ABSTRACT
Granulocyte-macrophage colony-stimulating factor (GM-CSF) is a hematopoietic growth factor that can stimulate a variety of cells, but its overexpression leads to excessive production and activation of granulocytes and macrophages with many pathogenic effects. This cytokine is a therapeutic target in inflammatory diseases, and several anti-GM-CSF antibodies have advanced to Phase 2 clinical trials in patients with such diseases, e.g., rheumatoid arthritis. GM-CSF is also an essential factor in preventing pulmonary alveolar proteinosis (PAP), a disease associated with GM-CSF malfunction arising most typically through the presence of GM-CSF neutralizing auto-antibodies. Understanding the mechanism of action for neutralizing antibodies that target GM-CSF is important for improving their specificity and affinity as therapeutics and, conversely, in devising strategies to reduce the effects of GM-CSF auto-antibodies in PAP. We have solved the crystal structures of human GM-CSF bound to antigen-binding fragments of two neutralizing antibodies, the human auto-antibody F1 and the mouse monoclonal antibody 4D4. Coordinates and structure factors of the crystal structures of the GM-CSF:F1 Fab and the GM-CSF:4D4 Fab complexes have been deposited in the RCSB Protein Data Bank under the accession numbers 6BFQ and 6BFS, respectively. The structures show that these antibodies bind to mutually exclusive epitopes on GM-CSF; however, both prevent the cytokine from interacting with its alpha receptor subunit and hence prevent receptor activation. Importantly, identification of the F1 epitope together with functional analyses highlighted modifications to GM-CSF that would abolish auto-antibody recognition whilst retaining GM-CSF function. These results provide a framework for developing novel GM-CSF molecules for PAP treatment and for optimizing current anti-GM-CSF antibodies for use in treating inflammatory disorders.
Keywords: Auto-antibodies, anti-GM-CSF therapeutics, Beta common cytokines, GM-CSF, GM-CSF mutations, pulmonary alveolar proteinosis, X-ray crystallography
Introduction
Granulocyte-macrophage colony-stimulating factor (GM-CSF) is a cytokine that was originally identified as a hematopoietic growth factor that stimulated proliferation of myeloid cells from bone marrow progenitors.1 It can be produced by a wide variety of cell types including activated T-cells, B-cells, macrophages, endothelial cells, fibroblasts and a number of tumor cell types (reviewed in Ref. 2–4). GM-CSF is now recognized as an immune modulatory cytokine due to its involvement in a range of pro-inflammatory functions, such as differentiation, adhesion, chemotaxis, and activation of multiple inflammatory and immune cells,5–7 such as monocytes, macrophages, neutrophils, microglia and dendritic cells. GM-CSF also plays a critical and non-redundant role in the maturation of alveolar macrophages in the lung.8–11
GM-CSF signals via a heterodimeric cell surface receptor that consists of a GM-CSF-specific alpha subunit, GMRα, and the major signaling subunit called beta common (βc) that is shared with the receptors for interleukin (IL) −3 and IL-5.2 Activation of the GM-CSF receptor is initiated when GM-CSF binds to GMRα (via the Site 1 interface) to form a low-affinity binary complex with a KD of ~ 10 nM.12 The binary complex then interacts with the βc subunit to form a high-affinity ternary complex with a KD of ~ 0.1 nM12,13 and a hexameric configuration (two GM-CSF:two GMRα:two βc monomers) that is a result of the symmetrical and homodimeric nature of βc.14,15 The active GM-CSF receptor complex further assembles into a dodecamer configuration, and possibly higher order complexes, for the initiation of receptor signaling through activation of the JAK-STAT pathway.13,16
While GM-CSF is not needed for steady-state myelopoiesis, overexpression of GM-CSF is associated with several human pathologies such as rheumatoid arthritis (RA), multiple sclerosis (MS), juvenile myelomonocytic leukemia and chronic myelomonocytic leukemia.2,3,17 As such, GM-CSF is a promising target for therapeutic intervention in a number of these conditions including RA, MS and asthma.18–24 GM-CSF activity can be inhibited by targeting GM-CSF itself or by targeting the GM-CSF receptor complex. Antibodies against relevant targets that are currently in clinical development include anti-GM-CSF antibodies (MOR103/GSK3196165, namilumab/IZN-101/MT203, lenzilumab/KB003, gimsilumab/MORAB-022)19–22,24 and mavrilimumab/CAM-3001 (specific for GMRα).25,26
The constitutive or sustained absence of GM-CSF activity results in defective maturation of alveolar macrophages and accumulation of surfactant in the alveolar spaces, a condition called pulmonary alveolar proteinosis (PAP). PAP patients suffer respiratory insufficiency and increased susceptibility to infections.10,11,27,28 Hereditary forms of PAP are associated with mutations in the genes encoding the GM-CSF receptor,8,9,29–33 while idiopathic autoimmune PAP is characterized by high levels of anti-GM-CSF auto-antibodies.34–36 Interestingly, the presence of single GM-CSF neutralizing monoclonal antibodies (mAbs) may not be harmful, and a recent study showed that, while single antibodies may potently neutralize GM-CSF activity in vitro, they may not be effective in reducing the amount of bioavailable GM-CSF in vivo.37 This study also showed that a combination of three or more non-cross competing antibodies completely neutralized GM-CSF activity in vitro and in vivo more efficiently than the single anti-GM-CSF antibodies that are currently being developed for treatment of autoimmune and inflammatory diseases. In order to identify potent, non-cross competing antibodies, it is necessary to determine the binding epitope and establish the molecular mechanism by which each antibody blocks GM-CSF bioactivity. Until recently the interaction epitopes of GM-CSF neutralizing antibodies were largely unknown, and their mechanism of action not resolved.
In 2012, Blech et. al., provided the first structural description of a high-affinity GM-CSF neutralizing auto-antibody, MB007, isolated from a patient with PAP.38 Their computational model of the GM-CSF:MB007 antigen-binding fragment (Fab) complex revealed only a small overlap between GMRα and the MB007 Fab when bound to GM-CSF. Microscale thermophoresis and surface plasmon resonance in vitro binding experiments as well as cell proliferation assays demonstrated that, although MB007 reduced the binding of GM-CSF to GMRα, MB007 did not prevent formation of a functional GM-CSF receptor complex.38 More recently, Eylenstein et al., published the first crystal structures of four related human anti-GM-CSF antibodies (the MOR series) in complex with GM-CSF. These antibodies were derived from a phage display library and subjected to in vitro affinity maturation to produce MOR103, a high-affinity neutralizing antibody against GM-CSF that is currently in clinical trials.39
In our previous study we characterized 19 mAbs against GM-CSF that were isolated from patients with PAP.36 All of these auto-antibodies specifically neutralized GM-CSF bioactivity and we identified five distinct groups of auto-antibodies with non-overlapping GM-CSF binding epitopes. Amongst these antibodies, the F1 auto-antibody was identified as the most effective at neutralizing GM-CSF bioactivity on a human erythroleukaemic cell line. In parallel, we also observed that the binding epitope of 4D4, a mouse mAb raised against human GM-CSF, overlapped with the binding epitope of several auto-antibodies isolated from PAP patients.36 To understand the mechanism of action of GM-CSF neutralizing antibodies and to identify their binding epitopes, we structurally characterized binary complexes of F1 and 4D4 Fabs bound to GM-CSF.
Our crystal structures of human GM-CSF in complex with the F1 and 4D4 Fabs reveal that these antibodies have non-overlapping binding sites on GM-CSF. While the 4D4 Fab binds at almost the same site as the MOR series of antibodies, the auto-antibody F1 Fab binds to a distinct GM-CSF epitope. Through functional analyses of GM-CSF residues forming the epitopes of these two antibodies, we found evidence for dissociation of GM-CSF residues required for antibody binding from those driving receptor binding and signaling. Thus, the structural characterization of GM-CSF:antibody complexes may assist in the development of GM-CSF variants that are partially, or completely, resistant to neutralizing auto-antibodies in patients with PAP.40 The data presented herein provides insight into the mechanism of action for anti-GM-CSF antibodies that are being developed to treat inflammatory disorders and provide a framework for developing novel GM-CSF molecules for the treatment of autoimmune PAP.
Results
Crystal structures of GM-CSF in complex with F1 and 4D4 Fabs
Binary complexes of the GM-CSF:F1 Fab and GM-CSF:4D4 Fab were purified, crystallized and the structure of the complexes determined (see Materials and Methods and Figs. S1–S3). The crystal structures of the GM-CSF:F1 and the GM-CSF:4D4 Fab complexes were solved at 2.6 Å and 2.0 Å resolution, respectively. The final data collection and refinement statistics are summarized in Table 1.
Table 1.
Summary of the data collection and refinement statistics.
| GM-CSF:F1 Fab complex |
GM-CSF:4D4 Fab complex |
|
|---|---|---|
| Data collection | Australian Synchrotron (MX2) | Australian Synchrotron (MX2) |
| Space group | P1 | P21 |
| Cell dimensions (Å; °) | 95.4,100.5,101.9; 91.1,117.9,108.6 |
70.5, 48.1, 77.6; 90.0, 99.9, 90.0 |
| Resolution (Å) | 50–2.6 (2.64–2.60)* | 40.0–2.0 (2.05–2.00)* |
| Rpim | 0.13 (0.82) | 0.045 (0.36) |
| I/σI | 39.0 (2.0) | 12.4 (2.3) |
| Completeness (%) | 98.7 (93.4) | 99.4 (99.0) |
| Redundancy | 4.1 (2.7) | 3.7 (3.8) |
| Wilson B factor (Å2) | 38.6 | 29.5 |
| CC ½ | 0.96 (0.34) | 0.99 (0.77) |
| Refinement | ||
| Resolution (Å) | 50–2.6 | 40–2.0 |
| Unique reflections | 94,668 (4471) | 34,696 (2557) |
| Rwork/Rfree | 0.23/0.26 | 0.18/0.24 |
| Total no. of atoms | ||
| Protein | 16,027 | 4128 |
| Water | 193 | 116 |
| Average B-factors | ||
| Protein | 48.9 | 35.5 |
| Water | 28.9 | 34.0 |
| R.M.S deviations | ||
| Bond lengths (Å) | 0.015 | 0.018 |
| Bond angles (°) | 1.70 | 1.82 |
*Values in parentheses are for highest resolution shell.
F1 Fab recognizes the β2-strand on GM-CSF
The crystal structure of the GM-CSF:F1 Fab complex (Figure 1 and Fig. S1A) was solved in the P1 space group with four copies of the antigen-antibody complex in the asymmetric unit. The four copies are almost identical, with the root-mean-square deviation (rmsd) over all Cα atoms < 0.2 Å. The overall fold of GM-CSF in the F1 Fab complex is almost identical (rmsd ~ < 0.6 Å) to the crystal structure of apo GM-CSF (PDB ID: 2GMF)41 and GM-CSF in a binary complex with its GMRα receptor subunit (PDB ID: 4RS1),12 indicating that binding of the auto-antibody F1 Fab does not result in any conformational changes in GM-CSF. The surface complementarity of the GM-CSF:F1 Fab interface (Sc = 0.77)42 and the relatively large total buried surface area of GM-CSF when complexed to the F1 Fab of 915 Å2 is consistent with the tight binding affinity of the complex (KD of 120 pM).36 Heavy chain interactions at the contact surface dominate, with the heavy chain contributing to a buried surface area of 641 Å2 and the light chain contributing to a buried surface area of 274 Å2. Electron density at the complementarity-determining region (CDR) loops was unambiguous for both the heavy and light chains of the F1 Fab.
Figure 1.
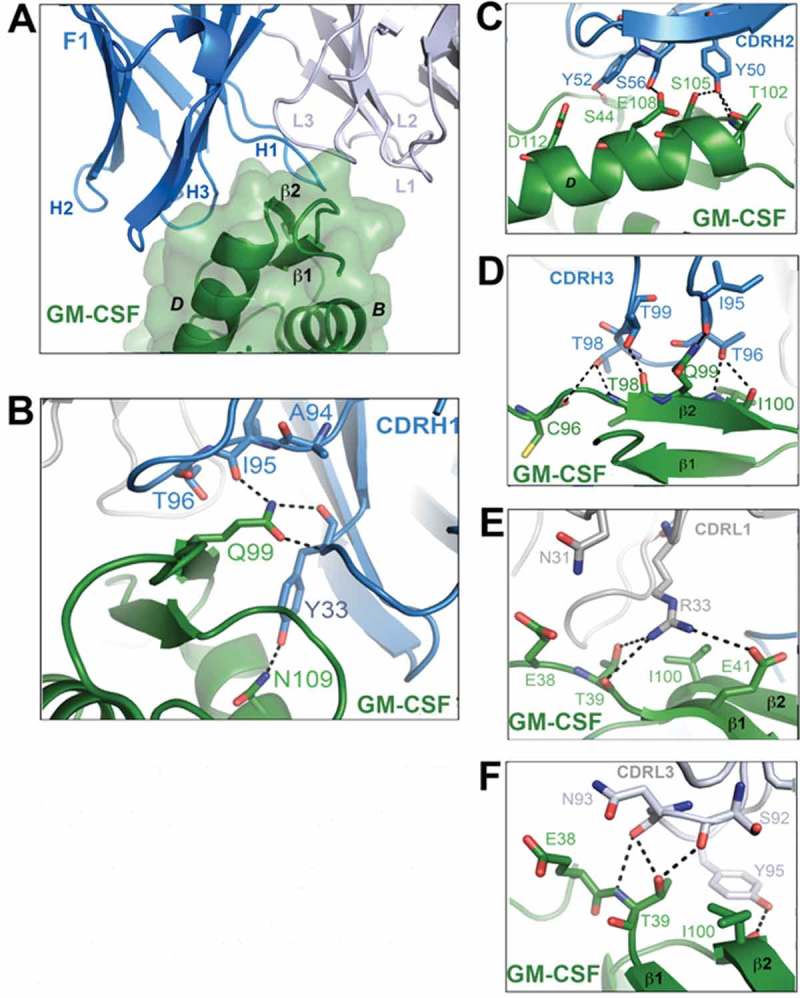
Crystal structure of the GM-CSF:F1 Fab complex. (A) A close-up view of the binding interface between GM-CSF and the F1 Fab. The F1 Fab molecule is shown in cartoon representation with the heavy chain colored blue and the light chain grey. GM-CSF is colored green and shown as a cartoon with a transparent molecular surface overlay. Key interactions made by the CDR loops of the F1 Fab with GM-CSF are shown in (B)–(F). Hydrogen bond interactions are depicted as black broken lines and the GM-CSF alpha helices are labeled in black type. For figure clarity, in (A) the six CDR loops are labeled H1, H2, H3, L1, L2 and L3. The complete view of the GM-CSF:F1 Fab complex is provided in Fig. S1A.
Based on the guidelines available for identifying antibody CDRs using the Kabat numbering system (http://www.bioinf.org.uk/abs/),43 the six F1 Fab CDR loops were identified to be: H1 (residues 26–33), H2 (residues 50–65), H3 (residues 95 −101), L1 (residues 25–35), L2 (residues 51–57) and L3 (residues 90–98) (Fig. S2). The crystal structure of the GM-CSF:F1 Fab complex reveals that the four helical (A-D) GM-CSF bundle engages within a broad groove along the CDR H1 and CDR H3 interface, primarily via insertion of the β2-strand of GM-CSF between the CDR H1 and CDR H3 loops (Figure 1A). Insertion of the GM-CSF β2-strand results in the projection of the GM-CSF Q99 side chain deep into the heavy chain CDR loops of the F1 Fab (Figure 1B, D). This single GM-CSF residue forms a network of hydrogen bonds and van der Waals interactions with residues I95, T96, T99 in the CDR H3 loop of the F1 Fab (Figure 1D), and Y32 and Y33 in the CDR H1 loop (Figure 1B). The CDR H2 loop residues Y50, Y52 and S56 make key interactions with GM-CSF residues T102, S105, S44 and E108 (Figure 1C). Among the light chain CDRs, residues in CDR L1 and CDR L3 form salt bridge/hydrogen bonding interactions with residues in GM-CSF while residues in CDR L2 do not make any contact with GM-CSF (details of the specific interactions are described in the Supplementary data section, Figure 1E,F). A total of 15 hydrogen bonds (3 from the light chain CDRs and 12 from the heavy chain CDRs) as well as a salt bridge are formed at the GM-CSF:F1 Fab interface (Figure 1B-F and Table 2).
Table 2.
Summary of the key interacting residues between GM-CSF and the heavy and light chain CDRs of the F1 and 4D4 Fabs.
| GM-CSF | F1 Heavy Chain | GM-CSF | 4D4 Heavy Chain |
|---|---|---|---|
| S44 | K31, Y52• | V16 | Y32 |
| E45 | K31, Y52 | Q20 | S28 |
| M46 | R30•, Y52, A53, S54 | R23 | N31• |
| C96 | T98• | L49 | Y33, W50, F52 |
| A97 | T98 | Q50 | W50, N56 |
| T98 | T97, G96, T98•, T99• | H83 | T97 |
| Q99 | Y32, Y33•, I95•, T96, T99 | H87 | T98 |
| I100 | T96•, G97 | L114 | N31 |
| I101 | Y33, Y50 | L115 | N31• |
| T102 | Y50•, F58 | V116 | F52 |
| E104 | S57 | I117 | N31• |
| S105 | Y50•, S56, S57 | P118 | N31•, Y33, F52 |
| E108 | S56• | F119 | S28, N31•, Y32 |
| N109 | Y33• | D120 | N31, Y32, Y33•, G96, T97 |
| D112 | S54 | C121 | Y33, K95•, T97•, T98 |
| W122 | Y33 | ||
| |
|
E123 |
K95••, W50 |
| GM-CSF |
F1 Light Chain |
GM-CSF |
4D4 Light Chain |
| N37 | N94 | Q86 | K50• |
| E38 | N31, N93 | C88 | W32 |
| T39 | S92•, N93•, Y95 | P89 | W32 |
| E41 | R33•• | E123 | G91, Y94• |
| I100 | R33, S92, Y95• | P124 | W32, G91, Q92 |
| I101 | Y95 | ||
| T102 | Y95 |
Residues making van der Waals contacts within 4 Å are listed in the table. Residues making hydrogen bonds are represented with a • symbol and those involved in salt bridge formation are marked with the •• symbol. GM-CSF residues that interact with GMRα are highlighted in grey. CDR H1 and CDR L1 residues are colored red, CDR H2 and CDR L2 residues blue, CDR H3 and CDR L3 residues black.
The heavy chain CDR loops drive the interaction between the 4D4 Fab and GM-CSF
The crystal structure of the GM-CSF:4D4 Fab complex (Figure 2 and Fig. S1B) was solved in the P21 space group with one copy of the antigen-antibody complex in the asymmetric unit. The electron density map at the GM-CSF:4D4 Fab interface is continuous and unambiguous. The total buried surface area of GM-CSF within the 4D4 Fab epitope is 881 Å2 and Sc is 0.72 for the GM-CSF:4D4 Fab interface. Like the auto-antibody F1 Fab, the 4D4 heavy and light chain contributions to the contact surface area are unequal: the heavy chain CDRs dominate interactions, contributing to a buried surface area of 614 Å2 and the light chain CDRs contributing to a buried surface area of 267 Å2. A structural comparison of GM-CSF from the 4D4 Fab complex with apo GM-CSF (PDB ID: 2GMF),41 GM-CSF in complex with GMRα (PDB ID: 4RS1)12 and GM-CSF from the auto-antibody F1 Fab complex (this work) showed that complexation with the 4D4 Fab did not result in any conformational change within the cytokine (rmsd < 0.7 Å over all Cα atoms).
Figure 2.
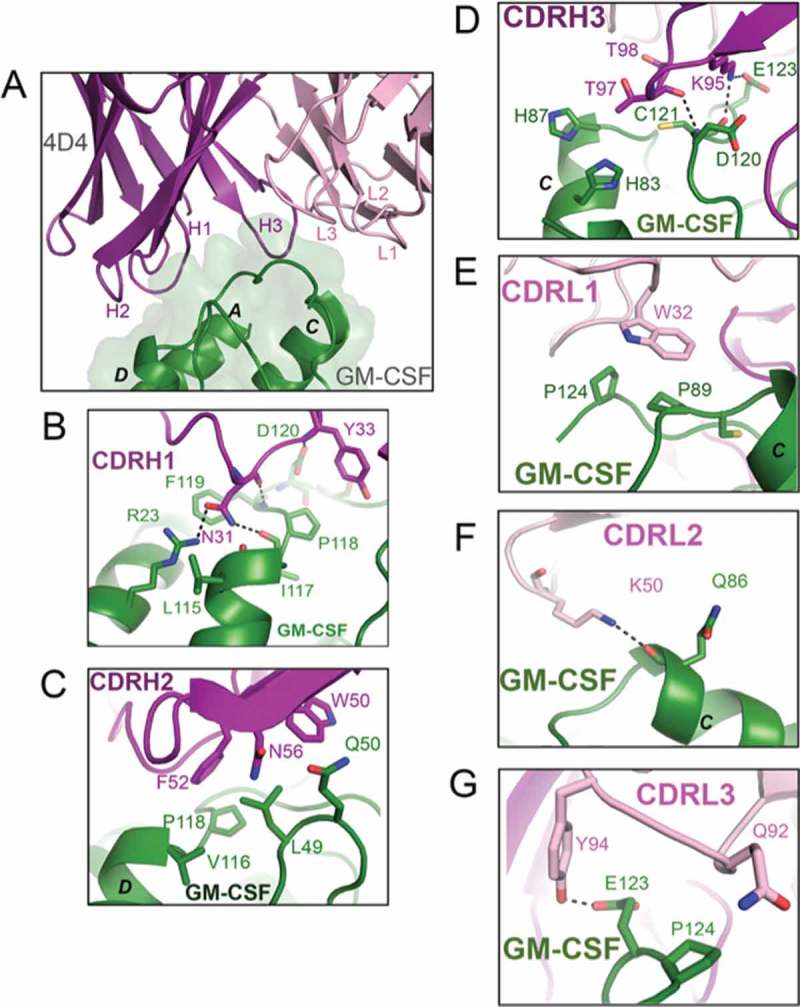
Crystal structure of the GM-CSF: 4D4 Fab complex. (A) A close-up view of the binding interface between GM-CSF and the 4D4 Fab. The 4D4 Fab molecule is shown in cartoon representation with the heavy chain colored purple and the light chain light pink. GM-CSF is colored green and shown as a cartoon with a transparent molecular surface overlay. Key interactions made by the six CDR loops of the 4D4 Fab with GM-CSF are shown in (B)–(G). Hydrogen bond interactions are depicted as black broken lines and the GM-CSF alpha helices are labeled in black type. For figure clarity, in (A) the six CDR loops are labeled H1, H2, H3, L1, L2 and L3. The complete view of the GM-CSF:4D4 Fab complex is provided in Fig. S1B.
The six 4D4 Fab CDR loops were identified, using the same guidelines described above, to be: H1 (residues 26–35), H2 (residues 50–61), H3 (residues 95–102), L1 (residues 24–34), L2 (residues 50–56) and L3 (residues 89–97). Compared to the F1 Fab, the 4D4 Fab makes fewer interactions with GM-CSF, while residues on CDR H2 and CDR L1 are predominantly involved in van der Waals interactions with residues in GM-CSF. Residues on the heavy chain CDR H1 and CDR H3, as well as light chain CDR L2 and CDR L3, contribute to a total of 10 hydrogen bonds (8 from the heavy chain and 2 from the light chain). Key GM-CSF residues involved in hydrogen bond interactions with the heavy chain CDRs in 4D4 Fab include, R23, L115, I117, P118, F119, D120, C121 and E123. GM-CSF residues Q86 and E123 are involved in hydrogen bond interactions with light chain CDRs in 4D4 Fab (Figure 2B-G). These interactions are described in detail in the Supplementary data section and listed in Table 2.
Structures reveal that both antibodies block GM-CSF interaction with GMRα but through distinct epitopes
Superimposition of the GM-CSF:F1 and GM-CSF:4D4 Fab complexes via the cytokine reveals that the binding epitopes of the two Fabs are distinct and have no overlap (Figure 3A). The binding epitope of the auto-antibody F1 Fab on GM-CSF covers residues from the AB loop (residues 37–39 and 44–46, Figure 1B, E, F), CD loop (residues 96–98, Figure 1D), helix D (residues 104, 105, 108, 109 and 112, Figure 1C) and the β2-strand (residues 99–102, Figure 1D, E, F) whereas the 4D4 Fab epitope predominantly covers residues on the C-terminal end of helix D (residues 114–124, Figure 2B, C, E, G) as well as residues on helix A (residues 16, 20 and 23, Figure 2B), the AB loop (residues 49 and 50, Figure 2C) and the CD loop (residues 83, 86–89, Figure 2D-F). We also compared the binding epitopes of the F1 and 4D4 Fabs with the four GM-CSF:MOR series Fab complex structures (PDB IDs: 5C7X, 5D70, 5D71 and 5D72).39 When aligned via GM-CSF, the binding epitopes of all four MOR series Fabs partially overlap with the binding epitope of the 4D4 Fab (Fig. S4A).
Figure 3.
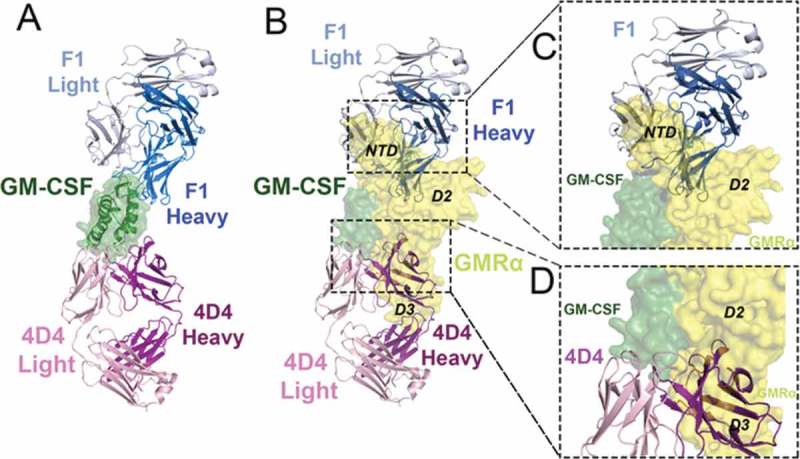
The F1 and 4D4 Fabs bind to opposed surfaces of GM-CSF but both disrupt GM-CSF interaction with GMRα. (A) Cartoon representation of the GM-CSF:F1 Fab and GM-CSF:4D4 Fab structures superimposed via GM-CSF (shown as a cartoon with a transparent molecular surface overlay). Molecules are colored; F1 Fab heavy chain (blue), F1 light chain (grey), 4D4 heavy chain (purple), 4D4 light chain (light pink) and GM-CSF (green). (B) The F1 and 4D4 Fab complexes from (A) were superimposed, via GM-CSF, with the GM-CSF:GMRα binary complex structure (PDB ID: 4RS1).10 The GM-CSF:GMRα binary complex is shown as a molecular surface, with GMRα in yellow. (C) Close-up view of the steric clash between the F1 Fab heavy and light chain variable domains with the GMRα NTD and D2 in the GM-CSF:GMRα binary complex. (D) Close-up view of the steric clash between the 4D4 Fab heavy chain variable domains with GMRα D2 and D3 in the GM-CSF:GMRα binary complex.
To gain an understanding of how these antibodies neutralize GM-CSF function, we aligned the structures of the GM-CSF:F1, GM-CSF:4D4 and the GM-CSF:MOR series Fab complexes to the GM-CSF binary complex (PDB ID: 4RS1),12 via the GM-CSF molecule. The alignment revealed extensive overlap between all six bound Fabs and GMRα (Figure 3B and Fig. S4B, C). The F1 Fab predominantly hinders the interaction between GM-CSF and the GMRα N-terminal domain (NTD) and, to a minor extent, the interaction with GMRα domain 2 (D2) (Figure 3B, C). In contrast, the 4D4 Fab and the MOR series of Fabs have extensive clashes with domain 3 of GMRα (D3) (Figure 3B, D and Fig. S4B, C). The steric clashes observed in these alignments indicates that the binding of Fabs F1, 4D4 and the MOR series are all likely to neutralize GM-CSF activity by preventing the cytokine from binding to GMRα, the initial step required for GM-CSF receptor activation.
Is the GMRα NTD interaction with GM-CSF a critical target for F1 Fab inhibition of GM-CSF function?
Structurally, the auto-antibody F1 Fab appears to block GM-CSF receptor activation by binding to the β2-strand of the cytokine, thereby preventing interaction with the GMRα NTD. In order to define the key interactions, we investigated the role of the GMRα NTD in GM-CSF target recognition by the F1 Fab. Although the GMRα NTD is required for maximal GM-CSF function, cells expressing GMRα lacking the NTD (GMRα ΔNTD) respond to GM-CSF, but with reduced potency.44 We speculated that cells expressing GMRα ∆NTD would be resistant to the neutralization of GM-CSF activity by the F1 Fab. We used CTL-EN/IL3Rα/βc cell lines transduced to express full-length GMRα or GMRα ∆NTD to assess the ability of the F1 Fab to block GM-CSF-mediated cell proliferation. GM-CSF stimulated proliferation of both cell types, but was considerably less active on cells expressing truncated GMRα (GMRα EC50 = 0.002 ± 0.001 ng/mL vs GMRα ∆NTD EC50 = 6.7 ± 1.5 ng/mL, Figure 4A). The CTL-EN/IL3Rα/βc cell lines also proliferate in response to IL-3 stimulation due to expression of the IL-3 receptor alpha-subunit (IL3Rα). In control experiments, IL-3-mediated proliferation of both cell types was identical (GMRα EC50 = 0.004 ± 0.001 ng/mL vs GMRα ∆NTD EC50 = 0.007 ± 0.003 ng/mL, Figure 4A). The F1 Fab was titrated against a near EC50 concentration of GM-CSF or IL-3 for each cell line, and cell proliferation was assessed. We observed robust inhibition of GM-CSF-mediated proliferation on cells expressing full-length GMRα (IC50 = 0.009 ± 0.006 μg/mL) and very weak, partial inhibition of GM-CSF-mediated proliferation on cells expressing GMRα ∆NTD (Figure 4B). There was no inhibition of IL-3-mediated proliferation for either cell line by F1 Fab (Figure 4B). Importantly, there was no statistically significant difference in GM-CSF- or IL-3-mediated proliferation of cells expressing GMRα ∆NTD in the presence of F1 Fab up to a concentration of 100 μg/mL. It is apparent from these experiments that the presence of the GMRα NTD is required to facilitate the blockade of GM-CSF-mediated function by the F1 Fab.
Figure 4.
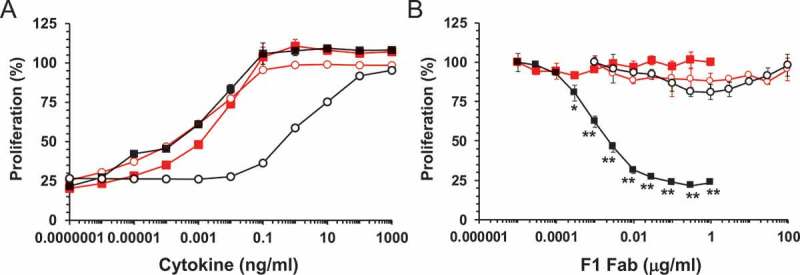
F1 disruption of functional GM-CSF interaction with GMRα. (A) CTL-EN cells stably expressing IL3Rα and βc with either wild type GMRα, CTL-EN/IL3Rα/βc/GMRα (■, ■) or the GMRα ∆NTD truncation mutant, CTL-EN/IL3Rα/βc/GMRα ∆NTD (○, ○) were stimulated with a titration of GM-CSF (black) or IL-3 (red) for 40 hours and cell proliferation determined. Data are plotted as a percentage of maximum proliferation in response to mIL-2 stimulation and are the mean of triplicate determinations from a representative experiment (n = 6). Error bars represent SEM. (B) As for (A) but cells were pre-incubated with titrations of F1 Fab for 1 hour followed by GM-CSF stimulation at 0.03 ng/mL (■) or 10 ng/mL (○) or IL-3 stimulation at 0.003 ng/mL (■, ○). Data are plotted as a percentage of maximum proliferation in the absence of F1 Fab and are the mean of triplicate determinations from a representative experiment (n = 4). Statistical significance of differences in functional response to F1 Fab treatment between GM-CSF and IL-3 stimulated CTL-EN/IL3Rα/βc/GMRα or CTL-EN/IL3Rα/βc/GMRα ∆NTD were determined by 2-way ANOVA with Sidak’s multiple comparisons test and are shown as asterisks, * p < 0.001 ** p < 0.0001.
Identification of GM-CSF residues in the F1 and 4D4 epitopes that are required for antibody neutralization
Although the F1 and 4D4 Fabs have distinct and non-overlapping binding sites on GM-CSF (Figure 3A), both of these antibodies will interfere with the cytokine’s engagement of GMRα (the interface between the cytokine and its alpha receptor subunit is defined as Site 1).3 We previously characterized the biological activity of GM-CSF variants that target GMRα Site 1,12 and a number of these GM-CSF residues, including E45, Q99, D112, L115 and F119, form part of the F1 or 4D4 Fab epitopes (Figure 5A, Table 2 and Table S1). To determine the contribution of these GM-CSF residues to the neutralizing activity of the F1 or 4D4 Fabs, we assessed the ability of the two antibodies to block TF-1 cell proliferation mediated by EC50 concentrations of wild type GM-CSF or selected Site 1 GM-CSF mutants (Figure 5B, C and Table S1).
Figure 5.

Disruption of F1 and 4D4 Fab epitope residues and the development of antibody resistance. (A) Cartoon representation of the GM-CSF:F1 Fab and GM-CSF:4D4 Fab structures that have been superimposed via GM-CSF (shown as a molecular surface). Molecules are colored: F1 Fab heavy chain (blue), F1 light chain (grey), 4D4 heavy chain (purple), 4D4 light chain (light pink), and GM-CSF (green). GM-CSF residues in the F1 and 4D4 Fab epitopes that contribute to GMRα interactions are labeled and shown in red while Q99 (shown in white and indicated by the arrow), does not contribute to GMRα interactions. (B) TF-1 cells were pre-incubated with the F1 Fab for 1 hour followed by stimulation with an EC50 concentration of wild type GM-CSF (0.02 ng/mL, ■) or the GM-CSF variants E45A (0.03 ng/mL, ○), E45K (0.05 ng/mL, ●), Q99A (0.019 ng/mL, ●), Q99E (0.018 ng/mL, ○), D112A (0.03 ng/mL, □) or D112K (0.17 ng/mL, ■). Data are plotted as a percentage of maximum proliferation in the absence of antibody and are the mean of triplicate determinations from a representative experiment (n = 3–4). Error bars represent SEM. (C) As for (B) but TF-1 cells were pre-incubated with intact 4D4 mAb for 1 hour followed by stimulation with an EC50 concentration of wild type GM-CSF (0.02 ng/mL, ■) or the GM-CSF variants L115A (0.02 ng/mL, ○), F119A (0.13 ng/mL, ■) or L115A, F119A (1.47 ng/mL, ○).
We observed that GM-CSF mutations E45K (IC50 = 0.09 ± 0.01 μg/mL) and D112K (IC50 = 0.26 ± 0.06 μg/mL) have a modest effect on the ability of the F1 Fab to block GM-CSF function (Figure 5B and Table S1) compared to wild type GM-CSF (IC50 = 0.033 ± 0.004 μg/mL). GM-CSF residues E45 and D112 make a significant contribution to GMRα binding affinity,12 but only a modest contribution to GM-CSF function (Table S1). In contrast, mutation of GM-CSF Q99 (i.e., Q99A and Q99E) completely abolished the neutralizing activity of the F1 Fab (Figure 5B and Table S1) at concentrations up to 100 μg/mL, which was the maximum concentration we were able to analyze in this assay. In the GM-CSF:F1 Fab complex, the Q99 side chain reaches between the CDR H1 and CDR H3 loops, forming three hydrogen bonds and stabilizing the conformation of these two loops (Figure 1B, D). Significantly, although GM-CSF Q99 is located at the Site 1 interface (Figs. 1B and 5A), mutation of this residue has a modest impact upon GMRα binding (2–7 fold reduction)12 and no impact on GM-CSF function (Table S1).
The neutralizing activity of Fab 4D4 is markedly inhibited by the GM-CSF F119A mutation (IC50 = 37.1 ± 5.4 μg/mL) compared to wild type GM-CSF (IC50 = 1.15 ± 0.17 μg/mL). Although F119A decreases GMRα binding affinity by at least two orders of magnitude,12 it only makes a modest contribution to GM-CSF function (Figure 5C and Table S1). In contrast, the GM-CSF L115A mutant also has significantly decreased GMRα binding affinity (by ~ two orders of magnitude),10 yet this mutation has no effect upon GM-CSF function or Fab 4D4 neutralizing activity (IC50 = 0.44 ± 0.09 μg/mL) (Figure 5C and Table S1). Thus, mutation of GM-CSF reveals residues required for optimal neutralization by the F1 and 4D4 antibodies, and yet not all of these residues play a significant role in receptor binding and/or signaling.
Discussion
To form an active signaling complex, GM-CSF initially binds to GMRα to form a low-affinity (KD ~ 10 nM) binary complex12 that is not considered capable of downstream JAK-STAT signaling. The binary complex then associates with βc to form a high-affinity (KD ~ 0.1 nM) ternary complex that assembles into higher-order complexes required for activation of the JAK-STAT pathway and initiation of the GM-CSF signaling cascade.12,13 Although the total surface area of GM-CSF buried within GMRα (1,175 Å2) in the binary complex is more expansive than the total surface area of GM-CSF buried in either the GM-CSF:F1 or GM-CSF:4D4 Fab complexes (915 Å2 and 881 Å2, respectively), both these Fabs inhibit GM-CSF bioactivity by obstructing formation of the binary complex, thereby effectively preventing the formation of a ternary complex and subsequent higher-order signaling complexes. Despite this, the 4D4 mAb is a relatively weak antagonist of GM-CSF function (IC50 = 1.15 ± 0.17 μg/mL) compared to the F1 Fab (IC50 = 0.033 ± 0.004 μg/mL), which is also known to bind GM-CSF with very high affinity (KD = 120 pM).36 These differences in potency may arise because the CDR loops of the 4D4 Fab make a limited number of hydrogen bonds and van der Waals interactions with GM-CSF compared to the interactions formed by the CDR loops of the F1 Fab.
The F1 and 4D4 Fabs predominantly block engagement of GM-CSF with GMRα; however our structural analysis of their binding epitopes suggests that they do so by binding to different sites on GM-CSF. Superimposition (via the cytokine) of the GM-CSF:F1 Fab complex and GM-CSF ternary complex (PDB ID: 4NKQ) suggested that the F1 Fab primarily blocks the interaction between GM-CSF and the GMRα NTD, and to some extent the interaction with GMRα D2 (Fig. S5). Superimposition of the GM-CSF:4D4 Fab complex and the GM-CSF ternary complex via the cytokine suggests that the 4D4 Fab predominantly blocks interaction between GM-CSF and GMRα D2 and D3 (Figure 3B, D and Fig. S5). Moreover, the presence of the 4D4 Fab will sterically interfere with the βc receptor subunit’s engagement of GM-CSF whereas the F1 Fab should not. Since the epitope of the 4D4 Fab partially overlaps with the epitope of the MOR series of Fabs,39 these antibodies are likely to have a similar mechanism of action (Fig. S4). Our results show that inhibition of GMRα binding is an effective mechanism of GM-CSF antagonism mediated by GM-CSF-specific antibodies. Other mechanisms of antagonism are also likely to be effective, including those that principally inhibit GM-CSF interactions with βc.45
The F1 and 4D4 antibodies were selected by nature or by experimentation, respectively, on the basis of their ability to inhibit GM-CSF function, but it is interesting to note that at least one other GM-CSF inhibitory molecule occurs in nature and has a completely distinct origin. The GM-CSF/IL-2 inhibition factor (GIF) is expressed by the orf virus, and it has been shown to antagonize both ovine GM-CSF and IL-2, although interestingly not their human orthologues,46 by binding to these cytokines through mutually exclusive binding sites and is likely to act as a competitive decoy receptor.47 Structural alignment (via the cytokine) of the ovine GM-CSF:GIF complex (PDB ID: 5D28)47 with the human GM-CSF:F1 Fab, GM-CSF:4D4 Fab and GM-CSF:GMRα (PDB ID: 4RS1) complexes reveals that the GIF homodimer is co-located with the heavy chain of the 4D4 Fab and with D2 and D3 of GMRα (Fig. S6). The alignment suggests that the binding of GIF to ovine GM-CSF would physically preclude ovine GMRα from binding the cytokine. Thus, GIF appears to act as a decoy ovine GMRα receptor subunit. GM-CSF interacts, via it’s A and D alpha helices, with both GIF and GMRα (D2 and D3 domains). Although human and ovine GM-CSF share 80% sequence identity,47 there are three non-conservative residue differences on the GIF/GMRα interacting face of GM-CSF helix A (Q20, R23 and R24 in human GM-CSF are replaced by K20, L23 and S24 in ovine GM-CSF) and two residues on helix D (human GM-CSF E104 and L115 are replaced by K104 and F115 in the ovine cytokine). Based on the structural alignment, these sequence differences in the human cytokine A and D alpha helices would likely generate steric clashes with GIF, preventing it from binding, and explain the species-restricted antagonism of GIF.
In addition to their prominent role in PAP, GM-CSF auto-antibodies have been observed in cases of cryptococcal meningitis and cryptococcal gatti48,49 and have been identified as a marker of aggressive Crohn’s disease.50 While commercially available recombinant human GM-CSF has been shown to have therapeutic activity in treating patients with PAP,51 GM-CSF variants that are resistant to neutralizing auto-antibodies may be even more effective. Surface-engineered GM-CSF variants have been proposed as a means of treating patients with Crohn’s disease, and in one study, mutations of H15 and R23 in helix A were shown to reduce functional GM-CSF neutralization by goat polyclonal antisera.40 The fact that individual PAP patients express multiple GM-CSF neutralizing mAbs with non-overlapping epitopes36 would appear to make the development of resistant GM-CSF variants a difficult task, unless dominant or conserved epitopes can be identified and suitable, broadly protected GM-CSF variants developed. The introduction of amino acid changes could also conceivably enhance the immunogenicity of GM-CSF or create novel epitopes and lead to the production of additional auto-antibodies. Our functional studies have identified GM-CSF residues within the distinct epitopes of F1 or 4D4 where mutation reduces GM-CSF function and/or binding affinity for GMRα12 while also reducing sensitivity to antibody neutralization (Figure 5). For example, GM-CSF E45 and D112 both influence F1 Fab function, whereas GM-CSF F119 influences 4D4 mAb function (Table S1). Interestingly, mutations of GM-CSF Q99 have only a small effect on GMRα binding affinity (2–7 fold)12 and no effect upon GM-CSF function (Table S1), but they abolish F1 Fab neutralization activity (Figure 5B and Table S1), and as such would represent ideal GM-CSF variants to treat patients expressing the F1 auto-antibody. Through our detailed knowledge of the GM-CSF receptor complex structure12 and characterization of the epitopes for multiple GM-CSF auto-antibodies, we anticipate it will be possible to identify multiple GM-CSF residues that can be mutated to disrupt common auto-antibody epitopes without compromising GM-CSF function. Additionally, a more comprehensive understanding of the mode of action of GM-CSF auto-antibodies and their epitopes may inform the development of new anti-GM-CSF antibodies for use in treating a range of inflammatory diseases.2,3,17–24
Materials and methods
Expression and purification of the GM-CSF:Fab binary complexes
Human GM-CSF (residues A1-E127 of the mature peptide) was expressed in E. coli and purified by anion exchange chromatography and reversed phase HPLC.52,53 Recombinant human Fab F1 was expressed in E. coli SHuffle T7 cells (New England Biolab, Cat. C3026J) using the pET-Duet (Millipore-Novagen, Cat. 71146–3) expression plasmid containing the genes for the light and heavy chains of F1 Fab. The cells were grown in Super Broth media with 0.1 mg/mL ampicillin at 37°C until early log phase (OD600 = 0.3–0.4). Isopropyl-β-D-thiogalactoside was added to 0.1 mM, the temperature was reduced to 18°C, and the culture was left to incubate for 44 hours. The cells were collected by centrifugation and lysed by sonication in TBS (50 mM Tris-HCl pH 7.6, 150 mM NaCl). The Fab was purified from the soluble cell extract using KappaSelect affinity media (GE Healthcare, Cat. 17–5458-12) followed by Mono S ion-exchange chromatography (GE Healthcare, Cat. 17–5168-01), then dialysed against 20 mM Tris-HCl (pH 8.0) and concentrated to 10 mg/mL for complex formation.54 The F1 Fab was also purified using KappaSelect affinity media followed by preparative size exclusion chromatography (SEC) as described below. The anti-GM-CSF mouse mAb 4D452,53 was digested for 24 hours at 37°C using immobilized Papain (Thermo Scientific, Cat. PIE44985) and the 4D4 Fab recovered in the flow-through after passing the digest reaction over Protein A Sepharose (GE Healthcare, Cat. 17–0780-01). Binary complexes consisting of GM-CSF and F1 Fab or GM-CSF and 4D4 Fab were isolated by SEC of a 2:1 mixture of GM-CSF:Fab using a Superdex 200 column (26 mm x 600 mm, GE Healthcare, Cat. 28–9893-36) operated at 2 mL/min at 24°C with 150 mM NaCl, 50 mM sodium phosphate pH 7.0, as running buffer (Figs. S2 and S3).
Cloning and sequencing of the 4D4 heavy and light chain
RNA was prepared from 4D4 hybridoma cells using an RNeasy plus micro kit (Qiagen) and total cDNA synthesized using SuperscriptIII (Invitrogen, Cat. 18080–051). The 4D4 immunoglobulin (Ig) heavy and light chains were PCR amplified using primers specific for the 5ʹ end of the variable domains from mouse Ig heavy or kappa light chain genes55 and an oligo-dT17 3ʹ primer. PCR products from primer combinations that generated an appropriate fragment (4D4H ~ 1.5kb, 4D4L ~ 0.9kb) were sequenced directly or cloned into the pSG5 plasmid (Agilent, Cat. 216201) using NotI sites incorporated in the PCR primers and the insert from at least six independent isolates sequenced.
Amino terminal peptide sequence and respective sense primers for 4D4 heavy chain PCR:
QV(K,Q)LQ(E,Q)SG 5ʹ aggt(gc)(ac)a(ga)ctgcag(gc)agtc(at)gg 3ʹ
EVQLQQ 5ʹ gaggtccagctgcagcagtc 3ʹ
QVQLKQ 5ʹ caggtgcagctgaagcagtc 3ʹ
Amino terminal peptide sequence and respective sense primer for 4D4 kappa chain PCR:
DIQLTQS 5ʹ gacatccagctgactcagtct 3ʹ
Functional studies with GMRα mutants
DNA fragments encoding wild type human GMRα or the GMRα ∆NTD variant, were generated by PCR, cloned into the retroviral expression vector pRufHygro and recombinant retrovirus generated by co-transfection of pRufHygro:GMRα or pRufHygro:GMRα ∆NTD with the pEQ-Eco packaging plasmid56 into HEK293-T cells using Lipofectamine 2000 (Invitrogen, Cat. 11668–019). Retrovirus was used to transduce CTL-EN/IL3Rα/βc13,57 cells as previously described.12 Cell surface expression of GMRα was assessed by flow cytometry using mAbs specific for GMRα, including 4H1,58 8G659 and mAb 1037 (Millipore, Cat. MAB1037) of which only mAb 1037 was able to bind the GMRα ∆NTD variant. Cell proliferation was assessed using CellTiter 96® AQueous (Promega, Cat. G358B) following the manufacturer’s protocol (Figure 4, 5). Where indicated, CTL-EN proliferation in response to GM-CSF and IL-3 stimulation was normalized relative to proliferation in response to 100 U/mL mIL-2. For figures, error bars are standard error of the mean (SEM). Data were analyzed using GraphPad Prism to determine EC50 values. A 2-way ANOVA with Sidak’s multiple comparisons test was used to calculate the significance of differences in F1 Fab functional studies.
Mutagenesis, purification and testing of GM-CSF mutants
A synthetic human GM-CSF cDNA cloned in the pIN-III-OmpA2 expression vector was used to produce soluble GM-CSF mutants from E. coli that were subsequently purified by immunoaffinity chromatography and reversed phase HPLC as previously described.12,52 Mutants were generated by PCR-mutagenesis of a NcoI/BamHI fragment of GM-CSF that encompasses residues E14-E127. The functional activity of wild type and mutant forms of GM-CSF was assayed by measuring proliferation of the human erythroleukemia cell line, TF-160 using CellTiter 96® AQueous (Promega, Cat. G358B) following the manufacturer’s protocol. For figures, error bars are SEM. Data were analyzed using GraphPad Prism to determine EC50 values.
Protein crystallization
Prior to setting up crystallization trials, a suitable buffer for the GM-CSF:F1 Fab complex was determined using differential scanning fluorimetry (DSF). The protein was buffer exchanged into 20 mM sodium citrate pH 6 and 500 mM NaCl and concentrated to 10 mg/mL for crystallization screens. Several commercial crystallization screens (including Hampton Crystal Screen, MCSG1 and JCSG+) were tested at 10°C and 21°C. Crystals were obtained from several hit conditions and these were tested at the Australian Synchrotron (Clayton, Victoria). Finally, diffraction data were collected on crystals that grew at 10°C in 0.16 M calcium acetate, 14.4% PEG 8000, 0.08 M sodium cacodylate pH 6.5 and 20% glycerol from the JCSG+ screen.
The GM-CSF:4D4 Fab complex was buffer optimized using DSF and buffer exchanged into a buffer containing 20 mM Tris pH 9 and 50 mM NaCl. The protein complex was concentrated to 13–20 mg/mL and crystallization screens were conducted using the Hampton Crystal Screen, PEG+ Suite and the JCSG+ screens at 10°C and 21°C. A hit was obtained in the JCSG+ screen condition consisting of 0.2 M NaCl, 20% PEG 8000 and 0.1 M phosphate citrate buffer pH 4.5 at 10°C. These crystals were cryo-protected in 20% ethylene glycol and data were collected at the Australian Synchrotron.
Data processing and structure determination
Data collection was controlled using Blue-Ice software for all work at the Australian Synchrotron.61 Due to radiation damage and the low symmetry space group of the GMCSF:F1 Fab crystals, three datasets were combined to maximize completeness and multiplicity for structure determination. The three datasets were processed individually in XDS62 and unmerged ASCII files were combined and scaled in Aimless63 to obtain a combined data set that was used for molecular replacement and refinement.
The components of the asymmetric unit were determined by the molecular replacement method using the existing crystal structure of GM-CSF (PDB ID: 2GMF)41 as one search model and PDB ID: 2JIX,64 with CDR loops removed, as the search model for the Fab component due to close sequence similarity with the F1 Fab. Matthew’s coefficient65 (3.53 Å 3/Da for four molecules, which corresponds to a solvent content of 65%) suggested that there were four or five molecules in the asymmetric unit cell and Phaser was used to search for up to five molecules of each of the components.66 Phaser could find four copies of the Fab but only two copies of GM-CSF, and refinement did not improve R factors. Trimming the N-terminal residues of GM-CSF led to a solution that placed the cytokine in an orientation where it bound to the CDR loops of the Fab heavy and light chains. The final model containing four molecules of the GM-CSF:F1 Fab complex was refined using Refmac 5 (CCP4)67,68 and Phenix.69 Residues forming the CDR loops of the F1 Fab model were built in manually and the structure was refined to a final Rwork of 0.23 and Rfree of 0.26. The overall stereochemical analysis reveals good geometry of the structure with two Ramachandran outliers, V51 (main-chain carbonyl involved in a hydrogen bond with a water molecule) and P148.
Diffraction data from the GM-CSF:4D4 Fab complex crystals were collected and processed in XDS.62 Matthews cell content analysis65 predicted one GM-CSF:4D4 Fab complex in the asymmetric unit cell, with a Matthews coefficient of 2.16 Å3/Da and a solvent content of 43%. The structure was solved by the molecular replacement method using the coordinates of GM-CSF (PDB ID: 2GMF)41 and a bactericidal Fab-H6831 (PDB ID: 1RJL)70 for the 4D4 Fab due to high sequence similarity. The structure was refined using Refmac 5 (CCP4)67,68 to final Rwork and Rfree of 0.18 and 0.24, respectively. Stereochemical analysis reveals good geometry of the structure with all residues in the allowed regions of the Ramachandran plot. Stereochemical analysis was carried out using the wwPDB Validation Service (https://validate-rcsb-1.wwpdb.org/). Data and refinement statistics are listed in Table 1. The PISA (Protein Interfaces, Surfaces and Assemblies) server http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html was used for all protein-ligand surface interaction calculations.
Accession numbers
Coordinates and structure factors of the crystal structures of the GM-CSF:F1 Fab and the GM-CSF:4D4 Fab complexes have been deposited in the RCSB Protein Data Bank under the accession numbers 6BFQ and 6BFS, respectively.
Abbreviations
- βc
beta common receptor subunit
- CDR
complementarity-determining region
- DSF
differential scanning fluorimetry
- EC50
half maximal effective concentration
- IC50
half maximal inhibitory concentration
- Fab
antigen-binding fragment
- GM-CSF
granulocyte-macrophage colony stimulating factor
- GMRα
GM-CSF-specific receptor alpha subunit
- IL-3
interleukin-3
- IL-5
interleukin-5
- IL3Rα
interleukin-3 specific receptor alpha subunit
- JAK
janus kinase
- mAb
monoclonal antibody
- MS
multiple sclerosis
- NTD
N-terminal domain
- PAP
pulmonary alveolar proteinosis
- RA
rheumatoid arthritis
- rmsd
root-mean-square deviation
- Sc
surface complimentarity
- STAT
signal transducer and activator of transcription
Funding Statement
This work was supported by the National Health and Medical Research Council of Australia (NHMRC) [1117183]; National Health and Medical Research Council of Australia (NHMRC) [1071897].
Acknowledgments
This research was partly undertaken on the MX2 beamline at the Australian Synchrotron, Victoria, Australia and we thank the beamline staff for their assistance. We thank Anna Sapa for technical assistance and we acknowledge the SA Pathology Detmold Family Cytometry Centre for use of flow cytometry facilities. We acknowledge the use of the C3 facility at CSIRO for optimisation of crystallisation conditions. This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRC) to M.W.P. and A.F.L., and from the Australian Cancer Research Foundation to M.W.P. Funding from the Victorian Government Operational Infrastructure Support Scheme to St Vincent’s Institute is acknowledged as is support by the Canadian Research Chairs program to E.F.P. and J.W.S. U.D. is supported by Cancer Council Victoria and M.W.P. is a NHMRC Research Fellow.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be access on the publisher’s website.
References
- 1.Burgess AW, Camakaris J, Metcalf D.. Purification and properties of colony-stimulating factor from mouse lung-conditioned medium. J Biol Chem. 1977;252:1998–2003. [PubMed] [Google Scholar]
- 2.Broughton SE, Dhagat U, Hercus TR, Nero TL, Grimbaldeston MA, Bonder CS, Lopez AF, Parker MW. The GM-CSF/IL-3/IL-5 cytokine receptor family: from ligand recognition to initiation of signaling. Immunol Rev. 2012;250:277–302. doi: 10.1111/j.1600-065X.2012.01164.x. [DOI] [PubMed] [Google Scholar]
- 3.Hercus TR, Dhagat U, Kan WL, Broughton SE, Nero TL, Perugini M, Sandow JJ, D’Andrea RJ, Ekert PG, Hughes T, et al. Signalling by the betac family of cytokines. Cytokine Growth Factor Rev. 2013;24:189–201. doi: 10.1016/j.cytogfr.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Metcalf D. Hematopoietic cytokines. Blood. 2008;111:485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelso A, Metcalf D. Clonal heterogeneity in colony stimulating factor production by murine T lymphocytes. J Cell Physiol. 1985;123:101–110. doi: 10.1002/jcp.1041230115. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 7.Fleetwood AJ, Cook AD, Hamilton JA. Functions of granulocyte-macrophage colony-stimulating factor. Crit Rev Immunol. 2005;25:405–428. 7afe009f5855b332,6288a0cd2796c22e [pii]. [DOI] [PubMed] [Google Scholar]
- 8.Dranoff G, Crawford AD, Sadelain M, Ream B, Rashid A, Bronson RT, Dickersin GR, Bachurski CJ, Mark EL, Whitsett JA, et al. Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science. 1994;264:713–716. [DOI] [PubMed] [Google Scholar]
- 9.Stanley E, Lieschke GJ, Grail D, Metcalf D, Hodgson G, Gall JAM, Maher DW, Cebon J, Sinickas V, Dunn AR. Granulocyte-macrophage colony-stimulating factor-deficient mice show no major pertubation of hematopoiesis but develop a characteristic pulmonary pathology. Proc Natl Acad Sci USA. 1994;91:5592–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitamura T, Tanaka N, Watanabe J, Uchida, Kanegasaki S, Yamada Y, Nakata K. Idiopathic pulmonary alveolar proteinosis as an autoimmune disease with neutralizing antibody against granulocyte/macrophage colony-stimulating factor. J Exp Med. 1999;190:875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue Y, Trapnell BC, Tazawa R, Arai T, Takada T, Hizawa N, Kasahara Y, Tatsumi K, Hojo M, Ichiwata T, et al. Characteristics of a large cohort of patients with autoimmune pulmonary alveolar proteinosis in Japan. Am J Respir Crit Care Med. 2008;177:752–762. doi: 10.1164/rccm.200708-1271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broughton SE, Hercus TR, Nero TL, Dottore M, McClure BJ, Dhagat U, Taing H, Gorman MA, King-Scott J, Lopez AF, et al. Conformational changes in the GM-CSF receptor suggest a molecular mechanism for affinity conversion and receptor signaling. Structure. 2016;24:1271–1281. doi: 10.1016/j.str.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134:496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 14.Carr PD, Conlan F, Ford S, Ollis DL, Young IG. An improved resolution structure of the human beta common receptor involved in IL-3, IL-5 and GM-CSF signalling which gives better definition of the high-affinity binding epitope. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:509–513. doi: 10.1107/S1744309106016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr PD, Gustin SE, Church AP, Murphy JM, Ford SC, Mann DA, Woltring DM, Walker I, Ollis DL, Young IG. Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell. 2001;104:291–300. [DOI] [PubMed] [Google Scholar]
- 16.Broxmeyer HE, Hoggatt J, O’Leary HA, Mantel C, Chitteti BR, Cooper S, Messina-Graham S, Hangoc G, Farag S, Rohrabaugh SL, et al. Dipeptidylpeptidase 4 negatively regulates colony-stimulating factor activity and stress hematopoiesis. Nat Med. 2012;18:1786–1796. doi: 10.1038/nm.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicks IP, Roberts AW. Targeting GM-CSF in inflammatory diseases. Nat Reviews Rheumatol. 2016;12:37–48. doi: 10.1038/nrrheum.2015.161. [DOI] [PubMed] [Google Scholar]
- 18.van Nieuwenhuijze A, Koenders M, Roeleveld D, Sleeman MA, van den Berg W, Wicks IP. GM-CSF as a therapeutic target in inflammatory diseases. Mol Immunol. 2013;56:675–682. doi: 10.1016/j.molimm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Behrens F, Tak PP, Ostergaard M, Stoilov R, Wiland P, Huizinga TW, Berenfus VY, Vladeva S, Rech J, Rubbert-Roth A, et al. MOR103, a human monoclonal antibody to granulocyte-macrophage colony-stimulating factor, in the treatment of patients with moderate rheumatoid arthritis: results of a phase Ib/IIa randomised, double-blind, placebo-controlled, dose-escalation trial. Ann Rheum Dis. 2015;74:1058–1064. doi: 10.1136/annrheumdis-2013-204816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Constantinescu CS, Asher A, Fryze W, Kozubski W, Wagner F, Aram J, Tanasescu R, Korolkiewicz RP, Dirnberger-Hertweck M, Steidl S, et al. Randomized phase 1b trial of MOR103, a human antibody to GM-CSF, in multiple sclerosis. Neurol(R) Neuroimm Neuroinflamm. 2015;2:e117. doi: 10.1212/NXI.0000000000000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krinner EM, Raum T, Petsch S, Bruckmaier S, Schuster I, Petersen L, Cierpka R, Abebe D, Molhoj M, Wolf A, et al. A human monoclonal IgG1 potently neutralizing the pro-inflammatory cytokine GM-CSF. Mol Immunol. 2007;44:916–925. doi: 10.1016/j.molimm.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Molfino NA, Kuna P, Leff JA, Oh CK, Singh D, Chernow M, Sutton B, Yarranton G. Phase 2, randomised placebo-controlled trial to evaluate the efficacy and safety of an anti-GM-CSF antibody (KB003) in patients with inadequately controlled asthma. BMJ Open. 2016;6:e007709. doi: 10.1136/bmjopen-2015-007709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton JA. GM-CSF as a target in inflammatory/autoimmune disease: current evidence and future therapeutic potential. Expert Rev Clin Immunol. 2015;11:457–465. doi: 10.1586/1744666X.2015.1024110. [DOI] [PubMed] [Google Scholar]
- 24.Huizinga TW, Batalov A, Stoilov R, Lloyd E, Wagner T, Saurigny D, Souberbielle B, Esfandiari E. Phase 1b randomized, double-blind study of namilumab, an anti-granulocyte macrophage colony-stimulating factor monoclonal antibody, in mild-to-moderate rheumatoid arthritis. Arthritis Res Ther. 2017;19:53. doi: 10.1186/s13075-017-1267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burmester GR, Weinblatt ME, McInnes IB, Porter D, Barbarash O, Vatutin M, Szombati I, Esfandiari E, Sleeman MA, Kane CD, et al. Efficacy and safety of mavrilimumab in subjects with rheumatoid arthritis. Ann Rheum Dis. 2013;72:1445–1452. doi: 10.1136/annrheumdis-2012-202450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crotti C, Raimondo MG, Becciolini A, Biggioggero M, Favalli EG. Spotlight on mavrilimumab for the treatment of rheumatoid arthritis: evidence to date. Drug Des Devel Ther. 2017;11:211–223. doi: 10.2147/DDDT.S104233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakagami T, Uchida K, Suzuki T, Carey BC, Wood RE, Wert SE, Whitsett JA, Trapnell BC, Luisetti M. Human GM-CSF autoantibodies and reproduction of pulmonary alveolar proteinosis. N Engl J Med. 2009;361:2679–2681. doi: 10.1056/NEJMc0904077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchida K, Beck DC, Yamamoto T, Berclaz PY, Abe S, Staudt MK, Carey BC, Filippi MD, Wert SE, Denson LA, et al. GM-CSF autoantibodies and neutrophil dysfunction in pulmonary alveolar proteinosis. N Engl J Med. 2007;356:567–579. doi: 10.1056/NEJMoa062505. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, Maranda B, Sakagami T, Catellier P, Couture CY, Carey BC, Chalk C, Trapnell BC. Hereditary pulmonary alveolar proteinosis caused by recessive CSF2RB mutations. Eur Respir J. 2011;37:201–204. doi: 10.1183/09031936.00090610. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki T, Sakagami T, Rubin BK, Nogee LM, Wood RE, Zimmerman SL, Smolarek T, Dishop MK, Wert SE, Whitsett JA, et al. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. J Exp Med. 2008;205:2703–2710. doi: 10.1084/jem.20080990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka T, Motoi N, Tsuchihashi Y, Tazawa R, Kaneko C, Nei T, Yamamoto T, Hayashi T, Tagawa T, Nagayasu T, et al. Adult-onset hereditary pulmonary alveolar proteinosis caused by a single-base deletion in CSF2RB. J Med Genet. 2011;48:205–209. doi: 10.1136/jmg.2010.082586. [DOI] [PubMed] [Google Scholar]
- 32.Griese M, Ripper J, Sibbersen A, Lohse P, Lohse P, Brasch F, Schams A, Pamir A, Schaub B, Muensterer OJ, et al. Long-term follow-up and treatment of congenital alveolar proteinosis. BMC Pediatr. 2011;11:72. doi: 10.1186/1471-2431-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Moczygemba M, Doan ML, Elidemir O, Fan LL, Cheung SW, Lei JT, Moore JP, Tavana G, Lewis LR, Zhu Y, et al. Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRalpha gene in the X chromosome pseudoautosomal region 1. J Exp Med. 2008;205:2711–2716. doi: 10.1084/jem.20080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uchida K, Nakata K, Suzuki T, Luisetti M, Watanabe M, Koch DE, Stevens CA, Beck DC, Denson LA, Carey BC, et al. Granulocyte/macrophage-colony-stimulating factor autoantibodies and myeloid cell immune functions in healthy subjects. Blood. 2009;113:2547–2556. doi: 10.1182/blood-2009-05-155689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe M, Uchida K, Nakagaki K, Kanazawa H, Trapnell BC, Hoshino Y, Kagamu H, Yoshizawa H, Keicho N, Goto H, et al. Anti-cytokine autoantibodies are ubiquitous in healthy individuals. FEBS Lett. 2007;581:2017–2021. doi: 10.1016/j.febslet.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Thomson CA, Allan LL, Jackson LM, Olson M, Hercus TR, Nero TL, Turner A, Parker MW, et al. Characterization of pathogenic human monoclonal autoantibodies against GM-CSF. Proc Natl Acad Sci U S A. 2013;110:7832–7837. doi: 10.1073/pnas.1216011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piccoli L, Campo I, Fregni CS, Rodriguez BM, Minola A, Sallusto F, Luisetti M, Corti D, Lanzavecchia A. Neutralization and clearance of GM-CSF by autoantibodies in pulmonary alveolar proteinosis. Nat Commun. 2015;6:7375. doi: 10.1038/ncomms8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blech M, Seeliger D, Kistler B, Bauer MM, Hafner M, Horer S, Zeeb M, Nar H, Park JE. Molecular structure of human GM-CSF in complex with a disease-associated anti-human GM-CSF autoantibody and its potential biological implications. Biochem J. 2012;447:205–215. doi: 10.1042/BJ20120884. [DOI] [PubMed] [Google Scholar]
- 39.Eylenstein R, Weinfurtner D, Hartle S, Strohner R, Bottcher J, Augustin M, Ostendorp R, Steidl S. Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody. MAbs. 2016;8:176–186. doi: 10.1080/19420862.2015.1099774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinzelman P, Carlson SJ, Cox GN. Cytokine refacing effect reduces granulocyte macrophage colony-stimulating factor susceptibility to antibody neutralization. Protein Eng Des Sel: PEDS. 2015;28:461–466. doi: 10.1093/protein/gzv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozwarski DA, Diederichs K, Hecht R, Boone T, Karplus PA. Refined crystal structure and mutagenesis of human granulocyte-macrophage colony-stimulating factor. Proteins. 1996;26:304–313. doi:. [DOI] [PubMed] [Google Scholar]
- 42.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 43.Abhinandan KR, Martin AC. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol Immunol. 2008;45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Mirza S, Walker A, Chen J, Murphy JM, Young IG. The Ig-like domain of human GM-CSF receptor alpha plays a critical role in cytokine binding and receptor activation. Biochem J. 2010;426:307–317. doi: 10.1042/BJ20091745. [DOI] [PubMed] [Google Scholar]
- 45.Panousis C, Dhagat U, Edwards KM, Rayzman V, Hardy MP, Braley H, Gauvreau GM, Hercus TR, Smith S, Sehmi R, et al. CSL311, a novel, potent, therapeutic monoclonal antibody for the treatment of diseases mediated by the common beta chain of the IL-3, GM-CSF and IL-5 receptors. MAbs. 2016;8:436–453. doi: 10.1080/19420862.2015.1119352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deane D, McInnes CJ, Percival A, Wood A, Thomson J, Lear A, Gilray J, Fleming S, Mercer A, Haig D. Orf virus encodes a novel secreted protein inhibitor of granulocyte-macrophage colony-stimulating factor and interleukin-2. J Virol. 2000;74:1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Felix J, Kandiah E, De Munck S, Bloch Y, van Zundert GC, Pauwels K, Dansercoer A, Novanska K, Read RJ, Bonvin AM, et al. Structural basis of GM-CSF and IL-2 sequestration by the viral decoy receptor GIF. Nat Commun. 2016;7:13228. doi: 10.1038/ncomms13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosen LB, Freeman AF, Yang LM, Jutivorakool K, Olivier KN, Angkasekwinai N, Suputtamongkol Y, Bennett JE, Pyrgos V, Williamson PR, et al. Anti-GM-CSF autoantibodies in patients with cryptococcal meningitis. J Immunol. 2013;190:3959–3966. doi: 10.4049/jimmunol.1202526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saijo T, Chen J, Chen SC, Rosen LB, Yi J, Sorrell TC, Bennett JE, Holland SM, Browne SK, Kwon-Chung KJ. Anti-granulocyte-macrophage colony-stimulating factor autoantibodies are a risk factor for central nervous system infection by Cryptococcus gattii in otherwise immunocompetent patients. MBio. 2014;5:e00912–14. doi: 10.1128/mBio.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gathungu G, Kim MO, Ferguson JP, Sharma Y, Zhang W, Ng SM, Bonkowski E, Ning K, Simms LA, Croft AR, et al. Granulocyte-macrophage colony-stimulating factor autoantibodies: a marker of aggressive Crohn’s disease. Inflamm Bowel Dis. 2013;19:1671–1680. doi: 10.1097/MIB.0b013e318281f506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seymour JF, Presneill JJ, Schoch OD, Downie GH, Moore PE, Doyle IR, Vincent JM, Nakata K, Kitamura T, Langton D, et al. Therapeutic efficacy of granulocyte-macrophage colony-stimulating factor in patients with idiopathic acquired alveolar proteinosis. Am J Respir Crit Care Med. 2001;163:524–531. doi: 10.1164/ajrccm.163.2.2003146. [DOI] [PubMed] [Google Scholar]
- 52.Hercus TR, Bagley CJ, Cambareri B, Dottore M, Woodcock JM, Vadas MA, Shannon MF, Lopez AF. Specific human granulocyte-macrophage colony-stimulating factor antagonists. Proc Natl Acad Sci U S A. 1994;91:5838–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hercus TR, Cambareri B, Dottore M, Woodcock JM, Bagley CJ, Vadas MA, Shannon MF, Lopez AF. Identification of residues in the first and fourth helices of human granulocyte-macrophage colony stimulating factor involved in binding to the α- and β- chains of the receptor. Blood. 1994;83:3500–3508. [PubMed] [Google Scholar]
- 54.Bryson S, Thomson CA, Risnes LF, Dasgupta S, Smith K, Schrader JW, Pai EF. Structures of preferred human IgV genes-based protective antibodies identify how conserved residues contact diverse antigens and assign source of specificity to CDR3 loop variation. J Immunol. 2016;196:4723–4730. doi: 10.4049/jimmunol.1402890. [DOI] [PubMed] [Google Scholar]
- 55.McCafferty J, Johnson KS. Construction and screening of antibody display libraries In: Kay B, Winter J, McCafferty J, editors. Phage display of peptides and proteins: A laboratory manual. San Diego (CA): Academic Press; 1996. p. 79–111. [Google Scholar]
- 56.Persons DA, Mehaffey MG, Kaleko M, Nienhuis AW, Vanin EF. An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol Dis. 1998;24:167–182. doi: 10.1006/bcmd.1998.0184. [DOI] [PubMed] [Google Scholar]
- 57.Jenkins BJ, Le F, Gonda TJ. A cell type-specific constitutive point mutant of the common β-subunit of the human granulocyte-macrophage colony-stimulating factor (GM-CSF), Interleukin (IL)-3, and IL-5 receptors requires the GM-CSF receptor α-subunit for activation. JBiolChem. 1999;274:8669–8677. [DOI] [PubMed] [Google Scholar]
- 58.Stomski FC, Woodcock JM, Zacharakis B, Bagley CJ, Sun Q, Lopez AF. Identification of a Cys motif in the common beta chain of the interleukin 3, granulocyte-macrophage colony-stimulating factor, and interleukin 5 receptors essential for disulfide-linked receptor heterodimerization and activation of all three receptors. J Biol Chem. 1998;273:1192–1199. [DOI] [PubMed] [Google Scholar]
- 59.Woodcock JM, Zacharakis B, Plaetinck G, Bagley CJ, Qiyu S, Hercus TR, Tavernier J, Lopez AF. Three residues in the common beta chain of the human GM-CSF, IL-3 and IL-5 receptors are essential for GM-CSF and IL-5 but not IL-3 high affinity binding and interact with Glu21 of GM-CSF. EMBO J. 1994;13:5176–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao YF, Miyazono K, Urabe A, Takaku F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 61.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, et al. Blu-ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/S0909049502015170. [DOI] [PubMed] [Google Scholar]
- 62.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Z, Stoll VS, Devries PJ, Jakob CG, Xie N, Simmer RL, Lacy SE, Egan DA, Harlan JE, Lesniewski RR, et al. A potent erythropoietin-mimicking human antibody interacts through a novel binding site. Blood. 2007;110:2408–2413. doi: 10.1182/blood-2007-04-083998. [DOI] [PubMed] [Google Scholar]
- 65.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. [DOI] [PubMed] [Google Scholar]
- 66.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collaborative Computational Project N The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 68.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becker M, Bunikis J, Lade BD, Dunn JJ, Barbour AG, Lawson CL. Structural investigation of Borrelia burgdorferi OspB, a bactericidal Fab target. J Biol Chem. 2005;280:17363–17370. doi: 10.1074/jbc.M412842200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


