Abstract
Insulin and insulin-like growth factor 1 (IGF-1) are closely related hormones involved in the regulation of metabolism and growth. They elicit their functions through activation of tyrosine kinase–type receptors: insulin receptors (IR-A and IR-B) and IGF-1 receptor (IGF-1R). Despite similarity in primary and three-dimensional structures, insulin and IGF-1 bind the noncognate receptor with substantially reduced affinity. We prepared [d-HisB24, GlyB31, TyrB32]-insulin, which binds all three receptors with high affinity (251 or 338% binding affinity to IR-A respectively to IR-B relative to insulin and 12.4% binding affinity to IGF-1R relative to IGF-1). We prepared other modified insulins with the aim of explaining the versatility of [d-HisB24, GlyB31, TyrB32]-insulin. Through structural, activity, and kinetic studies of these insulin analogs, we concluded that the ability of [d-HisB24, GlyB31, TyrB32]-insulin to stimulate all three receptors is provided by structural changes caused by a reversed chirality at the B24 combined with the extension of the C terminus of the B chain by two extra residues. We assume that the structural changes allow the directing of the B chain C terminus to some extra interactions with the receptors. These unusual interactions lead to a decrease of dissociation rate from the IR and conversely enable easier association with IGF-1R. All of the structural changes were made at the hormones' Site 1, which is thought to interact with the Site 1 of the receptors. The results of the study suggest that merely modifications of Site 1 of the hormone are sufficient to change the receptor specificity of insulin.
Keywords: insulin, insulin-like growth factor (IGF), structure-function, insulin receptor, protein design, kinetics, binding, Site 1
Introduction
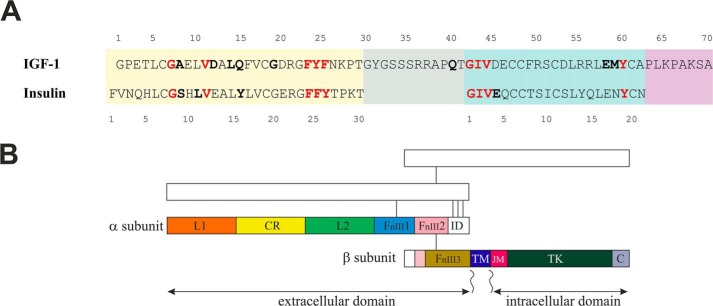
Insulin and insulin-like growth factors 1 and 2 (IGF-1 and IGF-2)3 are closely related protein hormones. Insulin is a key modulator of metabolism, whereas IGFs are factors indispensable for growth and development (1). They elicit their functions through activation of tyrosine kinase–type receptors (insulin receptor isoforms A and B (IR-A and IR-B), IGF-1 receptor (IGF-1R), or their hybrid forms) in cytoplasmic membranes of cells (2, 3). Malfunction of these hormones' complex signaling systems leads to both types of diabetes mellitus, increased cancer risk, and other life-threatening disorders (3). Amino acid sequences of insulin and IGF-1 and schematic organization of IR and IGF-1R domains are shown in (Fig. 1). Insulin is a two-chain molecule, where A and B chains are connected by two cysteine bridges, and the third disulfide bridge is in the A chain. IGF-1 is formed by a single chain organized into A, B, C, and D domains, where A and B domains are highly homologous to the A and B chains of insulin and share the same three-dimensional structure. The C terminus of the B domain in IGF-1 is connected to the N terminus of the A domain by a C domain. The D domain extends the C terminus of the A domain.
Figure 1.
Amino acid sequences of human insulin and IGF-1 (A) and schematic organization of IR and IGF-1R domains (B). A, insulin B chain and corresponding B domain in IGF-1 are highlighted by a yellow background. The green background indicates A chain and A domain. IGF-1 C and D domains are shown in gray or in violet. Amino acid residues depicted in boldface type were shown to contact the cognate receptor Site 1 in the crystal structures (6, 11–14). Red, residues found to interact with the receptors in the same fashion. B, structural domains of the receptor (αβ) dimer are marked on one half of the dimer, the second part is sketched. Subunits are disulfide-linked, as indicated by connecting lines. Domains are named as follows: leucine-rich (L), cysteine-rich (CR), fibronectin type III (FnIII), insert (ID), transmembrane (TM), juxtamembrane (JM), tyrosine kinase (TK), and C-terminal (C). For a review, see Belfiore et al. (2, 3)
The receptors are members of the receptor tyrosine kinase family. They are disulfide-linked (αβ)2 homodimers. Ligand binding to the receptor exhibits complex kinetics, characterized by a curvilinear Scatchard plot and negative cooperativity (4, 5). The commonly accepted assumption is that two distinct binding sites (Site 1 and Site 2) on a ligand interact with two receptor sites (Site 1 and Site 2, respectively) located on separate α subunits to create a high-affinity binding complex that is necessary for activation of the tyrosine kinase. The binding is asymmetrical; thus, only one ligand is bound per homodimer in the high-affinity complex (5–7).
Despite similarity in their primary and three-dimensional structures, insulin and IGF-1 bind the noncognate receptor with substantially reduced affinity. The structural basis for this discrimination is as yet unclear (6, 8). Insulin and IGF-1 receptors have a similar binding Site 1 that can accommodate both hormones. The difference in affinity of insulin and IGF-1 for the receptors results from different residues interacting with specificity-conferring regions on the two receptors. The studies of chimeric insulin/IGF-1 receptors ascribed insulin specificity mainly to the N-terminal sequence 1–68 of the leucine-rich (L1) domain of IR and IGF-1 specificity to the sequence 191–290 of the cysteine-rich (CR) domain of the IGF-1R (Fig. 1) (9, 10).
Major progress in the understanding of insulin and IGF-1 binding to their receptors was achieved through the recent solving of structures of a few ligand–receptor complexes that were engineered for protein crystallography (6, 11–14). These structures provided a wealth of information about the receptor's Site 1 interactions. However, the exact nature of interactions during Site 2 binding and the structural changes of the receptor leading to the tyrosine kinase activation are still not completely understood. Conformational change required for bridging of the receptor's Sites 1 and 2 is supposed to initiate signal transmission to the kinase region. Constraints holding transmembrane regions apart are thus released, and receptor kinase autophosphorylation is triggered (11, 15). The extent of conformational change was recently visualized using single-particle cryo-EM of full-length human IR reconstituted into lipid nanodiscs (16). The location of Site 2 in insulin and the amino acids important for the interaction were deduced from mutagenesis studies. Site 2 on the receptor was mapped near the junction of fibronectin type III domains 1 and 2 (FnIII1-FnIII2) of the α subunit (17). Recently, a structure of IR ectodomain with bound insulin, obtained using single-particle cryo-EM, was published (18). It positioned the Site 2 location within the first fibronectin-like domain (FnIII1). Rather different and much more restricted Site 2 interacting residues in insulin sequence (virtually CysA7 and ThrA8 only) were detected in the cryo-EM analysis, compared with the mutagenesis studies (ThrA8, IleA10, SerA12, LeuA13, GluA17, HisB10, GluB13, and LeuB17) (8, 19). Moreover, the IR–ectodomain dimer identified by cryo-EM did not resemble the crystallographic symmetry–generated dimer. Issues arising from these findings will need further clarification.
Concerning the IGF-1 binding, it is not clear to what extent Site 2 is important (19). It remains possible that the receptor's Site 2 is located in different parts of IGF-1R compared with IR (12). It was also proposed that Site 1 of IGF-1 is extended to the IGF-1 C domain and interacts also with the CR domain of IGF-1R (20). Unfortunately, the last three residues at the C termini of the B chain/domain of insulin and IGF-1 and the C and D domains of IGF-1 were not traceable in any of the structures of hormone–receptor complexes solved so far.
A number of studies have supported a critical role of insulin residues B24 and B25 in receptor binding (8). On the other hand, residues B26–B30 are not required for IR binding. Des-(B26–B30)-pentapeptide-B25-carboxamide insulin had full potency (21). In the past, we prepared a series of des-(B27-B30)-tetrapeptide-B26-carboxamide insulin (-DTI-NH2) analogs with a modified B26 position that have severalfold increased binding to the IR (22, 23). However, the residues B26–B30 of insulin are crucial for the formation of insulin dimers, conferring thermodynamic stability and self-assembly of insulin (24, 25), and TyrB26 was proposed as playing a role in the negative cooperativity of insulin (4).
Conversely, Slieker et al. (26) reported sensitivity of IGF-1R to structural changes in the C-terminal portion of the B chain of insulin. They prepared a series of insulin analogs, modified at B28-B29 positions that were approximately equipotent to insulin in binding to the IR but showed varying affinity to the IGF-1R. Basic amino acid residues increased, whereas acidic residues reduced relative affinity to the IGF-1R. Multiple basic residues in IGF-1 D and C domains were suggested as modulating IGF-1R specificity (27) and interacting with the CR region of IGF-1R that has negative surface electrostatic potential (28).
An insulin analog with three modifications in the C-terminal part of the B chain and with exceptionally increased (about 150-fold) affinity for IGF-1R is described in this paper. At the same time, the analog is also markedly more potent than human insulin in binding to both IR isoforms. We have designed a series of new analogs and performed several kinetic and biochemical experiments to explain this observation.
Results
Analog design
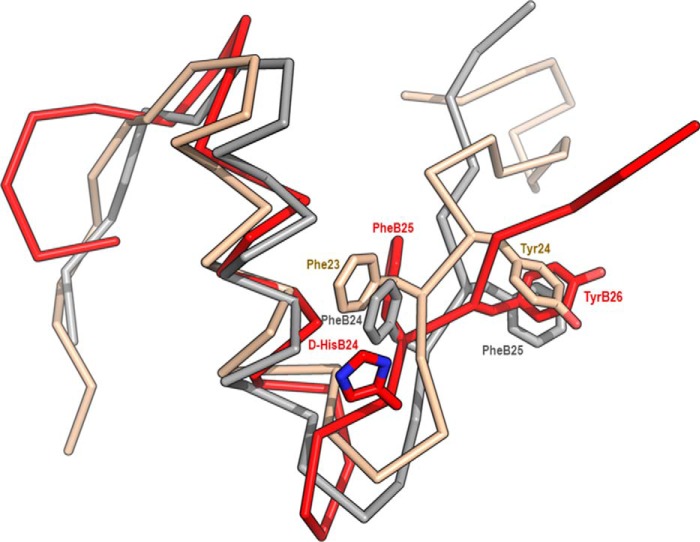
Previously synthetized analog [d-HisB24]-insulin (29) binds both IR-A and IR-B with higher affinity than human insulin (Table 1). NMR structure was also previously determined (29). The main structural feature of this analog was caused by the reverse chirality of the B24Cα atom that swayed the d-HisB24 side chain into the solvent. The pocket vacated by PheB24 was filled by PheB25, which mimicked the PheB24 side and main chains. The PheB25 downshift to the PheB24 position resulted in a subsequent downshift of TyrB26 into the B25 site and the departure of B26–B30 residues away from the insulin core (Fig. 2). In a course of routine testing of our analogs, we detected unexpectedly high stimulation of IGF-1R by [d-HisB24]-insulin. We designed a series of new analogs to explain this observation. A schematic illustration of the analogs is shown in Fig. 3.
Table 1.
Receptor binding affinities of human insulin, IGF-1, IGF-2, and insulin analogs
The Kd values were obtained from at least three measurements. n is the number of replicates. Asterisks indicate that binding of the ligand to a particular receptor differs significantly from that of insulin (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
| Analog | IR-A |
IR-B |
IGF-1R |
|||
|---|---|---|---|---|---|---|
| Kd ± S.D. (n) | Relativea binding affinity | Kd ± S.D. (n) | Relativea binding affinity | Kd ± S.D. (n) | Relativea binding affinity | |
| nm | % | nm | % | nm | % | |
| Insulinb | 0.36 ± 0.14# (5) | 100 | 0.55 ± 0.27# (4) | 100 | 292 ± 54.30€ (3) | 0.08 |
| 0.45 ± 0.11$ (6) | 0.37 ± 0.11$ (4) | |||||
| 0.27 ± 0.02∧ (5) | 0.39 ± 0.14∧ (4) | |||||
| 0.25 ± 0.05≈ (5) | 0.55 ± 0.11€ (6) | |||||
| 0.35 ± 0.06€ (4) | ||||||
| IGF-1b | 31.50 ± 5.63# (4)*** | 1.1 | 223.9 ± 32.9# (4)*** | 0.2 | 0.25 ± 0.03# (4) | 100 |
| 0.11 ± 0.05$ (5) | ||||||
| 0.12 ± 0.02∧ (5) | ||||||
| 0.24 ± 0.10€ (5) | ||||||
| IGF-2 | 2.92 ± 0.24≈ (3)*** | 8.4 | 35.45 ± 11.22# (4)*** | 1.6 | 2.32 ± 1.24# (3)*** | 10.7 |
| [d-HisB24]-DTI-NH2 | 0.18 ± 0.02# (3) | 204 | 0.30 ± 0.15# (4) | 183 | 288 ± 18.61# (3) | 0.09 |
| [d-HisB24]-insulin | 0.126 ± 0.015€ (3)** | 280 | 0.22 ± 0.05€ (3)** | 251 | 24.41 ± 10.59# (3)** | 1.02 |
| Des-PheB24-insulin | 0.12 ± 0.01# (4)* | 305 | 0.42 ± 0.11# (4) | 132 | 77.82 ± 20.71# (3)** | 0.32 |
| [TyrB25, PheB26, AsnB27, LysB28, ProB29]-insulin | 0.31 ± 0.07∧ (3) | 86 | 0.53 ± 0.21∧ (3) | 75 | 143.60 ± 54.67∧ (5) | 0.08 |
| [TyrB25, PheB26, AsnB27, LysB28, ProB29, GlyB31, TyrB32]-insulin | 0.21 ± 0.06∧ (3) | 131 | 0.19 ± 0.03∧ (3) | 207 | 16.01 ± 6.24∧ (5)*** | 0.77 |
| [GlyB31, TyrB32]-insulin | 0.17 ± 0.07∧ (3)* | 162 | 0.13 ± 0.05∧ (3)* | 310 | 45.66 ± 17.72∧ (4)** | 0.27 |
| [d-HisB24, GlyB31, TyrB32]-insulin | 0.18 ± 0.02$ (3)** | 251 | 0.12 ± 0.03$ (3)* | 338 | 0.89 ± 0.20$ (6)*** | 12.4 |
a Relative binding affinity is defined as (Kd of human insulin or IGF-1/Kd of analog) × 100 (%).
b The Kd of human insulin for IR-A was determined in five independent measurements (#, $, ∧, ≈, and €) and for IR-B in four measurements (#, $, ∧, and €), and Kd of human IGF-1 for IGF-1R was determined in four measurements (#, $, ∧, and €). The individual values of Kd of a particular ligand are relative to a corresponding insulin or IGF-1 Kd values (e.g. # to #, etc.).
Figure 2.
An overlay of the B chains of human insulin with human IGF-1 and [d-HisB24]-insulin. Insulin (PDB code 1MSO, crystal structure) is shown in gray, IGF-1 (PDB code 1GZR, crystal structure) is ocher, and [d-HisB24]-insulin (PDB code 2M2P, NMR structure with the lowest energy at pH 8) is red. Positions of downshifted d-HisB24, PheB25, and TyrB26 in [d-HisB24]-insulin are shown together with corresponding residues in insulin (PheB24 and PheB25) and IGF-1 (Phe23 and Tyr24).
Figure 3.
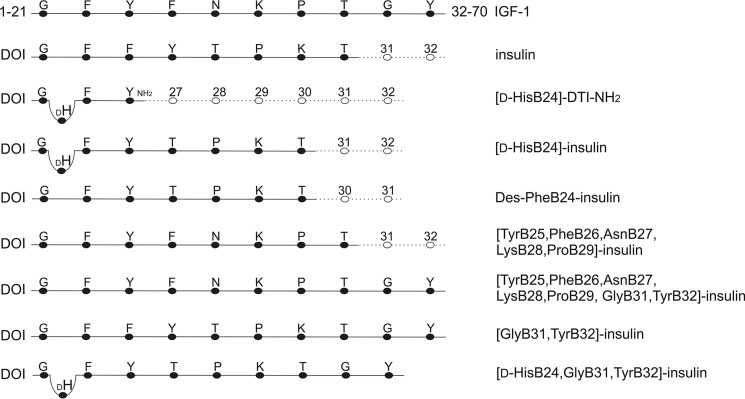
Schematic illustration of insulin analogs and comparison of the sequences with human insulin and IGF-1. Single-letter codes of amino acids are used. Vacant residues in insulin B chain are shown as empty circles; numbers represent their positions in the insulin B chain sequence. The respective sequence of human IGF-1 and its location in the amino acid chain is shown. Expected downshift of the B25 residue (and residues upward) due to the presence of d-His at B24 (DH) is indicated by bending of the connecting line. DOI, des-(B23-B30)-octapeptide-insulin. -DTI-NH2, des-(B27-B30)-tetrapeptide-B26-carboxamide insulin.
First, we aimed to check whether the d-His residue has any influence on IGF-1 receptor binding (in [d-HisB24]-DTI-NH2). Next, we examined the relevance of a downshift of PheB25 to the position of PheB24 and, in parallel, the switch of LysB29 and ProB28, which corresponds to the sequence in IGF-1 (in des-PheB24-insulin). We assumed that [d-HisB24]-insulin can adopt structural features mimicking respective parts of the IGF-1 B and C domains. Thus, we employed the B domain C-terminal sequence of IGF-1 (in [TyrB25, PheB26, AsnB27, LysB28, ProB29]-insulin) and also extended the C terminus of insulin analogs with GlyB31-TyrB32 residues, because Tyr31 in IGF-1 was shown to be important for IGF-1 receptor selectivity and activation (30) (in [TyrB25, PheB26, AsnB27, LysB28, ProB29, GlyB31, TyrB32]-insulin and in [GlyB31, TyrB32]-insulin). Finally, [d-HisB24, GlyB31, TyrB32]-insulin was designed to combine several tested features.
NMR spectroscopy and structure of [d-HisB24, GlyB31, TyrB32]-insulin
The NMR spectra of [d-HisB24, GlyB31, TyrB32]-insulin were acquired under the same conditions as used for [d-HisB24]-insulin described previously (29). The comparison of proton NMR data for both insulin analogs showed nearly the same values of chemical shifts (Δδ < 0.05 ppm) for protons of all residues except LysB29 and ThrB30. This is not surprising because they are the last two B chain residues in [d-HisB24]-insulin, whereas in the [d-HisB24, GlyB31, TyrB32]-insulin, the chemical shifts of LysB29 and ThrB30 are somewhat influenced by subsequent Gly31 and Tyr32. A comparison of the chemical shifts of backbone NH and Hα protons is presented in Table S2 and graphically demonstrated in Fig. S1. Based on these data, we can conclude that solution NMR structure of [d-HisB24, GlyB31, TyrB32]-insulin, except residues B29–B32, does not differ from the structure of [d-HisB24]-insulin described previously. The hydrophobic pocket of PheB24 was filled with PheB25, and d-HisB24 was left protruding from the structure. TyrB26 replaced the position of PheB25, and residues B27–B32 departed from the insulin core, thus mimicking the previously observed downshift in [d-HisB24]-insulin. The same conclusions for [d-HisB24, GlyB31, TyrB32] are supported by the presence of NOE cross-peaks TyrB16Hα/PheB25Hδ, Hϵ, Hζ as well as PheB25Hϵ/TyrB16Hδ, Hϵ cross-peaks. The absence of any medium- or long-range NOEs indicates a high degree of flexibility of the B26–B32 part.
Binding and stimulation of IR-A and IR-B
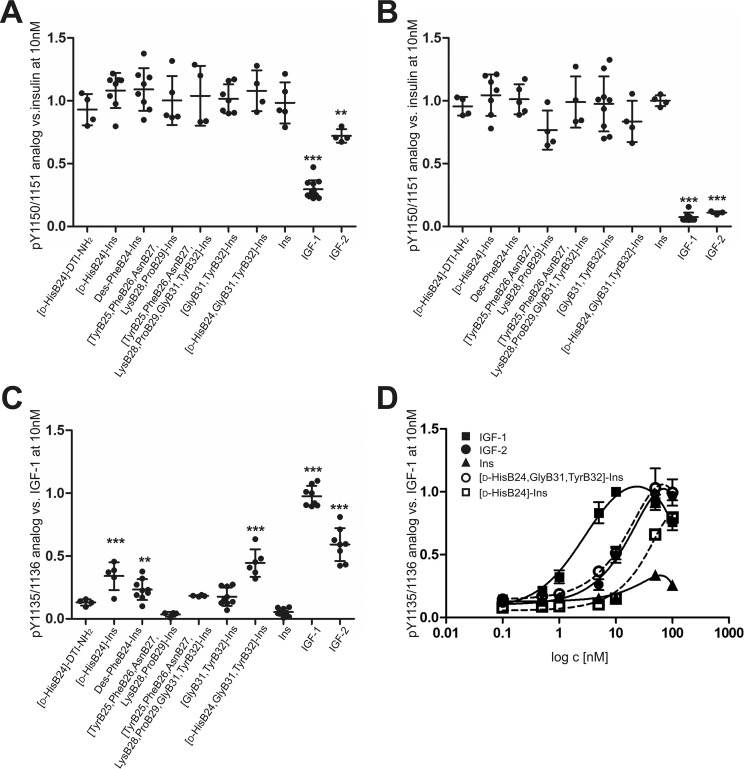
Five of seven analogs bound both IR-A and IR-B with increased affinity compared with human insulin (Table 1 and Figs. S2–S4). [TyrB25, PheB26, AsnB27, LysB28, ProB29]-insulin and [TyrB25, PheB26, AsnB27, LysB28, ProB29, GlyB31, TyrB32]-insulin, which bears the C-terminal sequence of the IGF-1 B domain, had affinity similar to insulin. The analogs with an extended C terminus with GlyB31–TyrB32 showed an increased ratio in IR-A/IR-B binding in favor of IR-B, whereas shortened analog des-PheB24-insulin showed the inverse ratio. We did not detect any deviations in the abilities of the analogs to stimulate the receptors compared with insulin (Fig. 4 (A and B) and Fig. S6).
Figure 4.
Relative abilities of human insulin, IGF-1, IGF-2, and insulin analogs to stimulate receptor phosphorylation. IR-A–transfected cells (A), IR-B–transfected cells (B), and IGF-1R–transfected cells (C) were stimulated with 10 nm ligands for 10 min. D, IGF-1R–transfected cells were stimulated with a 0.1–100 nm concentration range of WT ligands (continuous lines) and selected analogs (dashed lines). The data (mean ± S.D. (error bars), n ≥ 4) were expressed as the contribution of phosphorylation relative to the signal of human insulin (IR-A, IR-B) or IGF-1 (IGF-1R) at 10 nm in the same experiment. Data in A–C are from immunoblotting, and data in D were obtained using the In-Cell Western assay. Representative blots are shown in supporting Figs. S6 and S7. Ins, human insulin. Asterisks indicate that phosphorylation of the receptor induced by a ligand differs significantly from that of insulin (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Binding and stimulation of IGF-1R
The binding affinities (Table 1) of two analogs toward IGF-1R as well as their ability to stimulate the receptorwere unchanged compared with insulin. [d-HisB24]-DTI-NH2 bound IGF-1R equally to insulin. Also, [TyrB25, PheB26, AsnB27, LysB28, ProB29]-insulin, which bears the C-terminal sequence of the IGF-1 B domain, showed no change.
The addition of GlyB31-TyrB32 to insulin increased binding to IGF-1R about 3-fold, and the change of the sequence B23-B32 to an IGF-1–like sequence even increased binding 9-fold, although both analogs stimulated autophosphorylation of IGF-1R at 10 nm concentration similarly (Fig. 4C). The stimulation was increased about 3-fold compared with insulin, but the change appeared not to be significant in the analysis of variance.
Des-PheB24-insulin, [d-HisB24]-insulin, and [d-HisB24, GlyB31,TyrB32]-insulin showed enhanced binding to IGF-1R, accompanied with increased stimulation of the receptor. Binding of [d-HisB24, GlyB31, TyrB32]-insulin to IGF-1R was increased about 150-fold compared with insulin and was comparable with the IGF-2 binding. The analogs [d-HisB24]-insulin and [d-HisB24, GlyB31, TyrB32]-insulin were able to stimulate the IGF-1R at a 10 nm concentration to an extent approximating IGF-2 (Fig. 4C and Fig. S6). The dose–response curves of IGF-1R stimulation (Fig. 4D and Fig. S7) showed that [d-HisB24, GlyB31, TyrB32]-insulin is equipotent to IGF-2 and that [d-HisB24]-insulin is a less effective activator of IGF-1R than IGF-2, but significantly stronger than human insulin. In our further experiments, we focused on the most interesting analog, [d-HisB24, GlyB31, TyrB32]-insulin.
Binding kinetics
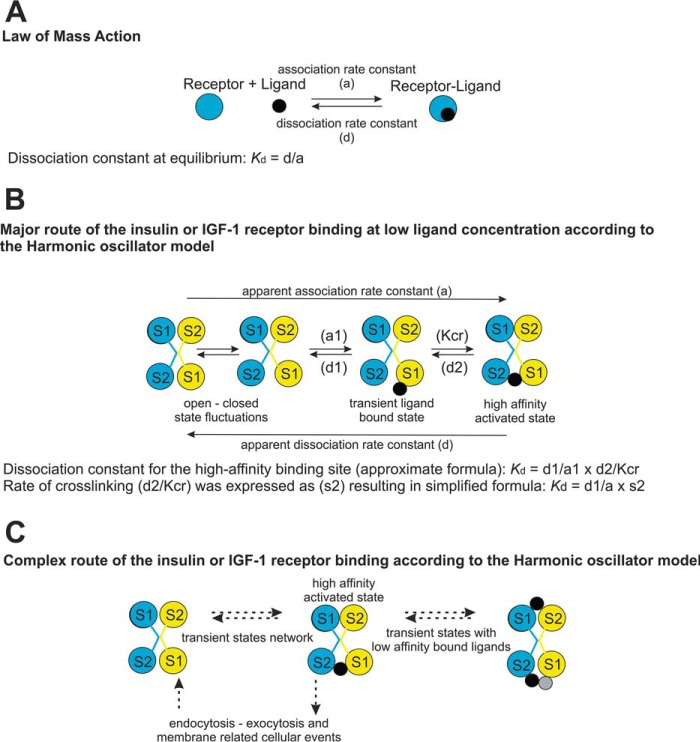
To interpret the data, we applied the presumptions and relations from the harmonic oscillator model (7, 31), as explained under “Experimental Procedures.” The model is schematically presented in Fig. 5. We considered only the major route (Fig. 5B) of a ligand cross-linking two binding sites on the receptor and ignored the other events, such as the rate of endocytosis and multiple low-affinity binding states (Fig. 5C).
Figure 5.
Simplified scheme of receptor–ligand interaction models used for interpretation of kinetic measurements. A, the law of mass action model; B, harmonic oscillator model, adapted from (6, 7) of insulin or IGF-1 binding at low ligand concentration, where a1 is the association and d1 is the dissociation coefficient of Site 1 (S1); d2 is the dissociation coefficient of Site 2 (S2), and Kcr is a constant characterizing the Site 1 and Site 2 assembly to form the high-affinity complex. C, model of insulin or IGF-1 receptor binding. Multiple factors influencing the kinetics are suggested. Receptors are represented by a blue circle or an assembly of blue and yellow circles showing binding sites on IR or IGF-1R homodimers. Black circles, ligands; gray circle, third ligand molecule bound to the IR, which is not allowed on IGF-1R.
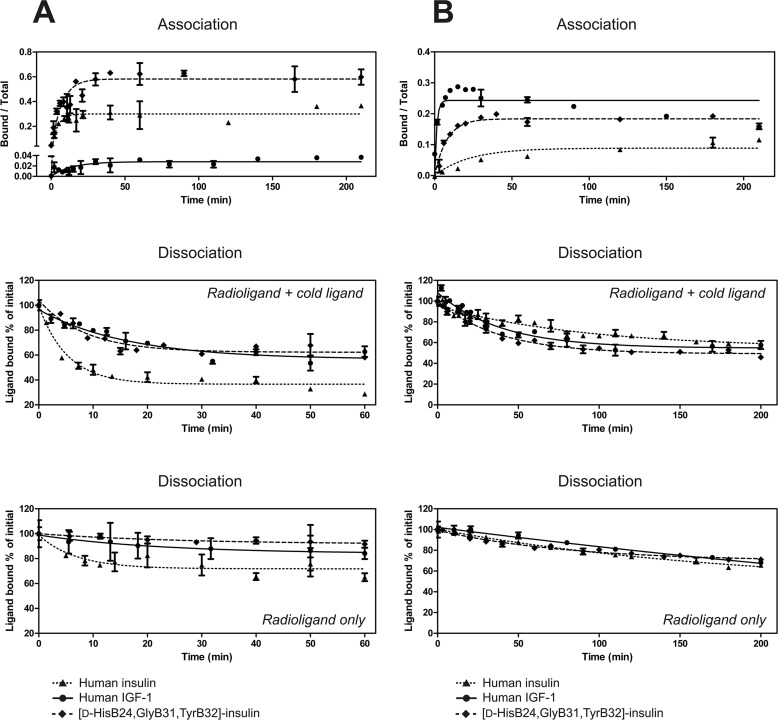
We measured association and dissociation kinetics (Fig. 6) and dose–response curves for accelerated dissociation of 125I-labeled insulin, IGF-1, and [d-HisB24, GlyB31, TyrB32]-insulin toward the IR-A and IGF-1R.
Figure 6.
Association and dissociation assays. A and B, association and dissociation of human 125I-insulin, 125I-IGF-1, and 125I-[d-HisB24, GlyB31, TyrB32]-insulin from IR-A in human IM-9 cells (A) and from IGF-1R in the CHO-K1 cell line stably transfected with human IGF-1R (B). Data are the mean ± S.D. (error bars) of duplicate values of a representative experiment. The results are expressed as the 125I-labeled ligand bound at the specific time point over total. Dissociation was measured in the presence of unlabeled ligand (for details, see “Experimental Procedures”) and with the radioligand only. Results are expressed as the percentage of 125I-labeled ligand bound at t = 0. Representative curves are shown.
Dose–response curves for accelerated dissociation had the same shape as reported previously (7) (i.e. bell-shaped for IR-A and sigmoid for IGF-1R in the case of all three ligands: human insulin, IGF-1, and [d-HisB24, GlyB31, TyrB32]-insulin) (Fig. S5).
To interpret the association and dissociation kinetics (Fig. 6), the measured Kd ratios (Table 1) were related to the ratios of dissociation coefficient at maximal acceleration, which should depend solely on Site 1 interactions (d1) (Fig. 5B) (31). In cases where the difference in interaction with receptor between ligands is dependent mainly on Site 1 interaction, the ratios of measured Kd and the d1 and a1 constants will follow a simple formula where X represents one ligand and Y is the other.
| (Eq. 1) |
We compared the measured apparent association constants (a) with theoretically expected values for Site 1 (a1) (Fig. 5B) and investigated whether the factors related to the Site 1–Site 2 (s2) cross-linking reaction must be encompassed by the following equation.
| (Eq. 2) |
The discussed ratios of the coefficients are shown in Table 2. The source data are in Table S1. Although the measurements showed high variability evident from the S.D. values (Table S1) and deviations from standard shape of the curves (Fig. 6B, IGF-1 association) we can draw the following conclusions.
Table 2.
Approximate ratios of kinetics factors for interaction of human insulin, IGF-1, and [d-HisB24, GlyB31, TyrB32]-insulin (An) with IR-A and IGF-1R
Ratios were calculated from mean values of the measured parameters. Relative S.D. values were in the range 10–50%; thus, the ratios must be considered as approximate. X and Y represent the values for the ligands listed in individual columns (insulin, IGF-1, and [d-HisB24, GlyB31, TyrB32]). For details, see “Experimental procedures” and Table S1. An, [d-HisB24, GlyB31, TyrB32]-insulin.
| Receptor type | Parameter | Insulin/IGF-1 | Insulin/An | IGF-1/An |
|---|---|---|---|---|
| IR-A | Kda | 1:88 | 2.5:1 | 221:1 |
| d1b | 6.4: 1 | 3.4: 1 | 1: 1.9 | |
| Theor. a1X/a1Y = | 563: 1 | 1.4: 1 | 1: 420 | |
| d1X/d1Y × KdY/KdXc | ||||
| ameas.d | 1.1: 1 | 1: 1.1 | 1: 1.3 | |
| aderived | 1.6: 1 | 1.2: 1 | 1: 1.3 | |
| Factor site 2 s2X/s2Y = | 1: 512 | 1: 1.5 | 323: 1 | |
| KdX/KdY × aX/aY × d1Y/d1Xe | 1: 352 | 1: 1.1 | 323: 1 | |
| IGF-1R | Kda | 1217: 1 | 150: 1 | 1: 8 |
| d1b | 1: 3 | 1: 3 | 1: 1 | |
| Theor. a1X/a1Y = | 1: 3651 | 1: 479 | 8: 1 | |
| d1X/d1Y × KdY/KdXc | ||||
| ameas.d | 1: 100 | 1: 15 | 6.7: 1 | |
| aderived | 1: 89 | 1: 14 | 6.4: 1 | |
| Factor site 2 s2X/s2Y = | 37: 1 | 30: 1 | 1: 1.2 | |
| KdX/KdY × aX/aY × d1Y/d1Xe | 41: 1 | 32: 1 | 1: 1.3 |
a Dissociation constants (Kd) are from receptor-binding assays.
b Ratios of dissociation factors for Site 1 (d1) were derived from the dissociation rate at maximal acceleration.
c Theoretical ratios of association constants (Theor. a1) were calculated as if the reaction followed the simple model of the law of mass action.
d Association factor (a) was derived from experimental curves, using the measured rate of dissociation of 125I-labeled ligand without the presence of cold ligand (ameas.), or constants calculated in the model for negative cooperativity (6) were applied (aderived).
e Factor Site 2 (s2) is an estimated value to complement the equation, using both association factors (ameas. upper value and aderived lower value).
Association of all three ligands (insulin, IGF-1, and [d-HisB24, GlyB31, TyrB32]-insulin with IR-A was very fast, whereas dissociation of IGF-1 and [d-HisB24, GlyB31, TyrB32]-insulin was markedly slower than that of insulin (Fig. 6A). In the case of [d-HisB24, GlyB31, TyrB32]-insulin, the decrease of the dissociation rate (d1) (3.4-fold) was reflected by an increase in Kd (2.5-fold) (Table 2). Our data confirm the presumption that the difference in Kd values between [d-HisB24, GlyB31, TyrB32]-insulin and insulin is related to the Site 1 interaction.
In the case of IGF-1 binding to IR-A, it was evident that factors other than simple interaction with Site 1 must play a role. If a simple model of law of mass action (Fig. 5A) is applied, the expected ratio of association coefficients (a) is 563:1 (association of IGF-1 with IR-A would be 563-fold slower than with insulin). However, the determined ratio was ∼1:1 (Table 2). The factors driving interaction with Site 2 (s2) are expected to play a major role in binding of insulin to IR-A, compared with binding of IGF-1.
The situation was opposite in the case of binding of the ligands to IGF-1R. Interaction of insulin with IGF-1R was characterized by a very slow association rate, whereas the dissociation rate was similar to that of IGF-1 (Fig. 6B). The factors driving interaction with Site 2 (s2) are also expected to be changed, but not to as great an extent as in the case of IGF-1 interacting with IR-A (∼1:400 in favor of insulin binding to IR-A, compared with 1:40 in favor of IGF-1 binding to IGF-1R) (Table 2). Increase in the association rate (about 15-fold) contributed substantially to the increase in Kd for IGF-1R of the [d-HisB24, GlyB31, TyrB32]-insulin compared with insulin (Fig. 6 and Table 2).
Discussion
Our laboratory is interested in the design of insulin and IGFs analogs, which should map the structure–activity relationship among ligands and their receptors and potentially serve medicinal purposes (22, 23, 29, 32–35). In the course of routine testing of our analogs, we have detected the unexpectedly high binding and stimulation of IGF-1R by [d-HisB24]-insulin (29). The binding and activation of IGF-1R was further dramatically accelerated by the addition of Gly31-Tyr32 to the C terminus of the B chain (251 or 338% binding affinity to IR-A respective to IR-B relative to insulin and 12.4% binding affinity to IGF-1R relative to IGF-1). To the best of our knowledge, the analog [d-HisB24, GlyB31, TyrB32]-insulin is one of the strongest insulin-like binders and activators of IGF-1R thus far reported and is a significantly more potent IGF-1R binder than the well-known “supermitogenic” analog X10 ([AspB10]-insulin) or a strong binder [ArgB31, ArgB32]-insulin (26, 36). Maybe only a chimera where insulin A and B chains are connected by the C-loop of IGF-1 is a stronger binder of IGF-1R (9). The chimera bound IR with 113% affinity relative to insulin and IGF-1R with 28% relative to IGF-1. However, [d-HisB24, GlyB31, TyrB32]-insulin differs from human insulin only in three positions but the chimera differs from human insulin in 12 residues (the whole extra C domain).
Among our analogs, [d-HisB24]-DTI-NH2 was primarily prepared to check whether the d-His residue exposed out of the insulin core has any influence on IGF-1R stimulation. Generally, des-(B27-B30)-B26-carboxamide insulins (-DTI-NH2) are characterized by high affinity to the IR (22, 23). This affinity has been explained by exposing the hydrophobic residues A1–A3 to direct interaction with the receptor. We tested the ability of other -DTI-NH2 insulin analogs to stimulate IGF-1R ([N-MeAlaB26]-DTI-NH2 and [d-ProB26]-DTI-NH2 (23); data not shown). The shortened analogs had characteristics comparable with the [d-HisB24]-DTI-NH2. The ability to bind and activate IGF-1R was similar to that of insulin. We concluded that the d-HisB24 residue and its structural effect on adjacent B25 and B26 positions makes no contribution to the increased IGF-1R binding and activation. Exposing the residues A1–A3 might have a certain impact on increased affinity to the IR, but not to the IGF-1R.
Other analogs [TyrB25, PheB26, AsnB27, LysB28, ProB29]-insulin, [TyrB25, PheB26, AsnB27, LysB28, ProB29, GlyB31, TyrB32]-insulin, and [GlyB31, TyrB32]-insulin were designed to probe whether the adoption of the IGF-1–like sequence in this portion of the molecule has any effect on binding and activation of the receptors. The [TyrB25, PheB26, AsnB27, LysB28, ProB29]-insulin did not acquire any new quality. The extension of the sequences by GlyB31-TyrB32 increased the affinity and activity of these analogs toward the IGF-1R. Surprisingly, the addition of GlyB31-TyrB32 also improved binding to IR-A and even more profoundly to IR-B. These results do not fit with the presumption that Tyr31 contributes to IGF-1R selectivity (30).
We also studied a previously reported des-PheB24-insulin (37). We synthetized this analog to prove the relevance of downshift of PheB25 to the position of PheB24. Our expectations were only partially complied with. This analog showed a high affinity to the IR-A and increased binding and activation of IGF-1R. The high affinity of des-PheB24-insulin to IR was explained analogously to the shortened -DTI-NH2- insulins and [d-HisB24]-insulin (i.e. by exposing A1-A3 residues, due to the relaxed structure at the C terminus of the B chain) (37). However, based on our data, the simple uncovering of the A1–A3 region is not the most likely driving force for an increased affinity of the analogs to the IGF-1R. We rather consider the idea that the specific change in the structure of the C-terminal part of the B chain in [d-HisB24, GlyB31, TyrB32]-insulin, resulting in its relaxation and redirection (in analogy with [d-HisB24]-insulin as confirmed here by NMR analysis), allows the residues to use a hidden potential of the receptor and bind it by means of some uncommon contacts.
To obtain a closer insight into the binding characteristics of this analog, we performed measurements of association and dissociation kinetics of insulin, IGF-1, and the [d-HisB24, GlyB31, TyrB32]-insulin toward the IR and IGF-1R (Fig. 6). We are aware that the presumptions used to interpret the data and our estimate calculations are roughly simplistic, but the conclusions seem to be eloquent. The presumption that IR and IGF-1R differ in their activation mechanism was supported. The difference between the receptors has already been implied from the shapes of the curves for negative cooperativity (7), from different binding characteristics of solubilized receptors compared with the membrane-bound receptors (5), and recently also from the structural studies (6, 12).
Based on our data, factors related to the formation of the high-affinity complex and dissociation kinetics seem to play the major role in the different affinities of the ligands (insulin, IGF-1, and the analog) for IR-A. The dissociation kinetics would probably be an even more distinguishing factor for IR-B affinity. We observed previously that specific extension of the C terminus of the insulin B chain can increase the affinity to IR-B (33). This is probably caused by additional contacts of the prolonged B chain of analogs with the receptor that slow down the dissociation from the receptor, similarly to our analogs with GlyB31-TyrB32.
Conversely, association kinetics seems to be the main factor in IGF-1R affinity. Site 1 on the receptors involves the N terminus of the L1 domain of one receptor α subunit and C-terminal residues of the insert domain of a second α subunit (αCT peptide). Site 1 on IR interacts with insulin amino acid residues: GlyA1, IleA2, ValA3, GluA4, TyrA19, GlyB8, SerB9, LeuB11, ValB12, TyrB16, PheB24, PheB25, and TyrB26 (8). Identical or homologous residues are present at the same positions in IGF-1 (Fig. 1) and were shown to interact with the IR as well as with IGF-1R in a similar manner (6, 12, 13). The only differences are in TyrB16 and SerB9 in insulin, which are replaced with Gln15 and Ala8 in IGF-1, respectively. In addition, Met59 in IGF-1, which does not have equivalent receptor-binding residue in insulin, was shown to interact with Arg704 IGF-1R αCT peptide, and its mutation abolishes receptor binding (34). Ala8 in IGF-1 was shown to interact with both the L1 domain (Glu91) and αCT peptide (Glu693) of IGF-1R, whereas insulin SerB9 interacts only with the IR αCT peptide (His710) (6, 8). Insulin TyrB16 interacts with Phe39 of IR L1 domain, which corresponds to Ser35 in IGF-1R, which is not involved in ligand binding (8, 28). Accordingly, the interaction of insulin TyrB16 with receptor Phe39 is crucial for specificity of insulin binding to IR (specificity-conferring region 1–68 IR) (9). We hypothesize that the lack of interactions provided by TyrB16 in insulin prevents anchoring of insulin on IGF-1R and accordingly slows down the association. We speculate that relaxation of the C terminus and the addition of GlyB31-TyrB32 in the [d-HisB24, GlyB31, TyrB32]-insulin provide additional contacts with the receptor, not necessarily the same as in IGF-1, and partially compensate for the missing anchor. We do not think that mimicking of structural features of the IGF-1 C-loop is the main reason for increased binding of [d-HisB24, GlyB31, TyrB32]-insulin to IGF-1R, because then also the other analogs with GlyB31-TyrB32 and not [d-HisB24]-insulin would have the same characteristics.
Phylogeny of insulin-like peptides and their interacting partners (receptors and binding proteins) dates to an outset of the Animalia kingdom. Insulin-like peptides can be found in primitive deuterostomes (38) as well as in insects (39). Whereas there is a vast diversity of insulin-like peptides sharing a similar fold of a compact three-dimensional structure, only one type of insulin-like receptor is found in the lower species. The separate IR and IGF-1R do not appear sooner than in vertebrates, and insulin receptor isoforms IR-A and IR-B exist only in mammals (2). The structural origins of the selectivity of the ligands (insulin, IGF-1, and IGF-2) to their cognate receptors still remain a mystery and a great example of natural selection. On the other hand, it is tempting to presume that the multiple insulin-like peptides found in invertebrate species should have the ability to exert specific functions, even though they act through the same receptor. Eliciting of different signaling and biological responses on the same receptor through the action of different ligands (insulin, IGFs or mimetic peptide S597) was reported (40, 41). The molecular mechanisms responsible for how different ligands activating the same receptor can initiate different biological responses in the same cell are not completely understood (42). It is thus possible that there is more than one way of activating the receptor. Our data with [d-HisB24, GlyB31, TyrB32]-insulin supports this assumption.
Based on the course of binding of insulin, IGF-1, and [d-HisB24, GlyB31, TyrB32]-insulin to the receptors, we offer insight into the factors contributing to the selectivity to the receptors. These discoveries can provide clues for the design of selective analogs and possibly antagonists of the receptors and demonstrate the power and effectivity of rational hormone engineering.
Experimental procedures
Synthesis of analogs
Previously described insulin analog [d-HisB24]-insulin (29) and five newly prepared analogs ([d-HisB24]-DTI-NH2, [TyrB25, PheB26, AsnB27, LysB28, ProB29]-insulin, [TyrB25, PheB26, AsnB27, LysB28, ProB29, GlyB31, TyrB32]-insulin, [GlyB31, TyrB32]-insulin, and [d-HisB24, GlyB31, TyrB32]-insulin), together with des-PheB24-insulin (37), were prepared by enzymatic semisynthesis of des-(B23-B30)-octapeptide insulin and corresponding tetra-, hepta-, octa-, and decapeptides. All semisynthetic procedures were described in detail previously (22), except that we used Fmoc-Lys(Pac)-OH, which was prepared by a modification (see supporting information) of a method described previously (43). The identities of peptides and insulin analogs were confirmed, using mass spectrometer LTQ-orbitrap XL (Thermo Fisher) or the TripleTOFTM 5600 system (AB SCIEX), and their purities (≥95%) were controlled by analytical HPLC. A schematic presentation of the analogs is shown in Fig. 3.
NMR spectroscopy
NMR spectra of 0.2 mm nonlabeled [d-HisB24, GlyB31, TyrB32]-insulin were acquired as a 0.4-ml solution in H2O + D2O (95:5) with 25 mm deuterated Tris buffer (pH 8.0) at 25 °C on a 600-MHz Bruker AVANCE spectrometer equipped with a triple-resonance cryoprobe. A series of 2D homonuclear spectra was recorded for structural assignment of proton signals: 2D TOCSY spectra with a mixing time 30, 60, and 90 ms and 2D NOESY spectra with mixing time 150, 200, and 300 ms. Proton NMR data are shown in Table S3.
Cell culture
Human IM-9 lymphocytes (ATCC) and mouse embryonic fibroblasts (IR-A, IR-B, and R+39) derived from IGF-1R knockout mice and stably transfected with the receptors IR-A, IR-B, and IGF-1R, kindly provided by A. Belfiore (Catanzarro, Italy) and R. Baserga (Philadelphia, PA), were grown as described previously (33, 42).
Chinese hamster ovary cell line CHO-K1 (ATCC) was stably transfected with pcDNA3-IGF-1R vector, kindly provided by R. O'Connor (Cork, Ireland), using Lipofectamine 2000 reagent and GeneticinTM (Thermo Fisher Scientific) as a selection antibiotic. A polyclonal population of cells stably expressing human IGF-1R was obtained (CCHO-R+). The cells were grown in Ham's F-12 medium, supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 0.5 mg/ml Geneticin, 100 units/ml penicillin, and 100 μg/ml streptomycin in humidified air with 5% CO2 at 37 °C. For receptor binding studies, the cells were trypsinized and transferred to an Erlenmeyer flask (5 × 106 cells/ml). They were maintained floating by constant agitation at 140 rpm at 37 °C overnight.
Receptor-binding studies
Human IM-9 lymphocytes (containing IR-A) and IGF-1R null mouse embryonic fibroblasts, stably transfected with either human IR-B or human IGF-1R, were employed for a whole-cell receptor-binding assay as described previously (32, 33, 42). For details, see supporting information. The binding curve of each analog was determined in duplicate, and the final dissociation constant (Kd) was calculated from at least three (n ≥ 3) binding curves. Significance of the changes in binding affinities in relation to insulin or IGF-1 was calculated using the two-tailed t test.
Dose–response curves for negative cooperativity (accelerated dissociation), association kinetics, and dissociation kinetics were measured for insulin, IGF-1, and [d-HisB24, GlyB31, TyrB32]-insulin toward the IR-A and IGF-1R. Basically, the procedures followed the published protocols (44). For details, see supporting information. Each curve was determined in duplicate, and experiments were repeated at least twice. IM-9 cells were used for IR-A measurements, and CHO-R+ cells were used for IGF-1R.
Labeled mono-125I-insulin and mono-125I-IGF-1 were purchased from PerkinElmer Life Sciences. Iodination of [d-HisB24, GlyB31, TyrB32]-insulin with 125I (Na[125I], product code I-RB-41, 0.1 mCi; Izotóp Intézet Kft. (Budapest, Hungary)) was performed, using the IODO-GENTM system (Pierce). The mono-iodinated ligand was separated on a Nucleosil 120 C18 column (5μ, 250 × 4.0 mm; Macherey Nagel). A detailed description of the iodination procedure is included in the supporting Methods.
For accelerated dissociation, the cells were preincubated with radiolabeled ligand for 2.5 h at 15 °C in all cases. For dissociation kinetics, the preincubation duration was estimated based on association kinetics (time after reaching the steady state). The cells were preincubated with radiolabeled ligand for 2.5 h in the case of insulin and analog binding to IR-A. In the case of IGF-1 binding to IR-A, the preincubation lasted 4 h. The period was 2.5 h for analog and IGF-1 binding to IGF-1R, and 7 h in the case of insulin binding to IGF-1R.
Dissociation kinetics of a radiolabeled ligand was determined both without and in the presence of cold ligand (170 nm for all of the experiments, except for insulin on IGF-1R, where a 17 μm solution of insulin was used).
Binding kinetics
To interpret the data obtained in the course of measuring the association and dissociation kinetics, we adopted some presumptions and relations from the harmonic oscillator model of insulin binding (7, 31) (Fig. 5).
First presumption
Kd value for the apparent high-affinity binding site can be calculated according to an approximate formula,
| (Eq. 3) |
where a1 is an association and d1 is a dissociation coefficient of Site 1; d2 is the dissociation coefficient of Site 2, and Kcr is a constant characterizing the Site 1 and Site 2 assembly to form the high-affinity complex.
Second presumption
Dissociation rate at maximum acceleration is proportional to the dissociation coefficient for Site 1 (d1).
Third presumption
If the difference in interaction with receptor between ligands is dependent solely on Site 1 interaction, then the ratios of measured Kd and d1 will be proportional to a ratio of constants (a1) of one ligand (X) to the other ligand (Y) as in Equation 1.
If the kinetics of the Site 1–Site 2 assembly is influencing the interaction, the found ratios of association coefficients will be disproportional and dependent on the Site 2 coefficients as follows.
| (Eq. 4) |
Because we cannot estimate the d2 and Kcr, we have simplified the formula to Equation 2, where s2 is a factor driven by the Site 2 interactions, and a is an apparent association coefficient determined from the experimental data.
The dissociation constants were calculated after fitting the experimental data to monoexponential decay in GraphPad Prism version 5. To fit experimental data to association kinetics in GraphPad Prism 5, it is mandatory to set the dissociation constant. However, assignment of this is ambiguous due to a multistep process of ligand binding and concentration dependence. Thus, the determined association and dissociation constants should be considered artificial. For calculations, we used the measured constants determined from dissociation in the absence of cold ligand. We also used constants calculated for first ligand dissociation (condition of low ligand concentration), derived from a recently refined model for negative cooperativity (6). The constants were adjusted to the respective analog according to the found d1 ratios. The data are shown in Table S1.
Receptor phosphorylation assay
Cell stimulation and detection of receptor phosphorylation were performed as described previously (33), using mouse fibroblasts (IR-A, IR-B, and R+39). For details, see supporting information. The cells were stimulated with 10 nm concentrations of the ligands for 10 min. Proteins were routinely analyzed using immunoblotting. The membranes were probed with anti-phospho-IGF-1Rβ (Tyr1135/1136)/IRβ (Tyr1150/1151) (Cell Signaling Technology). Each experiment was repeated four times. The data were expressed as the contribution of phosphorylation relative to the human insulin (IR-A, IR-B) or IGF-1 (IGF-1R) signal in the same experiment. Mean ± S.D. (n ≥ 4) values were calculated. The significance of the changes in stimulation of phosphorylation in relation to insulin was calculated, using one-way analysis of variance with Dunnett's test comparing all analogs versus control (i.e. insulin).
Ligand–dose response IGF-1R autophosphorylation levels for selected analogs were determined, using the In-Cell Western assay adapted for chemiluminescence as described (34). Data were subtracted from background values and expressed as the contribution of phosphorylation relative to the 10 nm IGF-1 signal. Experiments were repeated at least four times. Log(agonist) versus response curve fitting of data was carried out with GraphPad Prism 5.
Author contributions
M. C., L. Ž., A. M., O. S., M. B., and I. S. data curation; M. C., L. Ž., M. H., J. P., K. M., and I. S. investigation; M. C., L. Ž., A. M., O. S., M. B., M. H., J. P., K. M., and I. S. methodology; J. J. and I. S. conceptualization; J. J. funding acquisition; J. J. and I. S. writing-original draft; I. S. supervision.
Supplementary Material
Acknowledgments
We thank Kateřina Nováková from IOCB for performing MS analysis on MALDI-TOF/TOF. Also, we thank Maja Jensen and Rita Slaaby from Novo Nordisk A/S (Maløv, Denmark) for providing protocols for kinetic studies.
This work was supported by Medical Research Council Grant MR/K000179/1 and partly also by Medical Research Council Grant MR/R009066/1. Institutional support was provided by Czech Academy of Sciences Project RVO 61388963 (for the Institute of Organic Chemistry and Biochemistry). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S7.
- IGF
- insulin-like growth factor
- IR
- insulin receptor
- CR
- cysteine-rich
- IGF-1R
- IGF-1 receptor
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- 2D
- two-dimensional
- PDB
- Protein Data Bank.
References
- 1. Siddle K. (2012) Molecular basis of signaling specificity of insulin and IGF receptors: neglected corners and recent advances. Front. Endocrinol. (Lausanne) 3, 34 10.3389/fendo.2012.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belfiore A., Frasca F., Pandini G., Sciacca L., and Vigneri R. (2009) Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 30, 586–623 10.1210/er.2008-0047 [DOI] [PubMed] [Google Scholar]
- 3. Belfiore A., Malaguarnera R., Vella V., Lawrence M. C., Sciacca L., Frasca F., Morrione A., and Vigneri R. (2017) Insulin receptor isoforms in physiology and disease: an updated view. Endocr. Rev. 38, 379–431 10.1210/er.2017-00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Meyts P., Van Obberghen E., and Roth J. (1978) Mapping of the residues responsible for the negative cooperativity of the receptor-binding region of insulin. Nature 273, 504–509 10.1038/273504a0 [DOI] [PubMed] [Google Scholar]
- 5. Schäffer L. (1994) A model for insulin binding to the insulin receptor. Eur. J. Biochem. 221, 1127–1132 10.1111/j.1432-1033.1994.tb18833.x [DOI] [PubMed] [Google Scholar]
- 6. Xu Y., Kong G. K., Menting J. G., Margetts M. B., Delaine C. A., Jenkin L. M., Kiselyov V. V., De Meyts P., Forbes B. E., and Lawrence M. C. (2018) How ligand binds to the type 1 insulin-like growth factor receptor. Nat. Commun. 9, 821 10.1038/s41467-018-03219-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kiselyov V. V., Versteyhe S., Gauguin L., and De Meyts P. (2009) Harmonic oscillator model of the insulin and IGF1 receptors' allosteric binding and activation. Mol. Syst. Biol. 5, 243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Meyts P. (2015) Insulin/receptor binding: the last piece of the puzzle? What recent progress on the structure of the insulin/receptor complex tells us (or not) about negative cooperativity and activation. Bioessays 37, 389–397 10.1002/bies.201400190 [DOI] [PubMed] [Google Scholar]
- 9. Kristensen C., Andersen A. S., Hach M., Wiberg F. C., Schäffer L., and Kjeldsen T. (1995) A single-chain insulin-like growth factor I/insulin hybrid binds with high affinity to the insulin receptor. Biochem. J. 305, 981–986 10.1042/bj3050981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schumacher R., Soos M. A., Schlessinger J., Brandenburg D., Siddle K., and Ullrich A. (1993) Signaling-competent receptor chimeras allow mapping of major insulin receptor binding domain determinants. J. Biol. Chem. 268, 1087–1094 [PubMed] [Google Scholar]
- 11. Menting J. G., Whittaker J., Margetts M. B., Whittaker L. J., Kong G. K., Smith B. J., Watson C. J., Záková L., Kletvíková E., Jiráček J., Chan S. J., Steiner D. F., Dodson G. G., Brzozowski A. M., Weiss M. A., et al. (2013) How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245 10.1038/nature11781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ward C. W., Menting J. G., and Lawrence M. C. (2013) The insulin receptor changes conformation in unforeseen ways on ligand binding: sharpening the picture of insulin receptor activation. Bioessays 35, 945–954 10.1002/bies.201300065 [DOI] [PubMed] [Google Scholar]
- 13. Menting J. G., Lawrence C. F., Kong G. K., Margetts M. B., Ward C. W., and Lawrence M. C. (2015) Structural congruency of ligand binding to the insulin and insulin/type 1 insulin-like growth factor hybrid receptors. Structure 23, 1271–1282 10.1016/j.str.2015.04.016 [DOI] [PubMed] [Google Scholar]
- 14. Menting J. G., Yang Y., Chan S. J., Phillips N. B., Smith B. J., Whittaker J., Wickramasinghe N. P., Whittaker L. J., Pandyarajan V., Wan Z. L., Yadav S. P., Carroll J. M., Strokes N., Roberts C. T., Ismail-Beigi F., et al. (2014) Protective hinge in insulin opens to enable its receptor engagement. Proc. Natl. Acad. Sci. U.S.A. 111, E3395–E3404 10.1073/pnas.1412897111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kavran J. M., McCabe J. M., Byrne P. O., Connacher M. K., Wang Z., Ramek A., Sarabipour S., Shan Y., Shaw D. E., Hristova K., Cole P. A., and Leahy D. J. (2014) How IGF-1 activates its receptor. Elife 3, e03772 10.7554/eLife.03772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gutmann T., Kim K. H., Grzybek M., Walz T., and Coskun Ü. (2018) Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J. Cell Biol. 217, 1643–1649 10.1083/jcb.201711047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence M. C., McKern N. M., and Ward C. W. (2007) Insulin receptor structure and its implications for the IGF-1 receptor. Curr. Opin. Struct. Biol. 17, 699–705 10.1016/j.sbi.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 18. Scapin G., Dandey V. P., Zhang Z., Prosise W., Hruza A., Kelly T., Mayhood T., Strickland C., Potter C. S., and Carragher B. (2018) Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 556, 122–125 10.1038/nature26153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gauguin L., Delaine C., Alvino C. L., McNeil K. A., Wallace J. C., Forbes B. E., and De Meyts P. (2008) Alanine scanning of a putative receptor binding surface of insulin-like growth factor-I. J. Biol. Chem. 283, 20821–20829 10.1074/jbc.M802620200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keyhanfar M., Booker G. W., Whittaker J., Wallace J. C., and Forbes B. E. (2007) Precise mapping of an IGF-I-binding site on the IGF-1R. Biochem. J. 401, 269–277 10.1042/BJ20060890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakagawa S. H., and Tager H. S. (1987) Role of the COOH-terminal B-chain domain in insulin-receptor interactions: identification of perturbations involving the insulin mainchain. J. Biol. Chem. 262, 12054–12058 [PubMed] [Google Scholar]
- 22. Záková L., Kazdová L., Hanclová I., Protivínská E., Sanda M., Budesínský M., and Jirácek J. (2008) Insulin analogues with modifications at position B26: divergence of binding affinity and biological activity. Biochemistry 47, 5858–5868 10.1021/bi702086w [DOI] [PubMed] [Google Scholar]
- 23. Jirácek J., Záková L., Antolíková E., Watson C. J., Turkenburg J. P., Dodson G. G., and Brzozowski A. M. (2010) Implications for the active form of human insulin based on the structural convergence of highly active hormone analogues. Proc. Natl. Acad. Sci. U.S.A. 107, 1966–1970 10.1073/pnas.0911785107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antolíková E., Žáková L., Turkenburg J. P., Watson C. J., Hančlová I., Šanda M., Cooper A., Kraus T., Brzozowski A. M., and Jiráček J. (2011) Non-equivalent role of inter- and intramolecular hydrogen bonds in the insulin dimer interface. J. Biol. Chem. 286, 36968–36977 10.1074/jbc.M111.265249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pandyarajan V., Phillips N. B., Rege N., Lawrence M. C., Whittaker J., and Weiss M. A. (2016) Contribution of TyrB26 to the function and stability of insulin: structure-activity relationships at a conserved hormone-receptor interface. J. Biol. Chem. 291, 12978–12990 10.1074/jbc.M115.708347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Slieker L. J., Brooke G. S., DiMarchi R. D., Flora D. B., Green L. K., Hoffmann J. A., Long H. B., Fan L., Shields J. E., Sundell K. L., Surface P. L., and Chance R. E. (1997) Modifications in the B10 and B26–B30 regions of the B chain of human insulin alter affinity for the human IGF-I receptor more than for the insulin receptor. Diabetologia 40, Suppl. 2, S54–S61 10.1007/s001250051402 [DOI] [PubMed] [Google Scholar]
- 27. Zhang W., Gustafson T. A., Rutter W. J., and Johnson J. D. (1994) Positively charged side chains in the insulin-like growth factor-1 C- and D-regions determine receptor binding specificity. J. Biol. Chem. 269, 10609–10613 [PubMed] [Google Scholar]
- 28. Lou M., Garrett T. P., McKern N. M., Hoyne P. A., Epa V. C., Bentley J. D., Lovrecz G. O., Cosgrove L. J., Frenkel M. J., and Ward C. W. (2006) The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc. Natl. Acad. Sci. U.S.A. 103, 12429–12434 10.1073/pnas.0605395103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Žáková L, Kletvíková E., Veverka V., Lepsík M., Watson C. J., Turkenburg J. P., Jirácek J., and Brzozowski A. M. (2013) Structural integrity of the B24 site in human insulin is important for hormone functionality. J. Biol. Chem. 288, 10230–10240 10.1074/jbc.M112.448050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bayne M. L., Applebaum J., Chicchi G. G., Miller R. E., and Cascieri M. A. (1990) The roles of tyrosines 24, 31, and 60 in the high affinity binding of insulin-like growth factor-I to the type 1 insulin-like growth factor receptor. J. Biol. Chem. 265, 15648–15652 [PubMed] [Google Scholar]
- 31. Knudsen L., De Meyts P., and Kiselyov V. V. (2011) Insight into the molecular basis for the kinetic differences between the two insulin receptor isoforms. Biochem. J. 440, 397–403 10.1042/BJ20110550 [DOI] [PubMed] [Google Scholar]
- 32. Záková L., Kletvíková E., Lepšík M., Collinsová M., Watson C. J., Turkenburg J. P., Jiráček J., and Brzozowski A. M. (2014) Human insulin analogues modified at the B26 site reveal a hormone conformation that is undetected in the receptor complex. Acta Crystallogr. D Biol. Crystallogr. 70, 2765–2774 10.1107/S1399004714017775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Křížková K., Chrudinová M., Povalová A., Selicharová I., Collinsová M., Vaněk V., Brzozowski A. M., Jiráček J., and Žáková L. (2016) Insulin-insulin-like growth factors hybrids as molecular probes of hormone:receptor binding specificity. Biochemistry 55, 2903–2913 10.1021/acs.biochem.6b00140 [DOI] [PubMed] [Google Scholar]
- 34. Macháčková K., Collinsová M., Chrudinová M., Selicharová I., Pícha J., Buděšínský M., Vaněk V., Žáková L., Brzozowski A. M., and Jiráček J. (2017) Insulin-like growth factor 1 analogs clicked in the C domain: chemical synthesis and biological activities. J. Med. Chem. 60, 10105–10117 10.1021/acs.jmedchem.7b01331 [DOI] [PubMed] [Google Scholar]
- 35. Macháčková K., Chrudinová M., Radosavljević J., Potalitsyn P., Křížková K., Fábry M., Selicharová I., Collinsová M., Brzozowski A. M., Žáková L., and Jiráček J. (2018) Converting insulin-like growth factors 1 and 2 into high-affinity ligands for insulin receptor isoform A by the introduction of an evolutionarily divergent mutation. Biochemistry 57, 2373–2382 10.1021/acs.biochem.7b01260 [DOI] [PubMed] [Google Scholar]
- 36. Kurtzhals P., Schäffer L., Sørensen A., Kristensen C., Jonassen I., Schmid C., and Trüb T. (2000) Correlations of receptor binding and metabolic and mitogenic potencies of insulin analogs designed for clinical use. Diabetes 49, 999–1005 10.2337/diabetes.49.6.999 [DOI] [PubMed] [Google Scholar]
- 37. Jørgensen A. M., Olsen H. B., Balschmidt P., and Led J. J. (1996) Solution structure of the superactive monomeric des-[Phe(B25)] human insulin mutant: elucidation of the structural basis for the monomerization of des-[Phe(B25)] insulin and the dimerization of native insulin. J. Mol. Biol. 257, 684–699 10.1006/jmbi.1996.0194 [DOI] [PubMed] [Google Scholar]
- 38. Perillo M., and Arnone M. I. (2014) Characterization of insulin-like peptides (ILPs) in the sea urchin Strongylocentrotus purpuratus: insights on the evolution of the insulin family. Gen. Comp. Endocrinol. 205, 68–79 10.1016/j.ygcen.2014.06.014 [DOI] [PubMed] [Google Scholar]
- 39. Mizoguchi A., and Okamoto N. (2013) Insulin-like and IGF-like peptides in the silkmoth Bombyx mori: discovery, structure, secretion, and function. Front. Physiol. 4, 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Versteyhe S., Klaproth B., Borup R., Palsgaard J., Jensen M., Gray S. G., and De Meyts P. (2013) IGF-I, IGF-II, and insulin stimulate different gene expression responses through binding to the IGF-I receptor. Front. Endocrinol. (Lausanne) 4, 98 10.3389/fendo.2013.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jensen M., Hansen B., De Meyts P., Schäffer L., and Ursø B. (2007) Activation of the insulin receptor by insulin and a synthetic peptide leads to divergent metabolic and mitogenic signaling and responses. J. Biol. Chem. 282, 35179–35186 10.1074/jbc.M704599200 [DOI] [PubMed] [Google Scholar]
- 42. Morcavallo A., Genua M., Palummo A., Kletvikova E., Jiracek J., Brzozowski A. M., Iozzo R. V., Belfiore A., and Morrione A. (2012) Insulin and insulin-like growth factor II differentially regulate endocytic sorting and stability of insulin receptor isoform A. J. Biol. Chem. 287, 11422–11436 10.1074/jbc.M111.252478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Záková L., Zyka D., Jezek J., Hanclová I., Sanda M., Brzozowski A. M., and Jirácek J. (2007) The use of Fmoc-Lys(Pac)-OH and penicillin G acylase in the preparation of novel semisynthetic insulin analogs. J. Pept. Sci. 13, 334–341 10.1002/psc.847 [DOI] [PubMed] [Google Scholar]
- 44. Sajid W., Holst P. A., Kiselyov V. V., Andersen A. S., Conlon J. M., Kristensen C., Kjeldsen T., Whittaker J., Chan S. J., and De Meyts P. (2009) Structural basis of the aberrant receptor binding properties of hagfish and lamprey insulins. Biochemistry 48, 11283–11295 10.1021/bi901269j [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.