Abstract
The human genome encodes 10 insulin-like genes, whereas the Caenorhabditis elegans genome remarkably encodes 40 insulin-like genes. Knockout strategies to determine the roles of all the insulin/insulin-like peptide ligands (INS) in C. elegans has been challenging due to functional redundancy. Here, we individually overexpressed each of the 40 ins genes pan-neuronally, and monitored multiple phenotypes including: L1 arrest life span, neuroblast divisions under L1 arrest, dauer formation, and fat accumulation, as readouts to characterize the functions of each INS in vivo. Of the 40 INS peptides, we found functions for 35 INS peptides and functionally categorized each as agonists, antagonists, or of pleiotropic function. In particular, we found that 9 of 16 agonistic INS peptides shortened L1 arrest life span and promoted neuroblast divisions during L1 arrest. Our study revealed that a subset of β-class INS peptides that contain a distinct F peptide sequence are agonists. Our work is the first to categorize the structures of INS peptides and relate these structures to the functions of all 40 INS peptides in vivo. Our findings will promote the study of insulin function on development, metabolism, and aging-related diseases.
Keywords: insulin, insulin-like growth factor (IGF), Caenorhabditis elegans (C. elegans), aging, cell division, C. elegans, dauer formation, fat accumulation, insulin like peptides, L1 arrest
Introduction
The Caenorhabditis elegans insulin/insulin-like growth factor signaling (IIS)2 pathway has been extensively studied and the IIS pathway components are evolutionary conserved in metazoans (1). Insulin-like (INS) peptides bind to and activate cell-surface receptors with intrinsic tyrosine kinase activity (2, 3). Autophosphorylation of the receptors promote the recruitment and activation of downstream components to initiate their biological effects (4). The insulin superfamily genes are ubiquitous and have been identified in all animals (5, 6). Compared with human and the fruit fly, Drosophila melanogaster, which have 10 and eight INS peptides, respectively, the C. elegans genome encodes 40 INS genes (5, 7), suggesting that there may be more functional diversity among the C. elegans INS peptides. Strikingly, there is only one INS receptor, DAF-2/INSR, which the 40 INS peptides are thought to bind as ligands. To date, no single loss of function mutation can fully recapitulate the phenotypes associated with loss of the daf-2 insulin receptor (8). The lack of loss-of-function phenotypes for many of the INS peptides suggests that some act redundantly in C. elegans, and this makes addressing INS functions by knockout strategies challenging and limited (8, 9).
In mammals (including humans), INS peptides and IIS signaling control glucose levels, hormone homeostasis, and metabolism (10). In C. elegans, IIS signaling controls aging, development, behavior, dauer formation, as well as fat accumulation (11). By using dauer formation as a readout phenotype, only INS-4, -6, and -9 and DAF-28 were identified as potential agonists, whereas INS-1, -17, and -18 were identified as potential antagonists (7, 9, 12–14). Some INS peptides were suggested to be potential agonists or antagonists based on mRNA expression dynamics between fed and starved conditions (15) and the INS peptides have been shown to regulate each other transcriptionally (9). However, the roles and functions of all 40 INS peptides remain unclear. INS peptides are expressed primarily in the nervous system (7). As such, we individually overexpressed each of the 40 INS peptides in all neurons using a pan-neuronal promoter to direct gene expression (16). Overexpression (oe) lines were then characterized based on phenotypes associated with abnormal IIS signaling in C. elegans. As an example, in the absence of food, C. elegans can arrest development in the first larval stage (L1 arrest), preventing further growth and development (17). The level of INS signaling can shorten or lengthen the life span of L1-arrested animals. Here, we assayed the contribution of individual INS peptides on phenotypes associated with IIS signaling that include alterations in L1 arrest life span, Q neuroblast divisions, dauer formation, and fat metabolism. Based on these assays, seven INS peptides (INS-3, -4, -6, -9, -19, and -32 and DAF-28) were categorized as strong agonists and three INS peptides (INS-17, -37, and -39) were strong antagonists of the IIS receptor in vivo. Nine INS peptides (INS-1, -2, -10, -11, -13, -20, -24, -29, and -35) were found to be weak agonists and five INS peptides (INS-15, -21, -22, -36, and -38) functioned as weak antagonists. Five INS peptides (INS-5, -23, -26, -27, and -33) did not exhibit any significant phenotype in vivo. Eleven of the 40 INS peptides have diverse roles serving as agonists or antagonists of IIS depending on the phenotypes scored. The 40 INS peptides in C. elegans have been grouped into three classes: α, β, and γ, on the basis of predicted arrangements of their disulfide bonds (7). Our work revealed that the β class INS peptides that contain a sequence known as the F peptide is a strong predictor of an INS with activation properties. The majority of β class INS function as agonistic ligands of IIS. Our INS overexpression work reveals the functional nature of signaling for each of the 40 INS in vivo, and promotes future studies on the functions of the entire C. elegans insulin gene family on aging, development, and metabolic diseases.
Results
Overexpressed INS affect L1 arrest life span
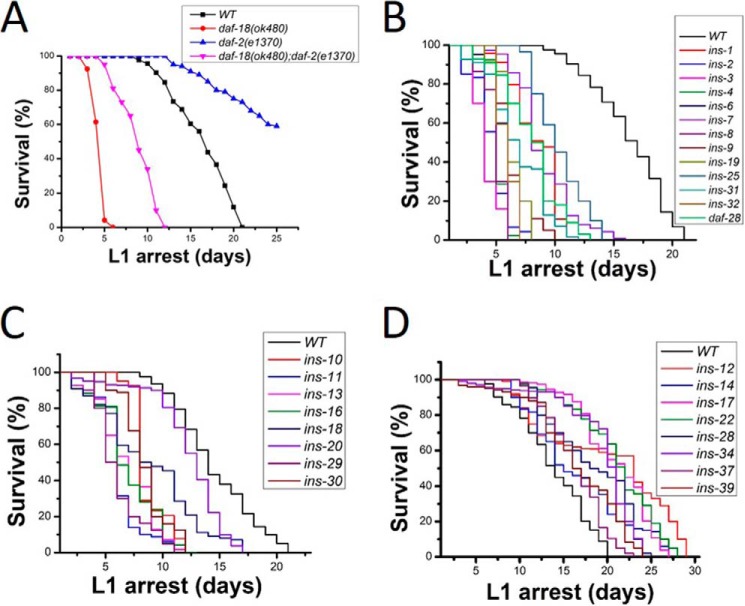
In the absence of food, newly hatched C. elegans larva (L1 stage) undergo a developmental quiescence called L1 arrest. Previously, we and others have shown that down-regulation of the IIS pathway is critical for L1 arrest survival (18, 19). WT L1 arrested worms live for a maximum of 21 days with a mean life span of 13 days when grown at 20 °C. Manipulation of IIS signaling can alter the normal 21 day survival period. DAF-18 is the worm orthologue of the human PTEN tumor suppressor. DAF-18/PTEN functions to inhibit the IIS pathway. Enhanced IIS caused by the loss of daf-18 resulted in shortened life span during L1 arrest (Fig. 1A). Blocking IIS by loss of the daf-2/INSR resulted in lengthened life span in L1-arrested worms (Fig. 1A). To categorize the functional role of INS peptides on the regulation of the IIS pathway, each of the 40 ins (oe) strains were scored for L1 arrest life span. We found that 21 (ins-1, -2, -3, -4, -6, -7, -8, -9, -10, -11, -13, -16, -18, -19, -20, -25, -29, -30, -31, -32 and daf-28) ins (oe) worms had significantly shorter life span when compared with WT worms during L1 arrest (Fig. 1, B and C). This suggests that these 21 INS peptides function as IIS agonists to activate the IIS pathway. Eight (ins-12, -14, -17, -22, -28, -34, -37, and -39) ins (oe) worms had significantly longer life span than WT L1 worms (Fig. 1D), suggesting that these INS function as IIS antagonists to shut down IIS. Eleven ins (oe) (ins-5, -15, -21, -23, -24, -26, -27, -33, -35, -36, and -38) strains had normal L1 arrest life span compared with WT worms (Table S2), suggesting that these INS peptides play neutral roles on L1 arrest life span.
Figure 1.
ins (oe) functions on L1 arrest life span. A, daf-18/pten mutants have shorter L1 arrest life span and the insulin receptor daf-2 mutants have a longer L1 arrest life span than WT worms. B and C, 21 ins (oe) strains have shorter L1 life span, suggesting these INS are IIS agonists. D, 8 ins (oe) strains have longer L1 life span, suggesting these INS peptides are IIS antagonists. Also see details in Table S2.
Agonistic INS peptides cause Q cell divisions during L1 arrest
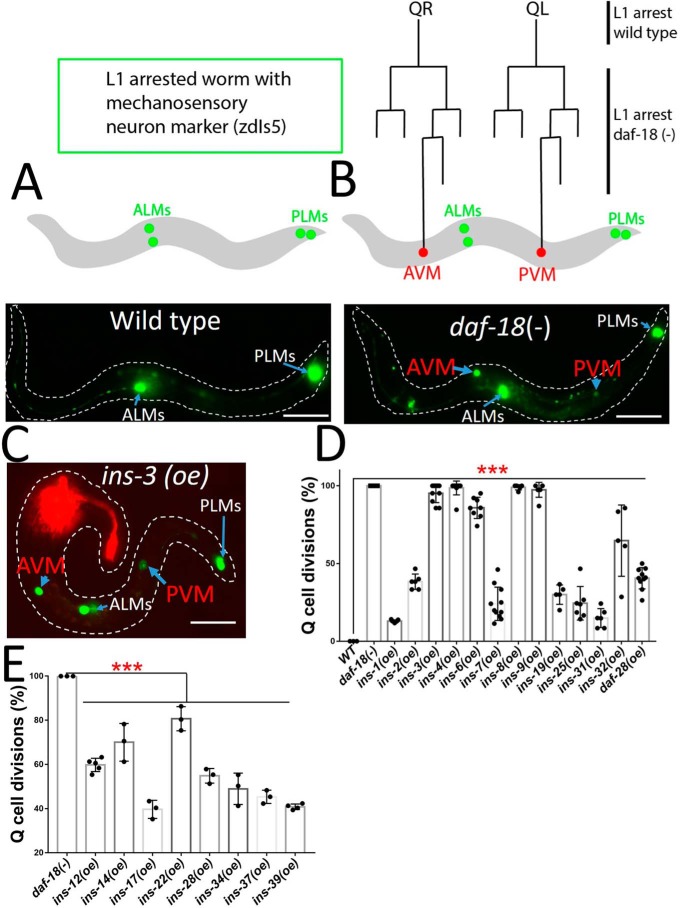
During L1 arrest all cell divisions are halted due, in part, to the shutdown of the IIS pathway. When the IIS pathway is activated during L1 arrest, we showed that the Q cell lineage undergoes cell divisions and movements.3 We asked whether INS (oe) could cause Q cell divisions during L1 arrest. To aid with the scoring of the Q cell divisions in L1 arrest, we scored the presence of the Q cell neuroblast descendants AVM and PVM. Normally, L1-arrested worms only have the embryonic mechanosensory neurons ALMs and PLMs (Fig. 2A). Our previous work found that AVM and PVM were present in L1-arrested daf-18 mutants (Fig. 2B), as loss of daf-18 enhances insulin signaling.3 Therefore in L1 arrest, if A/PVM are present it tells us the Q cell lineage has undergone its terminal divisions and can be used as a new readout to analyze the functions of the INS ligands.
Figure 2.
ins (oe) functions on L1 arrest Q cell divisions. A, WT worms stop development at L1 arrest. The WT worms with touch neuron marker (zdIs5) only have embryonic ALMs and PLMs. B, daf-18 (−) L1 arrest mutants have two terminal Q cell descendants AVM and PVM. QR/L are embryonic Q right (R) and left (L) neuroblasts. C, a representative ins-3 (oe). The red fluorescence is the AWC neuron from the odr-1::rfp transgenic marker for ins (oe) lines. D, 13 ins (oe) strains show L1 arrest Q cell divisions, suggesting these are IIS agonists. D and E, 8 ins (oe) antagonists, which have longer L1 arrest life span (Fig. 1D), can suppress the L1 arrest Q cell divisions in daf-18 (−) mutants, suggesting these INS are IIS antagonists for Q cell divisions. Scale bars represent 50 μm. Error bars represent the S.D. ***, p value (t test) versus control <0.001. Also see details in Table S3.
We found that 13 INS (oe) strains (ins-1, -2, -3, -4, -6, -7, -8, -9, -19, -25, -31, -32 and daf-28) caused Q cell divisions in L1-arrested worms (Fig. 2, C and D). Our results suggested that these INS peptides are strong IIS agonists. To provide evidence whether these INS peptides have a physiologically relevant role in L1 arrest Q cell divisions, we predicted that a loss of function mutation in these agonistic INS may suppress the daf-18 mutant L1 arrest Q cell divisions. We tested an ins-4 ins-6 double mutant (13) and indeed this double mutant did show suppression (Fig. S3). The suppression was not complete suggesting the other INS may also have roles for L1 arrest Q cell divisions. The Q cell divisions during L1 arrest is an excellent readout for INS peptides that activate the receptor. However, this phenotype on its own is not sufficient to determine INS peptides that are inhibitory to DAF-2 as the loss of DAF-2 is the same as WT (i.e. no Q cell divisions during L1 arrest). To examine INS peptides found to be inhibitory based on L1 arrest life span, we used daf-18 mutant worms (i.e. increased IIS signaling) and asked whether the inhibitory INS peptides could suppress the daf-18 Q L1 arrest cell divisions. As predicted, we found that all the ins (oe) strains, which could make L1-arrested worms live longer (Fig. 1D) could also significantly suppress the daf-18 Q cell divisions (Fig. 2E). These results confirm that the eight INS peptides (INS-12, -14, -17, -22, -28, -34, -37, and -39) function as IIS antagonists in both L1 arrest life span and Q cell divisions. All INS peptides that could induce L1 arrest Q cell divisions also shortened L1 arrest lifespan. However, not all the ins genes, which shorten L1 arrest life span can induce L1 arrest Q cell divisions (Fig. 1C), suggesting that these INS peptides have specific activities in controlling L1 arrest life span and cell divisions.
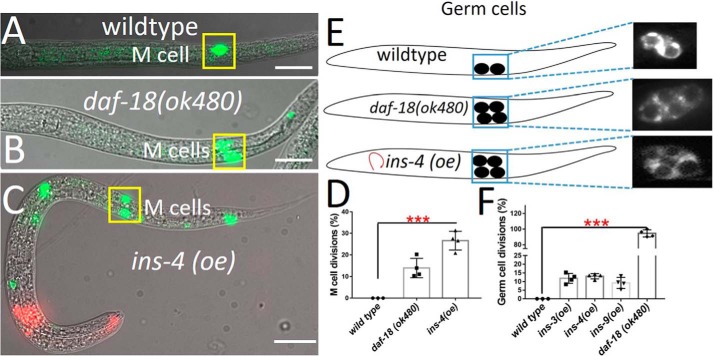
To ensure that INS peptides overexpressed in the nervous system were dependent on normal peptide processing, we tested whether the proprotein convertase-deficient animal (egl-3 mutant) (13) could suppress the function of agonistic INS peptides on L1 arrest Q cell divisions. We found that L1-arrested Q cell divisions in two strong ins (oe) worms (INS-3 and INS-4) were almost completely suppressed by egl-3 (Fig. S4A) suggesting that these insulin peptides need to be processed properly for their function. In addition, if these agonists INS peptides bind to and activate the DAF-2/INSR then daf-2 mutants should suppress the ins (oe) L1 arrest Q cell divisions. We found that each ins (oe) strain that induced L1-arrested Q cell divisions was completely suppressed in the daf-2 (e1370) mutant background (Fig. S4B). Previous work showed that the activated IIS pathway also induced germ cell and M cell divisions in L1-arrested worms (15, 20), therefore we determined whether these overexpressed INS peptides would induce L1 arrest germ cell divisions and/or M cell divisions. Indeed, the overexpressed agonistic INS peptides were able to act cell nonautonomously, resulting in M cell (INS-4) and germ cell divisions (INS-3, -4, and -9) during L1 arrest (Fig. 3).
Figure 3.
Pan-neuronal INS overexpression acts cell nonautonomously to induce nonneuronal cell divisions during L1 arrest. A, L1 arrest WT, only one M cell is observed (ayIs6). B–D, daf-18(−) and pan-neuronal INS overexpression cause the M cell to divide in L1 arrest. E and F, germ cells (Z2/Z3) in L1-arrested worms. daf-18(−) mutants and pan-neuronal INS overexpression induces germ cell divisions. Red fluorescence in the head is the (odr-1::rfp): transgenic marker. Data represent the average of at least 3 independent experiments from at least two stable transgenic lines. Scale bars represent 50 μm. Error bars represent the S.D. ***, p value (t test) versus control <0.001.
INS peptide function on dauer formation
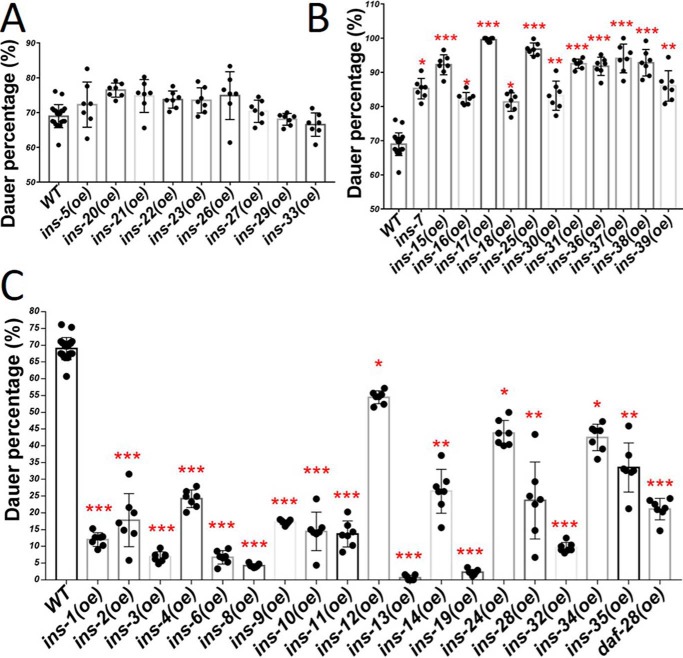
In C. elegans, animals in the second larval stage can enter a dauer diapause phase under adverse environmental conditions. Mutants that reduced IIS (such as daf-2 INSR (lf) or age-1 PI3K (lf)) have a dauer-constitutive (Daf-c) phenotype. Yet, individual INS knockout mutants only show a very weak dauer phenotype, which may imply functional redundancy (9). Functional redundancy was supported by creating a fully penetrant Daf-c phenotype by simultaneous removal of ins-4, ins-6, and daf-28 (13). However, the functions of the 40 ins genes on dauer formation are still not well-addressed. Here, we tested the function of individual pan-neuronal INS on dauer formation. Previous work showed that WT C. elegans can go into dauer under high temperatures even in the presence of food or noncrowding conditions (21). At 29 °C we found 19 INS peptides (INS-1, -2, -3, -4, -6, -8, -9, -10, -11, -12, -13, -14, -19, -24, -28, -32, -34, -35 and DAF-28) significantly reduced dauer formation (Fig. 4C), consistent with these INS peptides acting as IIS agonists. Twelve INS peptides (INS-7, -15, -16, -17, -18, -25, -30, -31, -36, -37, -38, and -39) caused higher dauer penetrance than WT (Fig. 4B), consistent with these INS peptides acting as IIS antagonists. Nine INS peptides (INS-5, -20, -21, -22, -23, -26, -27, -29, and -33) had no significant functions on dauer formation (Fig. 4A). Interestingly, 10 INS peptides (INS-1, -2, -3, -4, -6, -8, -9, -19, -32 and daf-28) overexpressed in worms showed shortened L1 arrest life span, promoted L1 arrest Q cell divisions, and reduced dauer formation, suggesting that these 10 INS can activate IIS consistently when scored by different phenotypes. Notably, not all the INS peptides, which extended L1 arrest life span enhanced dauer formation, suggesting that these INS peptides have different functions to control L1 arrest and dauer. For example, INS-12, -14, -22, -28, and -34 acted like IIS antagonists as they increased L1 life span, whereas they acted like agonists or had no function in dauer formation. INS-7, -16, -18, -25, -30, and -31 acted as IIS agonists as they reduced L1 arrest life span, but acted as IIS antagonists by increasing dauer formation.
Figure 4.
ins (oe) strains on dauer formation at high temperature. A, 9 ins (oe) worms have no significant functions on dauer formation. B, 12 ins (oe) worms (antagonists) induce more dauer than WT worms. C, 19 ins (oe) worms (agonists) have less dauer than WT worms. Error bars represent the S.D., *, p value (t test) versus control <0.05; **, p value (t test) versus control <0.01; ***, p value (t test) versus control <0.001. Also see details in Table S4.
INS peptide function on fat accumulation in adult worms
The IIS pathway plays an important role in controlling fat accumulation (22, 23). The daf-2/INSR (lf) causes fat accumulation in adults (24–27). Food cues are sensed by an olfactory receptor in the amphidal sensory neurons and this, in turn, is relayed to the IIS pathway to control fat metabolism (28, 29). However, the functions of all INS peptides on fat accumulation have not been identified. We studied the role of each of the 40 INS on fat accumulation in adult worms and found that 13 INS peptides (INS-3, -4, -6, -9, -11, -16, -18, -19, -25, -29, -30, -32 and DAF-28) had lower fat levels compared with WT (Fig. 5, A and C). Seven INS peptides (INS-8, -12, -17, -21, -28, -37, and -39) elevated fat levels compared with WT worms (Fig. 5B). Our results suggest that most of the INS that act as agonists had reduced fat staining, whereas most of the antagonists had increased fat staining. Of all the 13 INS (oe) that could induce L1 arrest Q cell divisions (Fig. 2D), most behaved as agonists for fat accumulation with the exception of INS-1, -2, -7, and -31, which had no effect, and INS-8, which acted as an antagonist for fat accumulation. Only INS-21 appeared to be specific for a role in fat accumulation exhibiting no effects on the other three phenotypes scored (Fig. 5B).
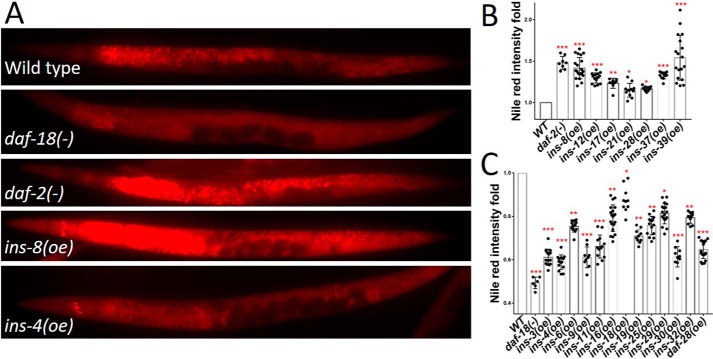
Figure 5.
ins (oe) functions on fat accumulation. A, fat accumulation analyzed by Nile Red staining in WT, daf-18(−), daf-2(−), and ins (oe) worms. The daf-2 and daf-18 mutants have higher and lower level fat accumulation than WT, respectively. B, 7 ins (oe) worms (antagonists) have higher fat accumulation than WT. C, 13 ins (oe) worms (agonists) have lower fat accumulation than WT. Error bars represent the S.D., *, p value (t test) versus control <0.05; **, p value (t test) versus control <0.01; ***, p value (t test) versus control <0.001. Also see details Table S5.
An F peptide and the β class INS act as agonists
According to our results, it is apparent that INS peptides that were structurally characterized as the β class and contain a sequence known as the F peptide are activators of the IIS (Fig. 6A). Nine of the β class INS peptides contain the F peptide (Fig. 6A). All of the β class INS behaved as agonists of IIS in our L1 arrest Q cell division assay, except for INS-10, which does not have an F peptide. We hypothesized that the F peptide contributes to the C. elegans INS activation in L1 arrest Q cell divisions. To test this hypothesis, we pan-neuronally expressed a strong agonist, INS-4 with a deletion of the F-peptide region or the F peptide alone and found that both failed to induce Q cell divisions during L1 arrest (Fig. 6, B and C). Pan-neuronal co-expression of the F peptide and the F peptide-lacking INS-4 resulted in the induction of Q cell divisions during L1 arrest (Fig. 6, B and C), showing that the F peptide can act in trans and is necessary for INS-4 activation of DAF-2/INSR. We then asked if co-expression of the F peptide with the β class INS-10, whose overexpression does not affect Q cell division, could induce L1 arrest Q cell divisions. The F peptide and INS-10 co-expression in trans failed to induce Q cell divisions during L1 arrest (Fig. 6C). This suggests that the F peptide functionally complements INS-4 (minus F peptide) activity in a peptide sequence-specific manner and/or that the pool of F peptides released upon processing of F peptide INS may not functionally complement INS peptides that lack an embedded F peptide sequence.
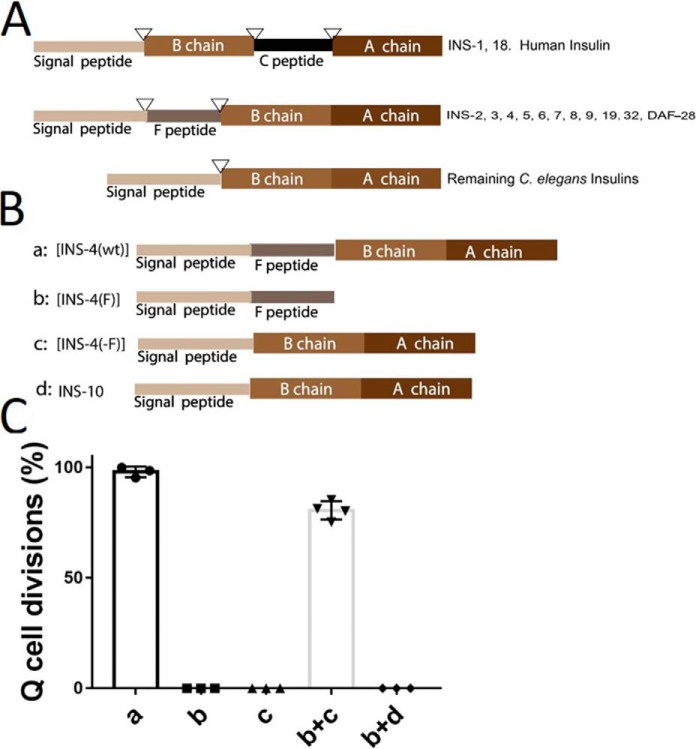
Figure 6.
The F peptide is needed for INS-4 activation. A, all INS contain at least a signal peptide, B chain, and A chain. Only INS-1 and INS-18 have a C peptide, like human insulin. An F peptide is present in INS-2 through INS-9 and DAF-28. Predicated cleavage sites for the proteolytic processing (triangles). B, variant of INS-4: a, WT ins-4 (INS-4(wt)); b, ins-4 F peptide only (INS-4(F)); c, ins-4 with F peptide deleted (INS-4(−F)). C, INS-4 with no F peptide (c) or F peptide alone (b) does not induce L1 arrest Q cell divisions. However, adding back both in trans (b+c) can induce L1 arrest Q cell divisions. INS-10 has a structure similar to INS-4 (β class), but INS-10 has no F peptide. INS-10 + F peptide from INS-4 (b) co-injection does not induce L1 arrest Q cell divisions.
Discussion
In this study, we created independent worm lines that overexpressed each of the 40 C. elegans INS peptides and assayed for IIS phenotypes to assign in vivo roles. Because this is an overexpression study, INS are not at their normal physiological levels (Table S1 and Fig. S2), and there are caveats to INS overexpression. For example, the observed phenotypes could be due to an artifact of the overexpressed INS affecting the processing of endogenous INS peptides. But, because we observed novel phenotypes for different INS peptides, we do not think this is likely. In addition, as Fernandes de Abreu et al. (9) reported there is INS-to-INS regulation, for example, INS-X may transcriptionally activate INS-Y, so in our study overexpressing INS-X may also (indirectly) overexpress endogenous INS-Y. Given these caveats, our overexpression study can identify physiologically relevant roles for the INS peptides. For example, INS-4 and INS-6 are likely endogenous INS for L1 arrest Q cell divisions as the double mutant can partially suppress daf-18 mutant L1 arrest Q cell divisions. Although the INS were expressed in the nervous system, we showed that these overexpressed INS peptides are processed by proprotein convertases, and are secreted as they can act cell nonautonomously on germline and M cells. Our study is the first to provide the functional data for all 40 INS on L1 arrest life span, Q cell divisions, heat-stress induced dauer formation, and fat accumulation. Our results are summarized in Table 1 and Fig. 7.
TABLE 1.
Data summary of functions of C. elegans INS peptides
Data are compared to wild type. +/−: with/without L1 arrest Q cell divisions. N: normal; L: low; H; high. See details in Supporting information for raw data. Color code: green indicates strong agonist; brown indicates agonist; red indicates strong antagonist; dark red indicates weak antagonist; and blue indacates neutral.
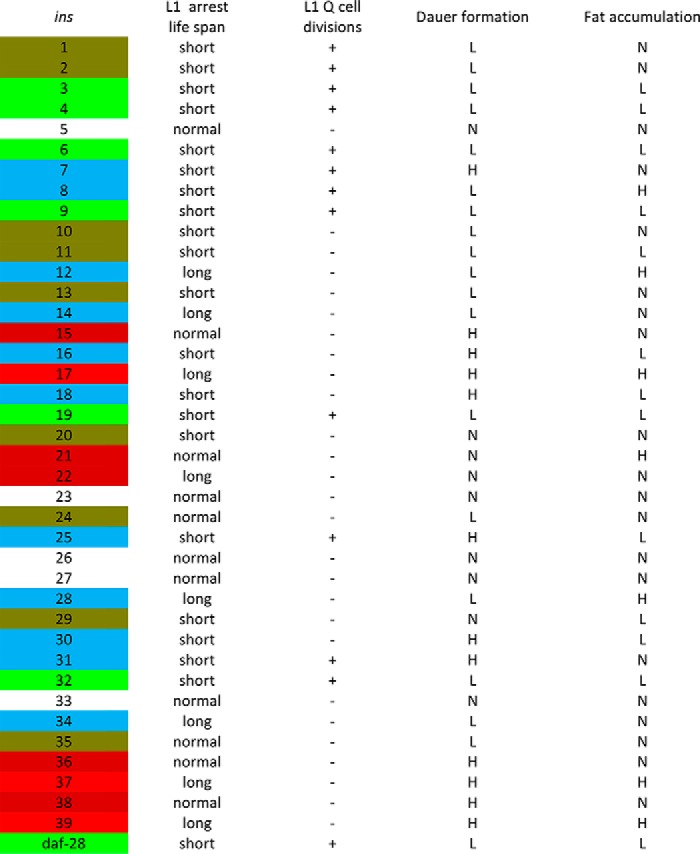
Figure 7.
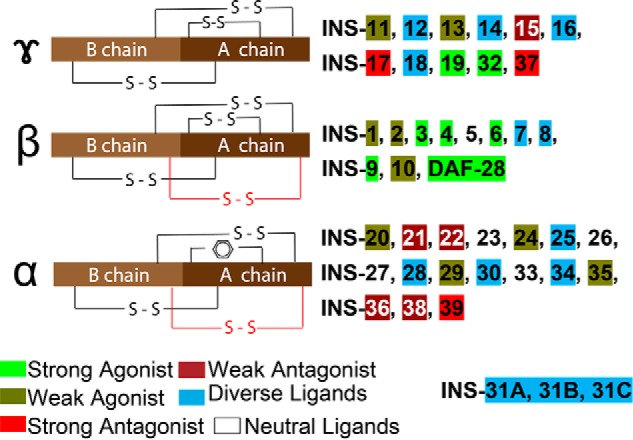
Insulin-like peptides in C. elegans. C. elegans INS can be classified into three types based on disulfide bonds (7). γ-Insulins have the arrangement of three disulfide bonds as found in vertebrates, whereas α- and β-insulins contain an additional intra-chain disulfide bond (red). α-Insulins lack the common intra-chain bond in the A chain, which is substituted by the interaction of aromatic amino acid side chains. INS-31 constitutes its own additional class with three repeats of B and A peptide chains. In our study we classified the 40 insulin ligands into 6 functional groups: strong agonist/antagonist: activity consistence within all tested phenotypes; weak agonist/antagonist: activity consistence within most tested phenotypes, but have no significant activity in other phenotypes; diverse, can have both agonist and antagonistic roles, and neutral ligands, no significant activity in all tested IIS assays.
Mutants with reduced IIS signaling have both a Daf-c phenotype and extended L1 arrest survival. We show that the INS-17, -37, and -39 are IIS antagonists exhibiting increased L1 arrest survival and increased dauer formation. INS-17 was previously reported to work as a IIS antagonist for dauer regulation (12), but this work is the first to assign INS-37 and -39 as antagonists. Our work also demonstrated that select INS peptides are IIS antagonists in controlling L1 arrest survival but are pleiotropic in their action in controlling dauer formation. For example, INS-12, -14, -28, and -34, which act as antagonists and can extend L1 arrest survival, can also act as agonists and have significantly lower dauer formation than WT worms. On the other hand, INS-7, -16, -18, -25, -30, -31, act as agonists in L1 arrest, but can have the opposite role in dauer, acting as an antagonist by increasing dauer formation. Our results suggest that INS function in L1-arrested worms may be different from that of controlling dauer formation because it is an alternative L3 development stage, and thus INS peptides have spatiotemporal compartmentalization with respect to their function. Our finding is consistent with studies that show that dauer arrest and adult lifespan regulation by IIS are also decoupled (30–33).
Pan-neuronal INS overexpression that caused L1 arrest Q cell divisions identified 13 INS peptides (INS-1, -2, -3, -4, -6, -7, -8, -9, -19, -25, -31, -32 and DAF-28) that act as agonists for the IIS. All 13 INS peptides also have short L1 arrest life span, suggesting that all 13 INS peptides behave as potential IIS agonists. Previous studies, based on differing assays assigned INS-3, -4, -6, -9, DAF-28 as potential agonist INS peptides (13–15, 34). These studies are consistent with our findings and support the reliability of our L1 arrest Q cell division readout as a means of categorizing the ins genes. INS-5 has been suggested to be an agonist (15), however, we did not find INS-5 to have any function in all of our assays, which is consistent with another report (13). INS-1 was shown to be an antagonistic peptide based on dauer formation (7, 13, 34). Overexpression of ins-1, enhances dauer arrest in weak daf-2 mutants, suggesting that INS-1 antagonizes insulin-like signaling. Also, INS-1 is antagonistic to DAF-2 for behavior (35). However, in our assays we found INS-1 to have weak activation properties. INS-1 may be a complex peptide as INS-1 acts as an agonist for IIS in salt chemotaxis learning (36).
We identified eight antagonistic INS peptides that could significantly extend L1 arrest life span and when overexpressed from the nervous system could suppress the daf-18/pten L1 arrest Q cell divisions. Thus, these INS peptides acted as therapeutic peptides for daf-18/pten worms. In humans, insulin and IGFs are thought to work as agonists and do not have antagonistic properties. Our work showed that C. elegans INS-6 is a strong agonist and INS-6 has been shown to bind and activate the human insulin receptor (37). It would be interesting to know whether the antagonistic INS peptides we have identified in this study can bind to and inhibit the human insulin or IGF-1 receptor, if so, these C. elegans INS peptides could be used as future therapeutics for hyperinsulinemia.
Our study revealed that INS-8 behaves as an agonist of IIS, because ins-8 (oe) shortens the life span of L1-arrested worms, has low penetrance dauer formation, and promotes L1 arrest Q cell divisions. A previous study suggested that INS-8 may work as an agonist (8). However, ins-8 (oe) worms behaved as an antagonist of IIS exhibiting higher fat accumulation. One study showed that ins-8 (oe) enhances ins-7 mutant life span, which would suggest that INS-8 is an antagonist (8). We suggest that the neuronal ins-8 (oe) is sufficient to work as an agonist to activate the IIS pathway, which in turn controls the L1 arrest life span and dauer formation, but in adult worms, it may work as an antagonist. This result with INS-8 is consistent with our finding that many INS peptides have distinct roles in mediating fat accumulation that is developmentally separate from its effects on dauer and L1 arrest life span. Insulin signaling temporally and in varying tissues of the body contributes differently to fat content (31).
Of the 40 INS peptides tested, eight appear to have specific functions in our phenotype assays. INS-15, INS-21, and INS-20 and -22 act as IIS antagonists specifically for regulating dauer, fat metabolism, and L1 arrest life span, respectively. Similarly, INS-24, -35, -36, and -38 only function in dauer formation.
To understand what makes an INS an activator we focused on the L1 arrest Q cell divisions as this assay determined with certainty which INS peptides acted as IIS activators. Our study revealed that the β class INS peptides, which contains the three canonical disulfide bonds as well as an additional inter-chain disulfide bond are good predictors of an INS peptide agonist. INS-1 to INS-10 and DAF-28 fall into this class (Fig. 7). Nine of the β class INS contain an F peptide (7), the exception is INS-10. The F peptide is processed at the N terminus by the signal peptidase cleavage site and at the C terminus by either the proprotein convertase enzymes EGL-3/PC2-like with cleavage sequence (RR or KR) or a KPC-1/PC1-like site (R-X-X-R) (13) (Fig. S1). INS-10 does have activation properties and reduces L1 arrest life span and dauer formation, but could not induce L1 arrest Q cell divisions. INS-5 was predicted to contain an F peptide (7), but upon further examination, INS-5 does not have a proprotein convertase site that would release the F peptide, but instead would be incorporated as the B chain (Fig. 7, Fig. S1). Thus, our results reveal a striking revelation that all INS peptides that are predicted to contain an F peptide should behave as agonists of the IIS (Fig. 7). We showed that the F peptide is indeed required for INS-4 to induce L1 arrest Q cell divisions and the F peptide can be added back in trans to restore INS-4 (minus F peptide) function. Note that the F peptide is not an absolute requirement for an INS to induce L1 arrest Q cell divisions as INS-1, -19, -25, -31, and -32 could induce L1 arrest Q cell divisions (albeit not as strong as other, e.g. INS-4). Interestingly, the predicted signal sequences for INS-32 was longer than average INS peptides and therefore may produce an F peptide. This prompted us to look more closely at the predicted peptides and using the SignalP 4.1 (38), we identified INS-32 as having potential F peptide (Fig. S1). In addition, we also propose that INS-19 has an F peptide as it has a potential proprotein convertase-cleavage site (Fig. S1).
Human insulin has a C peptide, and human IGF-1 and IGF-2 have E peptides that are cleaved during processing analogous to the F peptides identified in C. elegans INS peptides. Our work on the F peptide should stimulate closer examination of peptides released upon processing of human insulin and IGF. For instance, IGF-1 is one of the key molecules in cancer biology, however, little is known about the role of the E peptide. The E peptide is thought to have functional properties as the release from IGF-1 is thought to induce cellular proliferation in the human prostate cancer (39). The C peptide of proinsulin is important in the processing of mature insulin and may have biological activity as a report suggests that it binds to a G protein–coupled surface receptor and activates Ca2+-dependent intracellular signaling pathways (40). Because we have provided evidence in C. elegans that the F peptide can work in trans with the INS-4 lacking an F peptide, the F peptide serves as a modulator of INS-4 to induce L1 arrest Q cell divisions.
Finally, of the 40 INS (oe) strains tested, INS-5, -23, -26, -27, and -33 were not functional in the selected assays. These INS peptides may have specific roles that have not been uncovered through the assays selected. The INS peptides are also thought to function in a combinatorial fashion and perhaps these single INS peptides have no function on their own and may participate with the other INS to exert their function (9). Alternatively, these INS may bind to receptors other than DAF-2/INSR. A report has suggested that additional insulin-like receptors have been identified in the C. elegans genome (41).
In conclusion, our work systematically tested the functions of each of 40 INS on dauer formation, L1 arrest life span, L1 arrest Q cell divisions, and fat accumulation phenotypes (Table 1). By using these IIS phenotypes as readouts of insulin peptide activity, we found that seven INS peptides (INS-3, -4, -6, -9, -19, -32 and DAF-28) were strong agonists and three INS peptides (INS-17, -37, and -39) were strong antagonists of IIS, because these INS peptides acted either as agonists or antagonists in all our tested phenotypes. Five INS peptides (INS-15, -21, -22, -36, and -38) were found to be weak antagonists; and nine INS peptides (INS-1, -2, -10, -11, -13, -20, -24, -29, and -35) were weak agonists. Five INS peptides (INS-5, -23, -26, -27, and -33) were neutral ligands. Eleven INS peptides (INS-7, -8, -12, -14, -16, -18, -25, -28, -30, -31, and -34) have different roles in different stress environments and developmental stages (Table 1 and Fig. 7). These diverse functions of INS may contribute to important influences on development, metabolism, and aging-related diseases.
Experimental procedures
Strains
Strains used in this study were acquired from the Caenorhabditis Genetics Center (CGC). Standard culture methods were used as previously described (42). Strains were grown on OP50 Escherichia coli and cultured at 20 °C unless otherwise indicated. Strains used in this study: CZ10175: zdIs5 [mec-4p::GFP + lin-15(+)] I; RB712: daf-18(ok480); CB1370: daf-2 (e1370); DR1942: daf-2(e979), VC671: egl-3 (ok979); and PD4666: ayIs6 [Phlh-8::gfp + dpy-20(+)].
Transgenic strains
For the ins overexpression strains, the insulin genomic sequences were amplified from a N2 genomic DNA and placed under control of the pan-neuronal promoter Prgef-1 by standard cloning procedures (16). A plasmid with the injection marker odr-1::rfp was injected into Pmec-4::GFP(zdIs5) (43) worms using standard microinjection methods (44). For the F peptide experiment: Q5 mutagenesis (New England Biolabs) using primers was used to delete the F peptide sequence from the Prgef-1::INS-4 plasmid. For these extrachromosomal arrays, each injected plasmid we established and scored at least 3 independent lines. For a list of strains and primer sequences please see Tables S6 and S7.
L1 arrest Q Cell divisions
Nonstarved well-maintained mixed stage worms were collected to prepare embryos, as described (45). In brief, embryos were maintained and hatched in sterile M9 and incubated at 20 °C with low speed rocking to initiate L1 arrest. The final Q cell descendants (A/PVM) were observed under an Axioplan fluorescent microscope (Zeiss, Germany) after 2 days or more in L1 arrest. 50–100 μl of M9 containing greater than 50 L1-arrested worms were removed from the culture. The total number of worms and the worms with A/PVM cell divisions were counted. For transgenic strains, only the worms with the injection marker were counted and analyzed. Similarly, M cell divisions were analyzed by using ayIs6 strains (46).
Antibody staining
Antibody staining was performed as previously described (47). Worms were fixed by using 1× witches brew and paraformaldehyde (2.5% final). To detect germline cells, rabbit anti-PGL-1 (P-granule component) (1:20,000) (a gift from Dr. Susan Strome) was used as the primary antibody, which was diluted in PBST-A. Worms were incubated at room temperature overnight. Detection was with a FITC-labeled goat anti-rabbit secondary antibody (1:100). For transgenic strains, only the worms with the injection marker were counted and analyzed. The total number of worms and the worms with germ-cell divisions were counted. Analysis of worms was using an Axioplan fluorescent microscope (Zeiss, Germany).
L1 arrest life span assays
Life span was assessed in liquid medium (19). L1 worms were cultured in 1 ml of M9, 50–100 μl was taken to ensure the sample size was larger than 50, and the worms were scored every day. We scored survival by counting the number of worms that were moving (alive) and then dividing that number by the total number of worms in the aliquot. To compare the survival rates between strains, the L1 arrests were carried out in triplicate with at least 100 L1s and the mean survival rate calculated by the Kaplan-Meier method (48), which is the fraction of living animals over a time course. The significance of difference in overall survival rate is performed using the log-rank test (49).
Fat staining
Synchronized eggs were cultured on OP50 plates with 25 ng/ml of Nile Red for 3 days at 20 °C, and then washed 3 times with M9, cultured on normal OP50 plates for 1 more day at 20 °C to eliminate the Nile Red OP50 background in the intestine. Worms were collected and washed in M9 3 times, then fixed in 40% isopropyl alcohol for 3 min. At least 30 animals were imaged in at least three separate experiments using a Zeiss Axioplan. The fluorescent intensity was captured by using the TurboRFP (573 nm) channel and was quantified by using ImageJ. The nontransgenic (WT) siblings' intensity was set to “1” and the transgenic worms were calculated as “fold-increased or decreased” compared with WT animals. Nile Red has been a controversial as a stain for fat accumulation (27), so we also tested some of INS overexpressing strains with Oil Red O and they were consistent with the results obtained with Nile Red (see Fig. S5).
Dauer formation at high temperature
We analyzed L2 dauer formation at 29 °C as synchronized zdIs5 worm eggs hatched at 29 °C presented a higher percentage dauer phenotype. The dauer, dauer-like, and adult worms with injection marker were counted. The dauer percentages were calculated. Three independent trials were performed for each strain, each sample size was greater than 50.
For Q cell divisions, dauer formation, and fat staining experiments, nontransgenic siblings were used as WT controls. All the scatter plot data presented in the figures were made using GraphPad Prism 7.
Author contributions
S. Z. data curation; S. Z., H. C., and T. P. software; S. Z. and H. C. investigation; S. Z. and J. B. methodology; S. Z. writing-original draft; S. Z., W. B., and I. C.-S. writing-review and editing; J. B. and T. P. resources; W. B. validation; I. C.-S. supervision; I. C.-S. funding acquisition; I. C.-S. project administration.
Supplementary Material
Acknowledgments
We are grateful to Caenorhabditis Genomic Center for providing strains, which is funded by National Institutes of Health Office of Research Infrastructure Program Grant P40OD010440.
This work was supported by Natural Sciences and Engineering Research Council of Canada Grant NSERC 249779 and Canadian Institutes of Health Research Grant CIHR 130541. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5 and Tables S1–S7.
S. Zheng, Z. Qu, M. Zanetti, B. Lam, and I. Chin-Sang, unpublished data.
- IIS
- insulin/insulin-like growth factor signaling
- INS
- insulin-like
- IGF
- insulin-like growth factor.
References
- 1. Piñero González J., Carrillo Farnés O., Vasconcelos A. T., and Goñzalez Pérez A. (2009) Conservation of key members in the course of the evolution of the insulin signaling pathway. Bio. Systems 95, 7–16 10.1016/j.biosystems.2008.06.003 [DOI] [PubMed] [Google Scholar]
- 2. Ullrich A., Bell J. R., Chen E. Y., Herrera R., Petruzzelli L. M., Dull T. J., Gray A., Coussens L., Liao Y. C., and Tsubokawa M. (1985) Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature 313, 756–761 10.1038/313756a0 [DOI] [PubMed] [Google Scholar]
- 3. Ullrich A., Gray A., Tam A. W., Yang-Feng T., Tsubokawa M., Collins C., Henzel W., Le Bon T., Kathuria S., Chen E., Jacobs S., Francke U., Ramachandran J., and Fujitayamaguchi Y. (1986) Insulin-like growth factor-I receptor primary structure: comparison with insulin-receptor suggests structural determinants that define functional specificity. EMBO J. 5, 2503–2512 10.1002/j.1460-2075.1986.tb04528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taniguchi C. M., Emanuelli B., and Kahn C. R. (2006) Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 10.1038/nrm1837 [DOI] [PubMed] [Google Scholar]
- 5. Duret L., Guex N., Peitsch M. C., and Bairoch A. (1998) New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res. 8, 348–353 10.1101/gr.8.4.348 [DOI] [PubMed] [Google Scholar]
- 6. Kawano T., Ito Y., Ishiguro M., Takuwa K., Nakajima T., and Kimura Y. (2000) Molecular cloning and characterization of a new insulin/IGF-like peptide of the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 273, 431–436 10.1006/bbrc.2000.2971 [DOI] [PubMed] [Google Scholar]
- 7. Pierce S. B., Costa M., Wisotzkey R., Devadhar S., Homburger S. A., Buchman A. R., Ferguson K. C., Heller J., Platt D. M., Pasquinelli A. A., Liu L. X., Doberstein S. K., and Ruvkun G. (2001) Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15, 672–686 10.1101/gad.867301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ritter A. D., Shen Y., Fuxman Bass J., Jeyaraj S., Deplancke B., Mukhopadhyay A., Xu J., Driscoll M., Tissenbaum H. A., and Walhout A. J. (2013) Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res. 23, 954–965 10.1101/gr.150466.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernandes de Abreu D. A., Caballero A., Fardel P., Stroustrup N., Chen Z., Lee K., Keyes W. D., Nash Z. M., López-Moyado I. F., Vaggi F., Cornils A., Regenass M., Neagu A., Ostojic I., Liu C., et al. (2014) An insulin-to-insulin regulatory network orchestrates phenotypic specificity in development and physiology. PLoS Genet. 10, e1004225 10.1371/journal.pgen.1004225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anisimov V. N., and Bartke A. (2013) The key role of growth hormone-insulin-IGF-1 signaling in aging and cancer. Crit. Rev. Oncol. Hematol. 87, 201–223 10.1016/j.critrevonc.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murphy C. T., and Hu P. J. (2013) Insulin/insulin-like growth factor signaling in C. elegans. WormBook Dec. 26, 1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsunaga Y., Nakajima K., Gengyo-Ando K., Mitani S., Iwasaki T., and Kawano T. (2012) A Caenorhabditis elegans insulin-like peptide, INS-17: its physiological function and expression pattern. Biosci. Biotechnol. Biochem. 76, 2168–2172 10.1271/bbb.120540 [DOI] [PubMed] [Google Scholar]
- 13. Hung W. L., Wang Y., Chitturi J., and Zhen M. (2014) A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development 141, 1767–1779 10.1242/dev.103846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delaney C. E., Chen A. T., Graniel J. V., Dumas K. J., and Hu P. J. (2017) A histone H4 lysine 20 methyltransferase couples environmental cues to sensory neuron control of developmental plasticity. Development 144, 1273–1282 10.1242/dev.145722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y., and Baugh L. R. (2014) Ins-4 and daf-28 function redundantly to regulate C. elegans L1 arrest. Dev. Biol. 394, 314–326 10.1016/j.ydbio.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 16. Altun-Gultekin Z., Andachi Y., Tsalik E. L., Pilgrim D., Kohara Y., and Hobert O. (2001) A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951–1969 [DOI] [PubMed] [Google Scholar]
- 17. Baugh L. R. (2013) To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194, 539–555 10.1534/genetics.113.150847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukuyama M., Sakuma K., Park R., Kasuga H., Nagaya R., Atsumi Y., Shimomura Y., Takahashi S., Kajiho H., Rougvie A., Kontani K., and Katada T. (2012) C. elegans AMPKs promote survival and arrest germline development during nutrient stress. Biol. Open 1, 929–936 10.1242/bio.2012836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng S., and Chin-Sang I. D. (2016) C. elegans Methods to Study PTEN. Methods Mol. Biol. 1388, 307–321 10.1007/978-1-4939-3299-3_17 [DOI] [PubMed] [Google Scholar]
- 20. Fukuyama M., Rougvie A. E., and Rothman J. H. (2006) C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16, 773–779 10.1016/j.cub.2006.02.073 [DOI] [PubMed] [Google Scholar]
- 21. Ailion M., and Thomas J. H. (2003) Isolation and characterization of high-temperature-induced Dauer formation mutants in Caenorhabditis elegans. Genetics 165, 127–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hou N. S., and Taubert S. (2012) Function and regulation of lipid biology in Caenorhabditis elegans aging. Front. Physiol. 3, 143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones K. T., and Ashrafi K. (2009) Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Dis. Models Mech. 2, 224–229 10.1242/dmm.001933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horikawa M., and Sakamoto K. (2010) Polyunsaturated fatty acids are involved in regulatory mechanism of fatty acid homeostasis via daf-2/insulin signaling in Caenorhabditis elegans. Mol. Cell. Endocrinol. 323, 183–192 10.1016/j.mce.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 25. Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., and Ruvkun G. (2003) Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 10.1038/nature01279 [DOI] [PubMed] [Google Scholar]
- 26. Ogg S., and Ruvkun G. (1998) The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2, 887–893 10.1016/S1097-2765(00)80303-2 [DOI] [PubMed] [Google Scholar]
- 27. O'Rourke E. J., Soukas A. A., Carr C. E., and Ruvkun G. (2009) C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 10, 430–435 10.1016/j.cmet.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W., Kennedy S. G., and Ruvkun G. (2003) daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17, 844–858 10.1101/gad.1066503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerisch B., Weitzel C., Kober-Eisermann C., Rottiers V., and Antebi A. (2001) A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1, 841–851 10.1016/S1534-5807(01)00085-5 [DOI] [PubMed] [Google Scholar]
- 30. Apfeld J., and Kenyon C. (1998) Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95, 199–210 10.1016/S0092-8674(00)81751-1 [DOI] [PubMed] [Google Scholar]
- 31. Wolkow C. A., Kimura K. D., Lee M. S., and Ruvkun G. (2000) Regulation of C. elegans life-span by insulin-like signaling in the nervous system. Science 290, 147–150 10.1126/science.290.5489.147 [DOI] [PubMed] [Google Scholar]
- 32. Libina N., Berman J. R., and Kenyon C. (2003) Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489–502 10.1016/S0092-8674(03)00889-4 [DOI] [PubMed] [Google Scholar]
- 33. Dillin A., Crawford D. K., and Kenyon C. (2002) Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830–834 10.1126/science.1074240 [DOI] [PubMed] [Google Scholar]
- 34. Cornils A., Gloeck M., Chen Z., Zhang Y., and Alcedo J. (2011) Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138, 1183–1193 10.1242/dev.060905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kodama E., Kuhara A., Mohri-Shiomi A., Kimura K. D., Okumura M., Tomioka M., Iino Y., and Mori I. (2006) Insulin-like signaling and the neural circuit for integrative behavior in C. elegans. Genes Dev. 20, 2955–2960 10.1101/gad.1479906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tomioka M., Adachi T., Suzuki H., Kunitomo H., Schafer W. R., and Iino Y. (2006) The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51, 613–625 10.1016/j.neuron.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 37. Hua Q. X., Nakagawa S. H., Wilken J., Ramos R. R., Jia W., Bass J., and Weiss M. A. (2003) A divergent INS protein in Caenorhabditis elegans structurally resembles human insulin and activates the human insulin receptor. Genes Dev. 17, 826–831 10.1101/gad.1058003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nielsen H. (2017) Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73 10.1007/978-1-4939-7015-5_6 [DOI] [PubMed] [Google Scholar]
- 39. Armakolas A., Kaparelou M., Dimakakos A., Papageorgiou E., Armakolas N., Antonopoulos A., Petraki C., Lekarakou M., Lelovas P., Stathaki M., Psarros C., Donta I., Galanos P. S., Msaouel P., Gorgoulis V. G., and Koutsilieris M. (2015) Oncogenic role of the Ec peptide of the IGF-1Ec isoform in prostate cancer. Mol. Med. 21, 167–179 10.1007/s00894-015-2708-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wahren J., Ekberg K., Johansson J., Henriksson M., Pramanik A., Johansson B. L., Rigler R., and Jörnvall H. (2000) Role of C-peptide in human physiology. Am. J. Physiol. Endocrinol. Metab. 278, E759–E768 10.1152/ajpendo.2000.278.5.E759 [DOI] [PubMed] [Google Scholar]
- 41. Dlakić M. (2002) A new family of putative insulin receptor-like proteins in C. elegans. Curr. Biol. 12, R155–R157 10.1016/S0960-9822(02)00729-7 [DOI] [PubMed] [Google Scholar]
- 42. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clark S. G., and Chiu C. (2003) C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development 130, 3781–3794 10.1242/dev.00571 [DOI] [PubMed] [Google Scholar]
- 44. Mello C. C., Kramer J. M., Stinchcomb D., and Ambros V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fukuyama M., Kontani K., Katada T., and Rougvie A. E. (2015) The C. elegans hypodermis couples progenitor cell quiescence to the dietary state. Curr. Biol. 25, 1241–1248 10.1016/j.cub.2015.03.016 [DOI] [PubMed] [Google Scholar]
- 46. Kaplan R. E., Chen Y., Moore B. T., Jordan J. M., Maxwell C. S., Schindler A. J., and Baugh L. R. (2015) dbl-1/TGF-β and daf-12/NHR signaling mediate cell-nonautonomous effects of daf-16/FOXO on starvation-induced developmental arrest. PLoS Genet. 11, e1005731 10.1371/journal.pgen.1005731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chin-Sang I. D., George S. E., Ding M., Moseley S. L., Lynch A. S., and Chisholm A. D. (1999) The ephrin VAB-2/EFN-1 functions in neuronal signaling to regulate epidermal morphogenesis in C. elegans. Cell 99, 781–790 10.1016/S0092-8674(00)81675-X [DOI] [PubMed] [Google Scholar]
- 48. Rich J. T., Neely J. G., Paniello R. C., Voelker C. C., Nussenbaum B., and Wang E. W. (2010) A practical guide to understanding Kaplan-Meier curves. Otolaryngol. Head Neck Surg. 143, 331–336 10.1016/j.otohns.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Evason K., Huang C., Yamben I., Covey D. F., and Kornfeld K. (2005) Anticonvulsant medications extend worm life-span. Science 307, 258–262 10.1126/science.1105299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.