Abstract
Human cytochrome P450 enzymes are membrane-bound heme-containing monooxygenases. As is the case for many heme-containing enzymes, substitution of the metal in the center of the heme can be useful for mechanistic and structural studies of P450 enzymes. For many heme proteins, the iron protoporphyrin prosthetic group can be extracted and replaced with protoporphyrin containing another metal, but human membrane P450 enzymes are not stable enough for this approach. The method reported herein was developed to endogenously produce human membrane P450 proteins with a nonnative metal in the heme. This approach involved coexpression of the P450 of interest, a heme uptake system, and a chaperone in Escherichia coli growing in iron-depleted minimal medium supplemented with the desired trans-metallated protoporphyrin. Using the steroidogenic P450 enzymes CYP17A1 and CYP21A2 and the drug-metabolizing CYP3A4, we demonstrate that this approach can be used with several human P450 enzymes and several different metals, resulting in fully folded proteins appropriate for mechanistic, functional, and structural studies including solution NMR.
Keywords: membrane enzyme, cytochrome P450, heme, metalloenzyme, metalloprotein, nuclear magnetic resonance (NMR), metal, porphyrin, protoporphyrin, membrane protein, metal substitution
Introduction
The heme prosthetic group plays an indispensable structural and functional role in a large variety of proteins, including the oxygen transport proteins hemoglobin and myoglobin, proteins of the electron transport chain, and enzymes such as peroxidases, catalases, and cytochrome P450. Direct involvement of the heme in protein and enzyme function has prompted substitution of the iron protoporphyrin (FePPIX)2 with protoporphyrins containing other metals. For decades, this approach has been used to obtain insight into enzyme mechanism, to generate novel enzyme activities (1–4), and to facilitate structural studies (5). Many native heme-containing proteins can be produced in Escherichia coli either by relying on bacterial heme biosynthesis, by adding heme precursors during recombinant expression, or by expressing the apoprotein and reconstituting with PPIX during purification. For intrinsically stable apoproteins, such as myoglobin, hemoglobin, horseradish peroxidase, and cytochrome c peroxidase, either nonnative cofactor incorporation into the purified apoprotein or a simple acid-extraction method (6) has efficiently permitted heme substitution. Whereas this method can also work well for the more stable soluble cytochrome P450 monooxygenases (7, 8), it does not work well for the significantly less stable, membrane-bound P450 enzymes that play key roles in human drug metabolism and endogenous pathways, including steroidogenesis, vitamin metabolism, and fatty acid oxidation. These membrane P450 holoenzymes typically precipitate under heme extraction conditions and do not rebind protoporphyrins to a significant extent, resulting in very poor yield and indicating a need for other approaches.
Membrane cytochrome P450 enzymes can often be expressed recombinantly in E. coli as the holoprotein with a proximal cysteine thiolate coordinating the Fe3+ of heme b or iron protoporphyrin IX. Endogenous E. coli heme biosynthesis is usually supported by supplementing growth medium with the nonmetal heme precursor δ-aminolevulinic acid. The δ-aminolevulinic acid is readily taken up by E. coli, where it is used in heme synthesis by bacterial enzymes, and the heme is endogenously inserted into a human P450 polypeptide to produce enzymatically active protein. In contrast, heme (FePPIX) is not readily taken up by E. coli, so providing this or other metal-substituted PPIX to the P450-expressing bacteria requires additional engineering. The soluble bacterial P450 BM3 has been generated with manganese protoporphyrin IX (MnPPIX) instead of FePPIX by supplementing induction medium with MnPPIX and coexpressing the heme transporter chuA to promote its uptake (9). Another soluble bacterial thermophilic P450 protein (CYP119) has been generated by limiting iron and providing cobalt in the induction medium, relying on intrinsic bacterial uptake of the metal and endogenous ferrocheletase incorporation of cobalt into the protoporphyrin framework (10).
Despite the significant potential contributions in probing human P450 function and structure, we have found no reports of successful generation of metal-substituted membrane cytochrome P450 enzymes, which even in their native FePPIX state are often quasi-stable. Herein, we describe a method for generating human membrane cytochrome P450 proteins containing metals other than iron in the protoporphyrin. To determine how broadly applicable this method might be, we have evaluated exogenous PPIX uptake and incorporation into both the steroidogenic P450 enzymes CYP17A1 and CYP21A2 and drug-metabolizing CYP3A4. More detailed studies with the least stable of these enzymes, CYP17A1, demonstrated the ability to incorporate other metals without significant structural changes. This method was developed specifically for application to solution NMR. The goal was to reduce the deleterious effect that the unpaired electron spin of the paramagnetic iron has on resonances for nearby amino acids and thus facilitate NMR studies. However, it has the potential for broad application in mechanistic studies and other structural and functional investigations.
Results and discussion
Generation of membrane P450 proteins using exogenous FePPIX uptake
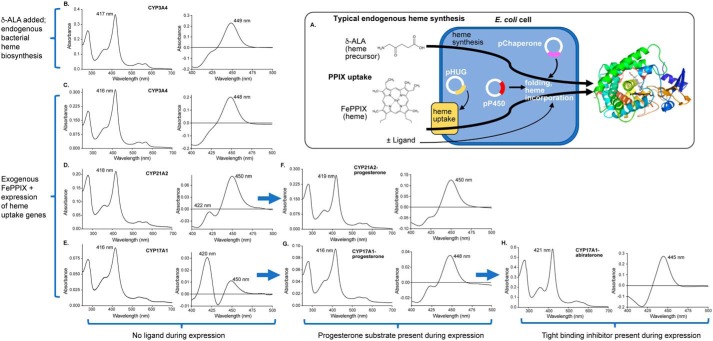
Initial trials reconfirmed that protoporphyrin extraction and reconstitution was not a feasible approach for substituting the prosthetic group in membrane P450 enzymes due to excessive precipitation of apo-P450 polypeptide and poor reincorporation of even the native FePPIX. To develop a method for inserting PPIX containing different metals during de novo P450 synthesis, the first step was to determine whether membrane P450 enzymes could be produced in suitable yields in minimal medium with PPIX uptake from the medium. Like many membrane proteins, human cytochrome P450 enzymes are notable for low recombinant expression even in rich medium, often Terrific Broth, but growth in minimal medium is necessary to limit the availability of iron and minimize endogenous bacterial FePPIX biosynthesis. To facilitate polypeptide folding, the GroEL/ES chaperone was coexpressed from a second plasmid with distinct antibiotic resistance and induction systems. This approach is frequently useful in increasing P450 protein yields even in rich medium, so its inclusion in minimal medium where lower yields were expected seemed logical. Additionally, because PPIX is not normally taken up into laboratory E. coli strains, a heme uptake system consisting of four additional proteins (hugA, tonB, exbBD, and hugBCD) from Plesiomonas shigelloides (11) was coexpressed. Induction of the heme uptake genes by the addition of an iron chelator further helped reduce adventitious iron. Thus, the method developed herein required E. coli to express the membrane P450 plus two chaperone proteins and four heme uptake proteins, while under selection with three different antibiotics, in iron-limited minimal medium (Fig. 1A). Initial trials were conducted for steroidogenic CYP21A2 and CYP17A1 and the drug-metabolizing CYP3A4 proteins in iron-depleted minimal medium supplemented with exogenous FePPIX. This was done to test not only PPIX uptake into cells, incorporation into P450, and the holoprotein yield, but also to permit determination of the catalytic activity of the resulting protein compared with a typical expression protocol involving endogenous bacterial FePPIX synthesis from the heme precursor δ-aminolevulinic acid, which is cell-permeable (Fig. 1A).
Figure 1.
Generation of three human membrane cytochrome P450 enzymes employing exogenous FePPIX. A, typical membrane P450 recombinant expression involves E. coli growth in rich medium supplemented by the heme precursor δ-aminolevulinic acid (δ-ALA), which is cell-permeable and supports endogenous bacterial heme synthesis for incorporation into the P450 polypeptide. The new method supports the possibility of P450 nonnative metal incorporation by growing E. coli in iron-depleted minimal medium supplemented with PPIX containing iron or other metals. Expression of four heme uptake genes from the pHUG plasmid facilitates uptake of this PPIX for incorporation into the P450 polypeptide. Both methods employ coexpression of chaperone proteins to facilitate folding and increase P450 yield. The usual endogenous heme biosynthesis method results in CYP3A4 protein (B) with a Soret peak consistent with water coordination to the heme iron (417 nm), whereas the reduced-carbon monoxide difference spectrum indicates mostly P450 and little P420. Very similar results are observed for CYP3A4 expressed with exogenous supplementation of FePPIX (C). However, the same exogenous FePPIX method results in purified CYP21A2 (D) and CYP17A1 (E) with normal absolute spectra (416–418 nm), but significant amounts of undesirable P420. The addition of the substrate progesterone in the medium during expression with exogenous FePPIX results in CYP21A2 (F) and CYP17A1 (G) in which the substrate has dissociated during purification (Soret at 416–419 nm) but which has increased yield (Table 1) and reduced amounts of P420. The addition of the tight-binding inhibitor abiraterone during expression of CYP17A1 with exogenous FePPIX (H) yields protein in which the abiraterone pyridine is still bound to the heme iron after purification (Soret at 421 nm), further increases yield (Table 1), and prevents P420 formation.
Unsurprisingly, E. coli cells grew very slowly under these conditions. Iron limitation was a significant factor in bacterial growth. As evaluated by the optical density at 600 nm, the growth rate was more than 3 times slower in terms of both the lag and initial exponential phases compared with the growth of E. coli containing the same constructs in iron-supplemented minimal medium. Otherwise, expression proceeded normally. Subsequent purification methods did not require optimization, proceeding with typical detergent extraction of E. coli membranes followed by metal-affinity and cation-exchange chromatography. During purification of proteins generated with exogenous FePPIX, the yields of the various steps were commensurate with parallel purifications of these proteins generated with endogenous FePPIX biosynthesis, without notable differences. For example, although expression levels differ for the various P450 proteins, a typical yield after ion exchange was 30–50% of the protein isolated from the metal-affinity chromatographic step. Following purification, the overall yields of holoprotein for the different membrane P450 enzymes generated in iron-depleted minimal medium with exogenous FePPIX (51, 37, and 6.8 nmol of P450 per liter of culture for CYP3A4, CYP21A2, and CYP17A1, respectively; Table 1) are 7–21% of the typical yields for the respective proteins expressed in Terrific Broth supplemented by δ-aminolevulinic acid (240, 350, and 100 nmol of P450 per liter of culture for CYP3A4, CYP21A1, and CYP17A1, respectively; Table 1).
Table 1.
Quantity and spectral properties of human P450 proteins expressed under different conditions
| Soret λmax | Total P450 from Soret peak | ASoreta/A280 | P450b | |
|---|---|---|---|---|
| nm | nmol/liter culture | % | ||
| CYP3A4 | ||||
| CYP3A4/chaperone expressed in Terrific Broth in the presence of: | ||||
| δ-Aminolevulinic acid/no ligand | 416 | 240 | 1.54 | 82 |
| CYP3A4/pHug/chaperone expressed in iron-depleted medium in the presence of: | ||||
| FePPIX/no ligand | 416 | 51 | 1.57 | 86 |
| CYP21A2 | ||||
| CYP21A1/chaperone expressed in Terrific Broth in the presence of: | ||||
| δ-Aminolevulinic acid/no ligand | 419 | 350 | 1.2 | 54 |
| CYP21A1/pHug/chaperone expressed in iron-depleted medium in the presence of: | ||||
| No PPIX/no ligand | 425 | 0.83 | 0.17 | 12 |
| FePPIX/no ligand | 418 | 37 | 1.07 | 81 |
| FePPIX/progesterone (substrate) | 419 | 83 | 1.18 | 89 |
| CYP17A1 | ||||
| CYP17A1/chaperone expressed in Terrific Broth in the presence of: | ||||
| δ-Aminolevulinic acid/no ligand | 417 | 100 | 0.9 | 74 |
| CYP17A1/pHug/chaperone expressed in iron-depleted medium in the presence of: | ||||
| No PPIX/no ligand | Tracec | Trace | Trace | Trace |
| FePPIX/no ligand | 416 | 6.8 | 1.25 | 39 |
| FePPIX/progesterone (substrate) | 416 | 23 | 1.28 | 75 |
| FePPIX/abiraterone (inhibitor) | 421 | 96 | 1.22 | 100 |
| CoPPIX/abiraterone (inhibitor) | 428 | 117 | 1.54 | No peak |
| GaPPIX/abiraterone (inhibitor)d | 421, 438 | 300 | 1.7 | No peak |
| InPPIX/abiraterone (inhibitor)d | 431 | 51 | 1.21 | 90 |
| CuPPIX/abiraterone (inhibitor)d | 422 | 40 | 1.04 | 100 |
| ZnPPIX/abiraterone (inhibitor)d | 426 | 108 | 1.4 | 100 |
| AgPPIX/abiraterone (inhibitor)d | 426 | 18 | 1.05 | 100 |
a The absorbance of the Soret peak (heme) to absorbance at 280 nm (total protein) is a relative measure of purity for an individual P450 enzyme.
b The percentage of P450 from the total of P450 and P420 species in the reduced, carbon monoxide difference spectrum reflects correct heme incorporation.
c Trace, trace amounts of protein too little to characterize.
d The CYP17A1 enzyme used was the A105L mutant, which has higher yields to help generate enough sample for NMR experiments.
To evaluate background levels of iron and possible endogenous FePPIX biosynthesis in iron-limited medium, the CYP17A1 and CYP21A2 were expressed under these iron-depleted conditions without FePPIX supplementation. This also resulted in very slow growth. The resulting protein was ≤2% of the yield compared with when exogenous FePPIX was supplied, was of very low purity, and had an unusual Soret peak for CYP21A2 (Table 1 and Fig. S1). This suggests that although trace amounts of iron may sometimes be present, the majority of the FePPIX-containing P450 protein was derived from supplemented FePPIX successfully imported from the medium into E. coli cells by the proteins of the heme utilization genes. Of course, some trace iron and/or FePPIX would be necessary to support the generation of essential bacterial respiratory proteins, catalases, peroxidases, and other heme-requiring proteins, so it is probably not beneficial to further limit iron sources. In addition, the current intended application of metal-substituted P450 enzymes is solution NMR, which would not be negatively affected by small amounts of P450 protein with FePPIX incorporation.
Characterization of P450 proteins generated from exogenous FePPIX uptake versus endogenous FePPIX biosynthesis
The quality of P450 proteins generated from exogenous FePPIX addition to the medium was compared with that of proteins generated by endogenous FePPIX synthesis. The latter is supported by the addition of cell-permeable, δ-aminolevulinic acid heme precursor to Terrific Broth medium just before P450 induction (Fig. 1A). Observations of metal incorporation and environment are facilitated by the electronic properties of PPIX, resulting in the Soret absorbance peak. The Soret peak shifts with changes in ligand binding to the metal, the identity of the metal, and the metal oxidation state. Endogenous FePPIX biosynthesis and expression of each of these P450 proteins typically result in a Soret peak at 417–418 nm (Table 1; shown for CYP3A4 only in Fig. 1B, left spectrum), consistent with water coordinated to the heme iron opposite the proximal Cys thiolate. The Soret peak for all three P450 enzymes expressed with exogenous FePPIX was located at 416–418 nm (Fig. 1 (C–E), left spectra). The ratio of the Soret absorbance maximum to absorbance at 280 nm is a relative measure of heme incorporation and purity for a specific P450 enzyme. These values for CYP3A4, CYP17A1, and CYP21A2 expressed with exogenous FePPIX (A416–418/A280 = 1.07–1.57) were similar to the purity for the same proteins generated using endogenous FePPIX synthesis (A416–418/A280 = 0.9–1.54; Table 1).
A second characteristic of a thiolate-coordinated heme iron is a peak at ∼450 nm for the reduced-carbon monoxide difference spectrum. This indicates that the iron of FePPIX is appropriately bonded to its proximal Cys thiolate. Observation of a peak at 420 nm after reduction and binding of carbon monoxide is instead consistent with incorrect interaction of the heme iron with the proximal polypeptide, proposed to be either the protonated thiol (12–14) or substitution by a nearby histidine (15, 16). This “P420” peak is a sensitive feature easily induced by many different environmental conditions and can often be readily observed to some extent for different P450 enzymes even under optimal expression conditions. When produced with endogenous FePPIX biosynthesis, typical percentages of P450 species for CYP3A4, CYP21A2, and CYP17A1 in the absence of ligand are 82% (Fig. 1B, right spectrum), 54%, and 74%, respectively. By comparison, the same enzymes generated with exogenous FePPIX supplementation demonstrated 86, 81, and 39% P450, respectively (Fig. 1 (C–E) and Table 1).
Because the presence of a ligand often stabilizes membrane P450 enzymes against P420 formation, the P450 enzymes with suboptimal amounts (>15%) of P420 (CYP21A2 and CYP17A1) were generated with the exogenous FePPIX system described above, but with the addition of their substrate progesterone during expression but not purification. The result was an increase in both protein quantity and quality. The Soret peak of the purified proteins indicated that the progesterone substrate had washed out during purification because the wavelength was consistent with water coordination to the heme (416 and 419 nm; Fig. 1 (F and G), left spectra). However, the presence of the ligand progesterone during expression resulted in a 2–3-fold increase in purified protein yield (Table 1) with similar purity as measured by the ratio of the Soret peak to the A280 for both proteins (Table 1). Even more importantly, the integrity of heme incorporation as measured by the reduced-carbon monoxide difference spectrum was increased from 81 to 89% for CYP21A1 and from 39 to 75% for CYP17A1 (Table 1 and Fig. 1 (F and G), right spectra).
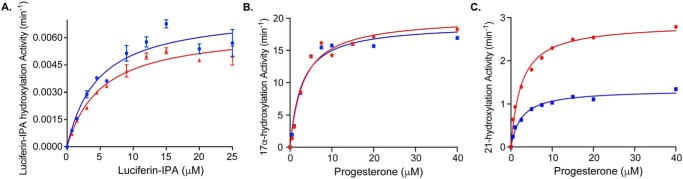
Finally, to further validate the general methodology, the catalytic activities of P450 enzymes expressed with exogenous FePPIX supplementation were compared with the same protein expressed with endogenous FePPIX biosynthesis. In each case, no ligand was included during expression or purification. CYP3A4 oxidation of luciferin-IPA proceeded similarly regardless of heme source (Fig. 2A and Table 2). Progesterone is a natural substrate for both CYP17A1 and CYP21A2, which perform hydroxylation at C17 and C21, respectively. Similar to CYP3A4, progesterone 21-hydroxylation by CYP21A2 generated with exogenous FePPIX yielded both Km and kcat values similar to that of CYP21A2 expressed with endogenous FePPIX biosynthesis (Fig. 2B and Table 2), and adding progesterone during expression did not significantly change the kinetic constants (Table 2). However, progesterone 17α-hydroxylation by CYP17A1 produced with exogenous FePPIX yielded a similar Km but lower kcat compared with CYP17A1 expressed with endogenous FePPIX synthesis (Fig. 2C and Table 2). In this case, adding progesterone during CYP17A1 expression with exogenous FePPIX supplementation resulted in a kcat more similar to CYP17A1 generated with endogenous FePPIX biosynthesis (Table 2).
Figure 2.
Enzyme kinetics of three human membrane P450 proteins produced with endogenous FePPIX synthesis (red) versus exogenous FePPIX supplementation (blue), both in the absence of ligand. CYP3A4 (A) and CYP21A2 (B) produced with exogenous FePPIX have Michaelis–Menten curves for their respective substrates similar to these enzymes produced using traditional endogenous bacterial FePPIX synthesis. C, CYP17A1 enzymes produced using the two methods have similar Km values, but the enzyme produced with exogenous FePPIX has an ∼2-fold lower kcat.
Table 2.
Comparison of kinetic constants for CYP3A4, CYP21A2, and CYP17A1 expressed with either bacterial endogenous FePPIX biosynthesis or the addition of exogenous FePPIX and heme uptake genes
CYP17A1 and CYP21A2 were evaluated using a progesterone C17- and C21-hydroxylation assay, respectively, whereas CYP3A4 was evaluated using a luciferin–isopropyl alcohol substrate. Values are reported for protein expressed in the absence or presence of the stabilizing substrate progesterone for the two steroidogenic enzymes.
| kcata | Km | kcat /Km | |
|---|---|---|---|
| min−1 | mm | min−1 mm−1 | |
| CYP3A4 expressed with: | |||
| Endogenous FePPIX | 0.0064 ± 0.0002 | 5.2 ± 0.6 | 0.0012 |
| Exogenous FePPIX | 0.0074 ± 0.0004# | 4.7 ± 0.7 | 0.0015 |
| CYP21A2 expressed with: | |||
| Endogenous FePPIX | 20 ± 1 | 3.2 ± 0.7 | 6.2 |
| Exogenous FePPIX | 19 ± 1 | 2.8 ± 0.6 | 6.9 |
| Endogenous FePPIX/progesterone | 20 ± 1 | 2.8 ± 0.7 | 7.2 |
| Exogenous FePPIX/progesterone | 20 ± 1 | 2.9 ± 0.7 | 6.7 |
| CYP17A1 expressed with: | |||
| Endogenous FePPIX | 2.9 ± 0.7 | 2.5 ± 0.3 | 1.1 |
| Exogenous FePPIX | 1.3 ± 0.4## | 2.5 ± 0.3 | 0.50 |
| Endogenous FePPIX/progesterone | 3.1 ± 0.1 | 2.2 ± 0.4 | 1.4 |
| Exogenous FePPIX/progesterone | 1.8 ± 0.1## | 2.9 ± 0.6 | 0.63 |
a #, p < 0.05; ##, p < 0.01 compared with the Km and kcat of the same enzyme expressed with endogenous FePPIX biosynthesis.
These results are consistent with different degrees of intrinsic stability for the different P450 enzymes generated either with endogenous FePPIX biosynthesis or with exogenousFePPIX supplementation. CYP3A4 did not require the presence of a ligand to yield a P450 spectrum similar to that with exogenous FePPIX biosynthesis. The addition of the substrate progesterone during expression improved the percentage of P450 for CYP21A2 to almost 90%. In contrast, although the addition of the substrate progesterone improved the percentage of P450 from 39 to 70% for CYP17A1, this was not sufficient to bring it to >85%, which was the target for structural studies. The progesterone substrate added to CYP17A1 and CYP21A2 increased the yield during expression with exogenous FePPIX, but was subsequently washed out during purification.
Optimization of ligand for CYP17A1 expression with exogenous FePPIX uptake
To further facilitate CYP17A1 stabilization, the inhibitor and prostate cancer drug abiraterone was used instead of the substrate progesterone because abiraterone binds more tightly (17). We previously established that abiraterone binds CYP17A1 with nanomolar affinity via coordination of its nitrogen heteroatom directly to the heme iron, as indicated by a red shift of the Soret peak and verified by the X-ray structure (17). The presence of abiraterone during expression with exogenous FePPIX, but not during purification, yielded purified CYP17A1, which retained the abiraterone inhibitor, as indicated by the Soret maximum at 421 nm (Fig. 1H, left spectrum). In addition, compared with expression with progesterone, expression with abiraterone resulted in a ∼4-fold higher yield after purification with similar purity and 100% P450 (Fig. 1H, right).
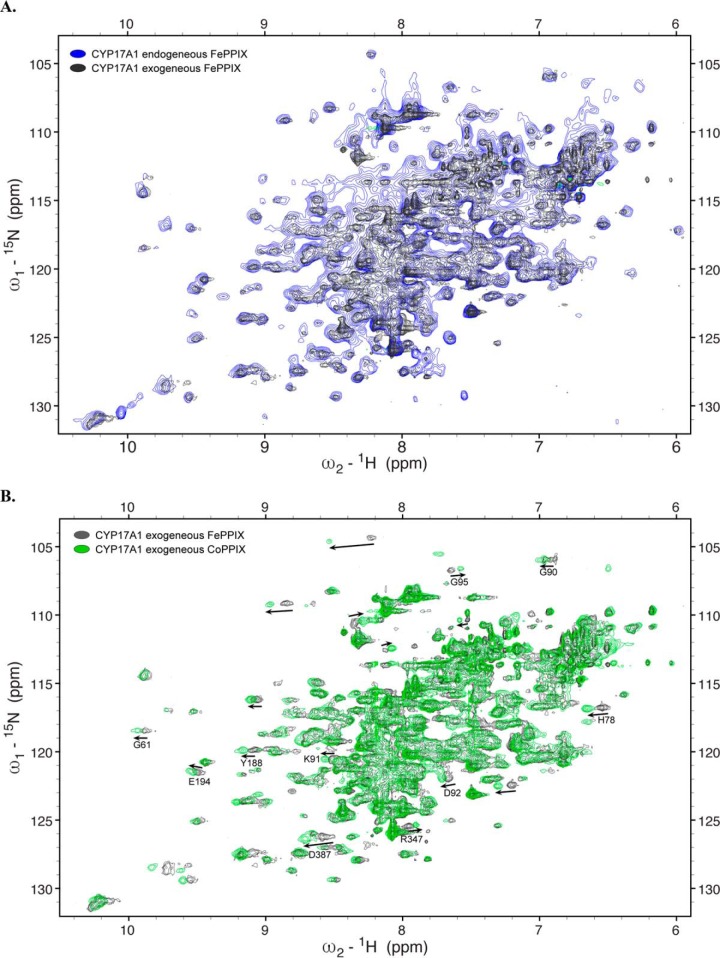
CYP17A1 is potently inhibited by the abiraterone inhibitor, which does not readily dissociate, so enzymatic activity could not be determined. However, solution NMR was used to identify any structural changes. Unlike CYP21A2 and CYP3A4, which are oligomers and too large for traditional solution NMR, CYP17A1 is monomeric, and its 55-kDa molecular mass is just small enough to yield a reasonable 2D 1H-15N spectrum. NMR is a highly sensitive, high-resolution approach that can readily be used to determine the similarities and differences between two versions of a protein, even without knowing the identity of the individual assignments. Thus, NMR permits evaluation of the entire CYP17A1 structure under different conditions, even if the spectrum is crowded by >500 amino acid resonances. For this purpose, CYP17A1 was generated in the presence of the abiraterone inhibitor with either exogenous PPIX supplementation or endogenous PPIX biosynthesis while substituting 15N-ammonium chloride for the unlabeled ammonium chloride in the minimal medium. The 2D 1H-15N TROSY-HSQC spectra revealed that 15N-CYP17A1 generated with exogenous FePPIX supplementation and endogenous FePPIX biosynthesis have a very similar overall distribution of resonances (Fig. 3A). This high similarity indicates very similar folds in both proteins with no major structural differences.
Figure 3.
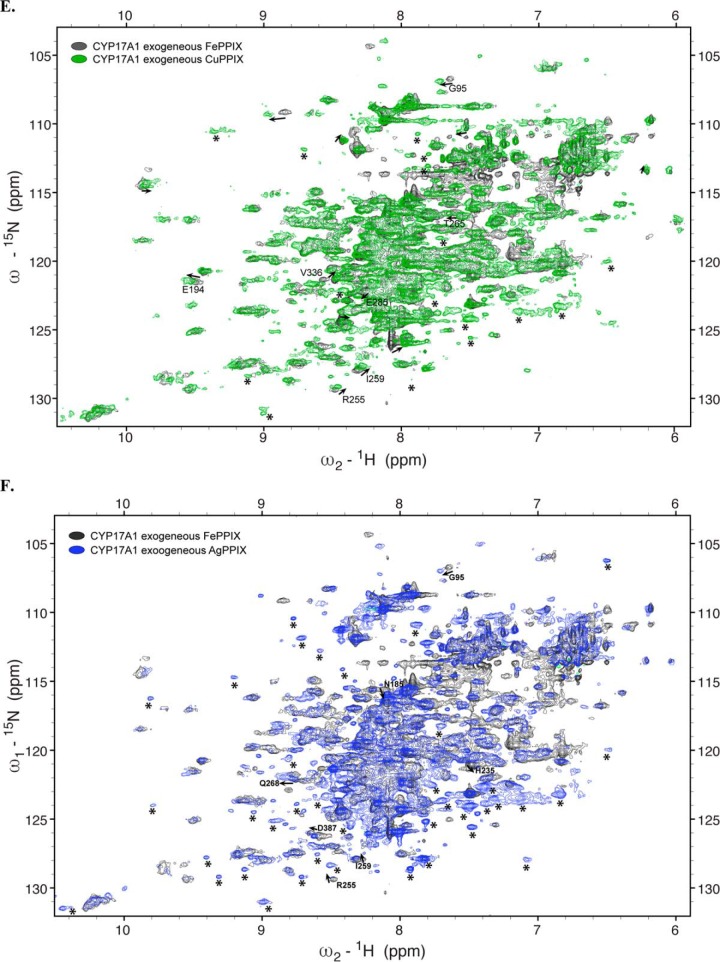
Overlay of 1H-15N TROSY-HSQC spectra of CYP17A1 expressed under various conditions. A–F, expression with exogenous FePPIX supplementation and pHUG genes (gray) versus δ-aminolevulinic supplementation supporting endogenous FePPIX biosynthesis (blue) (A), CoPPIX (green) (B), GaPPIX (red) (C), ZnPPIX (magenta) (D), CuPPIX (green) (E), or AgPPIX (blue) (F). Arrows, shifted resonances. Asterisks, peaks that can be detected in metal-substituted samples but not in the native FePPIX samples.
Overall, we were able to obtain acceptable yields (50–100 nmol) of purified membrane P450 enzymes at >85% P450 for CYP3A4, CYP21A2, and CYP17A1 by employing exogenous FePPIX supplementation in concert with the heme uptake genes. Thus, the general methodology for getting exogenous PPIX into cells and incorporated into these quasi-stable membrane proteins was validated.
Generation of CYP17A1 protein with conservative substitution of exogenous cobalt-substituted heme
The next challenge was to apply this approach to determine how well membrane P450 enzymes would incorporate PPIX containing nonnative metals. CYP17A1 with the inhibitor abiraterone was used for all subsequent studies because this was the most challenging protein to produce with exogenousFePPIX supplementation and because solution NMR was a comprehensive viable method for readily determining any changes in the overall structure that might result from metal substitution. Cobalt-containing PPIX (CoPPIX) was the first metal substitution employed because it demonstrated retention of the overall structure upon incorporation of CoPPIX into myoglobin (18), the ability to coordinate to a Cys side chain as observed for FePPIX in native P450 enzymes (19), and similarity to iron in being able to bind H2O (20), O2 (21), and nitrogen heterocycles (20) like that of the CYP17A1 abiraterone inhibitor.
The resulting purified CYP17A1/CoPPIX protein was obtained with a slightly higher yield (117 nmol/liter culture) than CYP17A1/FePPIX under the same conditions (96 nmol/liter culture) with similar purity (Table 1). CYP17A1/CoPPIX displayed a Soret peak at 429 nm, shifted from the 421-nm Soret peak of the same enzyme produced the same way with FePPIX (Fig. S2, A and B). This is consistent with metal substitution. A similar red shift was observed for the soluble bacterial P450 BM3 upon substitution of FePPIX with CoPPIX (9). In contrast to CYP17A1/FePPIX, CYP17A1/CoPPIX did not have a peak in the reduced-carbon monoxide difference spectrum (Fig. S2, A and B). This could be because carbon monoxide does not bind to CoPPIX or because the abiraterone bound so tightly to the CoPPIX that no displacement of the abiraterone pyridine occurred to permit CO binding. Others have reported that pyridine has a much lower dissociation and larger equilibrium constant for CoPPIX than FePPIX, at least in the context of the myoglobin polypeptide (20).
CoPPIX incorporation was evaluated using inductively coupled plasma MS (ICP-MS). As expected, CYP17A1 expressed with either endogenous FePPIX synthesis or exogenous FePPIX had very high iron content and less than 5% cobalt. However, CYP17A1 expressed with exogenous CoPPIX was 78% cobalt. Although no iron or FePPIX was added at any stage, iron is a nearly universal contaminant of water, glassware, and many reagents. Thus, the iron that was detected may be either free iron contaminant in the final sample or could have been traces in the growth medium that were incorporated into PPIX endogenously. Free iron in growth medium could be further reduced by filtering the minimal medium and buffers through Chelex 100, but as stated above, more severe iron depletion from growth medium is likely to be toxic to E. coli cell growth. However, by simply omitting iron from the minimal medium and growing with low levels of the iron chelator, the E. coli can grow slowly, likely employing most trace iron in their own biochemistry, before induction of P450, which largely utilizes the readily available metal protoporphyrin provided in the medium.
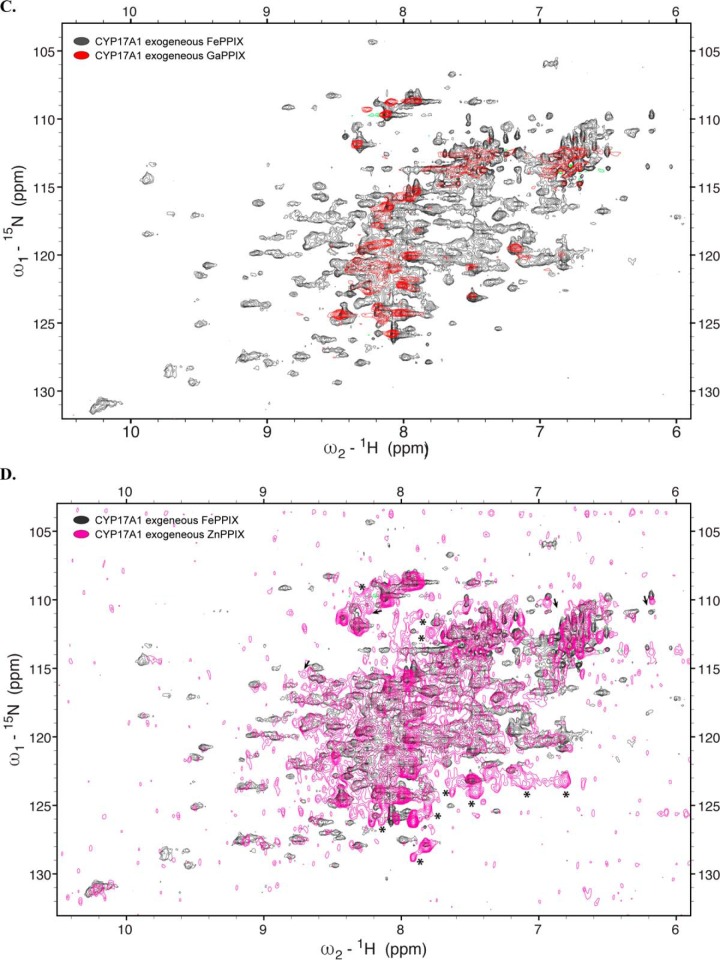
To evaluate any structural differences upon incorporation of exogenously supplemented CoPPIX, solution NMR was again employed. The 1H-15N TROSY-HSQC spectrum of 15N-CYP17A1/CoPPIX/abiraterone revealed a structure with a very similar overall fold and distribution of resonance peaks to CYP17A1/FePPIX/abiraterone (Fig. 3B). A subset of peaks displayed mostly minor and mostly downfield chemical shifts. This may indicate some small structural changes due to the metal substitution, but the overall success of this experiment led to exploration of additional metals.
Generation of CYP17A1 with diamagnetic metals
Development of this method was driven by the desire to replace FePPIX to reduce line broadening of NMR signals for residues in the proximity of paramagnetic metals like iron (and cobalt). Substitution with diamagnetic metals should reduce line broadening, resulting in the ability to detect resonances corresponding to amino acids in the proximity of the metal, in this case those lining the active site, and thus allow their assignment. Thus, the next step in method development was to try different diamagnetic metal protoporphyrins which might be successfully incorporated, result in negligible structural changes compared with protein containing FePPIX, and determine whether this would reveal new resonance peaks in the NMR spectra.
In moving away from iron and cobalt as the central metal, it was likely that at least a portion of the stabilization caused by the ligand nitrogen heterocycle interaction with the metal would be lost. For example, as stated above, abiraterone binds to CYP17A1 with FePPIX so tightly that when it is added during expression, one can purify the enzyme through multiple columns without further abiraterone addition and end up with abiraterone-saturated protein, as validated by the wavelength of the Soret peak (Fig. 1H). The Kd and IC50 values for abiraterone are in the nanomolar range (17, 22), but an abiraterone analog without the nitrogen that coordinates iron has an IC50 increased by over 3 orders of magnitude (22). Thus, two changes were made to compensate for the likely reduction in loss of abiraterone affinity to the various diamagnetic metals. First, abiraterone was included in all buffers throughout purification. Second, the A105L mutation of CYP17A1 was used. This mutation has been shown to increase the affinity for multiple steroidal compounds (23), likely by favoring a closed state for the active site, without changing the structure of the CYP17A1/abiraterone complex as observed by X-ray crystallography (23). Thus, from this point onward, CYP17A1 with an A105L substitution was expressed and purified in the presence of abiraterone with the commercially available exogenous metal protoporphyrins containing gallium (GaPPIX), indium (InPPIX), copper (CuPPIX), zinc (ZnPPIX), or silver (AgPPIX) added during P450 expression. In each case, ICP-MS confirmed in the respective purified proteins that the desired metal was increased by hundreds to thousands of times the normal background levels.
GaPPIX incorporation resulted in poor yield and purity. After the two purification columns, the yield was 30 nmol/liter of culture, which is similar to the use of no ligand, and the ratio of the Soret absorbance to A280 was significantly decreased, consistent with very poor purity. The Soret band was at 421 nm with a significant shoulder at 438 nm. (Fig. S2C). This is significantly different from the absorbance spectrum for GaPPIX alone in buffer. Pyridine is known to coordinate the gallium center of PPIX (24), but the affinity is unknown. It is possible that these two peaks represent abiraterone-bound and abiraterone-free populations. The same Soret shift was observed for CYP3A4 expressed with exogenous GaPPIX in the presence of a nitrogenous inhibitor (not shown). No P450 peak was observed in the reduced-carbon monoxide difference spectrum. Evaluation of the CYP17A1/GaPPIX/abiraterone sample by NMR indicates only a few resonances, mostly in the center of the spectrum (Fig. 3C), which is consistent with unfolded or disordered protein and a significant amount of protein precipitation during the NMR experiment, so this metal was not examined further.
Incorporation of InPPIX yielded slightly more protein (51 nmol/liter of culture), which could be purified with purity similar to FePPIX samples. The Soret peak was at 431 nm, consistent with InPPIX incorporation (Fig. S2C, left). Additionally, the reduced-carbon monoxide difference spectrum revealed a small peak at 445 nm and no peak at 420 nm (Fig. S2C, right). Although the 1H-15N TROSY-HSQC NMR spectrum for the InPPIX sample (Fig. S3) revealed more resonances and more dispersion than the GaPPIX sample, again resonances were mostly limited to the middle of the spectrum, suggesting substantial structural changes that were undesirable. In addition, significant protein precipitation was also observed over the course of the NMR experiment, suggesting low protein stability. Overall, this suggests that InPPIX, like GaPPIX, may not be a good diamagnetic replacement for FePPIX in membrane P450 enzymes.
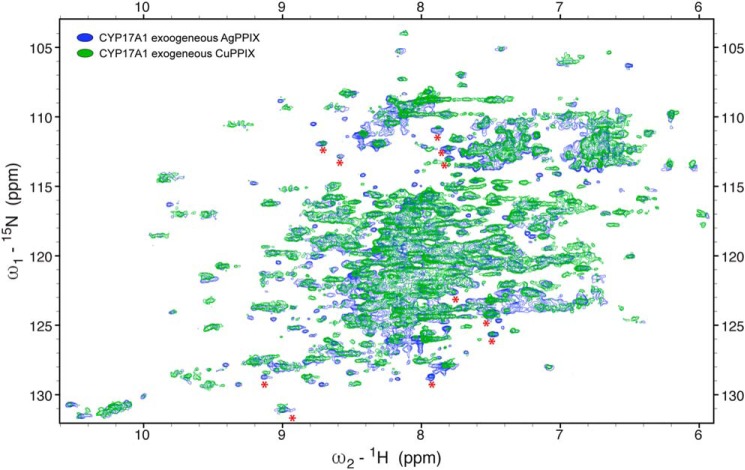
In contrast, ZnPPIX, CuPPIX, and AgPPIX were more successful in retaining the native structure of CYP17A1. The Soret peaks were 422 nm (CuPPIX) and 426 nm (ZnPPIX and AgPPIX) and, like InPPIX, resulted in reduced-carbon monoxide difference spectra with peaks consistently at 444–445 nm and no evidence of P420 (Fig. S2, E–G). Purified protein yields varied from 18 nmol/liter of culture (AgPPIX) to 40 nmol/liter of culture (CuPPIX) to 108 nmol/liter of culture (ZnPPIX) with purities similar to most other samples (Table 1). In contrast to the InPPIX and GaPPIX samples, 1H-15N TROSY-HSQC NMR spectra were consistent with well-folded protein with dispersed resonances and generally good overlap with the resonances of the FePPIX samples (Fig. 3, D–F), although all three showed some minor shifts for a few peaks. The ZnPPIX sample had a few new resonances (Fig. 3D), whereas the spectra for the CuPPIX and AgPPIX samples (Fig. 3, E and F) had many more resonances that had not been detectable with FePPIX. In addition, resonances for CYP17A1 with these latter two metals were sharper compared with the resonances with ZnPPIX, and this will facilitate the resolution of individual peaks, particularly in crowded parts of the spectra. Overall, this validates the utility of replacing the paramagnetic heme iron in order to be able to detect resonances near the heme active site. Many of these new resonances agree between the AgPPIX and ZnPPIX samples (Fig. 4). Although the newly detected resonances for CYP17A1 incorporating AgPPIX are stronger than when CuPPIX is incorporated, this serves as critical cross-validation of the data.
Figure 4.
Comparison of the two best 1H-15N TROSY-HSQC spectra with diamagnetic metals (CuPPIX in green and AgPPIX in blue) demonstrates that many of the resonance peaks that can now be detected are in agreement, although the signals from CYP17A1 with AgPPIX incorporated are stronger than the corresponding resonances when CuPPIX is incorporated.
Overall, the present results indicate that exogenous protoporphyrins can be taken up and correctly incorporated into even quasi-stable human membrane cytochrome P450 enzymes during recombinant expression. When FePPIX is exogenously supplemented, CYP3A4, CYP21A2, and CYP17A1 can be generated with normal spectroscopic features and often identical steady-state kinetic constants for their respective substrate oxidations. Using this methodology, other exogenous metal protoporphyrins could be readily incorporated into CYP17A1, often without significant perturbation in the overall protein structure as evaluated by 2D NMR. This was especially true if one took advantage of stabilizing factors, such as ligands and a mutation. The success with CYP17A1, the least stable of the three human P450 enzymes examined, suggests that the method is likely applicable to other human P450 enzymes, perhaps with additional stabilization. As a result of the incorporation of diamagnetic metals into CYP17A1, new NMR resonances were detected, which will assist in the assignment of amino acids otherwise broadened beyond detection by the nearby paramagnetic iron. In addition to the biophysical application in NMR discussed herein, other structural and mechanistic studies can profit from having control of the metal center at the heart of membrane P450 catalysis driving human drug metabolism and key endogenous and disease-related pathways such as steroidogenesis.
Materials and methods
Plasmid constructs
Recombinantly expressed human cytochrome P450 enzymes CYP17A1, CYP21A2, and CYP3A4 were used in this study. The gene for each P450 was constructed synthetically with codon optimization for expression in E. coli and modified to omit the single, N-terminal transmembrane helix and add a C-terminal histidine tag. Specifically, the 17A1Δ19H construct is as described (17). The CYP21A2dH construct omitted the N-terminal 2–19 residues, substituted the sequence coding for 20NWWKLRSLH28 with nucleotide coding for AKKTSSKGK, and added codons for a C-terminal four-histidine tag immediately before the stop codon (22). The CYP3A4Δ23H construct deleted N-terminal residues 3–23 (25) and similarly added a C-terminal four-histidine tag. All three genes were insertedin the pCWori+ vector, which confers carbenicillin resistance. The chloramphenicol-resistant pHUG21.1 plasmid containing the coding sequence for the heme utilization genes (hugA, tonB, exbBD, and hugBCD) from P. shigelloides under the control of the fur promoter was a gift from J. S. Olson (Rice University, Houston, TX). The pGro12 (26) plasmid containing the chaperone groES and groEL genes under the control of the araB promoter and including kanamycin resistance was a gift from Rita Bernhardt (Saarland University).
Protein expression and purification
E. coli DH5α cells were sequentially transformed with pHUG21.1, pGro12, and then either pCW17A1Δ19H, pCW21A2dH, or pCW3A4Δ23H plasmid. Cells transformed with all three plasmids were grown on lysogeny broth agar plates containing chloramphenicol (20 μg/ml), kanamycin (33 μg/ml), and carbenicillin (100 μg/ml) to select for the respective plasmids. Single colonies were first grown in 5 ml of lysogeny broth medium at 37 °C, 250 rpm in the presence of all three antibiotics until A600 = 0.6. Then 500 μl of this culture was inoculated into 10 ml of M9 minimal medium and grown for ∼16 h at 37 °C, 250 rpm. Iron trace metal (FeCl3) was omitted from the M9 minimal medium used from this point on. Of this 10-ml culture, 2.5 ml of culture was used to inoculate 50 ml of minimal medium, which was grown at 37 °C, 250 rpm overnight to acclimatize the cells to low-iron conditions. From this, 25 ml was inoculated in 1 liter of minimal medium and grown at 37 °C, 250 rpm until A600 reached 0.4. At this point, expression of heme utilization proteins and chaperone was induced by the addition of 10 mg of the iron chelator ethylenediamine-N,N′-bis((2-hydroxyphenyl)acetic acid) (EDDHA) and 500 mg of l-arabinose, respectively. The temperature was reduced to 27 °C, and cultures continued growing until A600 reached 0.8. At this point, P450 protein was induced with 0.8 mm isopropyl 1-thio-β-d-galactopyranoside and either 10 mm Fe(III)PPIX, Co(III)PPIX, Ga(III)PPIX, In(III)PPIX, Cu(II)PPIX, Zn(II)PPIX, or Ag(II)PPIX added into the 1-liter culture. Temperature and shaking were reduced to 25 °C and 200 rpm, respectively. The same amount of metal protoporphyrin was again added 24 h after induction. Stocks of metal protoporphyrin IX were prepared in 0.1 n NaOH. Cultures were harvested at 48 h, and cells were collected by centrifugation at 8000 × g for 10 min. Cells were resuspended in buffer (50 mm Tris-HCl, pH 7.4, 20% (v/v) glycerol, and 300 mm NaCl for CYP17A1; 100 mm potassium phosphate, pH 6.8, 20% glycerol, and 300 mm NaCl for CYP21A2; 100 mm potassium phosphate, pH 7.4, 20% glycerol, and 500 mm NaCl for CYP3A4) and stored at −80 °C until purification. CYP17A1 used in this study was expressed either without ligand or in the presence of its substrate progesterone (50 μm) or the inhibitor abiraterone (20 μm). CYP21A2 was expressed without adding ligand or in the presence of its substrate progesterone (50 μm). CYP3A4 was only expressed without adding a ligand. Ligands were added only during protein expression unless otherwise specified. For comparison, each P450 protein (CYP17A1, CYP21A2, and CYP3A4) was also expressed without PPIX addition but with the addition of heme precursor δ-aminolevulinic acid in Terrific Broth to support endogenous heme generation in the culture during expression. To evaluate background, CYP17A1 and CYP21A2 were also expressed without the addition of any PPIX or δ-aminolevulinic acid in iron-depleted minimal medium.
Human P450 enzymes were purified using metal-affinity and ion-exchange chromatography. CYP17A1 protein was purified as described (23), except that a French press was used for cell disruption. Final CYP17A1 protein was in 50 mm Tris, pH 7.4, 100 mm glycine, 500 mm NaCl, and 20% glycerol buffer. Purification of CYP21A2 was as described (22) with the final protein in 50 mm potassium phosphate, pH 6.8, 350 mm NaCl, and 20% (v/v) glycerol. CYP3A4 was purified similarly to CYP21A2 except that 1) 10–14 mm CHAPS was used as the detergent instead of Emulgen-913 and 2) potassium phosphate buffers were at pH 7.4 instead of pH 6.8. Purified CYP3A4 was in 50 mm potassium phosphate, pH 7.4, 250 mm NaCl, 20% (v/v) glycerol. The final quantity and purity of all P450 proteins were assessed by evaluating the UV-visible absorbance spectrum and SDS-PAGE. The final quality of purified proteins was evaluated by the reduced-carbon monoxide difference spectrum.
Functional assays
For assays, the P450 concentration was determined from the reduced-carbon monoxide difference spectrum either in the presence of progesterone (CYP21A2 and CYP17A1) or without ligand (CYP3A4).
CYP17A1 or CYP21A2 expressed with either endogenous heme production or supplementation of exogenous FePPIX was evaluated to determine its ability to hydroxylate progesterone using an end point assay. Progesterone hydroxylation was performed for CYP17A1 as described (23), and a similar method was followed using CYP21A2. Briefly, progesterone (0–40 μm) was co-incubated with CYP17A1 (25 pmol) and rat cytochrome P450 reductase (100 pmol) or CYP21A2 (10 pmol) and rat cytochrome P450 reductase (40 pmol). Reactions were started with the addition of 1 μm NADPH and allowed to proceed at 37 °C for 10 and 8 min for CYP17A1 and CYP21A2, respectively, before the addition of stop reagent (20% TCA). Precipitated protein was removed from each sample by centrifugation at 5000 rpm for 10 min, and supernatant was loaded onto an HPLC Luna C18 column (Phenomenex) with 40% acetonitrile and 0.2% acetic acid isocratic flow. The CYP17A1 17α-hydroxyprogesterone product and the CYP21A2 21-hydroxyprogesterone product were detected at 248 nm with retention times of ∼5.5 and ∼4.1 min, respectively.
CYP3A4 enzymatic activity was evaluated for protein expressed with endogenous heme synthesis or the exogenous addition of FePPIX using a luminescent end point assay. CYP3A4 (10 pmol) was mixed with 40 pmol of rat cytochrome P450 reductase and 10 pmol of human cytochrome b5 and a 0–25 μm concentration of the substrate luciferin-IPA (Promega). The reaction was initiated by the addition of 100 μm NADPH and proceeded for 30 min at 37 °C before it was stopped with esterase-containing luciferin detection reagent. Luminescence was recorded using a plate reader. d-Luciferin was used to generate a standard curve for converting luminescence to the amount of product formed by CYP3A4.
NMR spectroscopy
Uniformly 15N-labeled CYP17A1 was generated with either endogenous heme biosynthesis or the addition of an exogenous metal PPIX as described above except that during expression, 15N-labeled ammonium chloride was substituted for unlabeled ammonium chloride as the sole nitrogen source in the minimal medium. These proteins were expressed in the presence ofthe stabilizing inhibitor abiraterone (50 μm) and purified as described above. An additional size-exclusion chromatography step was employed for the CoPPIX and GaPPIX samples because the total amount of protein available was sufficient to do so. In these cases, separation was performed on a Superdex 200 column (GE Healthcare) in NMR buffer (50 mm potassium phosphate, pH 6.5, 50 mm NaCl). Proteins were concentrated to 250–300 μm, and 10% D2O was added before data collection.
NMR spectra (1H-15N TROSY-HSQC) were collected on either a Bruker Avance III HD 850 MHz equipped with a triple-resonance inverse TCI cryoprobe at the Ohio State University Campus Chemical Instrument Center or a Bruker Avance III HD 800 MHz with TXI cryoprobe with z-gradients at the University of Michigan Biomolecular NMR facility. Spectra were processed with Bruker Topspin version 4.0 and analyzed using NMRFAM-SPARKY (27).
Metal content analysis by ICP-MS
Incorporation of different metal protoporphyrins was verified using ICP-MS. All proteins (20 μm in 0.5 ml) were initially exchanged into buffer A (50 mm Tris, pH 7.4, 500 mm NaCl, 20% glycerol, 1 mm EDTA, 1 mm EDDHA) to chelate any free iron contaminant present in the sample. Proteins were then exchanged into buffer B (50 mm Tris, pH 7.4, 500 mm NaCl, 20% glycerol) to remove the EDDHA and EDTA. Both buffer A and B were passed through Chelex 100 resins (Bio-Rad) three times before use to avoid introducing transition metal ions to the protein samples. Finally, each protein in buffer B was subjected to nitric acid digestion before analysis by ICP-MS at the Elemental Analysis Core Laboratory at Oregon Health and Science University.
Author contributions
R. Y. and E. E. S. data curation; R. Y. and E. E. S. formal analysis; R. Y. validation; R. Y. investigation; R. Y. and E. E. S. visualization; R. Y. and E. E. S. methodology; R. Y. and E. E. S. writing-original draft; R. Y. and E. E. S. writing-review and editing; E. E. S. conceptualization; E. E. S. supervision; E. E. S. funding acquisition; E. E. S. project administration.
Supplementary Material
Acknowledgments
The pHUG plasmid for the heme uptake system was a generous gift from the laboratory of John Olson (Rice University, Houston, TX). The pGro12 plasmid containing the chaperone groES and groEL genes was a gift from Rita Bernhardt (Saarland University, Saarbrücken, Germany). ICP-MS measurements were taken in the Oregon Health and Science University Elemental Analysis Core with partial support from National Institutes of Health Core Grant S10RR025512. NMR spectra were collected either at the Ohio State University Campus Chemical Instrument Center or the University of Michigan BioNMR Core Laboratory.
This work was supported by National Institutes of Health Grant R37 GM076343 (to E. E. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
- PPIX
- protoporphyrin IX
- P450
- cytochrome P450
- P420
- P450 enzyme with maximal absorbance at 420 nm in the reduced-carbon monoxide difference spectrum
- 2D
- two-dimensional
- ICP-MS
- inductively coupled plasma MS
- EDDHA
- ethylenediamine-N,N′-bis((2-hydroxyphenyl)acetic acid).
References
- 1. Key H. M., Dydio P., Clark D. S., and Hartwig J. F. (2016) Abiological catalysis by artificial haem proteins containing noble metals in place of iron. Nature 534, 534–537 10.1038/nature17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gelb M. H., Toscano W. A. Jr, and Sligar S. G. (1982) Chemical mechanisms for cytochrome P-450 oxidation: spectral and catalytic properties of a manganese-substituted protein. Proc. Natl. Acad. Sci. U.S.A. 79, 5758–5762 10.1073/pnas.79.19.5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Furukawa Y., Ishimori K., and Morishima I. (2000) Electron transfer reactions in Zn-substituted cytochrome P450cam. Biochemistry 39, 10996–11004 10.1021/bi000874y [DOI] [PubMed] [Google Scholar]
- 4. Stone K. L., and Ahmed S. M. (2016) Advances in engineered hemoproteins that promote biocatalysis. Inorganics 4, 12 10.3390/inorganics4020012 [DOI] [Google Scholar]
- 5. Kazanis S., Pochapsky T. C., Barnhart T. M., Penner-Hahn J. E., Mirza U. A., and Chait B. T. (1995) Conversion of a Fe2S2 ferredoxin into a Ga3+ rubredoxin. J. Am. Chem. Soc. 117, 6625–6626 10.1021/ja00129a040 [DOI] [Google Scholar]
- 6. Teale F. W. (1959) Cleavage of the haem-protein link by acid methylethylketone. Biochim. Biophys. Acta 35, 543 10.1016/0006-3002(59)90407-X [DOI] [PubMed] [Google Scholar]
- 7. Wagner G. C., Gunsalus I. C., Wang M. Y., and Hoffman B. M. (1981) Cobalt-substituted cytochrome P-450cam. J. Biol. Chem. 256, 6266–6273 [PubMed] [Google Scholar]
- 8. Wagner G. C., Perez M., Toscano W. A. Jr., and Gunsalus I. C. (1981) Apoprotein formation and heme reconstitution of cytochrome P-450cam. J. Biol. Chem. 256, 6262–6265 [PubMed] [Google Scholar]
- 9. Lelyveld V. S., Brustad E., Arnold F. H., and Jasanoff A. (2011) Metal-substituted protein MRI contrast agents engineered for enhanced relaxivity and ligand sensitivity. J. Am. Chem. Soc. 133, 649–651 10.1021/ja107936d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Straub W. E., Nishida C. R., and de Montellano P. R. (2013) Expression in Escherichia coli of a cytochrome P450 enzyme with a cobalt protoporphyrin IX prosthetic group. Methods Mol. Biol. 987, 107–113 10.1007/978-1-62703-321-3_9 [DOI] [PubMed] [Google Scholar]
- 11. Graves P. E., Henderson D. P., Horstman M. J., Solomon B. J., and Olson J. S. (2008) Enhancing stability and expression of recombinant human hemoglobin in E. coli: progress in the development of a recombinant HBOC source. Biochim. Biophys. Acta 1784, 1471–1479 10.1016/j.bbapap.2008.04.012 [DOI] [PubMed] [Google Scholar]
- 12. Martinis S. A., Blanke S. R., Hager L. P., Sligar S. G., Hoa G. H., Rux J. J., and Dawson J. H. (1996) Probing the heme iron coordination structure of pressure-induced cytochrome P420cam. Biochemistry 35, 14530–14536 10.1021/bi961511u [DOI] [PubMed] [Google Scholar]
- 13. Perera R., Sono M., Sigman J. A., Pfister T. D., Lu Y., and Dawson J. H. (2003) Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. U.S.A. 100, 3641–3646 10.1073/pnas.0737142100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sabat J., Stuehr D. J., Yeh S. R., and Rousseau D. L. (2009) Characterization of the proximal ligand in the P420 form of inducible nitric oxide synthase. J. Am. Chem. Soc. 131, 12186–12192 10.1021/ja901016a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Y., Zeng W., Benabbas A., Ye X., Denisov I., Sligar S. G., Du J., Dawson J. H., and Champion P. M. (2013) Investigations of heme ligation and ligand switching in cytochromes P450 and P420. Biochemistry 52, 5941–5951 10.1021/bi400541v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wells A. V., Li P., Champion P. M., Martinis S. A., and Sligar S. G. (1992) Resonance Raman investigations of Escherichia coli-expressed Pseudomonas putida cytochrome P450 and P420. Biochemistry 31, 4384–4393 10.1021/bi00133a002 [DOI] [PubMed] [Google Scholar]
- 17. DeVore N. M., and Scott E. E. (2012) Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature 482, 116–119 10.1038/nature10743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brucker E. A., Olson J. S., Phillips G. N. Jr., Dou Y., and Ikeda-Saito M. (1996) High resolution crystal structures of the deoxy, oxy, and aquomet forms of cobalt myoglobin. J. Biol. Chem. 271, 25419–25422 10.1074/jbc.271.41.25419 [DOI] [PubMed] [Google Scholar]
- 19. Matta-Camacho E., Banerjee S., Hughes T. S., Solt L. A., Wang Y., Burris T. P., and Kojetin D. J. (2014) Structure of REV-ERBβ ligand-binding domain bound to a porphyrin antagonist. J. Biol. Chem. 289, 20054–20066 10.1074/jbc.M113.545111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neya S., Suzuki M., Hoshino T., and Kawaguchi A. T. (2013) Relaxation analysis of ligand binding to the myoglobin reconstituted with cobaltic heme. Inorg. Chem. 52, 7387–7393 10.1021/ic400078w [DOI] [PubMed] [Google Scholar]
- 21. Hoffman B. M., and Petering D. H. (1970) Coboglobins: oxygen-carrying cobalt-reconstituted hemoglobin and myoglobin. Proc. Natl. Acad. Sci. U.S.A. 67, 637–643 10.1073/pnas.67.2.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fehl C., Vogt C. D., Yadav R., Li K., Scott E. E., and Aubé J. (2018) Structure-based design of inhibitors with improved selectivity for steroidogenic cytochrome P450 17A1 over cytochrome P450 21A2. J. Med. Chem. 61, 4946–4960 10.1021/acs.jmedchem.8b00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petrunak E. M., DeVore N. M., Porubsky P. R., and Scott E. E. (2014) Structures of human steroidogenic cytochrome P450 17A1 with substrates. J. Biol. Chem. 289, 32952–32964 10.1074/jbc.M114.610998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pinter T. B., Dodd E. L., Bohle D. S., and Stillman M. J. (2012) Spectroscopic and theoretical studies of Ga(III)protoporphyrin-IX and its reactions with myoglobin. Inorg. Chem. 51, 3743–3753 10.1021/ic202731g [DOI] [PubMed] [Google Scholar]
- 25. Yano J. K., Wester M. R., Schoch G. A., Griffin K. J., Stout C. D., and Johnson E. F. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-Å resolution. J. Biol. Chem. 279, 38091–38094 10.1074/jbc.C400293200 [DOI] [PubMed] [Google Scholar]
- 26. Nishihara K., Kanemori M., Kitagawa M., Yanagi H., and Yura T. (1998) Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 64, 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee W., Tonelli M., and Markley J. L. (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.