined the role of LLO, InlA, and InlB in L. monocytogenes uptake by human cytotrophoblast-like BeWo cells. Cytotrophoblasts are cells of fetal origin located at the interface between maternal and fetal tissues. Invasion of the placenta requires traversal of the cytotrophoblast barrier. No role for LLO in L. monocytogenes association and entry was detected in BeWo cells. Two other cytotrophoblast-like cells, Jeg-3 and JAR, were also tested, leading to the same conclusion (data not shown). Only InlA plays a major role in L. monocytogenes association with BeWo cells, but it does not affect the efficiency of internalization (Fig. 6). Finally, no role for InlB was observed in the invasion of BeWo cells, as previously reported by others using the same bacterial strain (59).
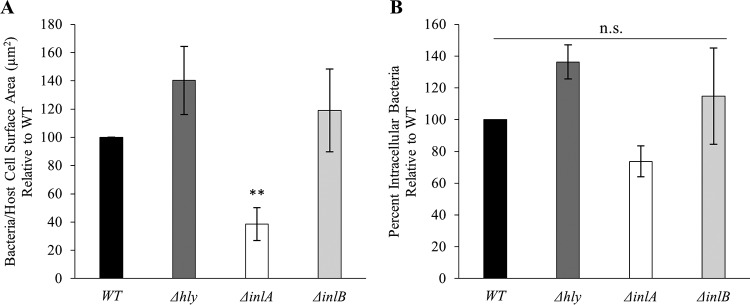
FIG 6.
Role of LLO, InlA, and InlB in L. monocytogenes invasion of human cytotrophoblasts. BeWo cells were infected with WT, LLO-deficient (Δhly), InlA-deficient (ΔinlA), or InlB-deficient (ΔinlB) bacteria (106 bacteria/well) for 30 min at 37°C. The cells were washed, fixed, and labeled with fluorescent antibodies and DAPI. (A) The bacterial association efficiency was calculated as the number of cell-associated bacteria per unit surface area (μm2). The average association for the WT strain before normalization was 0.0015 bacteria/μm2. (B) The bacterial internalization efficiency was measured as the percentage of intracellular bacteria. The average internalization for the WT strain before normalization was 13.82%. The average number of WT bacteria counted per experimental condition was 5,000, with a minimum count of 100 bacteria being required for any mutant with reduced association efficiency. Results are expressed as the mean ± SEM relative to the WT (n ≥ 3). Statistical analyses compared each strain to the WT strain and were performed on raw data before normalization (*, P < 0.01; **, P < 0.001; n.s., nonsignificant).
Uptake of L. monocytogenes by HUVECs is independent of the three invasion factors.
We next assessed the role of LLO, InlA, and InlB in the uptake of L. monocytogenes by human umbilical vein endothelial cells (HUVECs). We used the low MOI of 5 because HUVECs are severely damaged at higher MOIs due to LLO activity, as we have observed and as recently reported (42). Our data showed no role for InlA, InlB, or LLO in the invasion of HUVECs (Fig. 7). This is congruent with the most recent report in the literature regarding L. monocytogenes strain 10403S and HUVECs that supports the notion that bacterial uptake is largely independent of InlA, InlB, and LLO (42).
FIG 7.
Absence of a role for LLO, InlA, and InlB in L. monocytogenes invasion of human endothelial cells. HUVECs were infected with WT, LLO-deficient (Δhly), InlA-deficient (ΔinlA), or InlB-deficient (ΔinlB) bacteria (MOI of 5) for 30 min at 37°C. Cells were washed, fixed, and labeled with fluorescent antibodies and DAPI. (A) The bacterial association efficiency was calculated as the number of cell-associated bacteria per human cell. The average association for the WT strain was 0.13 bacteria/host cell. (B) The bacterial internalization efficiency was measured as the percentage of intracellular bacteria. The average internalization efficiency for the WT strain was 13.23%. The average number of WT bacteria counted per experiment was 500, and a minimum of 2,000 host cells were counted per condition. Results are expressed as the mean ± SEM relative to the WT (n ≥ 3). Statistical analyses compared each strain to the WT strain and were performed on raw data before normalization (n.s., nonsignificant).
Establishing cooperation between LLO and InlA in L. monocytogenes invasion of hepatocytes.
Hepatocyte infection data indicated an important role for both LLO and InlA in L. monocytogenes host cell invasion. This infection model was therefore appropriate for establishing whether LLO and InlA cooperate to potentiate the efficiency of host cell invasion. The biological expectation for positive cooperation between the two proteins, also referred to as synergism, is that the biological response when both proteins are expressed (when both genes are present) will be greater than the sum of their individual responses (when one of the corresponding genes is deleted) (60). To establish if InlA and LLO display positive cooperation in bacterial association with host cells on entry into host cells, we established four groups: InlA and LLO are both expressed (WT strain), LLO is expressed alone (ΔinlA strain), InlA is expressed alone (Δhly strain), and neither of the two proteins is expressed (ΔinlA Δhly double deletion mutant). A linear mixed-effects model was used to test this hypothesis: (μboth − μneither) > [(μA − μneither) + (μB − μneither)], i.e., μboth − μA − μB + μneither > 0, where both is the WT, neither is the double deletion mutant, and μ is the mean outcome for each group (60). If the P value for this test is significant, we claim that there is significant synergistic interaction (positive cooperation) between the two proteins. We used this analytical method to test whether InlA and LLO work synergistically to affect bacterial association and internalization of Listeria monocytogenes. Similar analyses were performed to test for potential positive cooperation between InlB and LLO and between InlB and InlA. Estimates and accompanying statistics are included in Table 1. In the process of bacterial association, no pattern of positive cooperation was observed (Table 1). This is consistent with InlA being the sole contributor to association among the tested invasins. In the process of bacterial internalization, no synergistic effect was observed between InlB and the two other invasins (Fig. S2 and Table 1), confirming that InlB does not affect the uptake of L. monocytogenes (strain 10403S) into human hepatocytes. Only LLO and InlA interact in a synergistic manner to potentiate L. monocytogenes internalization into HepG2 and PLC5 cells.
TABLE 1.
Invasion factor cooperation analysisa
| Invasion factor combination tested | Cell line | Cooperation in internalization |
Cooperation in association |
||||
|---|---|---|---|---|---|---|---|
| Estimate | SE | P value | Estimate | SE | P value | ||
| InlA/LLO | HepG2 | 13.996 | 5.9587 | 0.0232 | −0.04043 | 0.0379 | 0.2917 |
| Hep3B | −16.8597 | 9.9539 | 0.0995 | −1.1548 | 0.6499 | 0.0848 | |
| PLC5 | 6.5958 | 2.9439 | 0.0332 | −0.3497 | 0.4422 | 0.4357 | |
| Huh7 | 7.0611 | 6.8443 | 0.3114 | 0.01674 | 0.2237 | 0.9409 | |
| InlA/InlB | HepG2 | 7.6389 | 6.0995 | 0.2168 | −0.02705 | 0.03911 | 0.4928 |
| Hep3B | −2.8172 | 10.1881 | 0.7838 | −0.4551 | 0.6875 | 0.5126 | |
| PLC5 | 3.645 | 3.3279 | 0.2827 | −0.6872 | 0.4973 | 0.1779 | |
| Huh7 | −1.0749 | 7.8543 | 0.8922 | −1.0749 | 7.8543 | 0.8922 | |
| InlB/LLO | HepG2 | 8.3178 | 5.4892 | 0.1365 | −0.07209 | 0.03502 | 0.0452 |
| Hep3B | −3.0958 | 9.4808 | 0.746 | 0.4169 | 0.6232 | 0.5082 | |
| PLC5 | 0.9946 | 2.9439 | 0.738 | 0.04456 | 0.4422 | 0.9205 | |
| Huh7 | 2.6579 | 7.1474 | 0.7129 | 2.6579 | 7.1474 | 0.7129 | |
The estimate is the result of the synergistic interaction tests described in Results. Statistically significant P values (<0.05) indicate positive (synergistic) cooperation. SE, standard error. Boldface indicates statistically significant P values.
DISCUSSION
This work focused on establishing the relative roles of LLO, InlA, and InlB in L. monocytogenes (strain 10403S) association with and internalization into normally nonphagocytic human cells. The data show that LLO activity is cell type dependent, as LLO plays a significant role in L. monocytogenes internalization into hepatocytes but not into cytotrophoblasts or endothelial cells. InlA and LLO are the two virulence factors that significantly contribute to the invasion of human hepatocytes, with InlA playing a significant role as an adhesin and LLO as an invasin. To our surprise, no role for InlB was detected unless the prfA gene was replaced by a constitutively active prfA* mutant, indicating that higher expression levels of InlB are required for InlB-mediated bacterial internalization.
Studies that identified the L. monocytogenes virulence factors controlling host cell invasion have traditionally used the gentamicin survival assay. This assay robustly measures bacterial intracellular survival but presents some limitations. First, it indiscriminately and collectively reports the efficiencies of bacterial association and internalization. Second, host cell perforation by LLO allows for diffusion of gentamicin and potential targeting of intracellular bacteria (35). Finally, this assay generally involves long incubation times, which can be sufficient for intracellular bacterial division or killing. Because of these limitations, we analyzed cells infected for only 30 min at a low MOI and in the absence of gentamicin, using a fluorescence microscopy approach (61). Microscope automation allows for rapid acquisitions of a high number of images, and software-assisted analytical tools considerably decrease the time for analysis. Importantly, this approach specifically quantifies with sensitivity and accuracy the efficiencies of bacterial attachment and association with host cells (61).
No role for InlB was initially detected in the present work. This result was unexpected, because numerous studies report that InlB promotes host cell invasion (15, 20, 40, 44, 48, 49). Using the hepatocyte model, we showed that the InlB receptor, c-Met, was expressed and functional. In addition, the amount of InlB produced by L. monocytogenes 10403S under our experimental conditions was sufficient to activate c-Met-dependent signaling but not bacterial internalization (Fig. 3 and 4). Furthermore, hepatocytes could massively internalize polystyrene beads coated with high concentrations of recombinant InlB (Fig. 4C). Studies that characterized the role of InlB in host cell invasion mostly used strain EGD, which carries a mutation in the gene coding for the master regulator of the virulence gene prfA (designated prfA*), leading to high expression levels of InlB among other virulence factors (47). Among all sequenced L. monocytogenes strains analyzed, the prfA* mutation is very rarely observed (47). When the prfA* mutation was introduced into EGD-e, inlB transcription was increased over 40-fold (47). This led us to hypothesize that the strain used in our study, 10403S, may not produce enough InlB for productive bacterial internalization. To test this hypothesis, we replaced the WT prfA allele with a prfA* allele in the 10403S background and consequently observed a marked increase in InlB production and a statistically significant role for InlB in bacterial internalization. Together, these data support the idea that the level of expression of InlB is critical for bacterial internalization. Therefore, it is reasonable to extrapolate that any conditions, including different bacterial cell growth conditions or environmental conditions, that substantially increase InlB expression would favor InlB-dependent internalization. For example, the transcription level of inlB in strain EGD-e is increased in human blood and the murine intestine (62). One should also consider that the role of InlB observed at later time points of infection may be related to bacterial intracellular survival and/or multiplication and not to bacterial internalization.
As expected, InlA promotes invasion of cells that express its receptor, E-cadherin (19). Importantly, the role of InlA was substantial even in strain 10403S expressing wild-type prfA. Few studies have focused on distinguishing the role of E-cadherin in anchoring the bacterium to the host cell surface from its role in stimulating bacterial internalization. It was initially proposed that the InlA–E-cadherin interaction promotes both anchoring and internalization, since the intracellular domain of E-cadherin and its association with the F-actin cytoskeleton were necessary for InlA-dependent L. monocytogenes uptake by fibroblasts (63). More recent work studying L. monocytogenes invasion of MDCK epithelial cells expressing wild-type E-cadherin or the glycosylphosphatidylinositol (GPI)-anchored extracellular domain of E-cadherin concluded that the InlA–E-cadherin interaction anchors the bacterium to the host cell surface but is dispensable for F-actin-dependent internalization of the bacterium (64). Our results are in accordance with both studies. We report that the primary function of the InlA–E-cadherin interaction is to anchor the bacterium to the host surface, but this interaction can also control the efficiency of bacterial internalization in some, but not all, cell lines.
LLO plays a critical role in L. monocytogenes internalization into hepatocytes. Other studies established that the formation of LLO pores on the plasma membrane activates the following signaling cascade: influx of extracellular Ca2+, activation of Ca2+-dependent conventional protein kinase C upstream from the Rho GTPase Rac1, and Arp2/3-dependent formation of F-actin-rich membrane projections that promote internalization of the bacterium (34, 35, 65). Because LLO targets all membranes that contain cholesterol, it was expected that LLO would activate bacterial internalization in all animal cells, including cytotrophoblasts and HUVECs, but to our surprise, this was not the case. However, hepatocytes are not the only cells thus far identified to undergo LLO-dependent L. monocytogenes internalization, as this was also reported in HeLa cells, Hep2 cells, human neutrophils, and macrophages (34). Furthermore, LLO-dependent internalization has been demonstrated for L. monocytogenes strains 10403S, L028, and EGD (33, 34). The difference in host cell response to LLO should be investigated further to understand what makes some cell types permissive to the LLO-dependent entry pathway. This would be useful for understanding how pathogens can generally take advantage of plasma membrane perforation to gain entry into host cells (66, 67).
We report that InlA and LLO cooperate in an additive or synergistic fashion depending on the cell line. Though the mechanism by which LLO and InlA cooperate is still unknown, two non-mutually exclusive hypotheses can be envisioned. First, by anchoring L. monocytogenes to the host cell, InlA increases local LLO concentration and thereby LLO-dependent internalization. Along this line, InlA likely served as the adhesin and LLO promoted the signaling cascade for bacterial internalization into MDCK cells expressing GPI-anchored E-cadherin (64). Second, LLO- and InlA-induced signaling cascades may potentiate the activation of common transducers for the remodeling of F-actin and bacterial engulfment (65, 68).
Most studies that addressed the roles of InlA, InlB, and LLO utilized the laboratory strain EGD, EGD-e, or 10403S, which all belong to serovar 1/2a. EGD is derived from the strain of L. monocytogenes isolated from guinea pigs in 1926 (69). EGD-e is thought to be a derivative of strain EGD (47, 70). 10403S is a derivative of strain 10403, a strain initially isolated from a human skin lesion (71). Of these strains, EGD-e is the most virulent in mice and has been shown to express high levels of some of the PrfA-stimulated genes despite the absence of the prfA* allele (47). L. monocytogenes strains associated with clinical cases and outbreaks of listeriosis belong predominantly to serovars 1/2a, 1/2b, and 4b, with greater than 50% of isolates belonging to serovar 4b (72, 73). Characterization of virulence factors in clinical strains seems to be lacking. A role for InlA in the invasion of Caco-2 cells has been demonstrated with a clinical isolate (Scott A, serotype 4b) from an outbreak of listeriosis in Massachusetts in 1983 (74, 75). One epidemiological study reported that 96% of clinical isolates, and only 65% of food isolates, express full-length InlA (76), and other studies have similarly found a higher prevalence of full-length InlA in strains associated with human and animal infections, with more strains expressing truncated InlA in food isolates (77–79). Other work has found that LLO- and InlB-encoding genes are highly prevalent in clinical strains (80). However, these studies emphasize the importance of InlA, InlB, and LLO as virulence factors but do not directly inform on their mechanism of action in vivo. In vivo studies using animal models also established a role for these three virulence factors. Of the three factors, LLO is the most important for virulence, as LLO-deficient strains are avirulent, so dissecting its role in vivo is challenging. In mice infected with 10403S or EGD-e, InlB does not affect liver and spleen colonization or the 50% lethal dose (LD50) (81, 82). One recent study infecting E-cadherin-humanized mice and gerbils with EGD (prfA*) showed that neither InlA nor InlB affected infection of the liver (7). The same study also showed that InlA is important for infection of the intestines, colon, and cecum and that both InlA and InlB contribute to infection of the placenta and fetus.
In conclusion, to successfully cross the host barriers and invade multiple tissues, L. monocytogenes uses a collection of virulence factors that collectively facilitate bacterial anchoring to host cells and successive internalization. It appears that InlA is the major adhesin, while InlA, LLO, and InlB can stimulate bacterial internalization alone or in concert with InlA. Collectively, the three factors are conserved among clinical strains, but their roles likely vary in a tissue- and strain-dependent fashion.
MATERIALS AND METHODS
Bacterial strains and culture.
Escherichia coli XL1-Blue and BL21(DE3) were grown in Luria-Bertani (LB) broth under agitation at 37°C. Plasmids were maintained with either ampicillin (50 μg/ml) or kanamycin (30 μg/ml), as indicated. Wild-type (WT) L. monocytogenes (EGD-e) was a gift from Pascale Cossart (Pasteur Institute, Paris, France) (Table 2). WT L. monocytogenes (10403S) and Δhly (DP-L2161), ΔinlA (DP-L4405), ΔinlB (DP-L4406), and ΔinlAB (DP-L4404) isogenic mutants were gifts from Daniel Portnoy (UC Berkeley, CA, USA). Strain 10403S, a member of lineage II and serotype 1/2a, is a streptomycin-resistant derivative of strain 10403 (47, 71), which was originally isolated from a human skin lesion in 1968 (83). The ΔinlAB Δhly triple deletion mutant was developed previously (34). ΔinlA Δhly and ΔinlB Δhly double deletion mutants were constructed using DP-L4405 and DP-L4406, respectively, by knocking out the hly gene via allelic exchange using the pKSV7 integration shuttle vector and primers listed in Table 3, as described previously (34, 84). The deletion of hly was confirmed by PCR using primers listed in Table 3. L. monocytogenes strains were grown overnight under agitation at 37°C in brain heart infusion (BHI) (BD Biosciences). For invasion assays, overnight cultures were diluted 20-fold in BHI and grown at 37°C until an optical density at 600 nm (OD600) of 0.7 to 0.8 was reached. Cells were washed three times in sterile, 37°C phosphate-buffered saline (PBS) and diluted to the indicated multiplicity of infection (MOI) in appropriate mammalian cell culture medium without serum or antibiotic.
TABLE 2.
L. monocytogenes strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| EGD-e | Wild type | 70 |
| 10403S | Wild type | 87 |
| DP-L2161 | 10403S Δhly | 88 |
| DP-L4405 | 10403S ΔinlA | 59 |
| DP-L4406 | 10403S ΔinlB | 59 |
| DP-L4404 | 10403S ΔinlAB | 59 |
| SL33 | 10403S ΔinlA Δhly | This study |
| SL40 | 10403S ΔinlB Δhly | This study |
| SL20 | 10403S ΔinlAB Δhly | This study |
| NF-L1177 | 10403S prfA G145S actA-gus-neo-plcB | 89 |
| SL64 | DP-L4406 prfA G145S actA-gus-neo-plc | This study |
TABLE 3.
Primers used in this studya
| Purpose of constructs | Oligonucleotide sequence (5′–3′) | Reference |
|---|---|---|
| Construction of Δhly strains | Forward: GGG AAT TCA ATT GTT GAT ACA ATG ACA TC | 88 |
| Reverse: GGC TGC AGG GTC TTT TTG GCT TGT GTA T | 88 | |
| Primers to amplify the hly ORF | Forward: CCG TCG GAT CCA TGA AAA AAA TAA TGC TAG TTT TTATTACAC | 88 |
| Reverse: ATC CGC GCT GCA GTT CGA TTG GAT TAT CTA CTT TAT TAC | 88 | |
| pET29b-inB6His (bp 106 to 1890) | Forward: AAC GTG CAT ATG GAG ACT ATC ACC GTG CCA ACG | This study |
| Reverse: ATT CTC GAG TTT CTG TGC CCT TAA ATT AGC TGC | This study | |
| Sequencing prfA mutants | Forward: CTA TCT GTT GCA GCT CTT CTT GG | This study |
| Reverse: CAG CTA ACA ATT GTT GTT ACT GCC | ||
| Confirm gus-neo insertion (prfA* mutants) | Forward: GCA GTC AAT TAA TAT GCC GAG CC | This study |
| Reverse: CGG ACC AAC TAA GTT TAT GTG G | This study | |
| Hydrolysis primers and probes for qPCR for gene target | ||
| inlA | Forward: GGC AAA GAA ACA ACC AAA GAA G | This study |
| Reverse: GGG CAT CAA ACC AAC CAA | This study | |
| Probe: AT TGA CTG AAC CAG CTA AGC CCG T | This study | |
| inlB | Forward: CCG AGC ACT TAA CAC ATT CTA C | This study |
| Reverse: TTA TCT GCT ACC GGG ACT TTA T | This study | |
| Probe: ATG TCA GCG CCA ATA AAG CTG GC | This study | |
| hly | Forward: CTG GTT TAG CTT GGG AAT GG | This study |
| Reverse: ATT TCG GAT AAA GCG TGG TG | This study | |
| Probe: TGA TGA CCG GAA CTT ACC ACT TGT GA | This study | |
| gap | Forward: TCA CAG CGC AAG ACA AAG | This study |
| Reverse: ACT GTT TCA GTT CCG TCT AAT G | This study | |
| Probe: TG TTA TCT CCG CTC CAG CAA CTG G | This study | |
| rpoB | Forward: TGT AAA ATA TGG ACG GCA TCG T | 90 |
| Reverse: GCT GTT TGA ATC TCA ATT AAG TTT GG | 90 | |
| Probe: CT GAT TCG CGC AAA ACT TCT ACG CG | 90 |
All probes have a 5' 6-FAM reporter dye and a 3' Iowa Black FQ quencher.
Transduction and prfA* mutant isolation.
U153 bacteriophage (85) was used to infect L. monocytogenes strain NF-L1177 (prfA* G145S actA-gus-neo-plcB), and the phages were recovered and used to transduce the prfA* (leading to G145S) actA-gus-neo-plcB to the target strains, WT 10403S and the ΔinlB mutant, as previously described (U153 bacteriophage and strain NF-L1177 were gifts from Nancy Freitag [University of Illinois, Chicago, IL]) (52, 58). Transductants were selected by plating the mixture of phage and bacteria on BHI/agar plates (5 μg/ml neomycin) for 2 days at 37°C. Neomycin-resistant mutants were further screened by plating on BHI/agar plates containing 5 μg/ml neomycin plus 50 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-gluc) to confirm the prfA* mutation and the downstream actA-gus-neo-plcB transcription fusion. The actA-gus-neo-plcB insertion was then confirmed by PCR, and the prfA G145S mutation was confirmed by sequencing using primers described in Table 3.
RNA purification, reverse transcription, and RT-qPCR.
For RNA purification, L. monocytogenes was cultured in BHI under agitation at 37°C to an OD600 of 0.7 to 0.8. RNA was purified from 109 bacteria and subsequently treated with RNase-free DNase as described previously (12). RNA concentration and purity were measured via a NanoDrop ND-1000 spectrophotometer. RNA integrity was determined on a 1.2% agarose gel. Reverse transcription was performed using a high-capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacturer's instructions. Duplicate reaction mixtures lacking the reverse transcriptase enzyme were performed in parallel, and these samples were used in RT-qPCR to test for residual DNA contamination. RT-qPCR was performed using a CFX96 real-time system and a C1000 thermal cycler (Bio-Rad). All reactions were performed in 96-well plates using 1.5 ng of converted cDNA, iQ Supermix (Bio-Rad), forward and reverse oligonucleotide primers, and hydrolysis probes (Table 3). No-reverse-transcriptase (NRT) samples were used as negative controls. inlA, inlB, and hly gene expression was normalized to housekeeping genes gap and rpoB. Fold changes in gene expression are relative to that of WT L. monocytogenes. Primer and probe concentrations were optimized by testing a concentration gradient of all oligonucleotides as described previously (86). All primer/probe sets yielded reaction efficiencies of ∼100%. All RT-qPCR hydrolysis probes include a 5' 6-FAM reporter dye and a 3' Iowa Black FQ quencher. Samples were analyzed in triplicate by RT-qPCR.
InlB purification and generation of anti-InlB rabbit polyclonal antibodies.
The inlB gene, excluding the signal sequence (bp 106 to 1890), was amplified from genomic DNA of L. monocytogenes strain EGD-e using primers (Table 3) that contain NdeI and XhoI restriction sites. This DNA fragment was ligated into the pET29b expression vector upstream of the C-terminal 6His tag sequence. The resulting expression vector, pET29b-inlB, was transformed into Escherichia coli strain BL21(DE3). For expression of recombinant protein, this strain was grown at 37°C until an OD600 of 0.6 was reached, and expression of recombinant InlB-6His was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (48). After 5 h of induction, the bacteria were pelleted and suspended in binding buffer (5 mM imidazole, 500 mM NaCl, and 50 mM HEPES, pH 7.9) and lysed with a French press. The crude lysate was centrifuged, and the supernatant was incubated with Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen). After washes, the protein was eluted and dialyzed overnight. Purified recombinant InlB was sent to GenScript (Piscataway, NJ, USA) to generate rabbit anti-InlB polyclonal antibodies. To immunize rabbits, recombinant InlB and complete Freund's adjuvant were administered via subcutaneous injection. After the primary immunization, three boosts were performed over the course of 66 days. InlB-specific IgG antibodies were purified from serum by affinity chromatography using a Sepharose 4B gel coupled to recombinant InlB. The specificity of the antibodies was ensured by Western blotting of WT and inlB deletion mutant L. monocytogenes strains (see Fig. S1 in the supplemental material).
Mammalian cell culture.
The human hepatocyte cell line HepG2 (HB-8065) was purchased from ATCC. The human hepatocyte cell lines Hep3B (HB-8064; ATCC), PLC5 (CRL-8024; ATCC), and Huh7 (Health Science Research Resources Bank, Osaka, Japan; JCRB0403) were gifts from Ching-Shih Chen (The Ohio State University, OH, USA). HepG2, Hep3B, and PLC5 cells were grown in minimal essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS; Atlanta Biologicals), 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Huh7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% HI-FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The human choriocarcinoma cell line BeWo (ATCC CCL-98) was a gift from John Mitchell Robinson (The Ohio State University, OH, USA). BeWo cells were grown in DMEM-F12 medium (1:1) supplemented with 10% HI-FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Human umbilical vein endothelial cells (HUVECs; ScienCell Research Laboratories, San Diego, CA, USA) were cultured in endothelial cell medium (ECM) with 5% HI-FBS, endothelial cell growth supplement (ECGS; ScienCell), 100 U/ml penicillin, and 100 μg/ml streptomycin. All plates and flasks used for HUVEC culture were coated with 2 μg/cm2 human fibronectin (BD Biosciences).
Western blotting (LLO, InlB, c-Met).
Bacterial lysates were loaded at several dilutions (8 × 107, 4 × 107, and 2 × 107 bacteria loaded for LLO, and 3.2 × 108, 1.6 × 108, and 8 × 107 bacteria loaded for InlB) and subjected to SDS-PAGE and Western blot analysis using polyvinylidene difluoride (PVDF) membranes and anti-LLO antibody (rabbit polyclonal from Abcam), anti-InlB antibody (rabbit polyclonal from Genscript), and secondary anti-rabbit IgG antibody conjugated to horseradish peroxidase (Cell Signaling). For detection of InlB production in prfA* mutants, 1.6 × 108 cells were used. We also probed for p60 as a loading control (Adipogen). Signal detection was performed using an Amersham ECL select reagent kit (GE Healthcare) and a ChemiDoc XRS imaging system (Bio-Rad). Densitometry analysis was performed by enclosing each protein band within a region of standard size, and the intensity of each band was measured using ImageJ gel analysis. Results were the average intensities calculated from three independent experiments. All intensities were set relative to that of WT bacterial lysates. For detection of c-MET, hepatocytes were grown to 80% confluence under the same experimental conditions as those used for invasion assays. Cell lysates were subjected to SDS-PAGE and Western blot analysis using PVDF membranes with anti-c-MET (4F8.2; Millipore) antibodies and secondary anti-mouse IgG antibodies conjugated to horseradish peroxidase (Cell Signaling). Signal detection was performed as described above.
Measuring bacterial association and internalization.
HepG2 (105 cells/well), Hep3B (0.75 × 105 cells/well), PLC5 (0.75 × 105 cells/well), Huh7 (0.75 × 105 cells/well), and HUVECs (2 × 104 cells/well) were cultured in 24-well tissue culture plates on glass coverslips at 37°C in a 5% CO2 atmosphere for 48 h before infection. BeWo cells (0.85 × 104 cells/well) were cultured in 24-well tissue culture plates on glass coverslips coated in 0.2% gelatin for 72 h before infection. The hepatocyte cell lines were infected with L. monocytogenes at an MOI of 20 and HUVECs at an MOI of 5; BeWo cells were infected with 106 bacteria/well. Infection of hepatocytes with prfA* bacterial strains was performed at an MOI of 5 to avoid toxicity of LLO in prfA* strains. Plates were centrifuged for 5 min (500 × g) at room temperature and incubated for 30 min at 37°C. Cells were washed three times with PBS, fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature, and blocked for 1 h in 0.1 M glycine and 10% HI-FBS in PBS, pH 7.4. Extracellular bacteria were labeled with anti-L. monocytogenes rabbit polyclonal antibodies (GeneTex) and with anti-rabbit secondary antibodies conjugated to Alexa Fluor 488 (Molecular Probes). Samples were permeabilized with 0.5% Triton X-100, and total (extracellular and intracellular) bacteria were labeled with anti-L. monocytogenes antibodies and secondary antibodies conjugated to Alexa Fluor 568 (Molecular Probes). Slides were mounted in ProLong gold antifade mountant containing DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes) to stain nuclei. To quantify the number of cells, images (phase contrast, DAPI, Alexa Fluor 488, and Alexa Fluor 568) were automatically acquired for each experimental condition using the 20× objective. MetaMorph analysis software was used to enumerate the total numbers of bacteria (Nt), extracellular bacteria (Ne), and mammalian cells (Nc) (61). The efficiency of bacterial internalization was calculated as follows: internalization = [(Nt – Ne)/Nt] × 100. The efficiency of bacterial association was calculated as follows: association = Nt/Nc. For each experimental condition, a minimum of 100 bacteria were counted (this applies to bacterial mutants with the lowest association efficiency) and a minimum of 150 mammalian cells (this applies to Hep3B, which are the largest cells and the cells with which L. monocytogenes associates the most effectively). The average numbers of WT bacteria and corresponding mammalian cells counted in each experiment are indicated in the figure legends. Because BeWo cells clustered in a fashion that made individual cell nuclei challenging to enumerate, we quantified the cell surface area by tracing plasma membrane outlines in MetaMorph and determined the surface area in μm2. We then calculated the efficiency of bacterial association as follows: association = Nt/cell surface area (μm2).
Polystyrene bead coating with recombinant InlB and invasion assay.
Blue fluorescent carboxylate-modified latex beads (1-μm diameter; Molecular Probes) were coated covalently with a mixture of recombinant InlB (5 mg/ml) and BSA (5 mg/ml) according to the manufacturer's instructions. Control, BSA-coated beads were prepared with 10 mg/ml BSA under the same conditions. The beads were then washed three times with 0.33× PBS, pH 7.4, and stored at 4°C. To assess the capacity for InlB-coated beads to be ingested by hepatocytes, HepG2 cells were seeded in 24-well plates on cover glasses for 48 h, as described for bacterial invasion assays. Cells were washed with MEM, and InlB/BSA- or BSA-coated beads were added to the wells at an MOI of 5. Plates were centrifuged for 3 min at 500 × g and incubated for 30 min at 37°C in 5% CO2. Cells were washed with PBS, fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature, and washed and blocked for 1 h in 0.1 M glycine and 5% blotting-grade blocker (Bio-Rad) in PBS, pH 7.4. Extracellular beads were labeled with rabbit anti-BSA antibodies (Sigma-Aldrich; B1520), followed by anti-rabbit secondary antibodies conjugated to Alexa Fluor 488. Slides were mounted in ProLong gold antifade mountant containing DAPI to stain the nuclei. The percentage of intracellular beads was determined by fluorescence microscopy. The percentage of intracellular beads was calculated as the number of intracellular beads divided by the total number of beads, multiplied by 100.
Live-cell imaging to assess hepatocyte response to InlB.
Hepatocytes were seeded (HepG2, 4 × 105 cells/dish; Hep3B, PLC5, and Huh7, 3 × 105 cells/dish) in 35-mm-diameter imaging dishes (Matek; P35G-1.5-10-C) and cultured at 37°C in 5% CO2 for 48 h. Cells were placed on the 37°C microscope stage and incubated with cell imaging medium without phenol red. Differential interference contrast (DIC) images were acquired with the 63× objective every 20 s for 15 min. At 5 min after the start of imaging, recombinant InlB was added to the cell culture medium to a final concentration of 1 nM. Under the control condition, the cells were imaged for 15 min without InlB.
Western blotting of Akt phosphorylation.
Hepatocytes were seeded (HepG2, 5 × 105 cells/dish; Hep3B, PLC5, and Huh7, 3 × 105 cells/dish) in 35-mm-diameter cell culture dishes and cultured for 48 h. For exposure to recombinant InlB, cells were washed and incubated for 30 min in serum-free medium and then incubated with or without 1.25 nM InlB for 5 min at 37°C. The cells were then washed with cold PBS and lysed with cold lysis buffer (150 mM NaCl, 20 mM Tris/HCl, 2 mM EDTA, 1% NP-40, 3 mM sodium orthovanadate, 50 mM sodium fluoride, and 1× EDTA-free protease inhibitor cocktail [Roche]). To assess the effect of InlB produced by L. monocytogenes, the cells were washed with medium without serum and infected with WT or InlB-deficient bacteria at an MOI of 20 for 30 min at 37°C (same experimental conditions as the invasion assay). The cells were then washed and lysed. Cell lysates were subjected to Western blot analysis using PVDF membranes and anti-Akt or anti-phospho-Akt (Ser473) antibodies (Cell Signaling) and secondary anti-rabbit IgG antibodies conjugated to horseradish peroxidase (Cell Signaling).
Microscope equipment.
Images were acquired on a motorized, inverted, wide-field fluorescence microscope (Axio Observer D1, TempModule S, heating unit XL S; Zeiss) equipped with a PZ-2000 XYZ automated stage, 20× Plan Neofluar (numerical aperture [NA] = 0.5), 40× Plan Neofluar (NA = 1.3), and 63× Plan Apochromat (NA = 1.4) objectives, a high-speed Xenon fluorescence emission device (Lambda DG-4, 300 W; Sutter Instrument Company), a Lambda 10-3 optical emission filter wheel for the fluorescence imaging, a SmartShutter to control the illumination for phase-contrast and DIC imaging (Sutter Instrument Company), a back-illuminated, frame-transfer electron-multiplying charge-coupled device (EMCCD) camera (Cascade II 512; Photometrics), and an ORCA-Flash 4.0 sCMOS camera (Hamamatsu). The filter sets for fluorescence were purchased from Chroma Technology Corporation and were as follows: DAPI (49000), Alexa Fluor 488 (49002), Alexa Fluor 568 (49005), and Cy5 (49006). Images were acquired and analyzed using MetaMorph imaging software (Molecular Devices).
Statistical methods.
All experimental work involved at least three biological replicates, each performed on different days. Data obtained each day include different treatment conditions, which are considered a cluster. Data within the same cluster are more correlated to each other than to data from clusters obtained on different days. Linear mixed-effects models were used to account for the correlation among observations from a same cluster. Linear mixed-effects models were used to analyze data from invasion assays (bacterial entry and association), studies of the interaction between invasion proteins, RT-qPCR, and quantitative Western blot analyses. For RT-qPCR and Western blot analyses, data were first normalized to internal controls or the loading standard to reduce variation before analysis. Holm's procedure was used to adjust for multiple comparisons such as comparisons of each L. monocytogenes deletion mutant to the WT. SAS 9.4 was used for all analyses (SAS Institute, Inc., NC). Although normalized data were presented in some figures for a clearer visualization of results, all statistical analyses were performed on raw data before normalization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Portnoy (University of California, Berkeley, USA) and Nancy Freitag (University of Illinois, Chicago, USA) for providing the bacterial strains used in this study. We thank Francis Repoila (French National Institute for Agricultural Research, Paris, France) for providing a protocol for L. monocytogenes RNA isolation.
Research reported in the article was supported by the NIAID of the National Institutes of Health under award number RO1AI107250 to Stephanie Seveau and by award number 1-T32-AI-1112542, an NRSA training grant administered by the Center for Microbial Interface Biology at The Ohio State University, to Sarika Pathak-Sharma.
The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00555-18.
REFERENCES
- 1.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Teberg AJ, Yonekura ML, Salminen C, Pavlova Z. 1987. Clinical manifestations of epidemic neonatal listeriosis. Pediatr Infect Dis J 6:817–820. [DOI] [PubMed] [Google Scholar]
- 1b.Mylonakis E, Paliou M, Hohmann EL, Calderwood SB, Wing EJ. 2002. Listeriosis during pregnancy: a case series and review of 222 cases. Medicine (Baltimore) 81:260–269. [DOI] [PubMed] [Google Scholar]
- 1c.Robbins JR, Bakardjiev AI. 2012. Pathogens and the placental fortress. Curr Opin Microbiol 15:36–43. doi: 10.1016/j.mib.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berche P. 1995. Bacteremia is required for invasion of the murine central nervous system by Listeria monocytogenes. Microb Pathog 18:323–336. doi: 10.1006/mpat.1995.0029. [DOI] [PubMed] [Google Scholar]
- 3.Disson O, Lecuit M. 2012. Targeting of the central nervous system by Listeria monocytogenes. Virulence 3:213–221. doi: 10.4161/viru.19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaillard JL, Jaubert F, Berche P. 1996. The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J Exp Med 183:359–369. doi: 10.1084/jem.183.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Nikitas G, Deschamps C, Disson O, Niault T, Cossart P, Lecuit M. 2011. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J Exp Med 208:2263–2277. doi: 10.1084/jem.20110560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Disson O, Grayo S, Huillet E, Nikitas G, Langa-Vives F, Dussurget O, Ragon M, Le Monnier A, Babinet C, Cossart P, Lecuit M. 2008. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 8.Leimeister-Wachter M, Haffner C, Domann E, Goebel W, Chakraborty T. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of listeria monocytogenes. Proc Natl Acad Sci U S A 87:8336–8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mengaud J, Dramsi S, Gouin E, Vazquez-Boland JA, Milon G, Cossart P. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol Microbiol 5:2273–2283. doi: 10.1111/j.1365-2958.1991.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 10.de las Heras A, Cain RJ, Bielecka MK, Vazquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr Opin Microbiol 14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551–561. doi: 10.1016/S0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 12.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 13.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol Microbiol 16:251–261. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127–1141. [DOI] [PubMed] [Google Scholar]
- 17.Dhar G, Faull KF, Schneewind O. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725–3733. doi: 10.1021/bi992347o. [DOI] [PubMed] [Google Scholar]
- 18.Jonquieres R, Bierne H, Fiedler F, Gounon P, Cossart P. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol Microbiol 34:902–914. doi: 10.1046/j.1365-2958.1999.01652.x. [DOI] [PubMed] [Google Scholar]
- 19.Mengaud J, Ohayon H, Gounon P, Mege RM, Cossart P. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923–932. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Naujokas M, Park M, Ireton K. 2000. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501–510. doi: 10.1016/S0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- 21.Braun L, Ghebrehiwet B, Cossart P. 2000. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J 19:1458–1466. doi: 10.1093/emboj/19.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonquieres R, Pizarro-Cerda J, Cossart P. 2001. Synergy between the N- and C-terminal domains of InlB for efficient invasion of non-phagocytic cells by Listeria monocytogenes. Mol Microbiol 42:955–965. doi: 10.1046/j.1365-2958.2001.02704.x. [DOI] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Disson O, Lecuit M. 2013. In vitro and in vivo models to study human listeriosis: mind the gap. Microbes Infect 15:971–980. doi: 10.1016/j.micinf.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Bergmann B, Raffelsbauer D, Kuhn M, Goetz M, Hom S, Goebel W. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol Microbiol 43:557–570. doi: 10.1046/j.1365-2958.2002.02767.x. [DOI] [PubMed] [Google Scholar]
- 26.Pentecost M, Kumaran J, Ghosh P, Amieva MR. 2010. Listeria monocytogenes internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog 6:e1000900. doi: 10.1371/journal.ppat.1000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry R, Shaughnessy L, Loessner MJ, Alberti-Segui C, Higgins DE, Swanson JA. 2006. Cytolysin-dependent delay of vacuole maturation in macrophages infected with Listeria monocytogenes. Cell Microbiol 8:107–119. doi: 10.1111/j.1462-5822.2005.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seveau S. 2014. Multifaceted activity of listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Subcell Biochem 80:161–195. doi: 10.1007/978-94-017-8881-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilney LG, Portnoy DA. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol 109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun 55:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnupf P, Portnoy DA. 2007. Listeriolysin O: a phagosome-specific lysin. Microbes Infect 9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Moors MA, Levitt B, Youngman P, Portnoy DA. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect Immun 67:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dramsi S, Cossart P. 2003. Listeriolysin O-mediated calcium influx potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect Immun 71:3614–3618. doi: 10.1128/IAI.71.6.3614-3618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vadia S, Arnett E, Haghighat AC, Wilson-Kubalek EM, Tweten RK, Seveau S. 2011. The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog 7:e1002356. doi: 10.1371/journal.ppat.1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vadia S, Seveau S. 2014. Fluxes of Ca2+ and K+ are required for the listeriolysin O-dependent internalization pathway of Listeria monocytogenes. Infect Immun 82:1084–1091. doi: 10.1128/IAI.01067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnett E, Vadia S, Nackerman CC, Oghumu S, Satoskar AR, McLeish KR, Uriarte SM, Seveau S. 2014. The pore-forming toxin listeriolysin O is degraded by neutrophil metalloproteinase-8 and fails to mediate Listeria monocytogenes intracellular survival in neutrophils. J Immunol 192:234–244. doi: 10.4049/jimmunol.1301302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czuczman MA, Fattouh R, van Rijn JM, Canadien V, Osborne S, Muise AM, Kuchroo VK, Higgins DE, Brumell JH. 2014. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature 509:230–234. doi: 10.1038/nature13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drevets DA, Sawyer RT, Potter TA, Campbell PA. 1995. Listeria monocytogenes infects human endothelial cells by two distinct mechanisms. Infect Immun 63:4268–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greiffenberg L, Goebel W, Kim KS, Weiglein I, Bubert A, Engelbrecht F, Stins M, Kuhn M. 1998. Interaction of Listeria monocytogenes with human brain microvascular endothelial cells: InlB-dependent invasion, long-term intracellular growth, and spread from macrophages to endothelial cells. Infect Immun 66:5260–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parida SK, Domann E, Rohde M, Muller S, Darji A, Hain T, Wehland J, Chakraborty T. 1998. Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol Microbiol 28:81–93. doi: 10.1046/j.1365-2958.1998.00776.x. [DOI] [PubMed] [Google Scholar]
- 41.Greiffenberg L, Sokolovic Z, Schnittler HJ, Spory A, Bockmann R, Goebel W, Kuhn M. 1997. Listeria monocytogenes-infected human umbilical vein endothelial cells: internalin-independent invasion, intracellular growth, movement, and host cell responses. FEMS Microbiol Lett 157:163–170. doi: 10.1111/j.1574-6968.1997.tb12768.x. [DOI] [PubMed] [Google Scholar]
- 42.Rengarajan M, Hayer A, Theriot JA. 2016. Endothelial cells use a formin-dependent phagocytosis-like process to internalize the bacterium Listeria monocytogenes. PLoS Pathog 12:e1005603. doi: 10.1371/journal.ppat.1005603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson SL, Drevets DA. 1998. Listeria monocytogenes infection and activation of human brain microvascular endothelial cells. J Infect Dis 178:1658–1666. doi: 10.1086/314490. [DOI] [PubMed] [Google Scholar]
- 44.Ireton K, Payrastre B, Cossart P. 1999. The Listeria monocytogenes protein InlB is an agonist of mammalian phosphoinositide 3-kinase. J Biol Chem 274:17025–17032. doi: 10.1074/jbc.274.24.17025. [DOI] [PubMed] [Google Scholar]
- 45.Mansell A, Khelef N, Cossart P, O'Neill LA. 2001. Internalin B activates nuclear factor-kappa B via Ras, phosphoinositide 3-kinase, and Akt. J Biol Chem 276:43597–43603. doi: 10.1074/jbc.M105202200. [DOI] [PubMed] [Google Scholar]
- 46.Bhalla M, Law D, Dowd GC, Ireton K. 2017. Host serine/threonine kinases mTOR and protein kinase C-alpha promote InlB-mediated entry of Listeria monocytogenes. Infect Immun 85:e00087-. doi: 10.1128/IAI.00087-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kuhbacher A, Brisse S, Pucciarelli MG, Garcia-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerda J, Moszer I, Bierne H, Cossart P. 2014. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio 5:e00969-. doi: 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. 1997. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol 25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 49.Jiwani S, Wang Y, Dowd GC, Gianfelice A, Pichestapong P, Gavicherla B, Vanbennekom N, Ireton K. 2012. Identification of components of the host type IA phosphoinositide 3-kinase pathway that promote internalization of Listeria monocytogenes. Infect Immun 80:1252–1266. doi: 10.1128/IAI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braun L, Ohayon H, Cossart P. 1998. The InIB protein of Listeria monocytogenes is sufficient to promote entry into mammalian cells. Mol Microbiol 27:1077–1087. doi: 10.1046/j.1365-2958.1998.00750.x. [DOI] [PubMed] [Google Scholar]
- 51.Van Ngo H, Bhalla M, Chen DY, Ireton K. November 2017. A role for host cell exocytosis in InlB-mediated internalisation of Listeria monocytogenes. Cell Microbiol doi: 10.1111/cmi.12768. [DOI] [PubMed] [Google Scholar]
- 52.Wong KK, Freitag NE. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J Bacteriol 186:6265–6276. doi: 10.1128/JB.186.18.6265-6276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shetron-Rama LM, Mueller K, Bravo JM, Bouwer HG, Way SS, Freitag NE. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol Microbiol 48:1537–1551. doi: 10.1046/j.1365-2958.2003.03534.x. [DOI] [PubMed] [Google Scholar]
- 54.Vega Y, Rauch M, Banfield MJ, Ermolaeva S, Scortti M, Goebel W, Vazquez-Boland JA. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol Microbiol 52:1553–1565. doi: 10.1111/j.1365-2958.2004.04052.x. [DOI] [PubMed] [Google Scholar]
- 55.Mueller KJ, Freitag NE. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect Immun 73:1917–1926. doi: 10.1128/IAI.73.4.1917-1926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miner MD, Port GC, Bouwer HG, Chang JC, Freitag NE. 2008. A novel prfA mutation that promotes Listeria monocytogenes cytosol entry but reduces bacterial spread and cytotoxicity. Microb Pathog 45:273–281. doi: 10.1016/j.micpath.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ripio MT, Dominguez-Bernal G, Lara M, Suarez M, Vazquez-Boland JA. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol 179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruno JC Jr, Freitag NE. 2010. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One 5:e15138. doi: 10.1371/journal.pone.0015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bakardjiev AI, Stacy BA, Fisher SJ, Portnoy DA. 2004. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect Immun 72:489–497. doi: 10.1128/IAI.72.1.489-497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slinker BK. 1998. The statistics of synergism. J Mol Cell Cardiol 30:723–731. doi: 10.1006/jmcc.1998.0655. [DOI] [PubMed] [Google Scholar]
- 61.Haghighat AC, Seveau S. 2010. Quantification of host-microbe interactions by automated fluorescence microscopy. J Immunol Methods 352:186–191. doi: 10.1016/j.jim.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 62.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 63.Lecuit M, Hurme R, Pizarro-Cerda J, Ohayon H, Geiger B, Cossart P. 2000. A role for alpha-and beta-catenins in bacterial uptake. Proc Natl Acad Sci U S A 97:10008–10013. doi: 10.1073/pnas.97.18.10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ortega FE, Rengarajan M, Chavez N, Radhakrishnan P, Gloerich M, Bianchini J, Siemers K, Luckett WS, Lauer P, Nelson WJ, Theriot JA. 2017. Adhesion to the host cell surface is sufficient to mediate Listeria monocytogenes entry into epithelial cells. Mol Biol Cell 28:2945–2957. doi: 10.1091/mbc.e16-12-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lam JGT, Vadia S, Pathak-Sharma S, McLaughlin E, Zhang X, Swanson J, Seveau S. 2018. Host cell perforation by listeriolysin O (LLO) activates a Ca2+-dependent cPKC/Rac1/Arp2/3 signaling pathway that promotes Listeria monocytogenes internalization independently of membrane resealing. Mol Biol Cell 29:270–284. doi: 10.1091/mbc.e17-09-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luisoni S, Suomalainen M, Boucke K, Tanner LB, Wenk MR, Guan XL, Grzybek M, Coskun U, Greber UF. 2015. Co-option of membrane wounding enables virus penetration into cells. Cell Host Microbe 18:75–85. doi: 10.1016/j.chom.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Fernandes MC, Cortez M, Flannery AR, Tam C, Mortara RA, Andrews NW. 2011. Trypanosoma cruzi subverts the sphingomyelinase-mediated plasma membrane repair pathway for cell invasion. J Exp Med 208:909–921. doi: 10.1084/jem.20102518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sousa S, Cabanes D, Bougneres L, Lecuit M, Sansonetti P, Tran-Van-Nhieu G, Cossart P. 2007. Src, cortactin and Arp2/3 complex are required for E-cadherin-mediated internalization of Listeria into cells. Cell Microbiol 9:2629–2643. doi: 10.1111/j.1462-5822.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- 69.Murray EGD, Webb RA, Swann MBR. 1926. A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes (n. sp.). J Pathol Bacteriol 29:407–439. doi: 10.1002/path.1700290409. [DOI] [Google Scholar]
- 70.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couve E, de Daruvar A, Dehoux P, Domann E, Dominguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Garcia-del Portillo F, Garrido P, Gautier L, Goebel W, Gomez-Lopez N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Perez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. [DOI] [PubMed] [Google Scholar]
- 71.Bishop DK, Hinrichs DJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J Immunol 139:2005–2009. [PubMed] [Google Scholar]
- 72.McLauchlin J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur J Clin Microbiol Infect Dis 9:210–213. doi: 10.1007/BF01963840. [DOI] [PubMed] [Google Scholar]
- 73.Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot 65:1811–1829. doi: 10.4315/0362-028X-65.11.1811. [DOI] [PubMed] [Google Scholar]
- 74.Fleming DW, Cochi SL, MacDonald KL, Brondum J, Hayes PS, Plikaytis BD, Holmes MB, Audurier A, Broome CV, Reingold AL. 1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N Engl J Med 312:404–407. doi: 10.1056/NEJM198502143120704. [DOI] [PubMed] [Google Scholar]
- 75.Olier M, Garmyn D, Rousseaux S, Lemaitre JP, Piveteau P, Guzzo J. 2005. Truncated internalin A and asymptomatic Listeria monocytogenes carriage: in vivo investigation by allelic exchange. Infect Immun 73:644–648. doi: 10.1128/IAI.73.1.644-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jacquet C, Doumith M, Gordon JI, Martin PM, Cossart P, Lecuit M. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J Infect Dis 189:2094–2100. doi: 10.1086/420853. [DOI] [PubMed] [Google Scholar]
- 77.Nightingale KK, Windham K, Martin KE, Yeung M, Wiedmann M. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl Environ Microbiol 71:8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jonquieres R, Bierne H, Mengaud J, Cossart P. 1998. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect Immun 66:3420–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manuel CS, Van Stelten A, Wiedmann M, Nightingale KK, Orsi RH. 2015. Prevalence and distribution of Listeria monocytogenes inlA alleles prone to phase variation and inlA alleles with premature stop codon mutations among human, food, animal, and environmental isolates. Appl Environ Microbiol 81:8339–8345. doi: 10.1128/AEM.02752-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacquet C, Gouin E, Jeannel D, Cossart P, Rocourt J. 2002. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl Environ Microbiol 68:616–622. doi: 10.1128/AEM.68.2.616-622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brockstedt DG, Giedlin MA, Leong ML, Bahjat KS, Gao Y, Luckett W, Liu W, Cook DN, Portnoy DA, Dubensky TW Jr. 2004. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc Natl Acad Sci U S A 101:13832–13837. doi: 10.1073/pnas.0406035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sobyanin KA, Sysolyatina EV, Chalenko YM, Kalinin EV, Ermolaeva SA. 2017. Route of injection affects the impact of InlB internalin domain variants on severity of Listeria monocytogenes infection in mice. Biomed Res Int 2017:2101575. doi: 10.1155/2017/2101575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edman DC, Pollock MB, Hall ER. 1968. Listeria monocytogenes L forms. I. Induction maintenance, and biological characteristics. J Bacteriol 96:352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Camilli A, Tilney LG, Portnoy DA. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol 8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hodgson DA. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol Microbiol 35:312–323. doi: 10.1046/j.1365-2958.2000.01643.x. [DOI] [PubMed] [Google Scholar]
- 86.Gavrilin MA, Bouakl IJ, Knatz NL, Duncan MD, Hall MW, Gunn JS, Wewers MD. 2006. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc Natl Acad Sci U S A 103:141–146. doi: 10.1073/pnas.0504271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jones S, Portnoy DA. 1994. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect Immun 62:5608–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miner MD, Port GC, Freitag NE. 2008. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology 154:3579–3589. doi: 10.1099/mic.0.2008/021063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sue D, Fink D, Wiedmann M, Boor KJ. 2004. SigmaB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843–3855. doi: 10.1099/mic.0.27257-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.