ABSTRACT
Ectomycorrhizal fungi improve tree phosphorus nutrition through transporters specifically localized at soil-hyphae and symbiotic interfaces. In the model symbiosis between the fungus Hebeloma cylindrosporum and the maritime pine (Pinus pinaster), several transporters possibly involved in phosphate fluxes were identified, including three H+:Pi transporters. Among these three, we recently unraveled the function of one of them, named HcPT2, in both pure culture and symbiotic interaction with P. pinaster. Here we investigated the transporter named HcPT1.2, by analyzing inorganic phosphate transport ability in a yeast complementation assay, assessing its expression in the fungus associated or not with the plant, and immunolocalizing the proteins in ectomycorrhizas. We also evaluated the effect of external Pi concentration on expression and localization of HcPT1.2. Our results revealed that HcPT1.2 is involved in Pi acquisition by H. cylindrosporum mycelium, irrespective of the external Pi concentrations.
KEYWORDS: Ectomycorrhizal symbiosis, H+:Pi transporter, Immunolocalization, Inorganic phosphate, RT-qPCR, Yeast complementation
TEXT
Phosphorus nutrition of trees from boreal and temperate forests is mostly ensured by the ectomycorrhizal (ECM) symbiosis.1 ECM fungi are able to cover a large portion of soil, and to solubilize and assimilate inorganic orthophosphate (Pi) through extra-radical and mantle hyphae. Pi is then transported towards a network of hyphae colonizing the host roots called the Hartig net, where it is released into a symbiotic interface and taken up by plant cortical cells. Molecular mechanisms facilitating Pi fluxes in ectomycorrhizas is still barely known.2,3 In the model ECM fungus Hebeloma cylindrosporum, two H+:Pi transporters were functionally characterized in heterologous systems so far.4 Among them, HcPT1.1 seems mainly involved in Pi acquisition from the soil under limiting conditions.5 However, HcPT1.1 proteins were also localized in Hartig net hyphae, suggesting a possible role at the symbiotic interface too. More recently, we revealed that the second identified H+:Pi transporter of H. cylindrosporum, HcPT2, seems having a dual function in both Pi acquisition from the soil and Pi release towards the host cortical cells.6 With the recent genome and transcriptome sequencing of H. cylindrosporum,7–9 we also identified a third transporter that we named HcPT1.2 because of a higher level of similarity shared with HcPT1.1 (65.7 %) than with HcPT2 (48 %). Recently, an updated phylogenetic analysis of selected phosphorus transporter classes using the predicted proteins (https://jgi.doe.gov/JGI) of 35 fungal species, including 14 ECM ones, showed that both HcPT1.1 and HcPT1.2 proteins belong to the same subgroup (Ia) of H+:Pi transporter.3 Here, we report (i) the functional characterization of HcPT1.2 in yeast using non-labelled Pi, (ii) the expression of HcPT1.2 in both axenic and symbiotic conditions under high and low Pi supply, and (iii) the protein immunolocalization in Pinus pinaster ectomycorrhizas.
The PHO84 deficient yeast strain MB192 (MATa pho3-1 pho84::HIS3 ade2 leu2-3, 112 his3-532, trp1-289 ura3-1,2 can1)10 was transformed with an empty vector (pFL61, carrying the ura gene) or with the pFL61 vector containing HcPT1.2 construct (Figure 1). To build this latter vector, the coding sequence of HcPT1.2 was amplified in two steps from H. cylindrosporum cDNA using two sets of primers (HcPT1.2-F1 – CCTTGCGGCCGCACTAGTATGCCGAGCATCATCGAGAG, HcPT1.2-R2 – ACTATCTCCCGCAGCACATC, HcPT1.2-F2 –GTCAGGCCGTTCTATCTGGA, and HcPT1.2-R2 – TCAAAAACCCCTGCGACGG). Both fragments were reassembled using BamHI restriction enzyme, and cloned into pFL61 using NotI restriction enzyme.11 Yeast were transformed and selected on YNB medium (1.7 g l−1; (NH4)2SO4, 5 g l−1; glucose, 20 g l−1; agar, 20 g l−1) supplemented with the appropriate amino acids (Adenine, Leucine, Tryptophan) for transformant selection as previously described.12 Presence of HcPT1.2 coding sequence was checked by PCR using HcPT1.2-F and HcPT1.2-R. Two independent strains that incorporated HcPT1.2 were selected randomly from a collection of 10 transgenic cells for the complementation, HcPT1.2-a and HcPT1.2-b (Figure 1). HcPT1.2-a, HcPT1.2-b, wild-type MB192 (WT), and pFL61 strains were incubated at O.D. = 0.1 in liquid synthetic medium (SM) containing 50 µM of Pi supplied as KH2PO4, and the decrease in Pi concentration in the medium was measured at 24, 30, and 42 hours. One liter of SM medium contained 20 g glycerol, 5 g glucose, 0.8 g KCl, 0.5 g MgSO4,7H2O, 0.5 g CaCl2, 1.08 g (NH4)2SO4, 4 ml of micro-element solution (containing per liter: 125 mg H3BO3, 67.5 mg CuSO4 5H2O, 25 mg KI, 500 mg MnSO4 H2O, 50 mg NaMoO4 2H2O, 650 mg Fe2(SO4)3, 100 mg ZnSO4 7H2O, 30 mg CoCl2 6H2O, 25 mg NiSO4 6H2O), 1 ml of vitamin solution (containing per liter: 5 g panthotenate, 20 g inositol, 2 g nicotinic acid, 250 mg pyridoxal hydrochloride, 250 mg thiamine hydrochloride, 10 mg of biotine) and auxotrophy factors. Just after the inoculation, we also measured the actual Pi concentration in the medium which was a bit higher than 50 µM due to the inoculum, but comparable between all strains (Figure 1). While WT strain consumed all available Pi in 24 hours, less than 20 % was taken up by the pFL61 strain, and only 50 % of initial Pi was remaining after 42 hours. HcPT1.2-a and HcPT1.2-b strains consumed around 50 % of Pi after 24 and 30 hours, respectively, and almost 100 % after 42 hours. We also checked the expression of HcPT1.2 in pure culture and in ectomycorrhizas under high and low P conditions by RT-qPCR with the internal marker HcTub, as recently described (Fig. 2).6 HcPT1.2-qPCR-F and HcPT1.2-qPCR-R primers (CATTATGGCCGGAGCGTAC and CTCCGAGAGCGATTTGGATC, respectively) were used for the amplification. In pure culture, expression of HcPT1.2 was determined in two-week-old mycelia grown in liquid medium supplemented with 1 mM of Pi, in mycelia transferred in a Pi-free medium for 24 hours (Figure 2a) or 5 days (Figure 2b), and in mycelia re-enriched with 1 mM of Pi for 24 hours. Ectomycorrhizas were produced in rhizoboxes on thin soil layers enriched or not in P as described previously (Fig. 3).6 Similar expression levels were observed in all tested conditions, indicating that external Pi availability did not affect the expression of HcPT1.2 in both axenic and symbiotic culture. Finally, we immunolocalized HcPT1.2 proteins in ectomycorrhizas produced on high and low P soil conditions using a recently detailed protocol.6 Briefly, images were obtained by confocal microscopy on 50 µm cross-sections labelled with HcPT1.2 primary antibodies and Dylight 405-conjugated secondary antibodies (Figure 3). HcPT1.2 proteins were detected in extra-radical and mantle hyphae, but not in the Hartig net (Figure 3(a,c)). Consistently with the expression level data described above, no difference in signal intensity was detected under high or low P supply.
Figure 1.
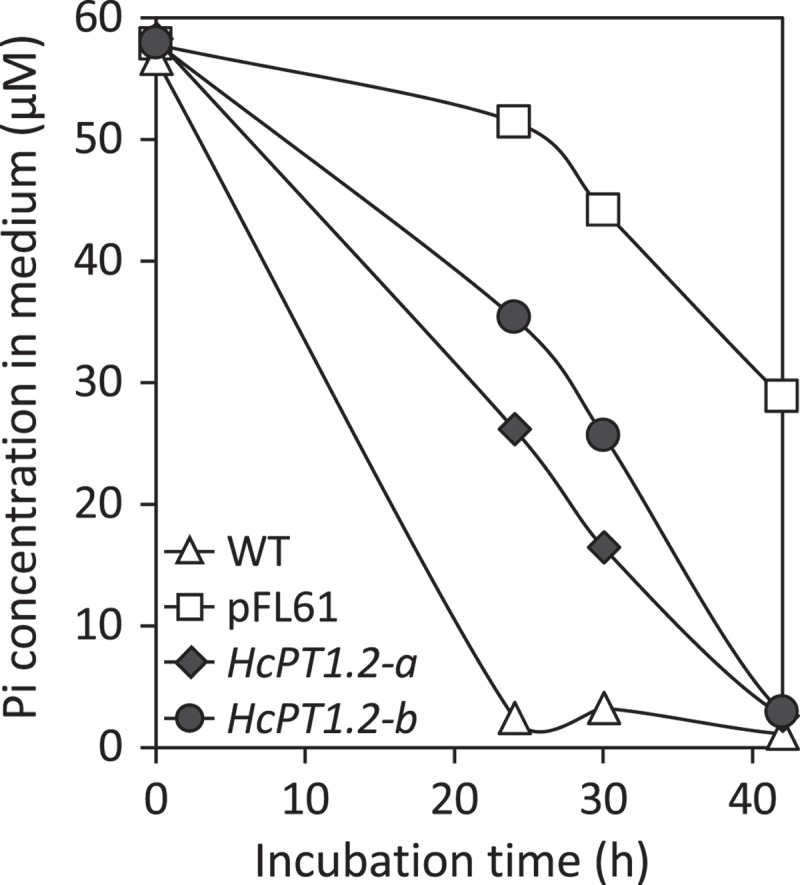
Functional complementation of PHO84 deficient yeast strain MB192 by HcPT1.2 Pi transporter of Hebeloma cylindrosporum. Phosphate absorption by wild-type (WT) or PHO84 deficient strains transformed with an empty vector (pFL61) or HcPT1.2 (HcPT1.2-a and HcPT1.2-b are two independent transformed strains) was determined by measuring Pi concentration directly in culture media at 24, 30 and 42 hours after inoculation. (n = 3).
Figure 2.
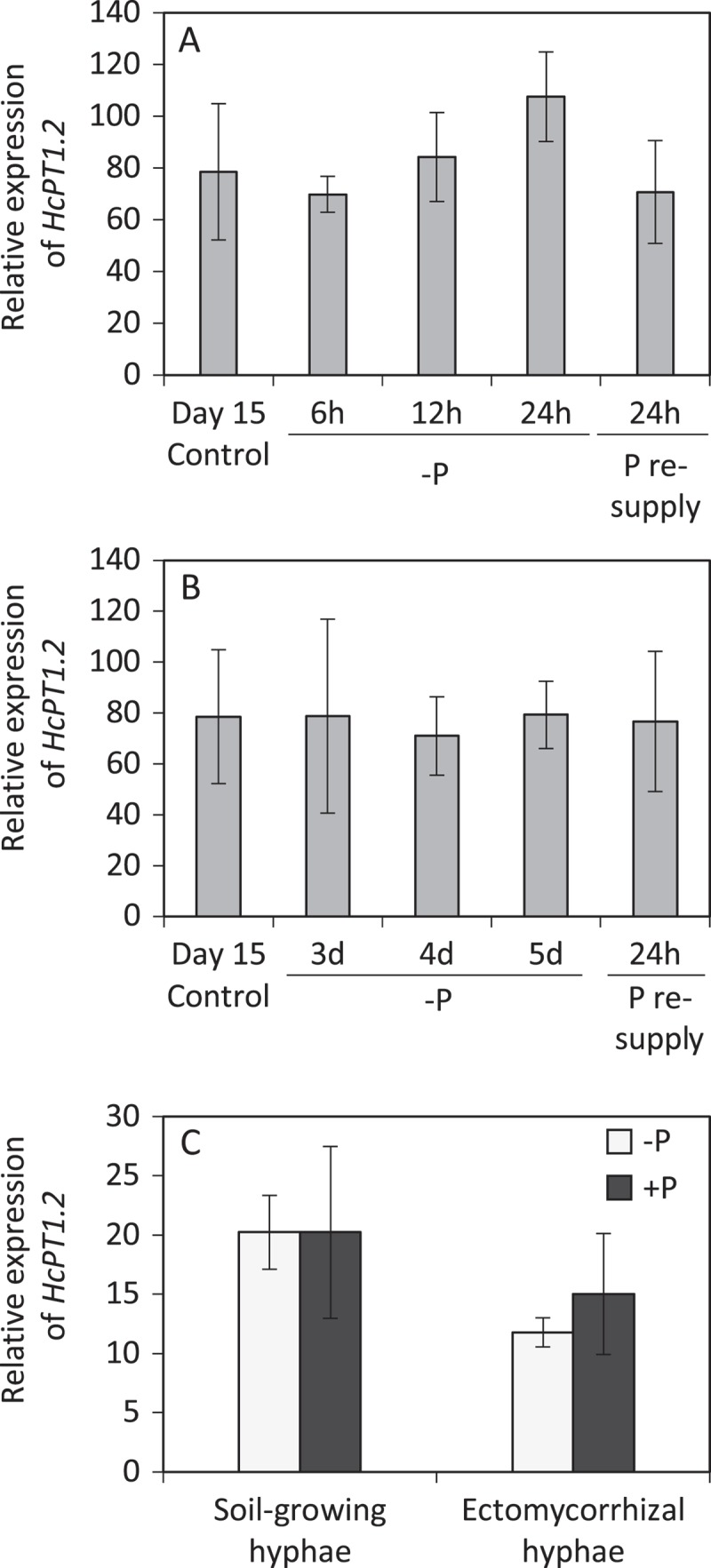
Expression of HcPT1.2 in pure culture and ectomycorrhizal symbiosis with Pinus pinaster under P-sufficient and -limiting conditions. (a,b) Expression of HcPT1.2 in pure culture was determined by RT-qPCR on two-week-old mycelia grown in liquid medium supplemented with 1 mM of Pi (Day 15 Control), and then in mycelia transferred in a Pi-free medium during 24h (a) or 5 days (b). Mycelia were finally transferred in a Pi-rich medium for 24h (1 mM, P resupply). (c) Expression of HcPT1.2 in symbiotic condition was determined in extra-radical (Soil-growing hyphae) or root-associated hyphae (Ectomycorrhizal hyphae), obtained in rhizoboxes on thin soil layers enriched in P (+ P) or not (-P). No difference was recorded in any conditions. Values were normalized with housekeeping gene (HcTub). Data are means ± standard deviation (n = 4).
Figure 3.
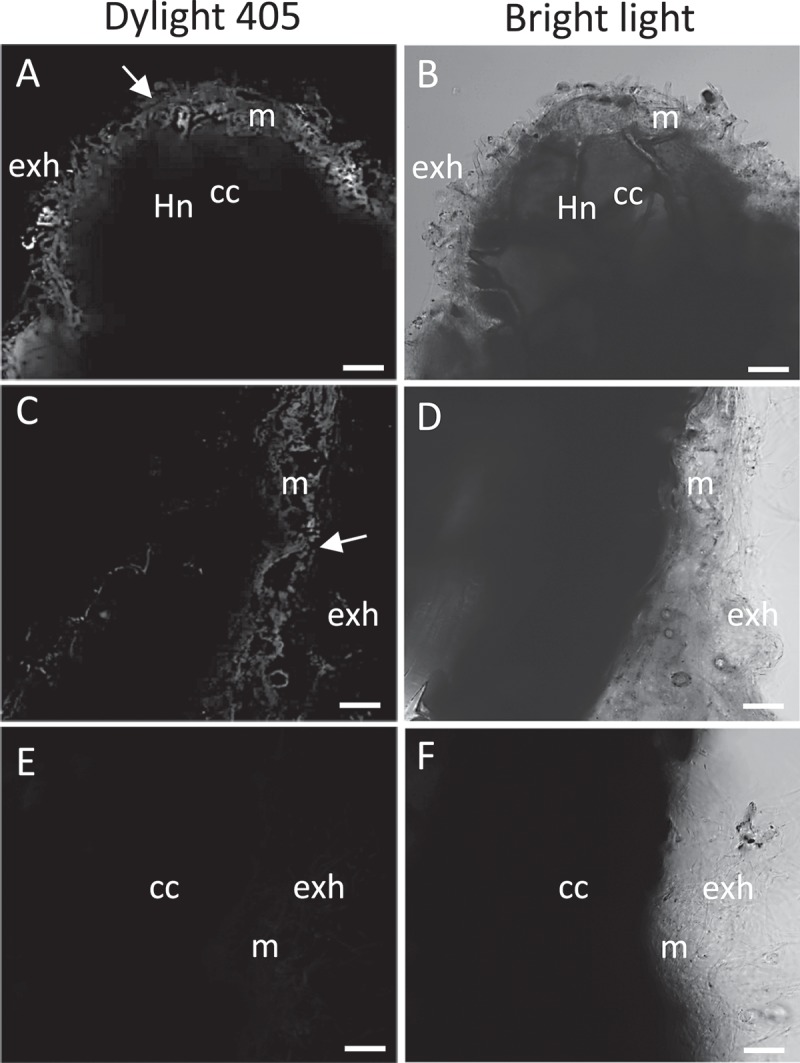
Immunolocalization of HcPT1.2 in Hebeloma cylindrosporum hyphae forming ectomycorrhizas with Pinus pinaster under P-sufficient and -limiting conditions. Ectomycorrhizas were obtained in rhizoboxes on thin soil layers enriched in P (a,b) or not (c,d). Cross-sections labelled with secondary antibodies only show no signal (E). Images were obtained by confocal microscopy on 50 µm cross-sections labelled with HcPT1.2 primary antibodies and Dylight 405-conjugated secondary antibodies. m, hyphal mantle; exh, extra-radical hyphae; Hn, Hartig net; cc, cortical cell.
Recently, we showed that transcript levels of HcPT1.2, the third H+:Pi transporter from H. cylindrosporum, were much lower in free-living mycelia and ectomycorrhizas than those of HcPT1.1 and HcPT2, but we did not described further this candidate.6,8 Here, we revealed its function, expression, and localization in the fungus in axenic and symbiotic conditions. Although we were not able to fully characterize HcPT1.2 because we did not use radio-labeled Pi, we have clearly demonstrated that HcPT1.2 proteins are able to partially complement the PHO84 deficient yeast strain MB192. This indicates that HcPT1.2 can participate in Pi acquisition in the ECM fungus H. cylindrosporum. We also did not observe a Pi-dependent regulation of HcPT1.2 expression and protein localization. This feature set HcPT1.2 apart from other H+:Pi fungal transporters characterized so far. Indeed, HcPT1.1 and HcPT2 transcripts are up-regulated by Pi limiting or sufficient conditions, respectively,4,5 and BePT, RlPT, and LbPT transcripts of Boletus edulis, Rhizopogon luteolus, and Leucocortinarius bulbiger, respectively, are up-regulated at low Pi.13,14 The absence of Pi-dependent regulation supposes that HcPT1.2 is more versatile than HcPT1.1 and HcPT2, and even if its expression is low it may play a critical role for a basal and stable Pi acquisition, particularly under fluctuating environmental conditions. However, we cannot exclude that P availability may impact the activity of HcPT1.2 proteins through post-translational modifications as described in plants and yeasts,15,16 and further experiments are still necessary to investigate this question. Localization of HcPT1.2 exclusively at the uptake sites of ectomycorrhizas (i.e. extra-radical hyphae and mantle) is consistent with its possible role in Pi acquisition from the soil. Once again, the absence of protein detection in the Hartig net sets HcPT1.2 apart from HcPT1.1 and HcPT2 since both of them localize in all hyphae of ectomycorrhizas.5,6 The generation of transgenic H. cylindrosporum strains up- or down-regulating the expression of candidate genes is a powerful approach to unravel their function in axenic and symbiotic conditions,6,17–20 and will help us to validate the Pi import role of HcPT1.2 in ectomycorrhizal symbiosis.
To conclude, although we have now started to investigate all H+:Pi transporters identified in H. cylindrosporum, important paths still remain unexplored. For example, efforts are now needed to unravel their shared and specific regulations by external P or unknown molecules originated from the host,6,21 the biosynthesis, movement, and degradation of polyphosphates,22 and the other putative candidates that might be involved in Pi and organic P transport we recently identified, e.g. HcPHO1 and HcPho90.6 Understanding the molecular mechanisms evolved by ECM fungi for P acquisition, transport, and delivery to the host is crucial to harness this symbiosis for a better and more sustainable production of crop trees.
Funding Statement
This work was supported by the Agence Nationale de la Recherche [ANR-10- LABX-04-01];Agence Nationale de la Recherche [“TRANSMUT” 2010 BLAN 1604 03];North Carolina Agriculture Research Service [Start-up funds].
Acknowledgments
A.B. was supported by an INRA CJS grant ‘Contrat Jeune Scientifique’. Microscopy observations were made at the Montpellier Rio Imaging facility (http://www.mri.cnrs.fr/). This work was supported by the ANR project “TRANSMUT” 2010 BLAN 1604 03 and by the program ‘Investments for the future’ (ANR-10- LABX-04-01) through the use of the Ecotrop platform from CeMEB labEx. K.G. acknowledges support of the North Carolina Agriculture Research Service (NCARS).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- 1.Becquer A, Trap J, Irshad U, Ali MA, Plassard C.. From soil to plant, the journey of P through trophic relationships and ectomycorrhizal association. Front Plant Sci. 2014;5:548. doi: 10.3389/fpls.2014.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia K, Doidy J, Zimmermann SD, Wipf D, Courty P-E.. Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 2016;21:937–950. doi: 10.1016/j.tplants.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Nehls U, Plassard C. Nitrogen and phosphate metabolism in ectomycorrhizas. New Phytol. 2018; doi: 10.1111/nph.15257. [DOI] [PubMed] [Google Scholar]
- 4.Tatry M-V, El Kassis E, Lambilliotte R, Corratgé C, van Aarle I, Amenc LK, Alary R, Zimmermann S, Sentenac H, Plassard C. Two differentially regulated phosphate transporters from the symbiotic fungus Hebeloma cylindrosporum and phosphorus acquisition by ectomycorrhizal Pinus pinaster. Plant J. 2009;57:1092–1102. doi: 10.1111/j.1365-313X.2008.03749.x. [DOI] [PubMed] [Google Scholar]
- 5.Garcia K, Haider MZ, Delteil A, Corratgé-Faillie C, Conéjero G, Tatry M-V, Becquer A, Amenc L, Sentenac H, Plassard C et al. Promoter-dependent expression of the fungal transporter HcPT1.1 under Pi shortage and its spatial localization in ectomycorrhiza. Fungal Genet Biol. 2013;58(59):53–61. doi: 10.1016/j.fgb.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Becquer A, Garcia K, Amenc L, Rivard C, Doré J, Trives-Segura C, Szponarski W, Russet S, Baeza Y, Lassalle-Kaiser B et al. The Hebeloma cylindrosporum HcPT2 Pi transporter plays a key role in ectomycorrhizal symbiosis. New Phytol. 2018; doi: 10.1111/nph.15281. [DOI] [PubMed] [Google Scholar]
- 7.Doré J, Perraud M, Dieryckx C, Kohler A, Morin E, Henrissat B, Lindquist E, Zimmermann SD, Girard V, Kuo A et al. Comparative genomics, proteomics and transcriptomics give new insight into the exoproteome of the basidiomycete Hebeloma cylindrosporum and its involvement in ectomycorrhizal symbiosis. New Phytol. 2015;208:1169–1187. doi: 10.1111/nph.13546. [DOI] [PubMed] [Google Scholar]
- 8.Doré J, Kohler A, Dubost A, Hundley H, Singan V, Peng Y, Kuo A, Grigoriev IV, Martin F, Marmeisse R et al. The ectomycorrhizal basidiomycete Hebeloma cylindrosporum undergoes early waves of transcriptional reprogramming prior to symbiotic structures differentiation. Environ Microbiol. 2017;19:1338–1354. doi: 10.1111/1462-2920.13670. [DOI] [PubMed] [Google Scholar]
- 9.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canback B, Choi C, Cichocki N, Clum A et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47:410–415. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 10.Bun-Ya M, Nishimura M, Harashima S, The OY. PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. PMID:2038328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minet M, Dufour M-E LF. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1046/j.1365-313X.1992.t01-38-00999.x. [DOI] [PubMed] [Google Scholar]
- 12.Dohmen RJ, Madura K, Bartel B, The VA. N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Li T, Wu X, Zhao Z. Molecular cloning and functional analysis of a H(+)-dependent phosphate transporter gene from the ectomycorrhizal fungus Boletus edulis in southwest China. Fungal Biol. 2014;118:453–461. doi: 10.1016/j.funbio.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Zheng R, Wang J, Liu M, Duan G, Gao X, Bai S, Han Y. Molecular cloning and functional analysis of two phosphate transporter genes from Rhizopogon luteolus and Leucocortinarius bulbiger, two ectomycorrhizal fungi of Pinus tabulaeformis. Mycorrhiza. 2016;26:633–644. doi: 10.1007/s00572-016-0702-7. [DOI] [PubMed] [Google Scholar]
- 15.Secco D, Wang C, Shou H, Whelan J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012;586:289–295. doi: 10.1016/j.febslet.2012.01.036. [DOI] [PubMed] [Google Scholar]
- 16.Gu M, Chen A, Sun S, Xu G. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Mol Plant. 2016;9:396–416. doi: 10.1016/j.molp.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Garcia K, Delteil A, Conéjéro G, Becquer A, Plassard C, Sentenac H, Zimmermann S. Potassium nutrition of ectomycorrhizal Pinus pinaster: overexpression of the Hebeloma cylindrosporum HcTrk1 transporter affects the translocation of both K(+) and phosphorus in the host plant. New Phytol. 2014;201:951–960. doi: 10.1111/nph.12603. [DOI] [PubMed] [Google Scholar]
- 18.Doré J, Marmeisse R, Combier J-P GG. A fungal conserved gene from the basidiomycete Hebeloma cylindrosporum is essential for efficient ectomycorrhiza formation. Mol Plant-Microbe Interact. 2014;27:1059–1069. doi: 10.1094/MPMI-03-14-0087-R. [DOI] [PubMed] [Google Scholar]
- 19.Guerrero‐Galán C, Delteil A, Garcia K, Houdinet G, Conéjéro G, Gaillard I, Sentenac H, Zimmermann SD. Plant potassium nutrition in ectomycorrhizal symbiosis: properties and roles of the three fungal TOK potassium channels in Hebeloma cylindrosporum. Environ Microbiol. 2018;20:1873–1887. doi: 10.1111/1462-2920.14122. [DOI] [PubMed] [Google Scholar]
- 20.Guerrero-Galán C, Garcia K, Houdinet G, Zimmermann SD. HcTOK1 participates in the maintenance of K+ homeostasis in the ectomycorrhizal fungus Hebeloma cylindrosporum, which is essential for the symbiotic K+ nutrition of Pinus pinaster. Plant Signal Behav. 2018; doi: 10.1080/15592324.2018.1480845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia K, Delaux P-M, Cope KR, Ané J-M. Molecular signals required for the establishment and maintenance of ectomycorrhizal symbioses. New Phytol. 2015;208:79–87. doi: 10.1111/nph.13423. [DOI] [PubMed] [Google Scholar]
- 22.Torres‐Aquino M, Becquer A, Le GC, Louche J, Amenc LK, Staunton S, Quiquampoix H, Plassard C. The host plant Pinus pinaster exerts specific effects on phosphate efflux and polyphosphate metabolism of the ectomycorrhizal fungus Hebeloma cylindrosporum: a radiotracer, cytological staining and 31P NMR spectroscopy study. Plant Cell Environ. 2017;40:190–202. doi: 10.1111/pce.12847. [DOI] [PubMed] [Google Scholar]


