We studied the effects of two Salmonella enterica serovar Typhimurium (host-adapted) strains (14028 and 4/74) and three S. Choleraesuis (non-host-adapted) strains (A50, A45, and B195) in human monocytes between 2 and 24 h postinfection (p.i.) to investigate whether differences in immune response may explain the much higher prevalence of sepsis in individuals infected with S.
KEYWORDS: salmonella, human, monocyte, sepsis
ABSTRACT
We studied the effects of two Salmonella enterica serovar Typhimurium (host-adapted) strains (14028 and 4/74) and three S. Choleraesuis (non-host-adapted) strains (A50, A45, and B195) in human monocytes between 2 and 24 h postinfection (p.i.) to investigate whether differences in immune response may explain the much higher prevalence of sepsis in individuals infected with S. Choleraesuis. Both serovars significantly increased the production of cytokines associated with acute sepsis (tumor necrosis factor alpha [TNF-α], interleukin β [IL-β], and IL-6), but temporal differences occurred between these serovars and between different S. Choleraesuis strains. Generally, all S. Choleraesuis strains induced significantly higher production of inflammatory cytokines than S. Typhimurium strains (P < 0.01 to 0.05). All S. Choleraesuis strains very significantly increased IL-10 production by monocytes at 6 and 24 h p.i. in comparison to S. Typhimurium strains (P < 0.01). In addition, ∼80% of monocytes were viable at 24 h p.i. with S. Choleraesuis A50, compared to only ∼40% following S. Typhimurium infection. Using S. Typhimurium 14028 and S. Choleraesuis A50 as examples of these two serovars, we also showed differential expression of genes within the Janus tyrosine kinase (JAK) and signal transducer and activator of transcription (STAT) (JAK/STAT) pathway via quantitative PCR (qPCR) microarray analysis. High serum IL-10 concentration and monocyte survival have been reported as markers of the development of human sepsis. We therefore conclude that high production of IL-10 by monocytes may, in part, explain the greater propensity for S. Choleraesuis to induce human sepsis and that this may be greater in strains such as A50, which induces both high IL-10 production and monocyte survival.
INTRODUCTION
Salmonella enterica serovar Typhimurium has a broad host range, and in humans, although it can cause bacteremia and sepsis, disease due to S. Typhimurium is usually seen as a self-limiting gastrointestinal infection. In contrast, Salmonella enterica serovar Choleraesuis is host adapted to pigs and in humans is most likely to cause bacteremia and sepsis with little gastrointestinal pathology (1). In humans, the mortality rate due to S. Choleraesuis infection may be as high as 30% (2), but predisposing factors such as underlying disease are also known to contribute to such a high rate (3). However, in comparison to S. Typhimurium, very little has been published regarding S. Choleraesuis pathogenesis in humans, and no comparisons have been reported regarding the effect of these Salmonella serovars in human immune cells.
The fact that S. Choleraesuis is much more likely to cause sepsis than S. Typhimurium may not be due simply to increased virulence and the ability to escape the intestinal barrier. For example, survival in blood cells may prolong infection, increasing the concentration of inflammatory cytokines produced by these cells and the development of the acute phase of sepsis.
Cytokine receptors which signal via the Janus tyrosine kinase/signal transducer and activator of transcription (JAK/STAT) pathway include gamma interferon receptor 1 (IFN-γR1), interleukin 6 receptor (IL-6R), and IL-10R (4), and the cytokines which signal via these receptors (IFN-α, IL-6, and IL-10) have been studied as prognostic markers during the systemic inflammatory response syndrome (SIRS) and compensatory anti-inflammatory response syndrome (CARS) phases of sepsis (5–7). The JAK/STAT pathway is not only activated by a range of different cytokines but is also responsible for the transcription of many different cytokine genes (4). The pathway includes three JAK proteins (JAK1 to JAK3), a JAK-related protein, tyrosine kinase 2 (TYK2), and seven STAT proteins (STAT1 to STAT4, STAT5a, STAT5b, and STAT6) (8). In postoperative human sepsis, mortality has been associated with monocyte resistance to IFN-γ, which fails to increase the release of crucial monocyte cytokines (9), and pneumonia-induced sepsis has been reported to be associated with single-nucleotide polymorphisms (SNPs) in IFNG genes (10). Many other experimental studies have now shown the essential requirement for IFN-γ in the killing of Salmonella by monocytes and macrophages, both in vitro and in vivo (11).
The aim of this study was to compare the pathogenesis of S. Typhimurium with that of S. Choleraesuis in human monocytes, with the goal of identifying possible differences that may explain the greater propensity of S. Choleraesuis to cause sepsis in humans.
RESULTS
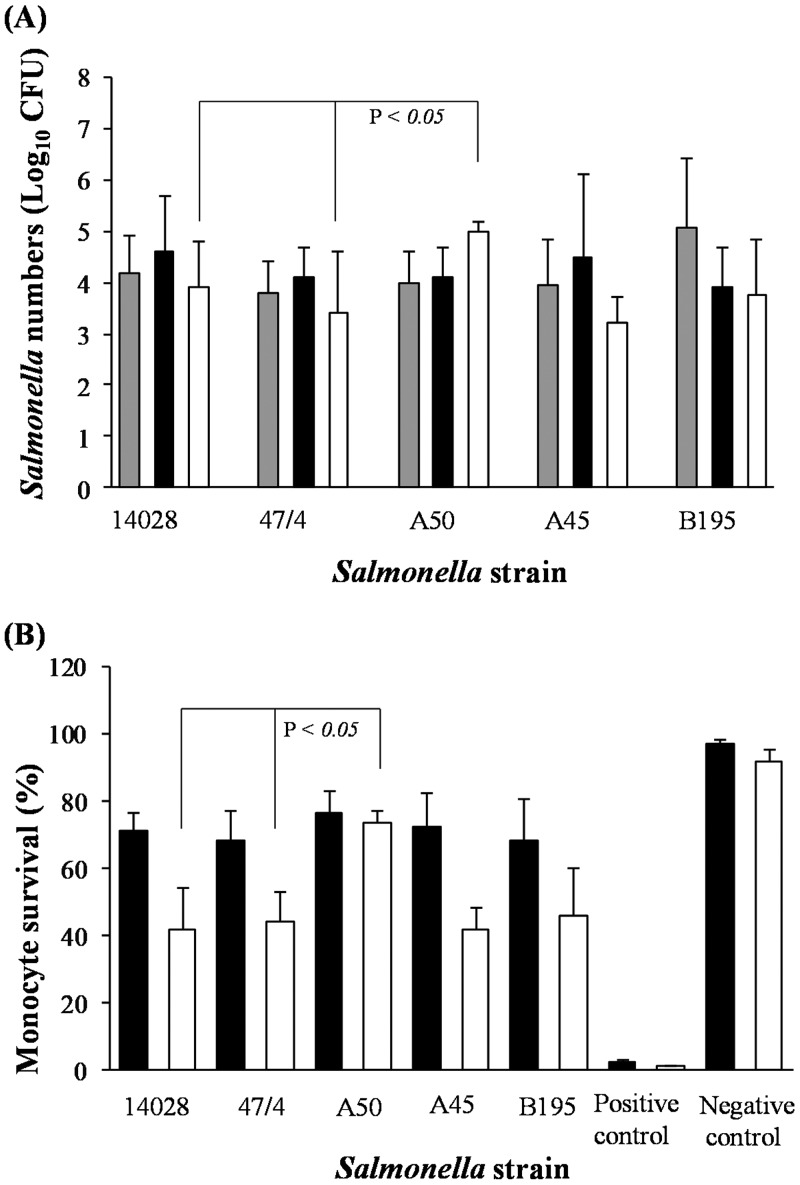
At 2 and 6 h postinfection (p.i.), comparable numbers of S. Typhimurium and S. Choleraesuis bacteria were isolated from monocytes (∼1 × 104 CFU/ml); although higher numbers of S. Choleraesuis B195 bacteria were isolated at 2 h p.i., the difference was not significant (Fig. 1A). At 24 h p.i., a significantly higher number of S. Choleraesuis A50 bacteria were isolated than either S. Typhimurium 14028 or 4/74 or other S. Choleraesuis strains (P < 0.05) (Fig. 1A). We then examined the effect of these bacteria on the monocytes themselves. At 6 h p.i., monocyte viability was ∼70% but fell to only ∼40% at 24 h p.i. when infected with S. Typhimurium 14028 or 4/74. In contrast, monocyte viability following S. Choleraesuis A50 infection was slightly higher, at ∼79%, at 6 h p.i. and was almost completely maintained (∼77%) at 24 h p.i. (Fig. 1B). This effect was not seen in monocytes infected with S. Choleraesuis A45 or B195, which had a viability similar to that of monocytes infected with S. Typhimurium (Fig. 1B).
FIG 1.
Salmonella bacterial numbers and human monocyte survival are increased following S. Choleraesuis A50 infection in comparison to S. Typhimurium infection. (A) Numbers of viable CFU of S. Choleraesuis (A50, A45, and B195) and S. Typhimurium (14028 and 4/74) recovered from monocytes at 2, 6, and 24 h p.i. (B) Monocyte viability was measured by propidium iodide uptake and FACS analyses following infection with S. Choleraesuis or S. Typhimurium strains after 6 and 24 h. Monocyte viability was compared with that of negative controls (uninfected monocytes cultured in medium for the same time period) and positive controls (monocytes immersed in ice-cold [−20°C] methanol for 30 min). Error bars represent standard deviations from the mean, and linkage bars at the top of the graph indicate significant differences (P = 0.05). Results shown are mean values calculated from triplicate experiments performed on five separate occasions. Gray bars, 2 h p.i.; black bars, 6 h p.i.; white bars, 24 h p.i.
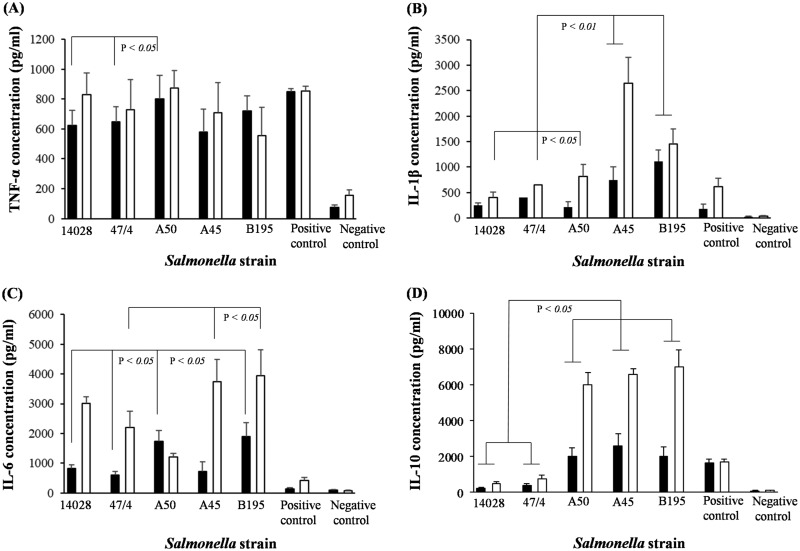
Only S. Choleraesuis A50 induced significantly higher production levels of tumor necrosis factor alpha (TNF-α) by monocytes than the levels induced by S. Typhimurium 14028 or 4/74 (6 h p.i.) (P < 0.05) (Fig. 2A). However, at 6 h p.i., S. Choleraesuis strains A45 and B195 induced significantly higher expression of IL-β (P < 0.05 and P < 0.01, respectively) than either S. Typhimurium serovar (Fig. 2B). At 24 h p.i., S. Choleraesuis A50 induced significantly higher IL-β production by monocytes than S. Typhimurium (P < 0.05), and this production level was further increased by S. Choleraesuis A45 or B195 (P < 0.01) (Fig. 2B).
FIG 2.
Hyperexpression of IL-10 by human monocytes is associated with S. Choleraesuis infection. ELISA analyses were performed to measure the production of proinflammatory cytokines TNF-α (A), IL-1β (B), and IL-6 (C) and anti-inflammatory IL-10 (D) by monocytes infected with S. Choleraesuis (A50, A45, and B195) or S. Typhimurium (14028 and 4/74). These production levels were compared with those of monocytes cultured with PMA (positive control) or of monocytes cultured only in medium for the same experimental time period. Error bars represent standard deviations from the mean, and linkage bars indicate significant differences (P = 0.05). Results shown are mean values calculated from triplicate experiments performed on five separate occasions. Black bars, 6 h p.i.; white bars, 24 h p.i.
The temporal production levels of IL-6 were also different in S. Typhimurium- and S. Choleraesuis-infected monocytes. Monocytes infected with S. Choleraesuis A50 or B195 produced significantly more IL-6 at 6 h p.i. (P < 0.05) than monocytes infected with S. Typhimurium 14028 or 4/74 (Fig. 2C). However, at 24 h p.i., monocytes infected with S. Choleraesuis A45 or B195 (but not A50) produced significantly more IL-6 (P < 0.05) than monocytes infected with S. Typhimurium 14028 or 4/74 (Fig. 2C).
The most striking difference in cytokine production that we measured was that of IL-10. At both 6 h and 24 h p.i., monocytes infected with S. Choleraesuis A50, A45, and B195 induced significantly (P < 0.01) more IL-10 than did those infected with S. Typhimurium 14028 or 4/74. However, it was the scale of difference that was most surprising, with around a 10-fold difference at 24 h p.i. (Fig. 2D).
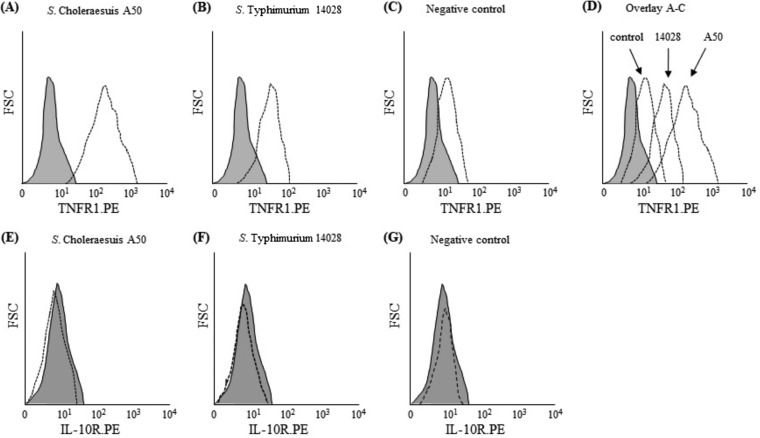
Using S. Choleraesuis A50 and S. Typhimurium 14028 as examples of these two serovars, we compared expression of TNFR1, IL-10R, and genes within the JAK/STAT pathway, all of which have been shown to be required for signaling during different phases of sepsis. Although both S. Typhimurium 14028 and S. Choleraesuis A50 increased TNFR1 protein expression on the surface of monocytes, this was much greater in S. Choleraesuis-infected monocytes (Fig. 3A to D). However, neither serovar affected the expression of IL-10R (Fig. 3E to G).
FIG 3.
S. Choleraesuis A50 increases TNFR1 protein on the surface of human monocytes in comparison to S. Typhimurium 14028. FACS analyses were used to measure TNFR1 and IL-10R proteins on the surface of monocytes infected with S. Choleraesuis A50 and S. Typhimurium 14028. (A) TNFR1 expression by S. Choleraesuis-infected monocytes; (B) TNFR1 expression by S. Typhimurium-infected monocytes; (C) TNFR1 expression by uninfected monocytes; (D) overlay of panels A to C showing increased TNFR1 expression in S. Choleraesuis-infected monocytes compared to S. Typhimurium-infected monocytes; (E) IL-10R expression by S. Choleraesuis-infected monocytes; (F) IL-10R expression by S. Typhimurium-infected monocytes; (G) IL-10R expression by uninfected monocytes. Broken lines represent cytokine receptor expression relative to that of isotype controls (gray-shaded area). Histograms are representative of those obtained in triplicate experiments performed on five separate occasions.
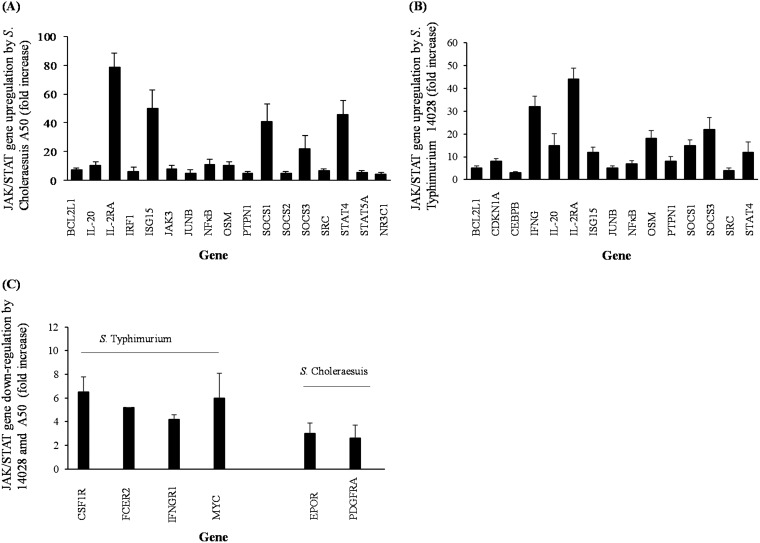
S. Choleraesuis A50 induced differential expression of 19 JAK/STAT genes, with 17 upregulated and 2 downregulated (Fig. 4A and C; Table 1). S. Typhimurium 14028 infection induced differential expression of 19 JAK/STAT genes, with 15 upregulated and 4 downregulated (Fig. 4B and C; Table 2). In monocytes infected with S. Typhimurium, expression of IFNG, which encodes IFN-γ protein, was significantly increased (P < 0.05) by >30-fold compared to that in uninfected cells (Fig. 4B). However, expression of the gene which encodes IFN-γ receptor 1 (IFNGR1) was significantly downregulated (P < 0.05) in S. Typhimurium-infected monocytes (Fig. 4C). No changes in expression of either IFNG or IFNGR1 was measured in S. Choleraesuis-infected monocytes, but S. Choleraesuis infection did significantly increase (P < 0.05) expression of the interferon regulatory factor 1 (IRF1) gene (Fig. 4A), which was not altered by S. Typhimurium infection. Both Salmonella serovars significantly upregulated (P < 0.05) IL-20 and IL-2RA genes, the latter of which encodes the IL-2 receptor and was upregulated >40-fold by S. Typhimurium but almost 80-fold by S. Choleraesuis (Fig. 4A and B).
FIG 4.
Differential expression of JAK/STAT genes by monocytes infected with S. Choleraesuis A50 and S. Typhimurium 14028. (A) Expression of JAK/STAT genes upregulated by S. Choleraesuis infection; (B) expression of JAK/STAT genes upregulated by S. Typhimurium infection; (C) expression of JAK/STAT genes downregulated by S. Typhimurium or S. Choleraesuis infection. Error bars represent standard deviations from the mean. Results shown are mean values calculated from triplicate experiments performed on three separate occasions. All bars indicate significant changes (P < 0.05) in threshold cycle (CT) values in comparison to gene expression in uninfected (control) monocytes.
TABLE 1.
Reputed functions of JAK/STAT genes differentially expressed in human monocytes infected with S. Choleraesuis A50a
| Gene | GenBank accession no. | Protein encoded | Gene function |
|---|---|---|---|
| Upregulated genes | |||
| BCL2L1 | NM_138578 | BCL2-like1 | Anti- or proapoptotic regulators that are involved in a wide variety of cellular activities |
| IL-20 | NM_018724 | Interleukin 20 | Encodes a cytokine that has been shown to transduce its signal through signal transducer and activator of transcription 3 (STAT3) in keratinocytes |
| IL2RA | NM_000417 | Interleukin 2 receptor alpha | Encodes IL-2 receptor protein |
| IRF1 | NM_002198 | Interferon regulatory factor 1 | Transcription factor which activates IFN-α and IFN-β |
| JAK3 | NM_000215 | Janus kinase 3 | Intracellular signaling molecule downstream of activated cytokine receptors |
| ISG15 | NM_005101 | ISG15 ubiquitin-like modifier | Encodes a ubiquitin-like protein that is conjugated to intracellular target proteins upon activation by interferon-alpha and interferon-beta |
| JUNB | NM_002229 | JunB proto-oncogene | Transcription factor involved in regulating gene activity following the primary growth factor response |
| NFκB | NM_003998 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 1 | Transcription regulator that is activated by various intra- and extracellular stimuli, such as cytokines, oxidant-free radicals, UV irradiation, and bacterial or viral products |
| OSM | NM_020530 | Oncostatin M | Encodes a growth regulator which inhibits the proliferation of a number of tumor cell lines |
| PTPN1 | NM_002827 | Protein tyrosine phosophatase, non-receptor type 1 | Encodes a PTP that was also reported to dephosphorylate JAK2 and TYK2 kinases, which implicated the role of this PTP in cell growth control and cell response to interferon stimulation |
| SOCS1 | NM_003745 | Suppressor of cytokine signaling 1 | SOCS1 functions downstream of cytokine receptors and takes part in a negative feedback loop to attenuate cytokine signaling |
| SOCS2 | NM_003877 | Suppressor of cytokine signaling 1 | Inhibitor of JAK/STAT pathway and IGF1R-mediated signaling |
| SOCS3 | NM_003955 | Suppressor of cytokine signaling 3 | Encodes a protein that can bind to JAK2 kinase and inhibit the activity of JAK2 kinase |
| SRC | NM_005417 | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | This proto-oncogene may play a role in the regulation of embryonic development and cell growth |
| STAT4 | NM_003151 | Signal transducer and activator of transcription 4 | Known to induce Th1 development and IFN-γ production following stimulation of Th0 cells with IL-12 |
| STAT5A | NM_003152 | Signal transducer and activator of transcription 4 | Transcriptional regulator of many different cytokine genes |
| NR3C1 | NM_000176 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Encodes glucocorticoid receptor proteins |
| Downregulated genes | |||
| EPOR | NM_000121 | Erythropoietin receptor | Encodes the erythropoietin receptor, a member of the cytokine receptor family; upon erythropoietin binding, this receptor activates Jak2 tyrosine kinase |
| PDGFRA | NM_006206 | Platelet-derived growth factor receptor, alpha polypeptide | Encodes a cell surface tyrosine kinase receptor for members of the platelet-derived growth factor family |
The protein encoded by each gene, the gene's known function, and the GenBank accession number are shown. Upregulated genes are genes that were upregulated by S. Choleraesuis infection. Downregulated genes are genes that were downregulated by S. Choleraesuis infection.
TABLE 2.
Reputed functions of JAK/STAT genes differentially expressed in human monocytes infected with S. Typhimurium 14028a
| Gene | GenBank accession no. | Protein encoded | Gene function |
|---|---|---|---|
| Upregulated genes | |||
| BCL2L1 | NM_138578 | BCL2-like 1 | Anti- or proapoptotic regulators that are involved in a wide variety of cellular activities |
| CDKN1A | NM_000389 | Cyclin D1 | Cyclins function as regulators of CDK kinases |
| CEBPB | NM_005194 | CCAAT/enhancer binding protein (C/EBP) beta | Encodes a protein whose activity is important in the regulation of genes involved in immune and inflammatory responses, among other processes |
| IFNG | NM_000619 | Interferon gamma | Encodes a soluble cytokine with antimicrobial, immunoregulatory and antitumor properties and is a potent activator of macrophages |
| IL-20 | NM_018724 | Interleukin 20 | Encodes a cytokine that has been shown to transduce its signal through signal transducer and activator of transcription 3 (STAT3) in keratinocytes |
| IL2RA | NM_000417 | Interleukin 2 receptor alpha | Encodes IL-2 receptor protein |
| ISG15 | NM_005101 | ISG15 ubiquitin-like modifier | Encodes a ubiquitin-like protein that is conjugated to intracellular target proteins upon activation by interferon-alpha and interferon-beta |
| JUNB | NM_002229 | Jun B proto-oncogene | Transcription factor involved in regulating gene activity following the primary growth factor response |
| NFκB | NM_003998 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 1 | NFκB is a transcription regulator that is activated by various intra- and extracellular stimuli, such as cytokines, oxidant-free radicals, UV irradiation, and bacterial or viral products |
| OSM | NM_020530 | Oncostatin M | Encodes a growth regulator which inhibits the proliferation of a number of tumor cell lines |
| PTPN1 | NM_002827 | Protein tyrosine phosphatase, non-receptor type 1 | Encodes a PTP that was also reported to dephosphorylate JAK2 and TYK2 kinases, which implicated the role of this PTP in cell growth control and cell response to interferon stimulation |
| SOCS1 | NM_003745 | Suppressor of cytokine signaling 1 | SOCS1 functions downstream of cytokine receptors and takes part in a negative feedback loop to attenuate cytokine signaling |
| SOCS3 | NM_003955 | Suppressor of cytokine signaling 3 | Encodes a protein that can bind to JAK2 kinase and inhibit the activity of JAK2 kinase |
| SRC | NM_005417 | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | This proto-oncogene may play a role in the regulation of embryonic development and cell growth |
| STAT4 | NM_003151 | Signal transducer and activator of transcription 4 | Known to induce Th1 development and IFN-γ production following stimulation of Th0 cells with IL-12 |
| Downregulated genes | |||
| CSF1R | NM_005211 | Colony-stimulating factor 1 receptor | Encodes a protein that is the receptor for colony-stimulating factor 1, a cytokine which controls many monocyte functions |
| FCER2 | NM_002002 | Fc fragment of IgE, low affinity II, receptor for CD23 | Encodes a protein that is a B cell-specific antigen and a low-affinity receptor for immunoglobulin E (IgE) |
| IFNGR1 | NM_000416 | Interferon gamma receptor 1 | The initial receptor subunit of IFN-γ receptor to bind IFN-γ |
| MYC | NM_002467 | V-myc myelocytomatosis viral oncogene homolog (avian) | Encodes a multifunctional, nuclear phosphoprotein that plays a role in apoptosis and cell cycle progression |
The protein encoded by each gene, the gene's known function, and the GenBank accession number are shown. Upregulated genes are genes that were upregulated by S. Typhimurium infection. Downregulated genes are genes that were downregulated by S. Typhimurium infection.
Genes which encode transcription factors of many different cytokines (JUNB, NF-κB, and CEBPD) were also upregulated by both S. Typhimurium and S. Choleraesuis (Fig. 4A and B). Both serovars also significantly upregulated (P < 0.05) the STAT4 gene, but this was most pronounced in S. Choleraesuis-infected monocytes, which showed a >40-fold increase (Fig. 4A), in comparison with about a 12-fold increase in S. Typhimurium-infected monocytes (Fig. 4B). In addition, S. Cholerasuis also significantly upregulated (P < 0.05) STAT5A and JAK3 genes (Fig. 4A), neither of which were changed by S. Typhimurium infection.
Differences were also measured in the expression of genes which encode suppressor of cytokine signaling 1 to 3 (SOCS1 to SOCS3). Both serovars induced significant increases in expression of SOCS1 and SOCS3 genes, and in the case of SOCS3, the expression levels were similar (∼20-fold increase); but, in the case of SOCS1, S. Choleraesuis induced double that induced by S. Typhimurium (Fig. 4A and B) and only S. Choleraesuis infection induced significant upregulation (P < 0.05) of SOCS2 (Fig. 4A). S. Choleraesuis infection also significantly upregulated the gene which encodes glucocorticoid receptor (NR3C1) (Fig. 4A), but neither serovar affected expression of the IL-10 receptor gene (IL-10RA).
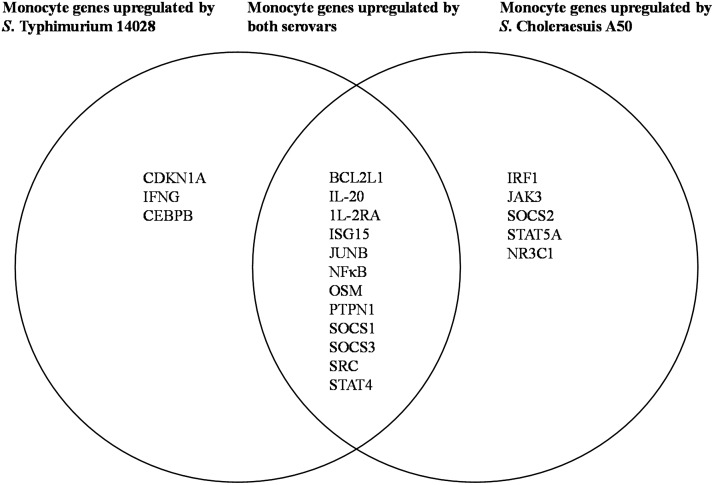
Expression of other genes within the JAK/STAT pathway, which may have a direct effect on the ability of Salmonella to survive in host cells, was differentially affected by Salmonella infection. These included genes which encode the interferon-stimulated gene 15 protein (ISG15), BCL2 protein (BCL2L1), and protein tyrosine-protein phosphatase nonreceptor type 1 (PTPN1). All three genes were significantly upregulated (P < 0.05) by S. Choleraesuis and S. Typhimurium (Fig. 4A and B) infection. In S. Typhimurium-infected monocytes, expression of the gene which encodes colony-stimulating factor 1 receptor (CSF1R) was downregulated by >6-fold (Fig. 4C) but was not changed during S. Choleraesuis infection. Differential upregulation of JAK/STAT gene expression due to S. Choleraesuis A50 and S. Typhimurium 14028 infection in human monocytes is shown as a Venn diagram in Fig. 5.
FIG 5.
Venn diagram to show JAK/STAT genes upregulated in human monocytes by infection with S. Choleraesuis A50, S. Typhimurium 14028, or both serovars. The overlapping portion shows JAK/STAT genes upregulated in human monocytes following infection by both S. Choleraesuis A50 and S. Typhimurium 14028. Nonoverlapping portions show differences in genes upregulated by each serovar.
DISCUSSION
We report differences in the immune responses to host-adapted S. Typhimurium and non-host-adapted S. Choleraesuis in human monocytes, which may have direct relevance to the greater propensity for S. Choleraesuis to cause human sepsis. Our study also indicates that some strains of S. Choleraesuis may be more likely to cause sepsis than others. First, S. Choleraesuis A50 infection prolonged monocyte viability much more so than infection with S. Typhimurium 14028 or 4/74 and at 24 h p.i., but this was not the case for S. Choleraesuis A45 or B195, which had results similar to S. Typhimurium. Previous studies have shown that increased apoptosis of blood monocytes correlates with increased survival of septic patients (12), and conversely, elevated monocyte numbers have been reported to correlate with sepsis (6). S. Typhimurium is known to induce apoptosis in human monocyte-derived macrophages (13) and, in a study by Watson et al. (14), S. Typhimurium bacteria damaged porcine macrophages that they invaded, but this was not the case for S. Choleraesuis. In pigs, as in humans, S. Choleraesuis causes a systemic infection and sepsis, and our results in combination with previously published work may indicate that in both humans and pigs, S. Choleraesuis A50-induced survival of monocytes may influence the development of sepsis. Susceptibility to the development of sepsis (and poor prognosis) in humans is also associated with a high serum IL-10 concentration (15–17). Our study shows that all strains of S. Choleraesuis induced ∼10-fold more IL-10 than S. Typhimurium. However, using S. Typhimurium 14028 and S. Choleraesuis A50 as examples, neither serovar increased IL-10R protein on the monocyte surface, which indicates that although S. Choleraesuis induces copious amounts of IL-10, this may not have an autocrine effect. High concentrations of IL-10, however, may inhibit the production of proinflammatory cytokines by Th1 lymphocytes (18) and/or induce peripheral tolerance via the differentiation of tolerogenic dendritic cells (19).
S. Choleraesuis A50 also induced a significant increase in TNF-α and IL-6 above that measured in S. Typhimurium 14028- or 4/74-infected monocytes after 6 h, but at 24 h p.i., TNF-α concentrations induced by the two serovars were comparable and the IL-6 concentration was significantly increased by S. Typhimurium infection above that induced by S. Choleraesuis A50 infection. Comparable levels of IL-1β were produced by monocytes infected with S. Choleraesuis A50 and both S. Typhimurium strains at 6 h p.i., but IL-1β production was increased at 24 h p.i. by S. Choleraesuis A50. However, S. Choleraesuis A45 and B195 infection induced the highest production levels of IL-6 and IL-1β by any Salmonella serovar or strain measured at 6 and 24 h p.i. Although all three cytokines are prominent in sepsis, TNF-α infusion in experimental animals induces septic shock and infusion of either TNF-α or IL-1β in humans induces symptoms very similar to those of the SIRS phase of acute sepsis (20). TNFR1 protein was increased on the surface of monocytes infected with either S. Choleraesuis A50 or S. Typhimurium 14028. Expression of TNFR1 was, however, much greater in S. Choleraesuis A50-infected monocytes, which may suggest that both infected monocyte populations are receptive to TNF-α but that this is possibly increased by S. Choleraesuis infection.
Analysis of gene expression in the JAK/STAT pathway also highlighted differences between S. Choleraesuis A50 and S. Typhimurium 14028. For example, although ISG15 expression was upregulated in monocytes infected with both serovars, S. Choleraesuis A50 induced a 50-fold increase compared to a 12-fold increase for S. Typhimurium 14028-infected monocytes. ISG15 has a number of different target proteins but is removed from these by ubiquitin-specific protease 18 (USP18) (21, 22), and USP18 knockout mice are susceptible to infection with both Salmonella and Mycobacterium and subsequently to Gram-negative sepsis (23, 24). We cannot say whether increased ISG15 expression was compensated for by a parallel increase in USP18, but it is possible that elevated ISG15 expression may be associated with increased numbers of S. Choleraesuis A50 bacteria measured in monocytes at 24 h p.i.
Expression of IFN-γ and IFN-γ receptor 1 genes (IFNG and IFNGR1, respectively) was also very different in monocytes infected with S. Choleraesuis A50 or S. Typhimurium 14028. S. Typhimurium 14028 upregulated expression of IFNG but downregulated IFNGR1, while neither gene was differentially expressed by S. Choleraesuis A50. In murine J774 macrophages, IFN-γ is required to kill wild-type S. Typhimurium 14028 (25), and human monocytes cultured with lipopolysaccharide (LPS) also produce IFN-γ (26), which may have an autocrine or paracrine effect on intracellular killing pathways.
However, viable bacteria are much more complex than LPS, since they express an array of different proteins required for intracellular infection and survival, and although we did not measure any additional effect due to IFN-γ in culture, the results may suggest that the effect of IFN-γ on intracellular S. Typhimurium may be reduced by the reduction in IFNGR1 expression in S. Typhimurium-infected monocytes and in S. Choleraesuis-infected monocytes in which neither IFNG nor IFNGR1 was differentially expressed.
In addition to a high level of IL-10 production, S. Choleraesuis A50-infected monocytes upregulated the expression of genes associated with immunosuppression. This included NR3C1, which encodes a glucocorticoid receptor, and SOCS1 to SOCS3, although S. Typhimurium 14028 also upregulated the expression of SOCS1 and SOCS2. SOCS proteins downregulate many inflammatory cytokines by influencing JAK/STAT signaling and downstream transcription (27), and overall, there was a lack of response in JAK and STAT genes in monocytes infected with either serovar. STAT4 genes were upregulated by both serovars, while S. Choleraesuis also induced upregulation of STAT5A and JAK3, but we do not know whether SOCS expression and production of SOCS proteins may have influenced JAK and STAT gene expression by these monocytes.
In conclusion, our study shows a differential immune phenotype expressed by human monocytes infected with either S. Choleraesuis or S. Typhimurium. Importantly, all strains of S. Choleraesuis stimulate much greater concentrations of IL-10 than S. Typhimurium, and one strain (S. Choleraesuis A50) also significantly prolonged monocyte survival. These traits are associated with human sepsis and may, in part, explain the much higher prevalence of sepsis in humans infected with S. Choleraesuis.
MATERIALS AND METHODS
Reagents.
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich, Poole, Dorset, United Kingdom. PCR and microarray reagents were purchased from Qiagen, Manchester, United Kingdom.
Bioethics.
Human blood was purchased from the blood transfusion service (Sheffield, United Kingdom). Blood used in this study was obtained with patient consent, and all studies were conducted following approval by local ethics committees.
Isolation of peripheral blood monocytes.
Whole blood was diluted with sterile phosphate-buffered saline (PBS) and then gently poured onto Histopaque-1077 prior to isolation of the buffy coat, as standard procedure. After appropriate washing steps, buffy coat supernatants were resuspended with appropriate amounts of cold MACS buffer and anti-CD14 antibody-coated micromagnetic beads (Miltenyi Biotech, Bisley, Surrey, United Kingdom) according to the manufacturer's instructions. The viability of isolated monocytes was assessed using trypan blue (10% vol/vol) and was found to be >90% prior to use. Experiments were performed in triplicate using blood from five individual (healthy) patients (five separate occasions).
Bacterial strains.
S. Typhimurium 14028 is virulent in mice (28) and in ex vivo human monocytes (29). S. Typhimurium 4/74 was originally isolated from a diseased calf and is highly virulent for pigs (30, 31) and in ex vivo human monocytes (32). S. Choleraesuis A50 was isolated from diseased pigs and is virulent in vivo (30) and in ex vivo systems (33). S. Choleraesuis A45 was isolated from diseased pigs but has not been previously reported. S. Choleraesuis B195 was originally isolated from diseased pigs but is virulent for many mammalian species (34). All strains were cultured for 24 h in 10 ml of Luria-Bertani broth (Difco Laboratories, Detroit, MI) in a shaking incubator (150 rpm) at 37°C. They generally reached densities of 3 × 109 to 5 × 109 cells/ml.
Salmonella invasion assays.
Monocytes were first washed with sterile PBS prior to the addition of S. Typhimurium or S. Choleraesuis at a multiplicity of infection (MOI) of 10:1 at 37°C, in 5% CO2, for 60 min. The cells were then washed and cultured with RPMI medium containing 100 μg/ml gentamicin and incubated for a further 60 min. The monocytes were washed again, and the medium was replaced with RPMI medium containing 25 μg/ml of gentamicin for a further 6 or 24 h in total. The cells were then washed three times with PBS at room temperature and then lysed using 1% Triton X (Fisher Scientific LTD, Loughborough, United Kingdom) for 15 min at 37°C. Intracellular bacterial counts were determined by serial dilution at different time points of 2, 6, and 24 h p.i. Viable bacterial cell counts were measured in CFU per ml. All counts were performed in triplicate on five separate occasions.
Monocyte survival assay.
The uptake of the fluorescent restriction dye propidium iodide (PI) was used to measure the viability of cells under the experimental procedures described above. At 6 and 24 h postculture, the monocytes were incubated in PBS containing PI (10 μg/ml) for 10 min. The number of nonviable cells (PI positive) was assessed using a FACSCanto II analyzer (Becton Dickinson, USA). Samples were acquired using BD FACSDiva (BD Biosciences, USA) and analyzed using CyFlogic 2.8 software, licensed to Nottingham University. Monocytes which had been immersed in ice-cold (−20°C) methanol for 30 min were used as a positive control, and monocytes cultured in medium only, for the same experimental time periods, were used as a negative control. All experiments were performed in triplicate on five separate occasions.
Cytokine concentration in monocyte supernatants.
An enzyme-linked immunosorbent assay (ELISA) (R&D Systems, Abingdon, United Kingdom) was used to determine the concentration of cytokines known to be highly important during the SIRS phase (IL-6, IL-1β, TNF-α) and the CARS phase (IL-10) of sepsis. Supernatants were analyzed (in accordance with the manufacturer's instructions) at 6 h and 24 h p.i. from S. Typhimurium- and S. Choleraesuis-infected monocytes and compared to uninfected (negative) monocytes and supernatants from monocytes cultured with phorbol myristate acetate (PMA; 10 μg/ml) (positive control).
Expression of cytokine receptors on monocyte membranes.
TNF-α and IL-10 have been proposed as possible biological markers of the SIRS and CARS phases of sepsis, respectively. Therefore, in addition to measuring TNF-α and IL-10 output, fluorescence-activated cell sorting (FACS) analyses were performed to compare the effects of S. Typhimurium and S. Choleraesuis infection on the expression of TNF-α receptor type 1 (TNFR1) and IL-10 receptor (IL-10R) protein on the surface of monocytes. Due to differential IL-10 profiles and monocyte survival associated with S. Choleraesuis A50 and S. Typhimurium, we chose S. Choleraesuis A50 and S. Typhimurium 14028 as examples of these two serovars. At 6 h p.i., 1 × 106 monocytes, from each group, were washed three times in FACS buffer (2 mM EDTA, 1% [wt/vol], bovine serum albumin [BSA]) at 300 × g for 10 min per wash step. The cell pellets were then incubated with human TruStain FcX (Biolegend, San Diego, CA, USA) for 15 min to block FC receptors. After washing three times in FACS buffer, the cell pellets were resuspended in FACS buffer containing rat anti-human IL-10R.PE (where PE is phycoerythrin; 0.5 μg/ml) or an isotype control (rabbit anti-rat Ig2a.PE) (Biolegend, USA) or containing mouse anti-human TNFR1.PE (0.5 μg/ml) or an isotype control (rat anti-mouse IgG2b.PE) (eBioscience, Cambridge). After antibody incubation, for 45 min in the dark at 4°C on an end-to-end shaker, the cells were washed three times in FACS buffer and then resuspended in 0.2 ml of FACS buffer prior to analysis on a FACSCanto II analyzer (BD Biosciences, USA). Samples were acquired using BD FACSDiva (BD Biosciences, USA) and analyzed using the CyFlogic 2.8 software. Each experiment was performed in triplicate on five separate occasions.
Multiplex PCR array analysis.
Following the results obtained for survival and cytokine production, we chose S. Choleraesuis strain A50 and S. Typhimurium strain 4/74 as examples of these two serovars to study gene regulation in the JAK/STAT pathway.
Monocytes were isolated at 6 h p.i. with S. Typhimurium 14028 or S. Choleraesuis A50 or unstimulated controls incubated with gentamicin for the same time period. The monocytes were washed briefly with ice-cold PBS, and the cell pellet was immediately harvested in 100 μl RNAprotect cell reagent, according to the manufacturer's recommendations. QIAshredder spin columns (Qiagen, Manchester, United Kingdom) were used to homogenize the cell lysate by centrifugation at 13,000 rpm (>10,000 × g) for 5 min. Total RNA was extracted using an RNAeasy plus minikit (Qiagen, United Kingdom). RNA quality was measured with a Nanodrop (ND 1000) spectrophotometer (Thermo Scientific, Loughborough, United Kingdom).
RT2 SYBR green master mix and an RT2 first-strand DNA kit were used, as per the manufacturer's instruction, to obtain monocyte DNA prior to multiplex PCR analysis using an RT2 profiler PCR array (Qiagen, United Kingdom). Changes in the expression of 84 genes of the JAK/STAT pathway were examined and compared to the mean expression of two reference genes, the beta-actin and glyceraldehyde 3-phosphate dehydrogenase genes. Mean fold changes in results from three replicate experiments performed on three separate occasions were recorded. Agarose gel electrophoresis was also used to verify quantitative PCR (qPCR) amplicon JAK/STAT products in infected monocytes.
Statistical analysis.
Analysis of variance (ANOVA) analyses with a one-way classification were performed to determine the significance between experimental groups. Tukey's post hoc test was used to determine significant differences between groups at the 95% confidence limit (P = 0.05).
ACKNOWLEDGMENTS
The work was funded by the Sudan University of Science and Technology and Faculty for the Future, the Gordon Memorial college scholarship (awarded to H.I.), and the University of Duhok/Kurdistan Region of Iraq scholarship (awarded to B.A.).
REFERENCES
- 1.Blaser MJ, Feldmann RA. 1981. Salmonella bacteremia: reports to the Centers for Disease Control, 1968–1979. J Infect Dis 143:743–746. doi: 10.1093/infdis/143.5.743. [DOI] [PubMed] [Google Scholar]
- 2.Chen P-L, Wu C-J, Chang C-M, Lee H-C, Lee N-Y, Shih H-I, Lee C-C, Ko N-Y, Wang L-R, Ko W-C. 2007. Extraintestinal focal infections in adults with Salmonella enterica serotype Choleraesuis bacteremia. J Microbiol Immunol Infect 40:240–247. [PubMed] [Google Scholar]
- 3.Chiu C, Su L, Chu C. 2004. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin Microbiol Rev 17:311–322. doi: 10.1128/CMR.17.2.311-322.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray P. 2007. The JAK-STAT signaling pathway: input and output integration. J Immunol 178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 5.Gogos CA, Drosou E, Bassaris HP, Skoutelis A. 2000. Pro- versus anti-inflammatory cytokine profile in patients with severe sepsis: a marker for prognosis and future therapeutic options. J Infect Dis 181:176–180. doi: 10.1086/315214. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J, Chaudhry H, Zhong Y, Ali MM, Perkins LA, Owens WB, Morales JE, McGuire FR, Zumbrun EE, Zhang J, Nagarkatti PS, Nagarkatti M. 2015. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine 71:89–100. doi: 10.1016/j.cyto.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong T-H, Chang C-H, Ko W-J, Lin C-F, Liu H-H, Chow L-P, Huang C-T, Yu S-L, Chen Y-S. 2014. Biomarkers of early sepsis may be correlated with outcome. J Transl Med 12:146. doi: 10.1186/1479-5876-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuai K, Lui B. 2003. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 9.Weighardt H, Heidecke CD, Emmanuilidis K, Maier S, Bartels H, Siewert JR, Holzmann B. 2000. Sepsis after major visceral surgery is associated with sustained and interferon-gamma-resistant defects of monocyte cytokine production. Surgery 127:309–315. doi: 10.1067/msy.2000.104118. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Zhong X, Huang D, Chen R, Bai G, Li Q, Yu B, Fan Y, Sun X. 2014. Functional polymorphisms of interferon-gamma affect pneumonia-induced sepsis. PLoS One 9:e87049. doi: 10.1371/journal.pone.0087049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon JJ, McSorley SJ. 2009. Tracking the dynamics of salmonella specific T cell responses. Curr Top Microbiol Immunol 334:179–198. doi: 10.1007/978-3-540-93864-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giamarellos-Bourboulis EJ, Routsi C, Plachouras D, Markaki V, Raftogiannis M, Zervakis D, Koussoulas V, Orfanos S, Kotanidou A, Armaganidis A, Roussos C, Giamarellou H. 2006. Early apoptosis of blood monocytes in the septic host: is it a mechanism of protection in the event of septic shock? Crit Care 10:R76. doi: 10.1186/cc4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Mantis N, Zhang X-R, Potoka DA, Watkins SC, Ford HR. 2000. Salmonella typhimurium induces apoptosis in human monocyte-derived macrophages. Microbiol Immunol 44:987–995. doi: 10.1111/j.1348-0421.2000.tb02594.x. [DOI] [PubMed] [Google Scholar]
- 14.Watson PR, Paulin SM, Jones PW, Wallis TS. 2000. Interaction of Salmonella serotypes with porcine macrophages in vitro does not correlate with virulence. Microbiology 146:1639–1649. doi: 10.1099/00221287-146-7-1639. [DOI] [PubMed] [Google Scholar]
- 15.Latifi SQ, O'Riordan MA, Levine AD. 2002. Interleukin-10 controls the onset of irreversible septic shock. Infect Immun 70:4441–4446. doi: 10.1128/IAI.70.8.4441-4446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanilova SA, Karakolev ZT, Dimov GS, Dobreva ZG, Miteva LD, Slavov ES, Stefanov CS, Stanilov NS. 2005. High interleukin 12 and low interleukin 10 production after in vitro stimulation detected in sepsis survivors. Intensive Care Med 31:401–407. doi: 10.1007/s00134-005-2575-7. [DOI] [PubMed] [Google Scholar]
- 17.Bozza F, Salluh J, Japiassu A, Soares M, Assis E, Gomes R, Bozza M, Castro-Faria-Neto H, Bozza P. 2007. Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit Care 11:R49. doi: 10.1186/cc5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiorentino DF, Zlotnik A, Vieira P, Mosmann TR, Howard M, Moore KW, O'Garra A. 1991. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J Immunol 146:3444–3451. [PubMed] [Google Scholar]
- 19.Morelli AE, Thomson AW. 2007. Tolerogenic dendritic cells and the quest for transplant tolerance. Nat Rev Immunol 7:610–621. doi: 10.1038/nri2132. [DOI] [PubMed] [Google Scholar]
- 20.Schulte W, Bernhagen J, Bucala R. 2013. Cytokines in sepsis: potent immunoregulators and potential therapeutic targets—an updated view. Mediators Inflamm 2013:165974. doi: 10.1155/2013/165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. 2006. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J 25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KI, Yan M, Malakhova O, Luo JK, Shen MF, Zou W, de la Torre JC, Zhang DE. 2006. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol Cell Biol 26:472–479. doi: 10.1128/MCB.26.2.472-479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richer E, Prendergast C, Zhang DE, Qureshi ST, Vidal SM, Malo D. 2010. N-ethyl-N-nitrosourea-induced mutation in ubiquitin-specific peptidase 18 causes hyperactivation of IFN-αβ signaling and suppresses STAT4-induced IFN-γ production, resulting in increased susceptibility to Salmonella typhimurium. J Immunol 185:3593–3601. doi: 10.4049/jimmunol.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dauphinee SM, Richer E, Eva MM, McIntosh F, Paquet M, Dangoor D, Burkart C, Zhang DE, Gruenheid S, Gros P, Behr M, Malo D. 2014. Contribution of increased ISG15, ISGylation and deregulated type I IFN signaling in Usp18 mutant mice during the course of bacterial infections. Genes Immun 15:282–292. doi: 10.1038/gene.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster N, Hulme SD, Barrow PA. 2003. Induction of antimicrobial pathways during early-phase immune response to Salmonella spp. in murine macrophages: gamma interferon (IFN-gamma) and upregulation of IFN-gamma receptor alpha expression are required for NADPH phagocytic oxidase gp91-stimulated oxidative burst and control of virulent Salmonella spp. Infect Immun 71:4733–4741. doi: 10.1128/IAI.71.8.4733-4741.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraaij MD, Vereyken EJ, Leenen PJ, van den Bosch TP, Rezaee F, Betjes MG, Baan CC, Rowshani AT. 2014. Human monocytes produce interferon-gamma upon stimulation with LPS. Cytokine 67:7–12. doi: 10.1016/j.cyto.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimura A, Naka T, Kubo M. 2007. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 28.Buchmeier N, Heffron F. 1991. Inhibition of phagosome-lysosome fusion by Salmonella typhimurium. Infect Immun 59:2232–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibrahim H, Askar B, Barrow P, Foster N. 2018. Dysregulation of JAK/STAT genes by vasoactive intestinal peptide (VIP) in Salmonella-infected monocytes may inhibit its therapeutic potential in human sepsis. Cytokine 105:49–56. doi: 10.1016/j.cyto.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Paulin S, Jagannathan A, Campbell J, Wallis T, Stevens M. 2007. Net replication of Salmonella enterica serovars Typhimurium and Choleraesuis in porcine intestinal mucosa and nodes is associated with their differential virulence. Infect Immun 75:3950–3960. doi: 10.1128/IAI.00366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster N, Richards L, Higgins J, Kanellos T, Barrow P. 2016. Oral vaccination with a rough attenuated mutant of S. Infantis increases post-wean weight gain and prevents clinical signs of salmonellosis in S. Typhimurium challenged pigs. Res Vet Sci 104:152–159. doi: 10.1016/j.rvsc.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Askar B, Ibrahim H, Barrow P. 2015. Vasoactive intestinal peptide (VIP) differentially affects inflammatory immune responses in human monocytes infected with viable Salmonella or stimulated with LPS. Peptides 71:188–195. doi: 10.1016/j.peptides.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Bolton A, Osborne M, Wallis T, Stephen J. 1999. Interaction of Salmonella choleraesuis, Salmonella dublin and Salmonella typhimurium with porcine and bovine terminal ileum in vivo. Microbiology 145:2431–2441. doi: 10.1099/00221287-145-9-2431. [DOI] [PubMed] [Google Scholar]
- 34.Barrow P, Huggins M, Lovell M. 1994. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect Immun 62:4602–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]