Abstract
The glycolipid transfer protein (GLTP) fold defines a superfamily of eukaryotic proteins that selectively transport sphingolipids (SLs) between membranes. However, the mechanisms determining the protein selectivity for specific glycosphingolipids (GSLs) are unclear. Here, we report the crystal structure of the GLTP homology (GLTPH) domain of human 4-phosphate adaptor protein 2 (FAPP2) bound with N-oleoyl-galactosylceramide. Using this domain, FAPP2 transports glucosylceramide from its cis-Golgi synthesis site to the trans-Golgi for conversion into complex GSLs. The FAPP2–GLTPH structure revealed an element, termed the ID loop, that controls specificity in the GLTP family. We found that, in accordance with FAPP2 preference for simple GSLs, the ID loop protrudes from behind the SL headgroup-recognition center to mitigate binding by complex GSLs. Mutational analyses including GLTP and FAPP2 chimeras with swapped ID loops supported the proposed restrictive role of the FAPP2 ID loop in GSL selectivity. Comparative analysis revealed distinctly designed ID loops in each GLTP family member. This analysis also disclosed a conserved H-bond triplet that “clasps” both ID-loop ends together to promote structural autonomy and rigidity. The findings indicated that various ID loops work in concert with conserved recognition centers to create different specificities among family members. We also observed four bulky, conserved hydrophobic residues involved in “sensor-like” interactions with lipid chains in protein hydrophobic pockets and FF motifs in GLTP and FAPP2, well-positioned to provide acyl chain–dependent SL selectivity for the hydrophobic pockets. In summary, our study provides mechanistic insights into sphingolipid recognition by the GLTP fold and uncovers the elements involved in this recognition.
Keywords: lipid-protein interaction, sphingolipid, transport, X-ray crystallography, protein complex, protein structure, 4-phosphate-adaptor-protein-2, FAPP2, nonvesicular lipid transport, protein family
Introduction
Human 4-phosphate-adaptor-protein-2 (FAPP2)4 is a Golgi-associated, two-domain protein consisting of 519 amino acids. The N-terminal pleckstrin homology domain (93 residues) that helps target the Golgi is connected by a 214-residue linker to a C-terminal glycolipid transfer protein homology (GLTPH) domain (212 residues) (1, 2). In vivo, FAPP2 transports glucosylceramides (GlcCer) from the synthesis site in the cis-Golgi to the trans-Golgi/TGN where conversion into more complex glycosphingolipids (GSLs) occurs via sequential sugar addition (1, 3). When docked with Golgi membranes via the pleckstrin homology domain, the long FAPP2 connecting linker enables the soluble GLTPH domain to move locally within the cytoplasm and approach the Golgi to acquire or release specific sphingolipids (SLs) from/to accessible membrane regions, i.e. act as a lipid-transfer protein (LTP) “on a leash.”
Structural homology modeling of the FAPP2–GLTPH domain suggests membership in the GLTP superfamily (1, 2, 4), a group of eukaryotic LTPs that selectively transfer SLs between membranes (5–11) and share a common protein fold first established from human GLTP crystal structure (12). The GLTP fold is composed of eight α-helices, organized as a curved two-layer “sandwich” that envelopes the SL aliphatic chains within an internal hydrophobic pocket that is accessed via a cleft-like gate, whereas the SL-specific headgroup-binding site includes a surface localized recognition center (12). Crystal structures of four other GLTP superfamily members (fungal heterokaryon incompatibility C2 protein, HET-C2, of Podosporaanserina; GLTP-like protein from the thermoacidophilic unicellular red alga, Galdieria sulfuraria; human ceramide-1-phosphate (C1P) transfer protein, CPTP; and accelerated cell death 11 protein (ACD11), an Arabidopsis CPTP homolog) reveal overall similarity with GLTP structure and nearly conserved arrangement of recognition center positions but with distinctions enabling selectivity of SLs with sugar (GSL) versus phosphate (C1P) headgroups (Refs. 10, 13, 14, and PDB code 2I3F), defining two subfamilies, GSL-specific and C1P-specific. Modeling of FAPP2–GLTPH relative to the GLTP structure shows conservation of residues required for sugar headgroup binding (2). However, FAPP2 prefers simple GSLs. The in vitro transfer of GlcCer and GalCer by FAPP2–GLTPH domain is equally efficient but is reduced by ∼50% with LacCer and is slower yet with complex GSLs, such as ganglioside GM1 that possess five sugars including N-acetylneuraminic acid, or sulfo-GalCer, sulfatide (SF) (2). By contrast, human GLTP efficiently transfers both complex and simple GSLs (6, 7, 9), as reviewed in Ref. 11. Crystal structures of different holo forms of human GLTP provide insights into the structural features responsible for the recognition of GalCer, GlcCer, LacCer, and SF. Also available are two apo structures of other GSL-specific members and different holo forms of two C1P-specific members, providing a potential source for comparative analyses. However, the basis for the focused FAPP2–GLTPH selectivity for simple GSLs has remained a matter of speculation because of the lack of structure determination for FAPP2. To address the issue, we present here the 1.45 Å resolution crystal structure of FAPP2–GLTPH domain (residues 308–519) complexed with the GSL, N-oleoyl-galactosylceramide (18:1-GalCer), and perform comparative analysis that includes other available structures of the GLTP superfamily members. The approach provides insights into the strict preference of FAPP2 for simple GSLs; reveals a unique element, termed the ID-loop, that distinguishes each particular protein of the family; and enables identification of previously unrecognized but important elements of the GLTP fold.
Results and discussion
Crystal structure of human FAPP2–GLTPH with bound N-oleoyl-galactosylceramide
After numerous crystallization trials were unsuccessful, we introduced the E377A/E378K double mutation far from functionally important regions (Fig. S1a) to reduce the excessive negative surface charge of FAPP2–GLTPH domain (residues 308–519) (2). This change did not affect transfer activity (Fig. S1b) but enabled a high-resolution crystal of the GLTPH domain complexed with N-oleoyl-galactosylceramide (18:1-GalCer) to form. Hereafter, “FAPP2–GLTPH” refers to the E377A/E378K mutant. The final model was refined to Rwork/Rfree values of 0.131/0.179 at 1.45 Å resolution. Table 1 summarizes the X-ray data collection and refinement statistics.
Table 1.
X-ray data collection and refinement statistics for human FAPP2–GLTPH·18:1-GalCer structure
The values in parentheses are for the highest-resolution shell. Rfree is calculated for 5% of randomly selected reflections excluded from refinement. RMSD, root-mean-square deviation from ideal values.
| Data collection | |
| Space group | P212121 |
| a (Å) | 66.24 |
| b (Å) | 74.61 |
| c (Å) | 93.59 |
| α = β = γ (°) | 90.0 |
| AU content | 2 molecules |
| Resolution (Å) | 58.34–1.45 (1.53–1.45) |
| Rmerge | 0.047 (0.754) |
| I/σI | 14.5 (2.0) |
| Completeness (%) | 99.7 (99.7) |
| Redundancy | 4.3 (4.3) |
| Refinement | |
| Resolution (Å) | 15.0–1.45 |
| No. of reflections | |
| Work | 78,300 |
| Free | 4122 |
| Rwork/Rfree | 0.131/0.179 |
| No. of atoms | |
| Protein | 3454 |
| Lipid | 102 |
| Water | 493 |
| B factors (Å2) | |
| Protein A/B | 26.6/29.4 |
| Lipid A/B | 36.0/34.6 |
| Water | 45.2 |
| RMSD | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.77 |
| PDB code | 5KDI |
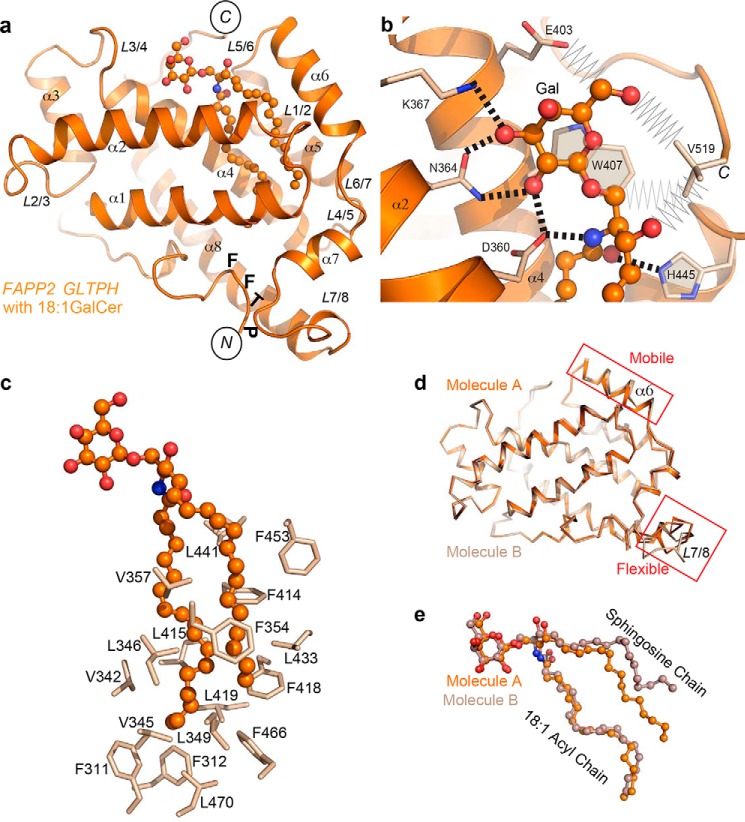
The overall structure of FAPP2–GLTPH with bound 18:1-GalCer (Fig. 1a) resembles GLTP complexed with 18:1 GlcCer (PDB code 3S0K) (15) with respect to lipid position and binding mode. The recognition center (Fig. 1b) displays a similar network of interactions as GLTP for anchoring the polar region of GalCer (16) with Trp407 serving as a stacking plate for the GSL sugar, whereas Asp360, Asn364, Lys367, and His445 form hydrogen bonds with the galactose-amide moiety. In addition, the C-terminal Val519 makes tight van der Waals contacts with two hydroxyls and the ceramide C1 atom, whereas residue Glu403 contacts with the sugar C6′ atom (Fig. 1b). Both nonpolar aliphatic chains are encapsulated by GLTPH via the so-called “sphingosine-in” binding mode (Fig. 1a) previously observed for human GLTP bound by GSLs containing an N-oleoyl-acyl chain (12, 15). Nonpolar residues such as Phe, Leu, and Val line the FAPP2 hydrophobic pocket, creating an appropriate environment for encapsulating the ceramide chains (Fig. 1c).
Figure 1.
Crystal structure of human FAPP2–GLTPH domain complexed with GSL, 18:1-GalCer. a, overall view of the complex showing labeled helices, loops, N and C termini, and PTFF sequence containing FF motif. b, protein recognition center interactions with galactose ring and ceramide amide moiety of bound GalCer, depicting hydrogen bonds (dashed lines) and the aromatic ring stacking over the glucose ring. Also depicted are van der Waals contacts (zig-zag lines) of the nonconserved residues Glu403 and C-terminal Val519. c, Phe, Leu, and Val residues lining the hydrophobic pocket that envelopes the lipid chains. d and e, superposition of two molecules of the complex occupying the asymmetric unit, showing either protein (d) or GSL (e). Molecules A and B are depicted in different shades of colors. In a–e, atoms are colored orange, red, and blue for carbon, oxygen, and nitrogen, respectively.
Comparison of the two molecules of GLTPH·18:1-GalCer complexes that occupy the asymmetric unit (AU) reveals moderate lateral mobility of α6-helix, noticeable flexibility of loop L7/8 (Fig. 1d), and different positions for the sphingosine chain of bound 18:1-GalCer in each molecule (Fig. 1e). Thus, high similarity exists with GLTP (12, 15, 16). However, compared with GLTP, FAPP2 allows different sphingosine chain positioning (Fig. 1e) within the same binding mode and for the same GSL molecule.
Overall comparison of GLTP family members
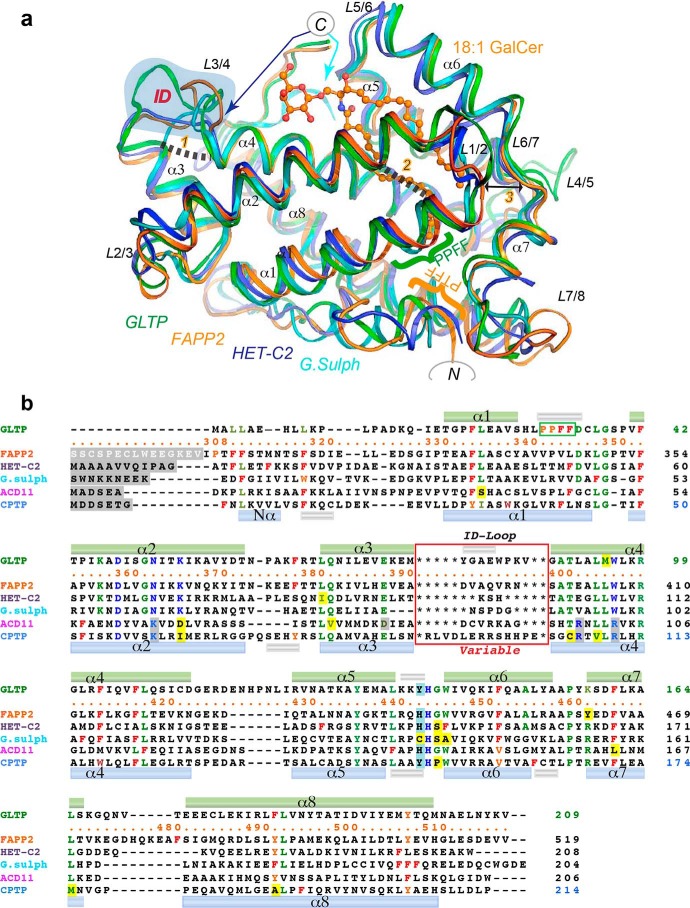
Fig. 2a shows the structure of FAPP2–GLTPH domain bound with 18:1-GalCer in superposition with other three members of the GSL-specific subfamily with known X-ray structures. Superimposition that includes C1P-specific members of the GLTP superfamily with known 3D structures is shown in Fig. S1a. Differences between ACD11 versus CPTP and CPTP versus FAPP2–GLTPH are highlighted in Fig. S1 (c and d). Poststructural sequence alignment of all six proteins (Fig. 2b) indicates conserved/semiconserved residues of the GLTP fold. Key recognition center residues (Fig. 2b, blue letters) identify five conserved positions in the GSL-specific subfamily and five conserved positions in the C1P-specific subfamily, four of which coincide in both subfamilies. Together they define the basic six-point pattern for the recognition center in the GLTP fold, with spatial templates DNKxWH and DKxRRH (where x indicates a nonconserved residue) characterizing the GSL-specific and C1P-specific families, respectively (Figs. 1b and 2b). The conserved recognition center templates for each subfamily suggest that specificity variations within a subfamily are controlled by other elements. Global comparisons (Fig. 2a and Fig. S1a) identify variable elements of the GLTP-fold including the “nonalignable” (Fig. 2b) loop L3/4 that differs among each member of the GLTP family. Also evident are three previously unrecognized but conserved interaction sites (Fig. 2a, labeled in orange as 1, 2, and 3) that are presumably important for GLTP-fold function.
Figure 2.
Comparison of FAPP2–GLTPH domain with other family members. a, structure superimposition of the GSL-specific proteins: human FAPP2–GLTPH domain (orange), human GLTP (green), fungal heterokaryon incompatibility C2 protein, HET-C2, of P. anserina (blue), and GLTP-like protein from the thermoacidophilic unicellular red alga, G. sulfuraria (cyan) (PDB codes 5KDI, 3S0K, 4KV0, and 2I3F, respectively). 18:1-GalCer is shown as bound to FAPP2–GLTPH. The blue-shaded area highlights the ID loops. The GLTP PPFF and FAPP2 PTFF sequences are indicated by green and orange braces, respectively. Newly identified conserved linkages 1 and 2 are indicated by dashed lines; the double-headed arrow 3 points to the strictly conserved Gly and Pro on opposite sides of the gate controlling the access to the hydrophobic pocket in the GLTP fold. Colored arrows point to the C-ends of HET-C2 (blue) and G. sulfuraria protein (cyan). Color codes for GSL atoms are defined in Fig. 1. b, poststructural sequence alignment of six proteins belonging to the GLTP family (see Fig. S1a). Color codes are blue for the recognition center residues, green for the conserved/semiconserved residues, and red and orange for Phe and similarly positioned Tyr/Trp, respectively. Recognition center residues of the C1P-specific family also are blue but shaded in gray when different from the GSL-specific family. Exceptions among conserved residues are shaded by yellow. The upper and lower cylinders (small silver for 310-helices; other for α-helices) indicate the locations of secondary structure elements found in GLTP and CPTP, respectively. Within the red rectangle are the ID-loop sequences that vary among each member.
GLTP-fold ID loop
The differing structural features of the L3/4 loop in FAPP2–GLTPH and other subfamily members are shown in Fig. 2a, whereas the variability of L3/4-loop length and sequence for each GLTP superfamily member can be appreciated in Fig. 2b (red box). Significant differences in L3/4-loop conformations in the GSL-specific subfamily and in the GLTP superfamily are highlighted in stereo mode in Fig. 3 (a and b, respectively). Because each L3/4 loop is unique, we refer to it as the “ID loop,” i.e. individual or identification loop.
Figure 3.
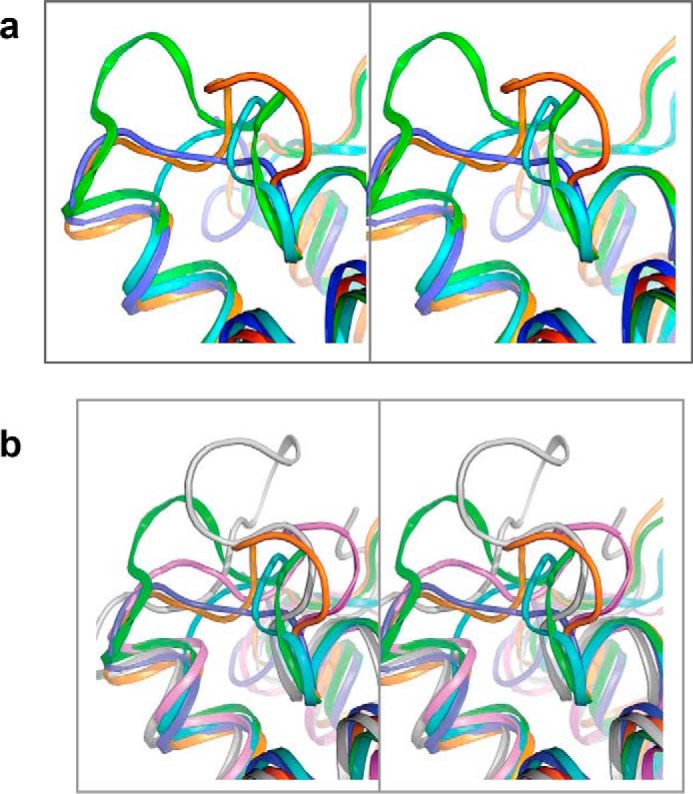
Comparative stereo view of the ID loops. a, based on Fig. 2a. b, based on Fig. S1a.
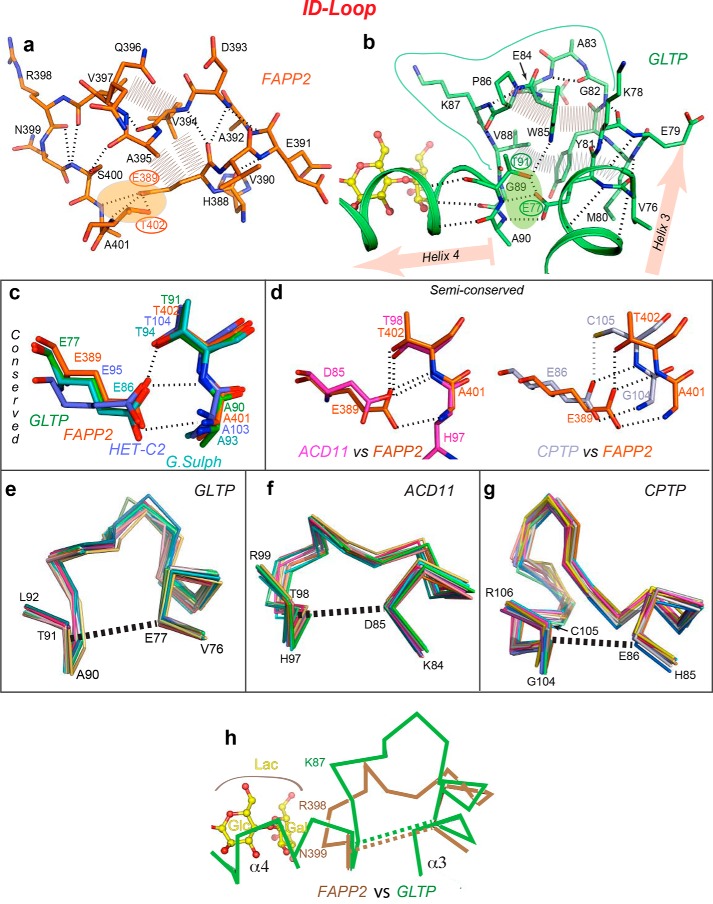
What makes the ID loop special compared with other helix–helix connectors is that each loop end interacts with each other to generate a pinched-together “clasp-like” structure (Fig. 2a, linkage 1). In FAPP2–GLTPH, this clasp is secured by an H-bond triplet (Fig. 4a, shaded area) formed by the side-chain carboxyl of Glu389, located in the C-terminal region of α3-helix, and two consecutive main-chain NH-groups (401 and 402) plus a side-chain hydroxyl (Thr402), grouped at the N terminus of α4-helix. Glu389 provides three acceptor vacancies for the triplet, whereas the partners from α4-helix provide three H-bond donors. The same H-bond triplet pattern, involving the same residues at the same locations, connects the ends of helices 3 and 4 in GLTP (Fig. 4b, shaded area) and is strictly conserved in other GSL-specific GLTP superfamily members (Fig. 4c). Moreover, as illustrated in Fig. 4d, the interaction that “locks” the ID loop persists in the C1P-specific subfamily, although Glu is replaced by Asp in ACD11 and Thr is replaced by Cys in CPTP. Regardless, the fastener for the ID loop remains intact, albeit with minor modifications (Fig. 4d). Thus, the ID-loop clasp is a conserved feature of the GLTP fold that endows the ID loop with relative structural autonomy and rigidity.
Figure 4.
Structural properties of the ID loop. a and b, intraloop interactions in human FAPP2–GLTPH (a) and human GLTP (b), depicting main-chain hydrogen bonds (dashed lines), side-chain van der Waals contacts (zig-zag lines), and orientation of flanking helices α3 and α4 (highlighted in b by pink arrows). Clasps formed by H-bond triplets that fasten the two ends of the ID loop to each other are shaded orange in a and green in b. c and d, the H-bond triplet interaction pattern in the GSL-specific family (c) and C1P-specific family (d) derived from global superposition of GLTP superfamily members in Fig. S1a. Two semiconserved patterns are compared with the H-bond triplet in FAPP2–GLTPH. e–g, ID-loop Cα backbones from superimposed multiple molecular structures of GLTP (e), ACD11 (f), and CPTP (g) showing the relative conformational rigidity of the ID loop in different GLTP superfamily members. Superimposed are 20, 12, and 15 molecular structures of GLTP, ACD11, and CPTP, respectively, with dashed lines indicating the approximate position of the H-bond triplets and the helical regions (of α3 and α4) adjacent the H-bond triplet ends. h, expected proximity differences in the location of bound LacCer headgroup (yellow) with respect to the ID-loop backbones in FAPP2 (beige) and GLTP (green).
Examination of the ID-loop structural details in FAPP2–GLTPH shows that the “loop body” is stabilized via a network of tight intraloop interactions involving main-chain hydrogen bonding and van der Waals contacts of side chains (Fig. 4a). The interactions span the entire loop and rigidify its conformation. In GLTP, the ID loop also maintains conformational rigidity by a system of intraloop interactions encompassing the entire loop body (Fig. 4b). Further support for the ID-loop rigidity arises from the very similar backbone conformations observed by superposition of different molecular structures available for GLTP (20 molecules; Fig. 4e), ACD11 (12 molecules; Fig. 4f), and CPTP (15 molecules; Fig. 4g). Although each protein backbone displays some conformational mobility (Fig. 4, e–g), the variations are less than those observed for most loops (e.g. see Fig. 7a). Even though the ID loops are longer, on average, in C1P-specific members ACD11 and CPTP than those in GSL-specific members (Fig. 2b), the similar conformations found in 12 different ACD11 molecules and 15 different CPTP molecules strongly support the relative rigidity of these ID loops (Fig. 4, f and g). We conclude that ID-loop conformations differ among superfamily members, but each ID loop is relatively rigid. Taken together, these data support the importance of the ID loop in the GLTP fold.
Figure 7.
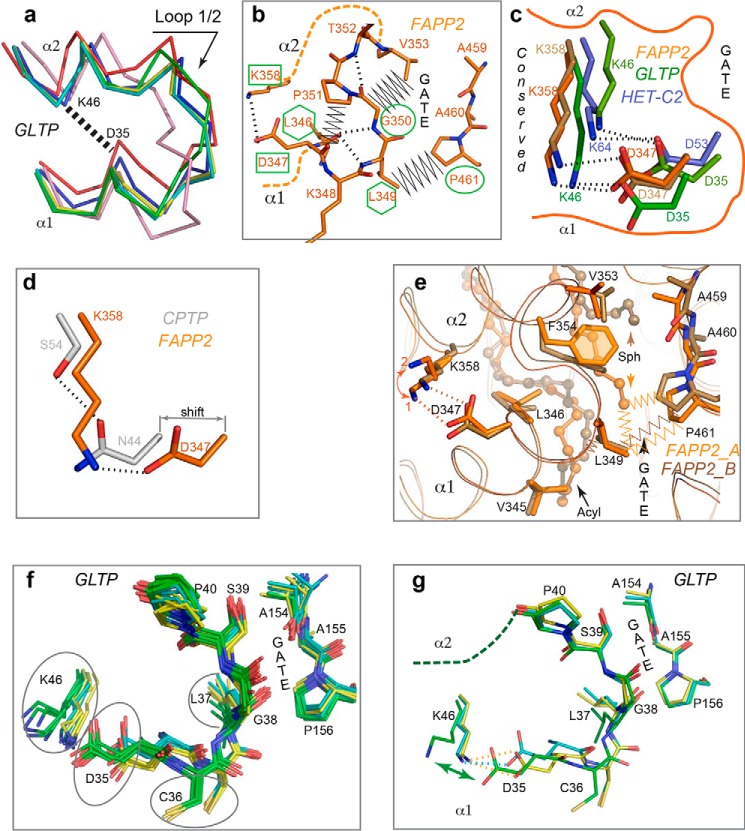
Adaptable clasp and other conserved elements involved in gate action. a, six superimposed molecular structures of human GLTP (PDB codes 1SX6, yellow; 1SWX, pink; 2EUK, cyan; 3RWA(a), red; 3RWA(b), blue; 3S0K, green) showing different conformations of one side of the gate, loop L1/2, adopted by the same protein. The dashed line indicates the approximate location of the conserved clasp for gate action. b, two opposing sides of the FAPP2–GLTPH gate depicting the strictly conserved Gly and Pro (outlined by ellipses), two conserved bulky-hydrophobic residues (outlined by hexagons), and the Lys358 and Asp347 residues (outlined by rectangles) of the conserved clasp for gate action. c, clasp connecting two ends of L1/2 loop of the gate and formed by the Asp347·Lys358 salt bridge (dashed lines) in both FAPP2–GLTPH molecules (different shades of orange) compared with human GLTP (different shades of green; PDB codes 1S0K and 1SWX) and HET-C2 (blue; PDB code 3KV0) illustrates bridge mobility and conservation in the GSL-specific family. d, substitution of the salt-bridged Asp·Lys pair in GSL-specific FAPP2–GLTPH for the H-bonded Asn·Ser pair in C1P-specific CPTP (PDB code 4K84) highlighting the preservation of the clasp (see Fig. S4e for unique Gly·Glu pair of ACD11). e, Φ1Φ2DXΦ3GX?XF motif conserved in the GSL-specific family (Φi = bulky hydrophobic residue; X = any residue; ? = X or none) with Φ1 (Val345), Φ2 (Leu346), Φ3 (Leu349), and Phe354 residues oriented toward the hydrophobic pocket and interacting with the acyl (Acyl) and sphingosine (Sph) chains. Note that the differing lipid conformations within the two protein molecules occupying AU (different shades of orange) correlate with conformational distinctions in their hydrophobic residues (mostly, in Phe354 and Leu349), as well as in their contacts (zig-zag lines). Remarkably, the Asp347·Lys358 pair also differs in two molecules. In FAPP2_A, Lys358 adopts the double conformation (orange 1 and 2), which either does not a salt bridge with Asp347 (orange 2) or results in a bridge that differs from that of in FAPP2_B (orange 1). f, superposition of 17 GLTP molecules from 12 PDB entries showing the gate-region with the Φ3-residue (Leu37) of the Φ1Φ2DXΦ3GX?XF motif that identifies three conformational groups for GLTP, colored in green, yellow, and cyan and three representative GLTP molecules (g) from visualizing the conformational distinctions between the three groups (f).
FAPP2 ID-loop limits selectivity to simple GSLs
Previous functional analyses show very different transfer activity by FAPP2–GLTPH for simple versus complex GSLs (2). Compared with GalCer and GlcCer, LacCer transfer efficiency declines by ∼50%, whereas transfer of SF and ganglioside GM1 is quite low. By contrast, GLTP efficiently transfers both simple and complex GSLs, including GalCer, GlcCer, LacCer, gangliosides, and sulfatides (17, 18).
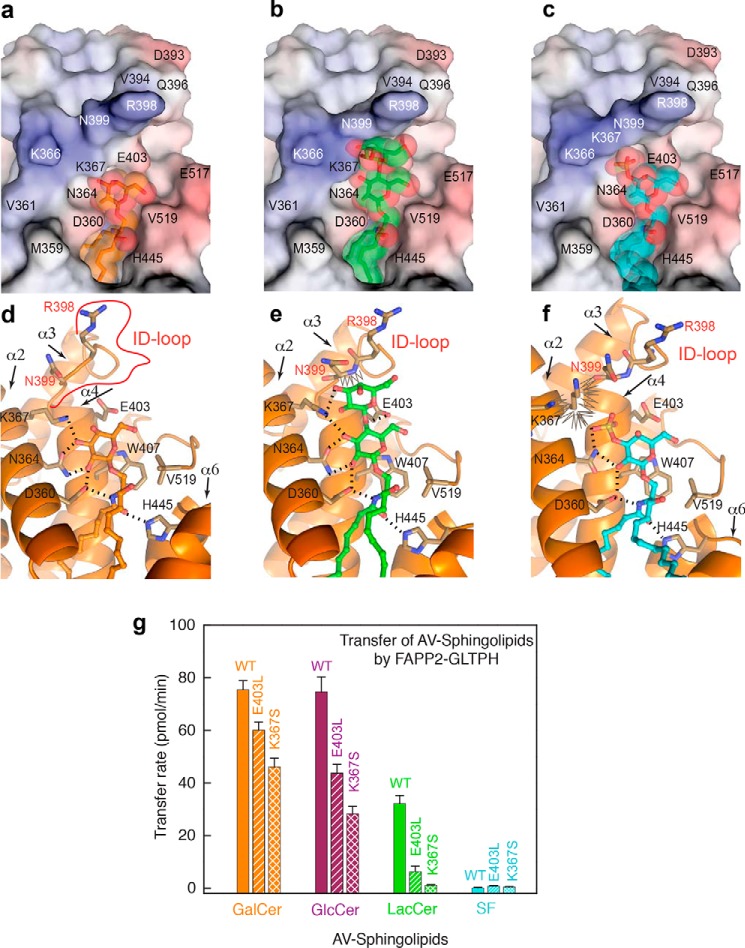
The ID-loop location behind the bound GSL headgroup (Figs. 2a and 5, a and d) suggests a potential regulatory role (negative or positive) in binding GSLs containing more than one sugar. Compared with GLTP, the main chain of the FAPP2 ID loop (especially Arg398 and Asn399) protrudes toward the GSL (Fig. 4h), where it can potentially interfere with complex headgroup binding, thereby limiting functional selectivity of FAPP2 to simple GSLs. To test the idea, we modeled the FAPP2·LacCer and FAPP2·SF complexes by superposition of the current crystal structure with GLTP·18:1-LacCer (PDB code 1SX6) and GLTP·3-O-SF (PDB code 3RZN and 4H2Z). In Fig. 5 (b, c, e, and f), the protein molecule is represented by FAPP2–GLTPH domain from the current crystal structure, but GalCer is replaced by LacCer or SF from the superimposed GLTP complexes. Because the ID loop is rigid, such models allow one to assess whether the loop and the GSL headgroup will collide in the potential complexes. For reference, the original structure of FAPP2–GLTPH·GalCer is shown in Fig. 5 (a and d). The protruding region of FAPP2 ID loop (R398 and N399 in Fig. 5, a–c) resembles a wall behind the GSL-sugars. The nearby Lys367 residue of α2-helix forms an H-bond with bound galactose (Fig. 5d). Accordingly, Lys367 projects inward in similar fashion to Lys55 in GLTP·GalCer complexes. The bound galactose is too far from the ID loop to be affected by nearby “wall” components, Asn399 and Arg398 (Fig. 5, a and d). However, in the modeled FAPP2–GLTPH·LacCer complex (Fig. 5, b and e), the second sugar ring of lactose moderately clashes against the wall. When GSLs have more than two headgroup sugars (e.g. gangliosides, globosides), serious interference by this wall effect is expected to significantly diminish GSL binding. This finding is consistent with functional data showing low transfer activity of FAPP2–GLTPH for ganglioside GM1 compared with GLTP (2).
Figure 5.
FAPP2 ID-loop regulation of GSL selectivity. a–c, electrostatics surface (blue, positive; red, negative; gray, neutral) of the FAPP2–GLTPH domain showing the ID loop, C terminus, and recognition center occupied by GalCer in the current crystal structure (a), compared with the partially impaired locations expected for LacCer (b), or sulfatide, SF (c), assessed by superposition with crystal structures of GLTP bound to LacCer (PDB code 1SX6) or 3-O-SF (PDB codes 3RZN and 4H2Z). GSL molecules are shown in stick representation, and GSL atoms are shown by spheres of van der Waals radii. GSL-atom color codes: blue for nitrogen; red for oxygen; and orange, green, and cyan for carbon in a, b, and c, respectively. d–f, protein·GSL interactions in the complexes shown in a–c, respectively, depicting hydrogen bonds (dashed lines) and van der Waals “clashes” (zig-zag lines). GSL atom color codes as in a–c. Lys367 and Asn399 conformations in b and e and Lys367 and Asn364 conformations in c and f were adapted to favorably interact with lipid headgroup and neighbor residues. For details, see text. g, FAPP2–GLTPH transfer activities for GalCer (orange), GlcCer (burgundy), acCer (green), and SF (cyan) by WT-GLTPH and E403L-GLTPH or K367S-GLTPH measured by FRET assay using AV lipids (see “Experimental procedures”).
Because the recognition center residues target mostly the initial sugar, binding of more complex heads is expected to involve additional nearby favorable interactions. Otherwise, the binding efficiency would decrease because of the increased size and complexity of the headgroup. Based on the favorable GM1 transfer by human GLTP (13), the complex double subloop architecture of the GLTP ID loop (Fig. 3a) appears likely to produce no local obstructions for the GM1 headgroup while promoting some additional interactions to tether the distal sugars to the protein surface.
In the modeled FAPP2–GLTPH·SF complex (Fig. 5, c and f), the proximity of the protruding ID loop to the potential 3-O-sulfo-group position (Fig. 5c) results in conformational restrictions, in particular for Lys367. Fig. 5 (c and f) illustrates the most favorable Lys367 conformation when the sulfo-group moderately clashes with the wall and Lys367 contacts too closely with a few surrounding atoms (zig-zag curve in Fig. 5f). Alternative conformations of Lys367 cause more noticeable clashes with either the sulfo-group or the wall indicating partial obstruction of SF binding to FAPP2–GLTPH because of limited space available for Lys367 in the complex. It is noteworthy that in human GLTP, the positively charged ϵ-amino group of Lys55 (FAPP2 Lys367 counterpart) plays only a secondary role in anchoring the negative sulfo-group to the protein. The primary role belongs to a conserved water molecule that shapes an adjustable complementary cavity on the protein surface (15). In all human GLTP crystal structures, this water molecule hydrogen bonds with the main-chain carbonyl at the ID/α4 junction and undergoes tight van der Waals contacts with certain atoms of ID loop and Leu92 (FAPP2 E403 counterpart) to not only favorably shape the surface cavity for complementary contacts with the 3-O-sulfo-group but also to directly bridge this group to the junction in the GLTP·SF complex (for details, see Fig. S2). Similarly, in GLTP·LacCer complexes, the same water molecule bridges one of the OH groups of the second sugar to the ID/α4 junction and makes favorable van der Waals contacts with other atoms of the ring. Thus, this conserved water (Wg in Fig. S2), found in all GLTP apo and holo forms, is a part of the protein structure, and its displacement from the protein surface is presumably associated with energy costs.
The finding of a similar water molecule, Wf, in the FAPP2 structure (Fig. S2, b and d) underscores its importance. However, the differently structured and protruding FAPP2 ID loop shifts Wf by ∼0.7 Å compared with Wg, a shift expected to generate a noticeable clash by Wf with the 3-O-sulfo-group in the potential FAPP2·SF complex (Fig. S2, b and d). To bind FAPP2, the 3-O-sulfo-group would need to displace Wf from the protein surface. Such displacement is not only energetically unfavorable but also would destroy the complementary support for the sulfo-group (for details, see Fig. S2, b and d). Thus, Wf in FAPP2 obstructs rather than supports SF binding via the protruding ID loop to interfere with transfer.
Point mutational analyses
To functionally evaluate the FAPP2–GLTPH ID loop regarding GSL-transfer selectivity while keeping the loop conformation intact, we point mutated residues Glu403 and Lys367 adjacent to the ID loop (Fig. 5a). Both residues contribute to the interaction network with the GSL headgroup (Fig. 1b) but are not primary recognition center residues. In Fig. 5g, FAPP2–GLTPH transfer activity for GalCer, GlcCer, LacCer, and 3-O-SF is compared with that of the E403L and K367S mutants using the well-established FRET approach for assessing transfer activity rates (14, 17, 19). Real-time kinetic traces for GSL intermembrane transfer by both mutants and wtFAPP2–GLTPH are shown in Fig. S3 (a–c). Transfer of GalCer and GlcCer still proceeded moderately well with the E403L and K367S mutants compared with the deleterious effect of both mutations on LacCer transfer (Fig. 5g). Remarkably, K367S substitution resulted in nearly complete loss of LacCer transfer by FAPP2—GLTPH, whereas GLTP-K55I (FAPP2-Lys367 counterpart) retained nearly full (90–97%) activity (12). It is noteworthy that neither FAPP2–GLTPH mutant can transfer SF (Fig. 5g), showing that removal of a negative (E403L) charge to reduce repulsion with the negatively charged sulfo-group or diminishing the crowding around it (K367S) did not improve SF transfer. This finding suggests that the protruding FAPP2 ID loop transforms the Wf water molecule from supporting 3-O-sulfo-group binding to hindering it.
Chimeric protein analyses
To further evaluate the role of FAPP2 ID loop (IDFAPP2) in GSL selectivity, we engineered two chimeras with swapped ID loops, IDFAPP2GLTP and IDGLTPFAPP2, and assessed ganglioside GM1 and sulfatide transfer. We acquired these data using surface plasmon resonance (SPR) because SPR can provide direct insights into both protein adsorption to the membrane as well as protein·GSL desorption from the membrane after GSL uptake within the same experiment (20–22). Fig. 6 shows the SPR response for the chimeras compared with control proteins, GLTP and FAPP2. The starting baseline in each sensorgram trace represents sensor chip saturation with immobilized 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) vesicles containing 0 (black), 5 (red), 10 (blue), or 20 mol % (green) of GM1 or sulfatide. Protein injections (red arrows) result in response increases that reflect protein adsorption onto the immobilized vesicles. Glycolipid uptake by proteins and departure from the immobilized vesicles is indicated by the signal decrease (relative to black trace) that occurs during the protein adsorption and generally requires ≥10 mol % glycolipid to clearly observe. The drops in the sensorgrams (black arrows) indicate a flow switch to buffer without protein. The magnitude of this drop is consistent with the relatively weak interactions of LTPs with membranes.
Figure 6.
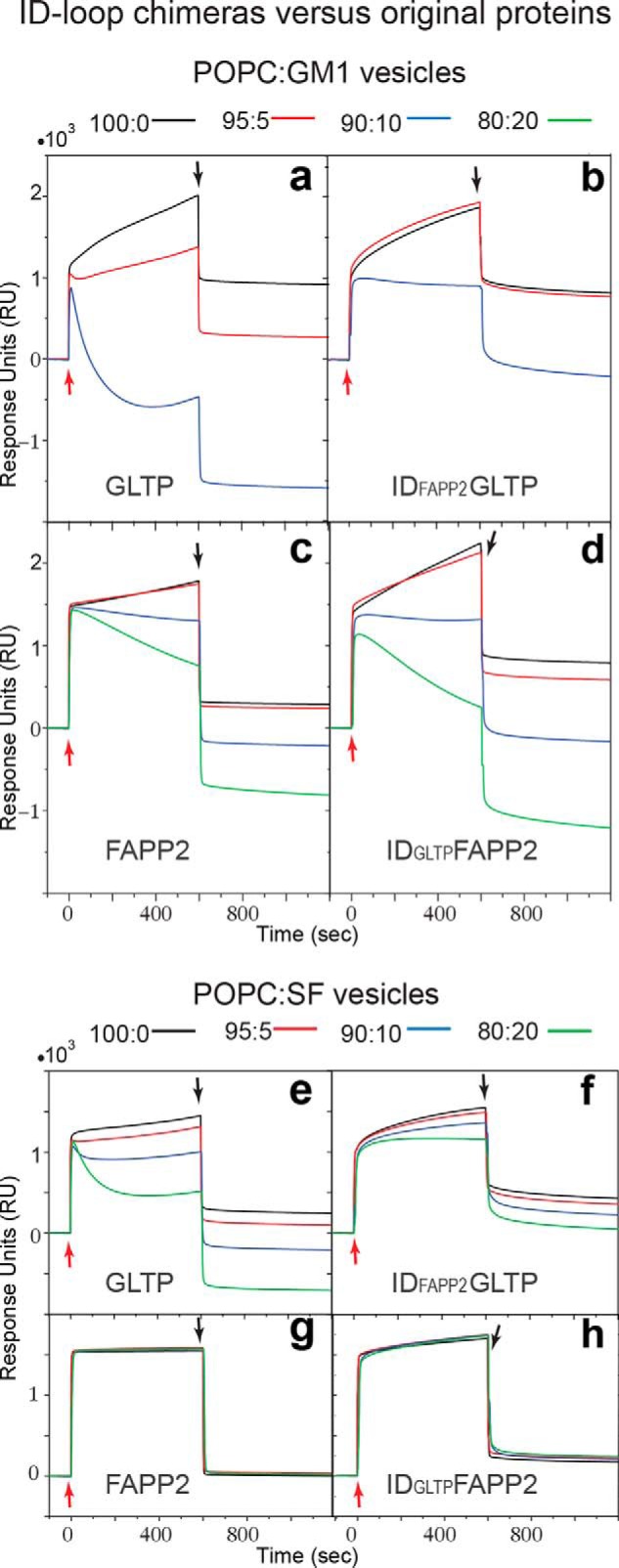
SPR sensorgrams of the interaction between lipid vesicles captured on a L1 chip and the engineered protein chimeras, IDFAPP2GLTP and IDGLTPFAPP2, versus GLTP and FAPP2–GLTPH. IDFAPP2 and IDGLTP are the ID loops of FAPP2 and GLTP, respectively. Vesicles are composed of POPC, 1-palmitoyl-2-oleoyl-sn-phosphatidylcholine, mixed with ganglioside GM1 (a–d) or 24:1 sulfatide, SF (e–h) in mol% ratios 100:0 (black), 95:5 (red), 90:10 (green), or 80:20 (blue). Proteins are dissolved at 0.1 mg/ml (5 μm) in running buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, pH 7.0). The flow rate is 5 μl/min. Red and black arrows indicate the start of protein injections and switching back to the buffer wash, respectively.
The GLTP data for GM1 (Fig. 6a) show that small GM1 amounts (5 mol %) alter the sensorgram shape and response magnitude as protein removes GM1 from vesicles (20, 21). At 10 mol % GM1 (blue curve), the response changes more drastically because of the highly efficient GM1 transfer by GLTP (2, 23). By contrast, the IDFAPP2GLTP chimera (Fig. 6b) triggers almost no change at 5% GM1, and the sensorgram at 10% GM1 (blue) is much less affected than those of GLTP, supporting the negative regulatory role of IDFAPP2 in GM1 transfer. Notably, the protein adsorption to immobilized vesicles is similar for both GLTP and the IDFAPP2GLTP chimera, indicating that the decreased GM1 transfer is not due to impaired protein–membrane interaction. With FAPP2 (Fig. 6c), vastly diminished GM1 uptake is observed, which is approximately nil at 5 mol % and moderately detectable at 10 and 20 mol % GM1. Uptake of GM1 by IDGLTPFAPP2 chimera (Fig. 6d) is better than for FAPP2, especially at higher GM1 levels, supporting a “gain-of-transfer function” associated with the GLTP ID loop.
SPR sensorgrams for POPC:sulfatide vesicles indicate efficient sulfatide uptake by GLTP (Fig. 6e) and significantly diminished uptake efficiency upon substitution of IDGLTP by IDFAPP2 (Fig. 6f), supporting the negative regulatory role of IDFAPP2 in sulfatide transfer. Puzzlingly, the complementary chimeric replacement (FAPP2 with IDGLTP) resulted in sensorgrams showing that neither FAPP2 (Fig. 6g) nor IDGLTPFAPP2 chimera (Fig. 6h) exhibit any ability to transfer sulfatide despite maintaining membrane interaction capacity.
Gating mechanism
As part of the SL transfer process, GLTP-family proteins capture the SL chain(s) via an undefined gating mechanism. The gate and associated cleft enable the lipid chains to access the hydrophobic pocket. The L1/2 loop and adjacent regions of α1 and α2 form one side of the gate (Fig. 2a), which is flexible, based on B-factor analysis (12) and different conformations adopted in same protein (Fig. 7a). The other side, which is composed of the L6/7 loop and adjacent regions of α6 and α7, appears to be more rigid based on its similar conformation in all GLTP superfamily members (Fig. 2a and Fig. S1a). Examination of the FAPP2 gate residues reveals that Gly350 (L1/2 loop) and Pro461 (L6/7 loop) are absolutely conserved in all members of the GLTP-family (Fig. 2b). Because Gly adds flexibility to a protein chain, and Pro usually forces a kink, their strict conservation in the gate (Fig. 2a, solid arrow) supports the importance of specific conformational properties for each side of the gate in the gating action. Fig. 7b illustrates the location of conserved Gly350 and Pro461 (outlined by green ellipses) in the FAPP2 gate.
Clasp control of gating action
A U-shaped arrangement for the α1- and α2-helices (Fig. 1a) results in an Asp347·Lys358 salt bridge (Fig. 7b, green rectangles) that connects the two helices near their junctions with the L1/2 loop (Fig. 2a, linkage 2). Fig. 2b shows conservation of the same Asp and Lys in the GSL-specific family. The Asp347·Lys358 salt bridges in FAPP2 are compared with GLTP and HET-C2 in Fig. 7c. In the C1P-specific family, different residues replace the positions of the conserved Asp and Lys in the GSL-specific family (Fig. 2b). Despite the changes, H-bond formation occurs between the Asn·Ser pair in most CPTP structures (Fig. 7d) as well as between the Gly·Glu pair in some ACD11 structures (Fig. S4e). The conserved location of the clasp residues near the gate in the GLTP fold suggests a role in maintaining gate functionality.
Hydrophobic sensor involvement in gate control
Inspection of the gate sequence (345VLDKLGPTVF354) encompassing the L1/2 loop and adjacent regions reveals a conserved Φ1Φ2DXΦ3GX?XF motif (where Φi is a bulky hydrophobic residue, X is any residue, and ? = X or none) (Fig. 2b) with four bulky hydrophobic residues, Φ1, Φ2, Φ3, and Phe. These residues potentially play a sensor role in gating regulation as shown by their locations and interactions with the bound aliphatic chains of 18:1-GalCer in the hydrophobic pocket (Fig. 7e). Val345 (Φ1) and Leu346 (Φ2) have contact with the acyl chain terminus and the C8–C12 methylene region, respectively, whereas conserved Phe354 interacts with the sphingoid chain. Leu349 (Φ3) contacts the sphingoid chain in one protein molecule and the acyl chain in the other. Leu349 also contacts the strictly conserved Pro of the L6/7 loop to firmly “latch” the gate at this location.
D·K bridging responds to sensor conformational changes
Fig. 7e also shows that sphingoid chain positional alterations are accompanied by “sensor” conformational change (mostly, Phe354 and Leu349 (Φ3)) as well as by variations in Asp347·Lys358 bridging. In FAPP2_A, Lys358 adopts a double-conformation (1 and 2), which either (conformer 2) does not contribute to the salt bridging with Asp347, or (conformer 1) results in a bridge that differs from that of in FAPP2_B. To address the issue of whether sensor conformations correlate with Asp·Lys bridging in the GLTP fold, we analyzed via superimposition of 17 molecular structures derived from 12 PDB entries (Fig. 7f). The side chains shown in Fig. 7f include the Φ3 residue, Leu37, and two salt bridge residues, Asp35 and Lys46. The superposition identifies three conformational groups of GLTP, colored cyan (five molecules), yellow (six molecules), and green (six molecules), while Fig. 7g emphasizes the distinctions between these three groups by showing three representative molecules from Fig. 7f. Two groups (cyan and yellow) share a similar Leu37 conformation (conformer 1) but differ by Asp35 position resulting in Asp oxygen “switching” in the Lys46·Asp35 salt bridge. The third group (green) displays a distinct Leu37 conformation (conformer 2) as well as changed Cys36 and Asp35 positions (Fig. 7g). The Asp35 carboxyl protrudes toward the Lys46 residue, which repositions to avoid clashing with Asp35. The resulting Lys46·Asp35 salt bridge distance increases from 2.6–3.2 Å (first two groups) to 3.1–4.0 Å (third group), indicating salt bridge weakening.
Bridge strength “recovery” is observed in the special conformation of the GLTP gate (PDB code 3S0K), where Asp35 protrudes even more toward Lys46 because of a conformational switching of the conserved Gly38 of the L1/2 loop (Fig. S4b). Remarkably, the Leu37 conformation reverts back to conformation 1, supporting the correlation with salt-bridge strength. The findings suggest that the conserved linkage 2 behaves as a “variable strength clasp” that is regulated by signals from the sensor residues of the hydrophobic pocket. It is noteworthy that GLTP structures that belong to the third conformational group are those of GSL-bound forms, in which the hydrophobic pocket is fully occupied by aliphatic chains including the bottom region designed for the terminal atoms of long acyl chains (12, 15, 16). Thus, the analyses suggest that the hydrophobic pocket occupancy influences the sensor conformations, whereas the clasp “senses” the alterations (Fig. 7, f and g) by receiving an integrated signal from the sensors.
Novel FF motif location in FAPP2–GLTPH
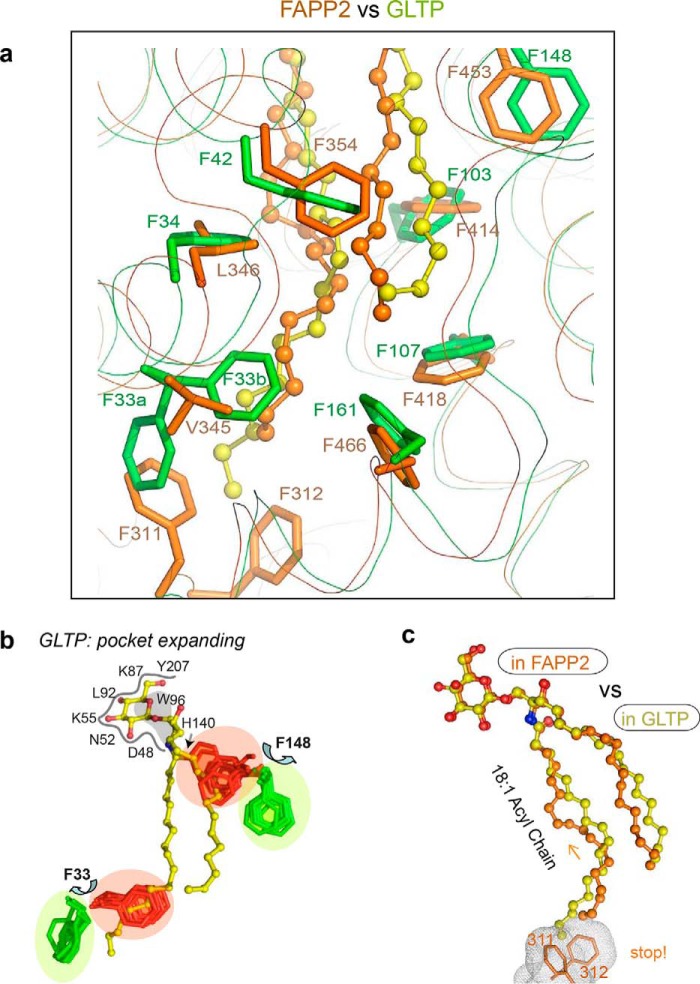
The hydrophobic pockets that encapsulate lipid chains are enriched in phenylalanines (10, 12, 14–16). Seven phenylalanines line the pockets of FAPP2 and GLTP (Fig. 8a), but only five are positionally conserved because of differently located FF motifs, both of which interact with the bound acyl chain, suggesting a possible functional role in regulating acyl chain accommodation in the pocket.
Figure 8.
Location of Phe and FF motifs in the hydrophobic pocket of FAPP2–GLTPH versus GLTP. a, superposition of FAPP2–GLTPH (orange) with GLTP (green) indicating five coinciding Phe positions in the hydrophobic pockets of both proteins but differently located FF motifs (Phe311–Phe312 in FAPP2 versus Phe33-Phe34 in GLTP; for details, see the text). b, two Phe conformations, the open-door (green) and closed-door (red), identified in human GLTP for Phe33 and Phe148 residues via superposition of all available GLTP structures. Phe functionality as doors for Phe33 and Phe148 regulates acyl and sphingosine chain access to the hydrophobic pocket of GLTP. c, 18:1-Acyl chain adaptation within the hydrophobic pockets of FAPP2–GLTPH (versus the pocket of GLTP) depicting the clashing role of FF motif that seals the bottom of the hydrophobic pocket in FAPP2.
In GLTP, the accessible volume of the hydrophobic pocket depends strongly on the conformation of Phe148 and Phe33 (Fig. 8b) (19). To collapse the pocket when unoccupied, both take the “closed-door” conformation (colored red). To accommodate the GSL chains, switching to the “open-door” conformations (colored green) is needed. The mechanism that controls the Phe148 closed-door/open-door transition appears to involve the synchronized action of three conserved residues, Tyr132, Phe42, and His140 (FAPP2 Tyr437, Phe354, and His445 counterparts), and requires GSL-headgroup anchoring by the recognition center to initiate the transition (15).
FAPP2 Phe453 (GLTP Phe148 counterpart) is maintained in the open-door conformation (Fig. 8a) by the same interaction pattern consistent with a similar transition mechanism for this Phe. By contrast, GLTP Phe33 is unique, because it belongs to the FF motif, in which two phenylalanines synchronize their action to help the pocket accommodate acyl chains of different lengths (16). The Phe33 closed-door/open-door transition (Fig. 8b) seems to help guide the terminal part of the long acyl chains to occupy the narrow bottom of the GLTP pocket rather than protruding outward.
The absence of a similarly located FF motif in FAPP2–GLTPH appears to be compensated by the differently located FAPP2 FF motif (Phe311–Phe312) that projects from the bottom of the hydrophobic pocket. The Phe311–Phe312 motif is held firmly in place by the tight contacts with α7- and α8-helices and seals the bottom of the pocket. Because the terminal part of the encapsulated acyl chain is surrounded by Val345, Leu346, Val342, Leu349, Leu415, Leu419, Leu349, and Leu470 (Fig. 1c), the chain must bend to avoid clashing with the FF motif. The conformational adaptation by the chain is evident by comparison with the 18:1 acyl chain of GlcCer bound to GLTP (Fig. 8c). Unlike FAPP2, GLTP has special space in its hydrophobic pocket for accommodating the terminal portions of long acyl chains (11, 16, 24). By sealing the bottom, the FAPP2 FF motif restricts the conformational space for GSLs with long acyl chains to perhaps change their binding mode with FAPP2.
Conclusions
The new data and analyses reported in this study lead to the conclusion that the all-α GLTP fold characterizing the GLTP superfamily is designed to recognize ceramide-based SLs with polar headgroups. The selectivity for the ceramide moiety originates from the highly conserved Asp and His residues of the recognition center (Fig. 2b) that bind the ceramide amide group in a strictly determinate manner (Fig. 1b). The requirement for a polar headgroup bonded to the ceramide O1 atom is evident from the existence of the recognition center (Fig. 2b).
The recognition center consists of six distinct residue positions grouped at one face of the protein. In the two known GSL- and C1P-specific GLTP subfamilies, the pattern consists of DNKxWH and DKxRRH. These spatial templates recognize the ceramide-amide group and initial ceramide linked sugar or phosphate, respectively.
The other key element used by the GLTP fold to control SL specificity is the ID loop. In each family member, the ID loop is distinct in sequence and length and locked via pinched together ends resulting in ID-loop structure that is relatively autonomous and rigid. In this way, ID-loop design is ideal for fine turning (enhancing or mitigating) specificity within a subfamily. Fig. 9 schematically depicts ID-loop action in GLTP and FAPP2 based on different transfer abilities for ganglioside GM1 observed in this study. Highly efficient GM1 transfer by human GLTP (Fig. 6a) leads us to conclude that the IDGLTP promotes additional (nonspecific) interactions that tether the distal GM1 sugar rings to the protein surface (Fig. 9a). By contrast, the FAPP2–GLTPH domain is much less efficient at transferring GM1 (Fig. 6c), because the IDFAPP2 creates a hindrance for the distal sugar rings, resulting in conformational stress upon the binding (Fig. 9b). The restrictive role of IDFAPP2 is supported by the loss of GM1 transfer efficiency by GLTP upon the substitution of IDGLTP by IDFAPP2 (Fig. 6b). Thus, we conclude that the recognition center works in concert with the ID loop to control the specificity in the GLTP family.
Figure 9.
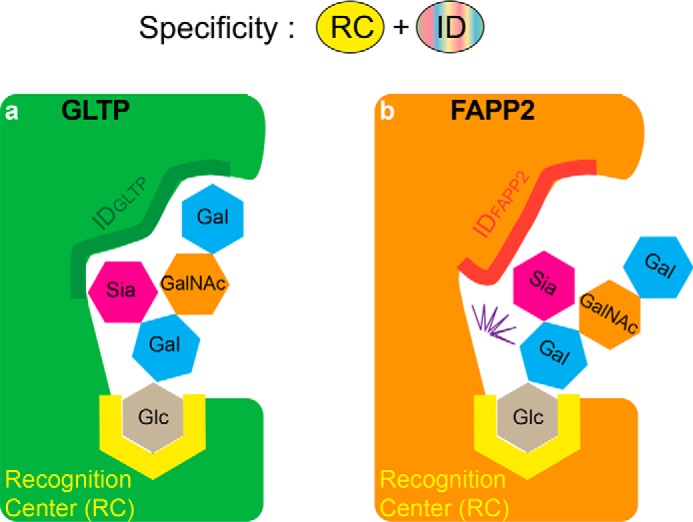
Schematic depiction of the recognition center and ID-loop elements controlling the SL specificity in the GLTP family. Conserved recognition center templates target the initial part of SL headgroup to designate binding of the SL class (GSL or C1P), whereas different ID loops interact with the distal parts of SL headgroups to select for certain species within an SL class (i.e. specificity “individualization” in subfamily members). GLTP and FAPP2–GLTPH are the two examples depicted for GSL-specific members of the GLTP family with ganglioside GM1 providing an example of a bulky headgroup GSL. a, supportive ID in GLTP facilitates interaction with the distal part of the SL headgroup to promote GM1 transfer activity by GLTP. b, obstructive ID in FAPP2 creates conformational stress (zig-zag) for GM1 headgroup to limit GM1 binding.
Among other findings, we also identified previously unrecognized and functionally important elements of the GLTP fold. They are (i) four hydrophobic bulky residues that play a sensor role in gating function; (ii) a conserved, adaptable, variable-strength clasp that maintains gate functionality and is controlled by sensors of the hydrophobic pocket occupancy; and (iii) an FF motif that seals the bottom of the hydrophobic pocket to restrict the conformational space for long acyl chains. Our structural data indicate that, in the absence of major conformational change, the FF motifs in the GLTP fold are unlikely to serve as the FFAT motifs (two phenylalanines in an acidic tract) that interact with VAP (vesicle-associated membrane protein-associated protein) (25, 26). However, the contributions of FF motifs to mechanisms that control the shaping and conformation of the hydrophobic pocket for lipid chains are evident.
Experimental procedures
Protein mutagenesis, expression, and purification
Human FAPP2 GLTPH-domain (C-terminal 212 residues = FAPP2-C212) was derived from PLEKHA8 ORF (519 residues; GenBankTM accession number MF465867) (2). Molecular cloning, expression, and initial purification of FAPP2–GLTPH, human GLTP (GenBankTM accession number AF209704), and all mutants involved fusion with the N-terminal His6–SUMO tag, which was then removed by proteolysis to achieve final purification (2). E377A/E378K double mutant and point mutations E403L and K367S were produced by QuikChange site-directed mutagenesis kit (Stratagene) and verified by sequencing. Cloning of chimeric human GLTP, IDFAPP2GLTP, and chimeric human FAPP2–GLTPH, IDGLTPFAPP2, containing swapped ID loops involved replacing the ID loop of GLTP (YGAEWPKVG) with that (DVAQVRNS) of FAPP2 and vice versa. Transformed BL21 (DE3) cells were grown in Luria–Bertani medium at 37 °C until the exponential growth phase, induced with 0.1 mm isopropyl β-d-thiogalactopyranoside, and then grown for 16–20 h at 20 °C. Ni2+–nitrilotriacetic acid affinity chromatography purification from soluble lysate of transformed cells was followed by overnight cleavage of His6-SUMO tag at 4 °C with SUMO-specific protease, Ulp1. Cleanup involved a nickel-chelating column followed by gel filtration chromatography purification using a HiLoad 16/60 Superdex-75 prep grade column (GE Healthcare) equilibrated in buffer containing 25 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 1 mm DTT. Protein purity was assessed by SDS–PAGE plus peptide fingerprinting for FAPP2.
Crystallization and X-ray data collection
E377A/E378K mutant of FAPP2–GLTPH domain displayed improved crystallizability. The protein–lipid complex was prepared by mixing the mutant and 18:1-GalCer (Avanti Polar Lipids, Alabaster, AL) at a 1:1 molar ratio in ethanol (∼40% EtOH). Crystals of the complex were grown by the hanging-drop vapor-diffusion method using PEG 8000 (13–15%) as precipitant and 100 mm MES, pH 6.5, as buffer. A single crystal was transferred into well solution containing 20% glycerol cryoprotectant, mounted in a fiber loop, and flash-frozen in a cold nitrogen stream. X-ray data were collected at λ = 0.97242 Å and 100 K to 1.45 Å resolution using Synchrotron radiation on Beamline ID 23-1 at the European Synchrotron Radiation Facility (Grenoble, France) equipped with Pilatus 6M-F detector. The data were processed, scaled, and divided into work (95%) and test (5%) subsets using the XDS program (27) and the Scala program (28) from the CCP4 suite.
Structure determination and refinement
Crystals of FAPP2–GLTPH complexed with 18:1-GalCer belonged to the P212121 space group and contained two molecules in the AU. The structure was determined by molecular replacement, using the MOLREP program (29) against X-ray data cut at 2.3 Å resolution and protein structure from the GLTP/24:1-GalCer complex (PDB code 2EUK) (16) as a search model. Found solution was prerefined with REFMAC (30) at 3.0 Å resolution to Rwork/Rfree = 0.53/0.55 and used as the initial model in ARP/wARP automated model building procedure intermittent with REFMAC refinement cycles (31). The model building performed against X-ray data cut at 1.8 Å resolution has been completed with protein chains built by 89% and refined to Rwork/Rfree of 0.246/0.294 for the resolution range 15.0–1.8 Å. Insertions of two lipid molecules and missing residues of polypeptide chains were made manually using electron density maps in COOT program. Further refinement to Rwork/Rfree of 0.131/0.179 was executed using a full data set up to 1.45 Å resolution with REFMAC alternating with minor model rebuildings in Coot. ARP/wARP was used to add solvent molecules (32). The final structure was validated by PROCHECKm and coordinates have been deposited in the Protein Data Bank (code 5KDI). Data collection and model refinement statistics, along with unit cell dimensions, are summarized in Table 1.
Fluorescent lipids
POPC was purchased from Avanti Polar Lipids. Lipid containing 3-perylenoyl (Per) or anthrylvinyl (AV) fluorophore (Per-PC, AV-GalCer, AV-GlcCer, AV-LacCer, and AV-sulfatide) and Me4-BODIPY-15-GalCer were synthesized as described in Refs. 33–35.
Vesicle preparation
Acceptor POPC vesicles and donor vesicles composed of POPC (98 mol %), AV-lipid (1 mol %), and Per-PC (1 mol %) or POPC (97.5 mol %), Me4-BODIPY-15-GalCer (1 mol %), and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylin-docarbocyanine perchlorate (1.5 mol %) were prepared as described in Ref. 36. Acceptor vesicle diameter averaged ∼25 nm. The final acceptor vesicle concentration in the FRET lipid transfer assay was ∼85 μm, which was 10–15-fold higher than the donor vesicle concentration. Buffer contained 10 mm potassium phosphate (pH 6.6), 150 mm NaCl, and 0.2% EDTA. POPC:GSL vesicles for SPR assays containing different amounts (0, 5, 10, or 20 mol%) of ganglioside GM1 or 24:1 sulfatide, SF, were prepared by brief sonication and purified as described in Refs. 21 and 22.
Fluorescent lipid transfer between membranes
Real-time intermembrane transfer rates of fluorescent glycolipids by FAPP2–GLTPH and all mutants were obtained by FRET using a SPEX FluoroLog3 spectrofluorimeter (Horiba Scientific), with excitation and emission bandpasses of 2 nm and a stirred (∼100 rpm), temperature-controlled (25 ± 0.1 °C) sample cuvette holder. All fluorescent lipids were localized initially to the donor vesicles, formed by rapid ethanol injection. Minimal emission by AV or BODIPY lipid occurred upon excitation (370 or 460 nm, respectively) because of resonance energy transfer to nearby Per-PC or C18-DiI. The addition of ∼10-fold excess of sonicated POPC acceptor vesicles produced minimal change in fluorescence signal (36). Protein addition triggered a sudden, exponential increase in AV or BODIPY emission intensity (415 or 503 nm, respectively) as FRET decreased because of protein transport of fluorescent glycolipids from donor vesicles to acceptor vesicles, creating separation from “nontransferable” Per-PC or C18-DiI lipids. The addition of Tween 20 detergent after extended incubation provided a measure of maximum intensity achievable by “infinite” fluorophore separation. Nonlinear regression analyses using ORIGIN 7.0 software enabled quantification of the initial lipid transfer rate, ν0, for the first-order exponential transfer process. Standard deviations were calculated at 95% confidence interval. R2 values for all estimates were >0.96.
Surface plasmon resonance
The assays were performed at 25 °C using a Biacore T200 system (GE Healthcare Bio-Sciences Corp). Vesicles (1 mm) in running buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, pH 7.0) were captured to a final surface density of 3000–6000 response units on a Series S Sensor Chip L1 to establish the baseline prior to protein addition. Injections of proteins (0.1 mg/ml) were performed for 10 min at 5 μl/min flow rates. Then the chip surface was washed for 10–15 min with running buffer and then was regenerated with CHAPS, as described in Refs. 20–22. The measurements were repeated twice for each sample.
Author contributions
B. O.-L., A. N. P., V. R. S., X. Z., and L. M. data curation; B. O.-L., Y.-G. G., X. Z., S. K. M., I. A. B., J. G. M., and D. K. S. investigation; B. O.-L., Y.-G. G., and X. Z. methodology; V. R. S., R. E. B, and L. M. validation; Y.-G. G., V. R. S., and X. Z. and L. M. formal analysis; D. J. P., R. E. B., and L. M. writing, reviewing, and editing; R. E. B. and L. M. conceptualization; R. E. B. and L. M. supervision; R. E. B. funding acquisition; L. M. writing-original draft; R. E. B. and L. M. project administration.
Supplementary Material
Acknowledgments
We thank the Proteomics Platform at CIC bioGUNE, a member of the Spanish ProteoRed-ISCIII Network and CIBERehd, for peptide fingerprint tests.
This work was supported in whole or part by National Institutes of Health Grants GM45928 from NIGMS, CA121493 from NCI, and HL125353 from NHLBI; Ministerio de Ciencia e Innovacion Grant BFU2010-17711; Russian Foundation for Basic Research Grant 14-04-01671; CIC bioGUNE funds; the Abby Rockefeller Mauze Trust; the Maloris and Hormel Foundations; and the Federal Agency of Scientific Organizations. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
The atomic coordinates and structure factors (code 5KDI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FAPP2
- phosphoinositol 4-phosphate adaptor protein 2
- GLTP
- glycolipid transfer protein
- GLTPH
- GLTP homology
- GLTP fold
- the fold first found in human GLTP
- SL
- sphingolipid
- GSL
- glycosphingolipid
- GlcCer
- glucosylceramide
- GalCer
- galactosylceramide
- LacCer
- lactosylceramide
- POPC
- 1-palmitoyl-2-oleyl phosphatidylcholine
- Per
- perylenoyl
- AV
- anthrylvinyl
- SF
- sulfatide
- PDB
- Protein Data Bank
- LTP
- lipid-transfer protein
- C1P
- ceramide-1-phosphate
- ACD11
- accelerated cell death 11 protein
- AU
- asymmetric unit
- SPR
- surface plasmon resonance
- SUMO
- small ubiquitin-like modifier.
References
- 1. D'Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C. C., van der Spoel A. C., Platt F. M., Hannun Y. A., Polishchuk R., Mattjus P., and De Matteis M. A. (2007) Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature 449, 62–67 [DOI] [PubMed] [Google Scholar]
- 2. Kamlekar R. K., Simanshu D. K., Gao Y. G., Kenoth R., Pike H. M., Prendergast F. G., Malinina L., Molotkovsky J. G., Venyaminov S. Y., Patel D. J., and Brown R. E. (2013) The glycolipid transfer protein (GLTP) domain of phosphoinositol 4-phosphate adaptor protein-2 (FAPP2): structure drives preference for simple neutral glycosphingolipids. Biochim. Biophys. Acta 1831, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Angelo G., Uemura T., Chuang C. C., Polishchuk E., Santoro M., Ohvo-Rekila H., Sato T., Di Tullio G., Varriale A., D'Auria S., Daniele T., Capuani F., Johannes L., Mattjus P., Monti M., Pucci P., Williams R. L., Burke J. E., Platt F. M., Harada A., and De Matteis M. A. (2013) Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature 501, 116–120 [DOI] [PubMed] [Google Scholar]
- 4. Mattjus P. (2009) Glycolipid transfer proteins and membrane interaction. Biochim. Biophys. Acta 1788, 267–272 [DOI] [PubMed] [Google Scholar]
- 5. Abe A., Yamada K., and Sasaki T. (1982) A protein purified from pig brain accelerates the inter-membranous translocation of mono- and dihexosylceramides, but not the translocation of phospholipids. Biochem. Biophys. Res. Commun. 104, 1386–1393 [DOI] [PubMed] [Google Scholar]
- 6. Abe A., and Sasaki T. (1985) Purification and some properties of the glycolipid transfer protein from pig brain. J. Biol. Chem. 260, 11231–11239 [PubMed] [Google Scholar]
- 7. Brown R. E., Stephenson F. A., Markello T., Barenholz Y., and Thompson T. E. (1985) Properties of a specific glycolipid transfer protein from bovine brain. Chem. Phys. Lipids 38, 79–93 [DOI] [PubMed] [Google Scholar]
- 8. Brown R. E., Jarvis K. L., and Hyland K. J. (1990) Purification and characterization of glycolipid transfer protein from bovine brain. Biochim. Biophys. Acta 1044, 77–83 [DOI] [PubMed] [Google Scholar]
- 9. Metz R. J., and Radin N. S. (1982) Purification and properties of a cerebroside transfer protein. J. Biol. Chem. 257, 12901–12907 [PubMed] [Google Scholar]
- 10. Simanshu D. K., Kamlekar R. K., Wijesinghe D. S., Zou X., Zhai X., Mishra S. K., Molotkovsky J. G., Malinina L., Hinchcliffe E. H., Chalfant C. E., Brown R. E., and Patel D. J. (2013) Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature 500, 463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malinina L., Simanshu D. K., Zhai X., Samygina V. R., Kamlekar R., Kenoth R., Ochoa-Lizarralde B., Malakhova M. L., Molotkovsky J. G., Patel D. J., and Brown R. E. (2015) Sphingolipid transfer proteins defined by the GLTP-fold. Q Rev. Biophys. 48, 281–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malinina L., Malakhova M. L., Teplov A., Brown R. E., and Patel D. J. (2004) Structural basis for glycosphingolipid transfer specificity. Nature 430, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kenoth R., Simanshu D. K., Kamlekar R. K., Pike H. M., Molotkovsky J. G., Benson L. M., Bergen H. R. 3rd, Prendergast F. G., Malinina L., Venyaminov S. Y., Patel D. J., and Brown R. E. (2010) Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold. J. Biol. Chem. 285, 13066–13078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simanshu D. K., Zhai X., Munch D., Hofius D., Markham J. E., Bielawski J., Bielawska A., Malinina L., Molotkovsky J. G., Mundy J. W., Patel D. J., and Brown R. E. (2014) Arabidopsis accelerated cell death 11, ACD11, is a ceramide-1-phosphate transfer protein and intermediary regulator of phytoceramide levels. Cell Rep. 6, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Samygina V. R., Popov A. N., Cabo-Bilbao A., Ochoa-Lizarralde B., Goni-de-Cerio F., Zhai X., Molotkovsky J. G., Patel D. J., Brown R. E., and Malinina L. (2011) Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein. Structure 19, 1644–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malinina L., Malakhova M. L., Kanack A. T., Lu M., Abagyan R., Brown R. E., and Patel D. J. (2006) The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure. PLos Biol. 4, e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown R. E., and Mattjus P. (2007) Glycolipid transfer proteins. Biochim. Biophys. Acta 1771, 746–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mattjus P. (2016) Specificity of the mammalian glycolipid transfer proteins. Chem. Phys. Lipids 194, 72–78 [DOI] [PubMed] [Google Scholar]
- 19. Samygina V. R., Ochoa-Lizarralde B., Popov A. N., Cabo-Bilbao A., Goni-de-Cerio F., Molotkovsky J. G., Patel D. J., Brown R. E., and Malinina L. (2013) Structural insights into lipid-dependent reversible dimerization of human GLTP. Acta Crystallogr. D Biol. Crystallogr. 69, 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sugiki T., Takahashi H., Nagasu M., Hanada K., and Shimada I. (2010) Real-time assay method of lipid extraction activity. Anal. Biochem. 399, 162–167 [DOI] [PubMed] [Google Scholar]
- 21. Ohvo-Rekila H., and Mattjus P. (2011) Monitoring glycolipid transfer protein activity and membrane interaction with the surface plasmon resonance technique. Biochim. Biophys. Acta 1808, 47–54 [DOI] [PubMed] [Google Scholar]
- 22. Zhai X., Gao Y. G., Mishra S. K., Simanshu D. K., Boldyrev I. A., Benson L. M., Bergen H. R. 3rd, Malinina L., Mundy J., Molotkovsky J. G., Patel D. J., and Brown R. E. (2017) Phosphatidylserine Stimulates Ceramide 1-Phosphate (C1P) Intermembrane Transfer by C1P Transfer Proteins. J. Biol. Chem. 292, 2531–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carton I., Malinina L., and Richter R. P. (2010) Dynamic modulation of the glycosphingolipid content in supported lipid bilayers by glycolipid transfer protein. Biophys. J. 99, 2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malinina L., Patel D. J., and Brown R. E. (2017) How alpha-Helical Motifs Form Functionally Diverse Lipid-Binding Compartments. Annu. Rev. Biochem. 86, 609–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tuuf J., Wistbacka L., and Mattjus P. (2009) The glycolipid transfer protein interacts with the vesicle-associated membrane protein-associated protein VAP-A. Biochem. Biophys. Res. Commun. 388, 395–399 [DOI] [PubMed] [Google Scholar]
- 26. Mikitova V., and Levine T. P. (2012) Analysis of the key elements of FFAT-like motifs identifies new proteins that potentially bind VAP on the ER, including two AKAPs and FAPP2. PLoS ONE 7, e30455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsh W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. D66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evans P. R. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. D62, 72–82 [DOI] [PubMed] [Google Scholar]
- 29. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31. Perrakis A., Morris R., and Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 32. Lamzin V. S., and Wilson K. S. (1993) Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 33. Molotkovsky J. G., and Bergelson L. D. (1982) Perylenoyl-labeled lipid-specific fluorescent probes. Bioorg. Khim. 8, 1256–1262 [Google Scholar]
- 34. Molotkovsky J. G., Mikhalyov I. I., Imbs A. B., and Bergelson L. D. (1991) Synthesis and characterization of new fluorescent glycolipid probes. Molecular organisation of glycosphingolipids in mixed-composition lipid bilayers. Chem. Phys. Lipids 58, 199–212 [Google Scholar]
- 35. Boldyrev I. A., Zhai X., Momsen M. M., Brockman H. L., Brown R. E., and Molotkovsky J. G. (2007) New BODIPY lipid probes for fluorescence studies of membranes. J. Lipid Res. 48, 1518–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mattjus P., Molotkovsky J. G., Smaby J. M., and Brown R. E. (1999) A fluorescence resonance energy transfer approach for monitoring protein-mediated glycolipid transfer between vesicle membranes. Anal. Biochem. 268, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.