ABSTRACT
The Arabidopsis thaliana Fasciclin like arabinogalactan protein 4 (FLA4) locus is required for normal root growth in a linear genetic pathway with the FEI1 and FEI2 loci coding for receptor-like kinases. The two Fas1 domains of FLA4 are conserved among angiosperms but only the C-terminal Fas1 domain is required for genetic function. We show that at low salt deletion of the N-terminal Fas1 domain of transgenic FLA4 leads to enhanced root elongation compared to the tandem Fas1 wild type version. Modeling the hypothetical interaction between FLA4 and FEI1 we show that the predicted interaction is predominantly involving the C-terminal Fas1 domain. Relative conformational mobility between the two FLA4 Fas1 domains might regulate the interaction with the FEI receptor kinases. We therefore speculate that the FLA4 FEI complex might be a sensor for environmental conditions in the apoplast.
KEYWORDS: Fasciclin like arabinogalactan proteins, receptor like kinase, homology modelling
Results and discussion
The fasciclin 1 (Fas1) domain is a structural feature of many extracellular proteins in all clades of life and has been implicated with adhesion to and signalling influenced by the extracellular matrix.1–5 Many eukaryotic Fas1 proteins, including several groups of fasciclin like arabinogalactan proteins (FLAs) in plants, contain two or more Fas1 domains arranged in tandem. However, the biological role of Fas1 tandems is elusive. The FLA4 protein encoded by the Arabidopsis thaliana Salt Overly Sensitive 5 (SOS5) locus is required for normal root growth and salt tolerance and acts in a linear genetic pathway with two leucine rich repeat receptor kinases encoding loci named FEI1 and FEI2.6–8 Sequence searches suggest that every angiosperm genome contains at least one putative orthologue of FLA4 containing both, the N-proximal Fas1-1 and the C-proximal Fas1–2 domain in tandem. Therefore, it was surprising that, in a study previously published in The Plant Journal,9 FLA4 exerted its role for root growth depending exclusively on Fas1-2 and did not require Fas1-1 for its function. Both full-length FLA4-citrin (F4C) constructs as well as a F4C construct lacking the Fas1-1 (F4C∆Fas1-1) restored normal root growth to sos5 mutants. In fact, in most experiments, when root growth on NaCl-free full-strength Murashige and Skoog medium (MS) was measured, independent UBQ:F4C∆Fas1-1 (sos5-1) lines not only complemented the growth defect of the sos5-1 mutant background, but slightly exceeded the values of UBQ:F4C(sos5-1) lines or the Col gl wildtype. The difference between UBQ:F4C(sos5-1) and UBQ:F4C∆Fas1-1 (sos5-1) lines, however, was significant (P < 0.01) after prolonged growth periods on half strength MS medium containing no or only low concentrations of NaCl (Figure 1). This suggests that at low salt concentrations, Fas1-1 plays a negative role for the dominant growth stimulatory role of FLA4 Fas1-2 in root growth. By contrast, when the NaCl concentration exceeded 50 mM, the full length UBQ:F4C(sos5-1) and the truncated UBQ:F4C∆Fas1-1 (sos5-1) lines behaved identically which indicates that under these conditions, the N-terminal Fas1-1 domain is neutral for the role of FLA4 in root growth.
Figure 1:
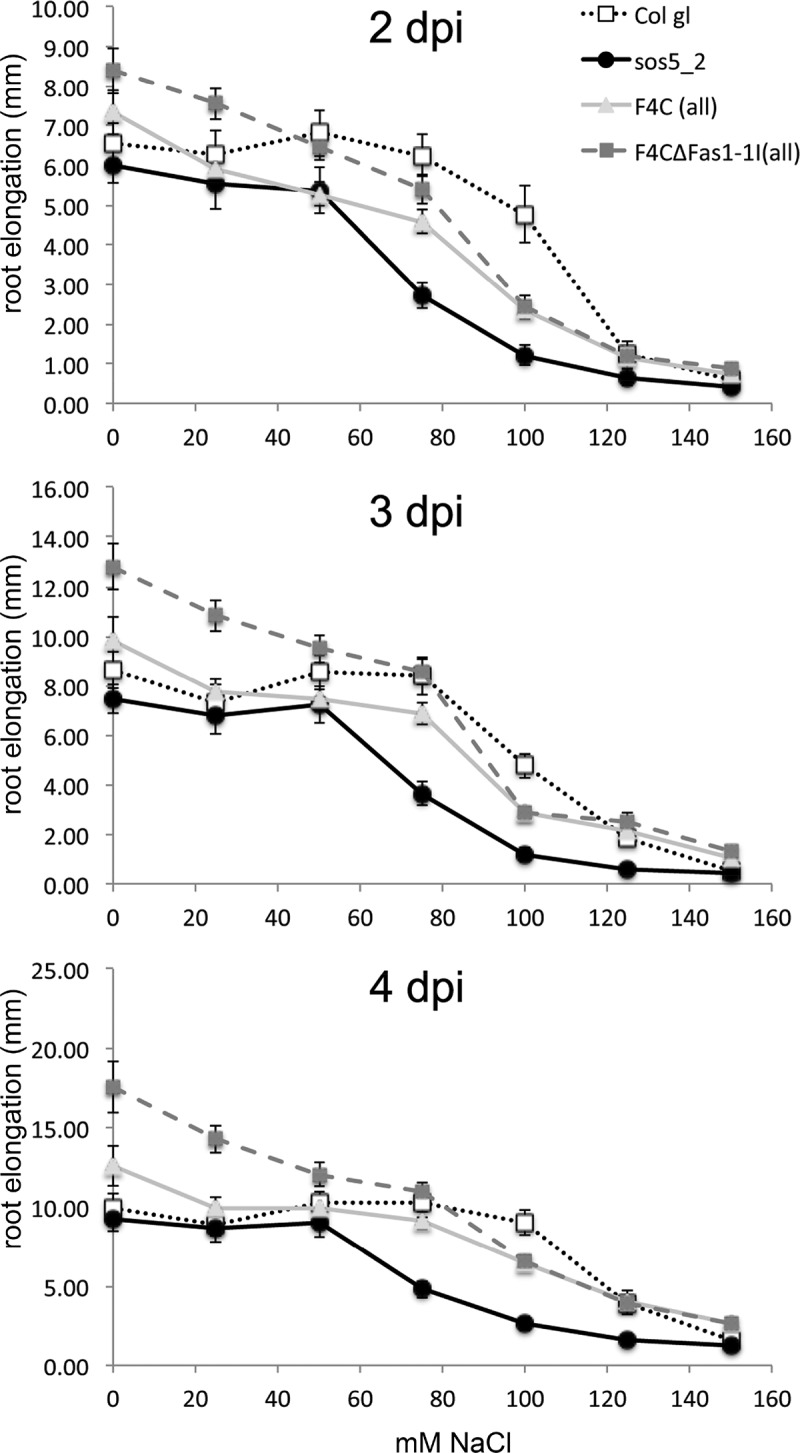
Root growth in Col gl (wild type), sos5-1 (mutant) and sos5-1 transformed with UBQ:F4C or UBQ:F4C∆Fas1-1 constructs. Seedlings were transferred to semi-solid half-strength MS medium containing 0.5% sucrose and varying concentrations of NaCl. Root growth was determined 2 to 4 days after transfer (dpi). 20 seedlings per genotype and three independent transformed lines for each construct were tested. Confidence margins for α = 5% are shown.
Is there a structural-mechanistic explanation for this observation? Because no crystal structures of FLA4 or FEI RLKs are available, we approached this question using molecular modelling. One of the crystal structures of Fas1 domains in the Protein Data Bank belongs to human transforming growth factor-beta-induced protein (TGFBIp) having four Fas1 domains (PDB ID 5NV6,10) and another one shows the Fas1-3 and Fas1-4 domains of Drosophila fasciclin I (PDB ID 1O70,11). We used SwissModel to make a homology model of FLA4 using these two crystal structures as a template.12–14 The FLA4 sequence has a higher identity with TGFBIp than Drosophila fasciclin I with 21.6% and 17.4%, respectively (the input sequence for the Swissprot query and a list of available structures including sequence identity and GMQE and is presented in the online supplement). Furthermore, although both models do not have a high global model quality estimation score (GMQE: 0.5), the model built with TGFBIp has a better agreement between the model structure and experimental structures. This ‘degree of nativeness’ is represented by the QMEAN score which is higher in the model based on TGFBIp with a score of −4.6 compared to model with Drosophila fasciclin I of −5.3. For these reasons, we continued with the model that was based on the TGFBIp structure. Next, a 3D molecular model of FLA4 with glycosylation at eight N-glycosylation sites was made. Representative glycan structures were added from a library created through enhanced sampling to get a large variety of conformers, as explained in reference.15 As a putative interaction partner, a homology model of the extracellular domain of FEI1 was modelled based on the structure of the extracellular domain of Brassinosteroid Insensitive 1- Associated Kinase 1 (BAK1, PDB code 4MN8;16 32.6% sequence identity with FEI1, corresponds to LRR 1–5 domains).
We next generated a model for the hypothetical protein-protein interaction between FEI1 and FLA47 using a knowledge based protein interaction prediction tool, PRISM.17 PRISM predictions use the observation that different protein structures can interact through shared motifs. First, PRISM predicts binding residues by using structural and evolutionary similarity to known protein interfaces as a template set, and then it performs a flexible docking with a fiberdock energy function, producing negative values for strong interactions and positive values for poor interactions.18 After the predicted structural similarity is compared with the geometrical alignment, a possible interaction is proposed. Conserved hot spots at the interface are identified as the residues having evolutionary similarity. Using our homology models for FLA4 and FEI1 as an input to predict possible interaction two predictions were suggested with two different interfaces; 1x77AB and 3shoBC. The first interaction model yielded −9.41 fiberdock score representing the larger interface between FEI1 and Fas 1-2 (Figure 2). The second interface related to the smaller interaction between FEI1 and Fas1-1 which corresponded to the beginning of the homology model of FEI1 that has no secondary structure and has only few hotspots on the interface. Hence we believe the Fas1-2 interaction to be the more likely one (Figure 2). The interface contacts for each residue are summarized in Table 1.
Figure 2:
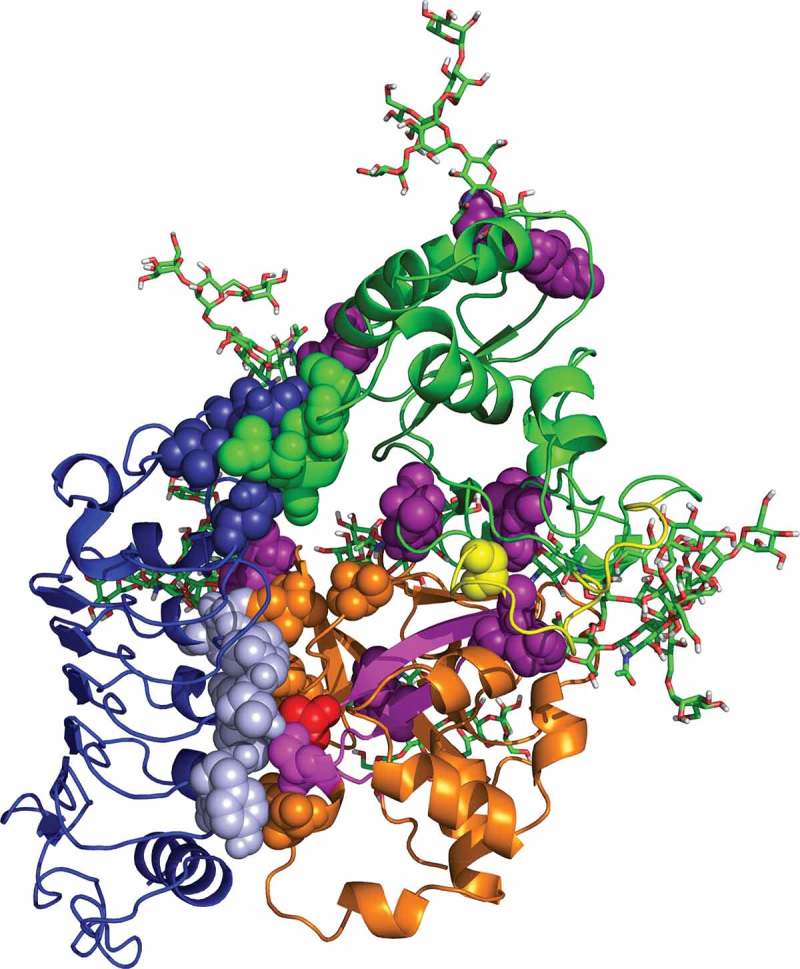
FLA4 – FEI1 interaction model. Homology model of FLA4 protein having flexible linker (coloured in yellow) connecting the Fas1-1 (green) and Fas1-2 (orange) domains. N- glycosylation was modelled at positions 30, 40, 135, 154, 167, 207, 312 and 317 using short high-mannose glycan structures for representation. Corresponding ASN residues colored in purple are represented as spheres. The predicted interaction face between FEI1 (coloured in blue) and Fas1-2 is shown as light blue and orange spheres, respectively. Interface residues between FEI1 and Fas1-1 domain are coloured in dark blue and green. Conserved residue SER348 coloured in red is substituted in the full loss of function allele sos5-1.6 Fas1-2 domain hotspots SER251, SER254, ASP255, VAL316, ASN317, ILE320, LEU329, ALA330, THR333, SER348 are shown as orange spheres. Their interaction partners on FEI1 PRO89, TYR111, GLY112, ALA113, THR116, THR135, GLY136, PRO137, ALA140, GLU141 are light blue spheres. Fas1-1 domain hotspots are PHE73, SER74, ALA76, SER77, LEU78 and THR81 shown as green spheres. Their interaction partners on FEI1 are ASN205, VAL213, CYS215 and GLN216 (dark blue spheres, this part of FEI1 corresponds to the beginning of the model, and shows no secondary structure in the model).
Table 1.
Interface residue contacts between FLA4 and FEI1. Two hydrogen bond pairs in the Fas1-2 domain are marked with asterisk.
| Fas1-1 Domain | FEI |
|---|---|
| PHE 73 | GLN 216 |
| SER74 | GLN 216, CYS 215 |
| ALA 76 | GLN 216 |
| SER 77 | ASN 205, GLN 216 |
| LEU 78 | ASN 205, GLN 216, VAL 213 |
| THR 81 |
GLN 216 |
|
Fas1-2 Domain |
FEI |
| SER 251* | THR 135*, GLY 112 |
| SER 254 | TYR 111 |
| ASP 255 | PRO 89, GLY 112 |
| VAL 316 | GLU 141 |
| ASN 317 | ALA 140 |
| ILE 320 | ALA 140 |
| LEU 329 | THR 116 |
| ALA 330* | THR 116* |
| THR 333 | GLY 136, PRO137 |
| SER 348 | ALA 113 |
The relatively weak support for an interaction with the Fas1-1 domain concurs with the recent observation that Fas1-1 was not essential for complementation.9 By contrast, the interaction model between FEI1 and Fas1-2 showed multiple interaction sites. To understand the driving force of the specific interaction, it is important to determine the characteristics of the protein interfaces. Physical and chemical features of the proposed interface were investigated in terms of hydrophobicity, electrostatic interactions, interface size and the occurrence of hydrogen bonds. In Table 1, residue contacts at the interface of the proposed complex between FLA4 and FEI1 is given for both FLA4 domains (Fas1-1 and Fas1-2) individually. As can be seen from both Figure 3 and Table 2, the interface between Fas1-2 and FEI1 is larger than the interface with the Fas1-1 domain. Furthermore, there are two hydrogen bond pairs at the Fas1-2 and FEI1 interface, reported in Table 1. The PDBePISA webserver19 was used to obtain the solvation free energy gain upon interface formation (ΔiG, in kcal/mol) reported in Table 2 along with the accessible and buried surface area of the interface. It can be seen from Table 2 that the Fas1-2 Domain and FEI1 interface has higher accessible surface area, which is an important structural characteristic during the formation of the protein-protein interaction. Also, it has ΔiG = −9.8 kcal/mol, indicating a hydrophobic interface.
Figure 3.
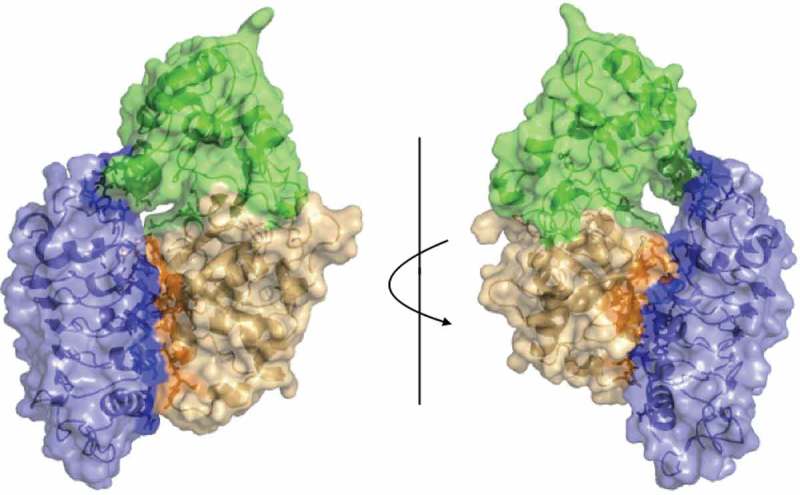
Surface representation of the proposed interaction model of FLA4-FEI (blue) from front view (left) and back view (right). Fas1-1 and Fas1-2 shown in green and orange respectively. Interface residues are shown darker with sticks.
Table 2.
Characteristics of the interface between proposed interaction of the FLA4 – FEI model.
| ASA* () | BSA* () | ΔiG (kcal/mol) | |
|---|---|---|---|
| Fas1-1 Domain & FEI1 | 7322 & 1700 | 284 & 281 | −2.3 |
| Fas1-2 Domain & FEI1 | 8086 & 7173 | 568 & 651 | −9.8 |
*ASA: Accessible Surface Area, BSA: Buried Surface Area, ΔiG: Solvation energy effect
While some LRR ligands such as flagellin bind to the ascending lateral (convex) surface of the LRR-domain,20 others such as the Spätzle morphogen21 or the follicle-stimulating hormone22 interact with the concave LRR surfaces of their respective receptors. In our model the convex surface of the FEI1 ectodomain binds to the surface of the Fas1-2 domain (Figure 3) that is formed by the outward bent β-sheet harbouring the Ser348 residue (marked red in Figure 2). Intriguingly, this residue is mutated to Phe in the strong sos5-1 allele,6 adding genetic support to our structural model.
We suggest that the Fas1-1 domain might negatively regulate FLA4 Fas1-2 binding to FEI1. This arrangement allows for regulatory flexibility. The relative conformation between Fas1-1 and Fas1-2 might be affected by post-translational modification (e.g. N-glycosylation seen in Figure 2) and their post-secretory remodelling, by interactions with other macromolecules or by apoplastic factors such as salinity or pH. In combination with FEI RLKs FLA4 might be an environmental sensor. Based independently on genetic evidence and structural predictions we speculate that the conformational changes of the tandem Fas1 protein FLA4 might regulate its interaction with FEI1 (and FEI2) LRR-RLKs to control root growth. Under all conditions Fas1-2 is predicted to interact with FEI1 which might in turn control growth by interacting with the biosynthesis and perception of growth regulators.7,23,24 Under some conditions such as low salt concentration, this interaction might be negatively modulated in an auto-inhibitory fashion by the Fas1-1 domain.
To gain an insight about the possible reconfiguration of FLA4, elastic network models (ENMs) were used. These models take the structure of a protein as a network taking atoms as nodes connected by springs. To predict the collective motion of FLA4, we used the Elastic Network Model server.25–27 The relative motion of the Fas1-1 and Fas1-2 domains are shown in the left panel of in Figure 4. Although the predicted motion is not causing a complete closing of the domains, rearrangement might affect the interaction of FEI as illustrated in the right panels of Figure 4.
Figure 4.
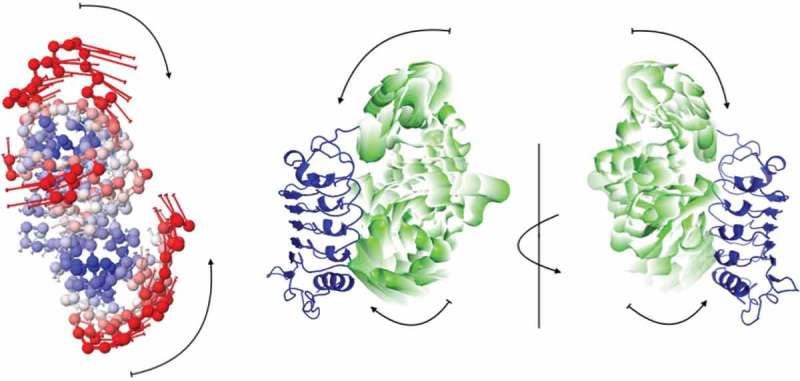
Predicted collective motion of FLA4 from Elastic Network Model. In the left panel, the FLA4 structure is colored according to the size of motions; red being most mobile and blue the most rigid. In the right panels, the motion is represented in the complex, where the transparency of the Fas1-1 and Fas1-2 domains (green) decreases, the closer to the FEI domain (blue) they are.
Alternatively, Fas1-1 might prevent FLA4 from ectopically interacting with other receptor kinases. An auto-regulatory role, previously suggested for human TGFBIp,2 might be the basis for the evolution of Fas1 tandems in eukaryotes and for the conservation of the tandem Fas1 architecture of FLA4 orthologues in angiosperms. Besides being fully consistent with available evidence, our speculative model provides testable predictions on the hypothetical physical interaction between these intriguing regulatory proteins.
Funding Statement
This work was supported by the Austrian Science Fund [W1224];Austrian Science Fund [I1182-B22].
Acknowledgements
Georg Seifert received support from the Austrian Science Fund FWF grant number I1182-B22. Aysegül Turupcu is supported by the FWF-funded doctoral programme “BioTechnology of Proteins (BioToP)”, grant number W1224.
References
- 1.Burroughs AM, Balaji S, Iyer LM, Aravind L.. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moody RG, Williamson MP.. Structure and function of a bacterial Fasciclin I Domain Protein elucidates function of related cell adhesion proteins such as TGFBIp and periostin. FEBS Open Bio. 2013;3:71–77. doi: 10.1016/j.fob.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mosher DF, Johansson MW, Gillis ME, Annis DS.. Periostin and TGF-beta-induced protein: two peas in a pod?. Critical Reviews in Biochemistry and Molecular Biology. 2015;50(5):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson KL, Jones BJ, Bacic A, Schultz CJ.. The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003;133(4):1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seifert GJ. Fascinating Fasciclins: A Surprisingly Widespread Family of Proteins that Mediate Interactions between the Cell Exterior and the Cell Surface. Int J Mol Sci. 2018;19(6). doi: 10.3390/ijms19061628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. The arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 2003;15(1):19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu SL, Rahman A, Baskin TI, Kieber JJ. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell. 2008;20(11):3065–3079. doi: 10.1105/tpc.108.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu D, Tian L, Debrosse T, Poirier E, Emch K, Herock H, Travers A, Am S. Glycosylation of a Fasciclin-Like Arabinogalactan-Protein (SOS5) Mediates Root Growth and Seed Mucilage Adherence via a Cell Wall Receptor-Like Kinase (FEI1/FEI2) Pathway in Arabidopsis. PLoS ONE. 2016;11(1):e0145092. doi: 10.1371/journal.pone.0145092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue H, Veit C, Abas L, Tryfona T, Maresch D, Ricardi MM, Estevez JM, Strasser R. Seifert GJ: Arabidopsis thaliana FLA4 functions as a glycan-stabilized soluble factor via its carboxy-proximal Fasciclin 1 domain. The Plant Journal: for Cell and Molecular Biology. 2017;91(4):613–630. doi: 10.1111/tpj.13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Castellanos R, Nielsen NS, Runager K, Thogersen IB, Lukassen MV, Poulsen ET, Goulas T, Enghild JJ, Gomis-Rüth FX. Structural and Functional Implications of Human Transforming Growth Factor beta-Induced Protein, TGFBIp, in Corneal Dystrophies. Structure. 2017;25(11):1740–1750. e1742 doi: 10.1016/j.str.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Clout NJ, Tisi D, Hohenester E. Novel fold revealed by the structure of a FAS1 domain pair from the insect cell adhesion molecule fasciclin I. Structure. 2003;11(2):197–203. [DOI] [PubMed] [Google Scholar]
- 12.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 13.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4(1):1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 14.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turupcu A, Oostenbrink C. Modeling of Oligosaccharides within Glycoproteins from Free-Energy Landscapes. J Chem Inf Model. 2017;57(9):2222–2236. doi: 10.1021/acs.jcim.7b00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342(6158):624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 17.MAPK group. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends in Plant Science. 2002;7(7):301–308. [DOI] [PubMed] [Google Scholar]
- 18.Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc. 2011;6(9):1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Ia W. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335(6070):859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parthier C, Stelter M, Ursel C, Fandrich U, Lilie H, Breithaupt C, Mt S. Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity. P Natl Acad Sci USA. 2014;111(17):6281–6286. doi: 10.1073/pnas.1320678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan QR, Wa H. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433(7023):269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seifert GJ, Xue H, Acet T. The Arabidopsis thaliana FASCICLIN LIKE ARABINOGALACTAN PROTEIN 4 gene acts synergistically with abscisic acid signalling to control root growth. Ann Bot. 2014;114(6):1125–1133. doi: 10.1093/aob/mcu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinwand BJ, Xu S, Polko JK, Doctor SM, Westafer M, Jj K. Alterations in auxin homeostasis suppress defects in cell wall function. PLoS ONE. 2014;9(5):e98193. doi: 10.1371/journal.pone.0098193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyal E, Yang LW, Bahar I. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006;22(21):2619–2627. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 26.Eyal E, Lum G, Bahar I. The anisotropic network model web server at 2015 (ANM 2.0). Bioinformatics. 2015;31(9):1487–1489. doi: 10.1093/bioinformatics/btu847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Chang YY, Lee JY, Bahar I, Lw Y. DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res. 2017;45(W1:W374–W380. [DOI] [PMC free article] [PubMed] [Google Scholar]


