Abstract
DNA-binding proteins from starved cells (Dps, EC: 1.16.3.1) have a variety of different biochemical activities such as DNA-binding, iron sequestration, and H2O2 detoxification. Most bacteria commonly feature one or two Dps enzymes, whereas the cyanobacterium Nostoc punctiforme displays an unusually high number of five Dps proteins (NpDps1–5). Our previous studies have indicated physiological differences, as well as cell-specific expression, among these five proteins. Three of the five NpDps proteins, NpDps1, -2, and -3, were classified as canonical Dps proteins. To further investigate their properties and possible importance for physiological function, here we characterized and compared them in vitro. Nondenaturing PAGE, gel filtration, and dynamic light-scattering experiments disclosed that the three NpDps proteins exist as multimeric protein species in the bacterial cell. We also demonstrate Dps-mediated iron oxidation catalysis in the presence of H2O2. However, no iron oxidation with O2 as the electron acceptor was detected under our experimental conditions. In modeled structures of NpDps1, -2, and -3, protein channels were identified that could serve as the entrance for ferrous iron into the dodecameric structures. Furthermore, we could demonstrate pH-dependent DNA-binding properties for NpDps2 and -3. This study adds critical insights into the functions and stabilities of the three canonical Dps proteins from N. punctiforme and suggests that each of the Dps proteins within this bacterium has a specific biochemical property and function.
Keywords: ferritin, DNA binding protein, iron, oxidative stress, reactive oxygen species (ROS), multifunctional protein, bacterial metabolism, cyanobacteria, metal homeostasis, DNA-binding proteins from starved cells
Introduction
Iron, the fourth most abundant element in the Earth's crust, plays an essential role in biological processes. A great variety of co-factors and prosthetic moieties that contain iron as a major constituent, e.g. iron-sulfur clusters and heme groups, can be found throughout all three domains of life (1). Iron is involved in, e.g. photosynthesis, respiration, N2-fixation, gene regulation, and DNA biosynthesis. The two physiologically relevant iron species, namely ferrous (Fe2+) and ferric iron (Fe3+), are interconvertible iron redox states and under an oxic environment, Fe2+ is usually oxidized to Fe3+, whose solubility is limited under physiological conditions (2). Free ferrous iron is also potentially dangerous because it can react with hydrogen peroxide, and the toxic reactive oxygen species (ROS)4 hydroxyl radical is formed by the Fenton reaction. Therefore, organisms have developed strategies to regulate the intracellular iron pool, for example, by storage inside “ferritin-like” proteins (2). This protein class consists of ferritins (Ftn), bacterioferritins (Bfr), and dodecameric mini-ferritins, known as DNA-binding proteins from starved cells (Dps). Aside from storing up to 500 iron ions (3), Dps proteins have been found to detoxify hydrogen peroxide (H2O2) and thereby protect cells in general, and DNA specifically, under different stressful conditions in which ROS are being formed (4–7).
Dps proteins can be found in prokaryotes that commonly accommodate one or two different dps genes in their genomes. However, some organisms, e.g. heterocyst-forming multicellular cyanobacteria, such as Nostoc punctiforme ATCC 29133 (8), are known to exhibit multiple Dps proteins. N. punctiforme has five genes annotated as Dps family proteins in its genome.
All the five dps-like genes are expressed in N. punctiforme, as shown by our earlier studies (8, 9), and are denoted NpDps1 (Npun_R3258), NpDps2 (Npun_F3730), NpDps3 (Npun_R5701), NpDps4 (Npun_R5799), and NpDps5 (Npun_F6212). Under certain conditions this cyanobacterium can form up to four different cell types. In the case of nitrogen depletion, some vegetative cells, which perform oxygenic photosynthesis (10), will differentiate into heterocysts that sustain a microoxic environment, in which oxygen-sensitive enzymes such as nitrogenase, the enzyme that fixates N2, can be active (11, 12).
The five NpDps proteins were, based on their predicted structural similarities to Ftns, Bfrs, and Dps proteins, clustered into specific clades. NpDps1–3 clustered together with typical Dps proteins and there was a strong predicted structural homology between NpDps1–3 and the Dps family prototype of Escherichia coli (EcDps). Therefore NpDps1–3 were classified as canonical Dps proteins. In contrast NpDps4 was clustered together with atypical Dps (8, 13) and NpDps5 shows predicted structural elements similar to Bfrs (8).
A functional characterization of the complete set of Dps proteins considering their individual functions and their possible interdependent physiological roles has not yet been addressed. A striking and fundamental question is: why would the cyanobacterium N. punctiforme need three different Dps proteins that all belong to the canonical group of Dps? Although NpDps1–3 have been predicted to be structurally similar, functional genetics studies have revealed that they are not redundant. NpDps2 is of core importance for H2O2 tolerance in N. punctiforme, and the lack of NpDps2 activity could not be compensated for by any of the other four Dps proteins or other H2O2 detoxification proteins in this cyanobacterium, such as catalases or peroxidases (8). The physiological functions of NpDps1 and NpDps3 are not resolved yet, but Npdps3, similar to Npdps2, is expressed in vegetative cells as well as in heterocysts (8). As yet there are limited studies on cyanobacterial Dps proteins from, e.g. Thermosynechococcus elongatus (13, 14), Synechococcus PCC 7942 (15, 16), Synechocystis PCC 6803 (17, 18), and Trichodesmium erythraeum (19). In particular, multiple Dps proteins within one cyanobacterium have only been studied in Nostoc sp. PCC 7120 (also known as Anabaena sp. PCC 7120) (20) and N. punctiforme (8, 21).
We present here in vitro and in silico characterizations of three individual canonical Dps proteins NpDps1, NpDps2, and NpDps3 within the one and the same cyanobacterium, N. punctiforme focusing on protein complex stability, DNA-binding properties, and iron incorporation/oxidation. The working hypothesis is that, based on results of our earlier studies (8), these multiple Dps proteins have different and specific functions of importance for cell homeostasis in vivo, and that these differences will be reflected in the biochemical properties of the individual Dps proteins. Such understanding of the biochemical potential will be of importance for future research on the individual physiological roles of the multiple Dps proteins in vivo in multicellular cyanobacteria.
Results
SDS-PAGE analysis of the purified proteins
To study the biochemical properties of the canonical Dps proteins from N. punctiforme we expressed and purified the NpDps1, NpDps2, and NpDps3 from E. coli cells via affinity chromatography utilizing a Strep-tag that was genetically introduced at the C terminus of each protein sequence. As a control for the different biophysical and biochemical characterizations, the Dps protein from E. coli was additionally expressed and purified. For each protein purification, SDS-PAGE analysis was used to ensure the purity of the Dps protein samples.
All the Dps proteins migrated through the polyacrylamide matrix close to the expected molecular mass of their monomers (Fig. 1) and all protein sequence identities were confirmed by MS. The band pattern occurring with EcDps was analyzed by MS confirming the protein sequence identity, accordingly. The double band pattern for EcDps has been described by Schmidt et al. (22) to be due to short N-terminal truncation inside E. coli. The EcDps sample contained an additional protein with approximately double the molecular mass of its monomer. This additional protein band (between 31.0 to 45.0 kDa) was also identified by MS to be the correct Dps sequence indicating the formation of dimers of this Dps protein. The persistence of dimeric structures in the gel after SDS and high temperature treatment might be due to high protein (thermo)stability. A band of a similar size from EcDps in SDS-PAGE has been observed earlier (22). The NpDps protein material was used for further analytical in vitro investigations.
Figure 1.
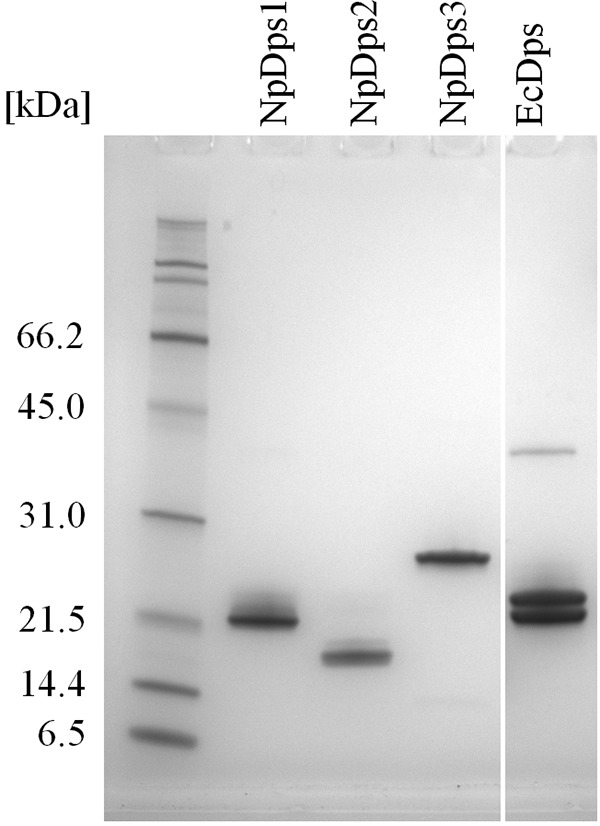
SDS-PAGE analysis on three recombinant Dps proteins from N. punctiforme and E. coli Dps (EcDps). 1 μg of each of the purified proteins was loaded on an SDS-PAGE gel. Expected molecular masses of the Dps monomers (including the alanine-alanine linker and the Strep(II)-tag sequence) were: 21.2 kDa for NpDps1, 19.5 kDa for NpDps2, 22.2 kDa for NpDps3, and 19.9 kDa for EcDps. SDS-PAGE broad range molecular weight standards (Bio-Rad) were used as molecular mass markers. The gel was stained with colloidal Coomassie.
DNA-binding properties
Dps proteins are often found to exhibit DNA-binding properties that might protect DNA against oxidative cleavage (6). To investigate whether the cyanobacterium N. punctiforme exhibits one or more Dps proteins that may protect DNA by forming DNA-Dps complexes, we conducted a series of electrophoretic mobility shift assays (EMSA) under different pH conditions. NpDps2 and NpDps3 formed DNA-Dps complexes at pH 6.0 as indicated by the DNA retardation in the gel (Fig. 2). At pH 7.0 the DNA-binding affinity for the two NpDps proteins, found to bind to DNA at pH 6.0, was completely abolished (Fig. 2). NpDps1 did not form a DNA-Dps complex, at either pH 6.0 or 7.0. In contrast to all NpDps proteins, EcDps bound to the complete amount of DNA at both pH 6.0 and 7.0, likewise demonstrated by an earlier study on EcDps (5). In EcDps DNA-binding is mediated by a lysine-rich N-terminal motif (5). Interestingly, similar motifs were found in the N-terminal and C-terminal protein sequence of NpDps2 and NpDps3, respectively, whereas no such motifs were found in NpDps1 (see Fig. S1 for further details).
Figure 2.
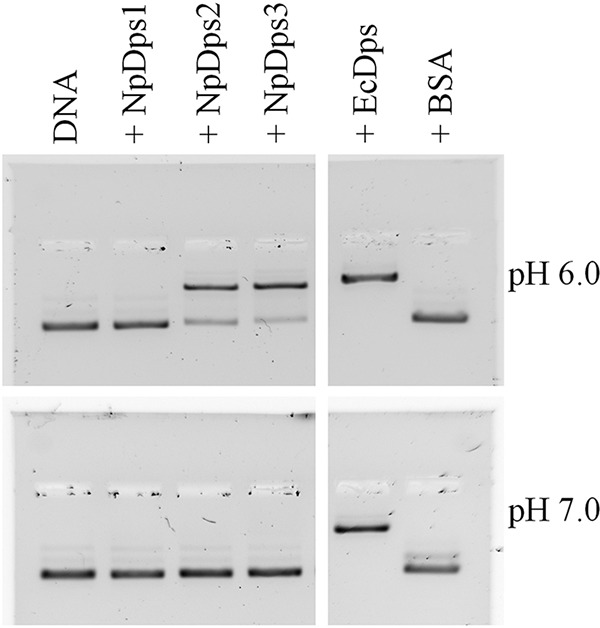
EMSA to analyze the DNA-binding properties of the NpDps proteins under different pH conditions. At pH 6.0 and 7.0, 125 ng of plasmid DNA (pSB1A3 vector) was incubated with 1 μg of each Dps protein, and separated on an agarose gel (1%). E. coli Dps (EcDps) and BSA served as a positive and negative control, respectively. Plasmid DNA is shown in lane DNA. Thiazole orange staining was performed after gel electrophoresis for DNA detection. The gel documentations are shown in inverted colors.
Dps multimerization
Members of the Dps protein family are generally expected to form dodecameric protein complexes that fulfil various functions such as iron incorporation and storage (6). To investigate whether the NpDps proteins were able to form such multimeric complexes, and if there was a correlation between the multimeric state of the proteins and DNA-binding properties, multiple strategies were followed.
Nondenaturing PAGE
Our first approach to identify possible multimers of the NpDps proteins was based on polyacrylamide gel electrophoresis (PAGE) under nondenaturing conditions. To estimate the approximate molecular masses of each investigated NpDps protein, the multimeric forms of bovine serum albumin (BSA) were used. Under nondenaturing conditions, BSA appeared to be present in its monomeric (66 kDa), dimeric (132 kDa), trimeric (198 kDa), and its tetrameric forms (264 kDa) (24) (Fig. 3). The three NpDps samples showed formation of different multimeric protein complexes when analyzed with nondenaturing PAGE at approximately pH 8 (Fig. 3). NpDps3 formed a large protein complex with comparable molecular mass to EcDps, which was included in this study as a control protein and was found to form a single multimeric protein complex. NpDps1 formed the lowest molecular mass complex of the three NpDps proteins with a band at ∼60 kDa, which might indicate its presence as a trimer (theoretical molecular mass 63.3 kDa). NpDps2 was found to have formed a putative pentamer of about 100 kDa (theoretical molecular mass 97 kDa). Even though NpDps1 and NpDps2 do not form protein complexes as large as NpDps3 under the gel electrophoresis conditions they both formed multimeric protein complexes. This provided the first indication of differential multimerization behavior among these NpDps proteins.
Figure 3.
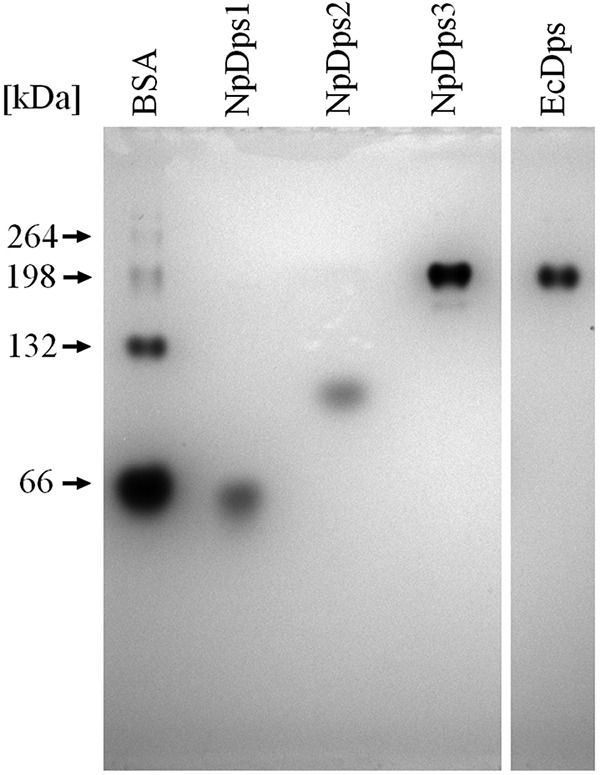
Multimerization of NpDps1, NpDps2, and NpDps3 and E. coli Dps (EcDps) analyzed by nondenaturing PAGE. The theoretical molecular masses of putative dodecamers were 253 kDa for NpDps1, 233 kDa for NpDps2, 265 kDa for NpDps3, and 236 kDa for EcDps. Additionally, the different native forms of BSA were used to estimate the approximate molecular masses. The pH of the gel was 8.5 and the pH of the running buffer was 8.0. The gel was stained with colloidal Coomassie.
Dynamic light scattering
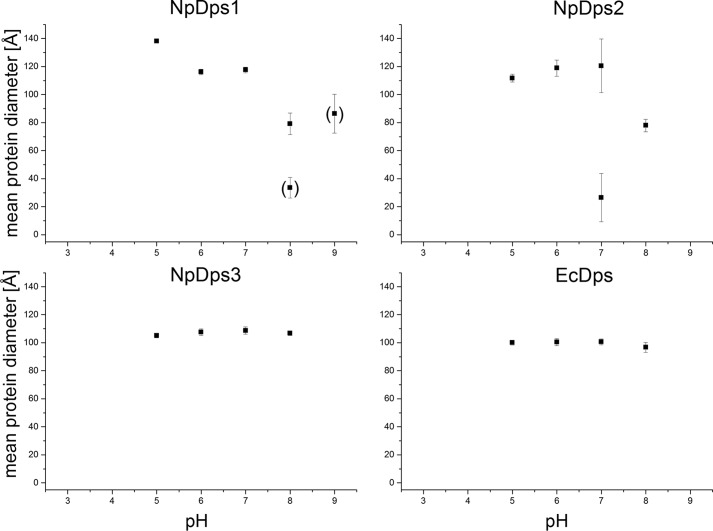
To expand our investigation of the multimeric Dps protein complexes from N. punctiforme, we conducted a series of dynamic light scattering (DLS) experiments, in solution, under different pH conditions. Our experimental design included the analysis of EcDps serving as a control protein to indicate the size of a Dps dodecamer, which has been earlier reported to be ∼101 Å in hydrodynamic diameter by DLS analysis (25). The Dps proteins were analyzed by DLS at pH 3.0, 5.0, 6.0, 7.0, 8.0, and 9.0 using different buffer systems (Fig. 4). There was a clear variation of pH-dependent stability between the individual NpDps. Nevertheless, at certain pH values, all three NpDps formed protein species with a hydrodynamic diameter larger than 99 Å indicating the presence of multimeric protein complexes.
Figure 4.
DLS on NpDps1, NpDps2, and NpDps3 at different pH conditions. The hydrodynamic diameters of all protein species are displayed as a mean of the average hydrodynamic diameter from each experimental series in Å at pH 3.0, 5.0, 6.0, 7.0, 8.0, and pH 9.0. Particles with a lower peak occurrence than 1% (number related) are not displayed, whereas particles with occurrence between 1 and 10% are shown in parentheses. The E. coli Dps (EcDps) served as a control. Mean ± S.E. particle diameters are given as error bars.
The proteins forming the most stable multimers with respect to varying pH conditions were NpDps3 and EcDps, where only large protein forms of similar hydrodynamic diameters (107 and 100 Å) were found at pH 5.0, 6.0, 7.0, and 8.0. At pH 9.0 and 3.0, no protein species were found for any of the Dps, which might be due to complete protein denaturation.
For NpDps1, a large protein species was found at pH 5.0, 6.0, and 7.0. However, at pH 5.0, this large species was about 21 Å larger than the species formed at pH 6.0 and 7.0, which had an average size of about 117 Å. At pH 8.0, no such large protein complex was observed, but two other smaller protein species were found with sizes of 81 and 28 Å. For both species, considerably larger mean ± S.E. were found, which may indicate the existence of intermediate protein species. At pH 9.0, only a protein species with a hydrodynamic diameter of 86 Å was observed, again, with large mean ± S.E. indicating more variable protein forms. Furthermore, the low peak occurrence for NpDps1 at pH 9.0 might be related to protein denaturation.
NpDps2 formed a large protein species (117 Å) at pH 5.0, 6.0, and 7.0. However, at pH 7.0 another smaller protein species of about 27 Å coexists with the large species. Large mean ± S.E. again implies partial decomposition of the large protein species into intermediate forms. At pH 8.0, only a medium-sized protein species of about 78 Å was observed for NpDps2. Note also that these DLS results correlate well with those from the nondenaturing PAGE analysis at pH ∼8, where NpDps3 formed the largest protein complex, whereas smaller complexes were observed for NpDps2 and NpDps1.
Size-exclusion chromatography
To clarify the overall oligomerization state of the three investigated NpDps proteins in solution at different pH (6.0 and 8.0), we performed a classic molecular mass determination using size-exclusion chromatography. EcDps was used as a control. For further chromatographic details, see Fig. S2. We found that all NpDps proteins could form a dodecamer, but lower multimeric states (n) calculated on the basis of protein mass standards could also be observed for NpDps1 and NpDps2. At pH 6.0, NpDps1 and NpDps3 are predominantly in their dodecameric state (n = 11.3 and 12.4, respectively), whereas NpDps2 was found to only partially form such state (n = 11.2), accompanied by a trimer (n = 2.63). Surprisingly, the mass of the control protein EcDps indicated an octamer (n = 8.06). For NpDps1 and NpDps2, there was a clear shift from a dodecameric state to lower multimeric form with an increase in pH from pH 6.0 to 8.0. By contrast, NpDps3 did not change its oligomerization state with pH, and remained predominantly a dodecamer even at pH 8.0.
Pores for iron attraction and entry
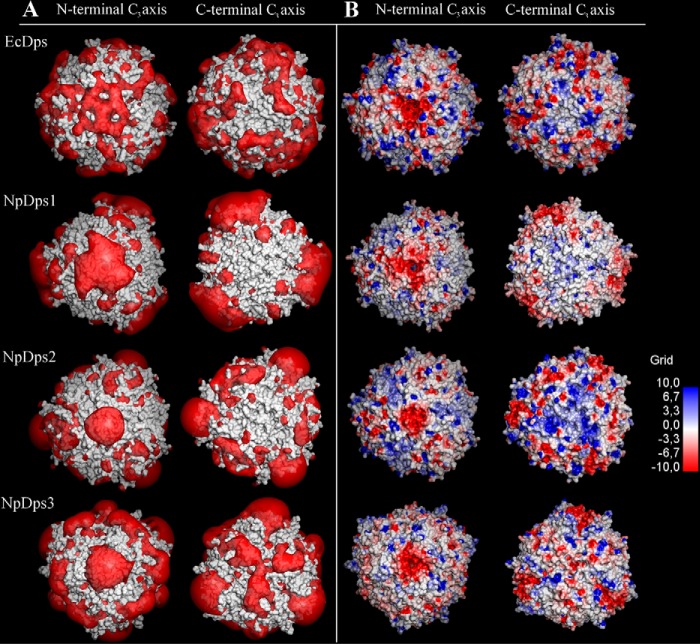
Besides the DNA-binding properties, another function that has been reported for Dps proteins is the incorporation of ferrous iron for its oxidation and storage inside the protein cavity. Two different sets of channel structures that link the protein interior to the exterior have been discovered, namely, the ferritin-like pore and the “Dps-type” pore (23). The ferritin-like pore has been demonstrated to contain several conserved negatively charged amino acids that line the path into the protein cavity, attracting metal ions and facilitating their incorporation into the protein interior (26). The ferritin-like pore is formed around a C3-symmetrical axis at the interaction interface of three Dps monomers close to their N-terminal random coil sequences (N-terminal C3 axis). Another pore structure, the Dps-type pore, is formed by three Dps monomers around a C3-symmetry axis, in the vicinity of their random coil C-terminal sequences (C-terminal C3 axis). In contrast to the ferritin-like pore, the Dps-type pore has been shown to be highly variable in terms of its pore size. There is also no general trend regarding its hydrophilicity or lipophilicity (14).
To investigate whether the NpDps proteins might form structures that could function as pores for incorporating metal ions such as ferrous iron, we first created model structures of NpDps1, NpDps2, and NpDps3 based on the crystal structure of the EcDps protein (PDB code 1DPS), which has been demonstrated to incorporate iron into its dodecameric structure. The electrostatic potentials of the predicted NpDps1, NpDps2, and NpDps3 structures were calculated with focus on the region around N- and C-terminal C3-axes (Fig. 5). Electrostatic potentials were also calculated for the EcDps for comparison. Note that because the structures of the random coil N and C termini could not easily be predicted for the dodecameric models, they were truncated prior to model building. Therefore, possible electrostatic effects of these tails on the pores were not included in this analysis. Examining the region around the N-terminal C3-axes (Fig. 5), it can be seen that the area around the ferritin-like pore here has a significantly negative electrostatic potential for all three NpDps models, as well as the EcDps reference. The isosurface visualization (Fig. 5A, left) highlights this and allows for comparison between the models at the same calculated negative potential value. This can also be seen in the visualization of the proteins' surfaces as a function of electrostatic potential, where the region surrounding the N-terminal pore have highly negative potentials for all four proteins (Fig. 5B, left). Analysis of the amino acids at these pores reveals that this is due to clusters of negatively charged aspartates in all these model structures. This correlates well with the earlier proposed functions of these N-terminal, ferritin-like pores in attracting and transporting positive ferrous ions into the interior of the dodecamer.
Figure 5.
Calculated electrostatic potential around the N- and C-terminal C3-axes in E. coli Dps (EcDps) and the model structures of NpDps1, NpDps2, and NpDps3. A, electrostatic potential isosurfaces (red; −1.0 kT/e) of the Dps proteins, superimposed onto the van der Waals surface of the proteins (gray). B, the van der Waals surfaces of the Dps proteins colored according to the calculated electrostatic potentials from electronegative (red) to electropositive (blue) by a continuous color gradient. The N-terminal C3-axes indicates the ferritin-like pore and the C-terminal C3-axes indicates the Dps-type pore.
The results for the Dps-type pore surrounding the C-terminal C3-symmetry axis were less uniform across these four models. For the EcDps, NpDps1, and NpDps2, the regions immediately surrounding the C-terminal pores did not have significantly negative electrostatic potentials (Fig. 5, A and B, right), although the results for the EcDps did show some patches of negative potential slightly further away from the actual pore. By contrast, it could be seen that there were regions with significantly negative electrostatic potentials for the NpDps3 around the corresponding C-terminal Dps-type pore (Fig. 5A, right). However, this was not as concentrated at the pore compared with the case of the N-terminal, ferritin-like pore discussed above, and inspection of the overall electrostatic potential map (Fig. 5B, right) also shows that the electrostatic potential values were not as negative either. For EcDps, NpDps1, and NpDps2, the C-terminal pore was essentially electrostatically neutral. Even patches of positive electrostatic potential were observed slightly further away from the pore, especially in the case of NpDps2. Given the calculated potentials obtained here, it seems unlikely that there would be incorporation of positively charged metal ions through the C-terminal pore. This is in contrast to NpDps3 that showed a more electrostatically negative pore. This correlates well with literature regarding the variability of similar Dps-type pores (14).
Iron content analysis of the purified NpDps protein material
To investigate whether Dps proteins from N. punctiforme had incorporated iron during heterologous expression in E. coli, a spectrophotometric iron quantification assay was performed on the purified NpDps proteins (Table 1).
Table 1.
Iron quantification, by a colorimetric assay, of the Dps proteins from N. punctiforme and E. coli (EcDps)
Hemoglobin served as a control. Standard errors were derived from four independent experiments.
| Protein sample | Fe2+ ions/protein dodecamer |
|---|---|
| NpDps1 | 2.5 ± 0.6 |
| NpDps2 | 0.7 ± 0.3 |
| NpDps3 | 15.1 ± 2.5 |
| EcDps | 11.0 ± 1.9 |
| Hemoglobin tetramer | 4.2 ± 0.1 |
The three investigated NpDps proteins were found to contain iron after heterologous expression in E. coli and purification. This was the case without any further addition of iron to the proteins. For NpDps the average amount of iron bound to the proteins was determined to be in range between 1 and 15 iron ions per protein (presumed as dodecamers). Analyses of the control protein, hemoglobin, showed that it contained about 4 iron ions per tetramer, which matched the iron content reported by the manufacturer (Sigma). Additionally, the EcDps, overexpressed in E. coli and purified in the same way as the three investigated NpDps proteins, was shown to contain 11 iron ions. BSA served as a negative control and its iron content was below the detection limit of the assay.
Iron oxidation activity
To investigate the enzymatic activity of the three canonical NpDps after recombinant expression and purification, kinetic Fe2+ oxidation experiments were performed at pH 6.0, a pH condition that was found to preserve higher multimeric states for all the three investigated NpDps (see DLS and gel filtration results). Iron oxidation was followed by absorption spectroscopy at 310 nm and no change in the absorbance could be observed when proteins were incubated for at least 10 min in the reaction buffer and subsequently mixed with Fe2+ under aerobic conditions (Fig. 6). Upon the addition of an excess amount of H2O2, the absorbance at 310 nm increased hyperbolically for NpDps1 and NpDps2 as well as for the protein-free control reaction, but with different kinetic profiles. The Fe2+ oxidation was fastest for NpDps1 and NpDps2, and the slowest reaction was found for the control. In contrast to the hyperbolic reaction trend, the reaction in the presence of NpDps3 appeared to be a two-phase reaction. The first phase of the reaction was too fast to be resolved in our experimental setup, therefore the maximum A310 nm was not identified. The second phase showed a slow decay that stabilized at approximately A310 nm of 0.050 after 10 min. For all reactions a further addition of H2O2 was performed to verify the completion of the oxidation, which proved to be the case for all reactions. Furthermore, the addition of H2O2 did not alter the absorption at 310 nm when Fe2+ was not added beforehand (data not shown).
Figure 6.
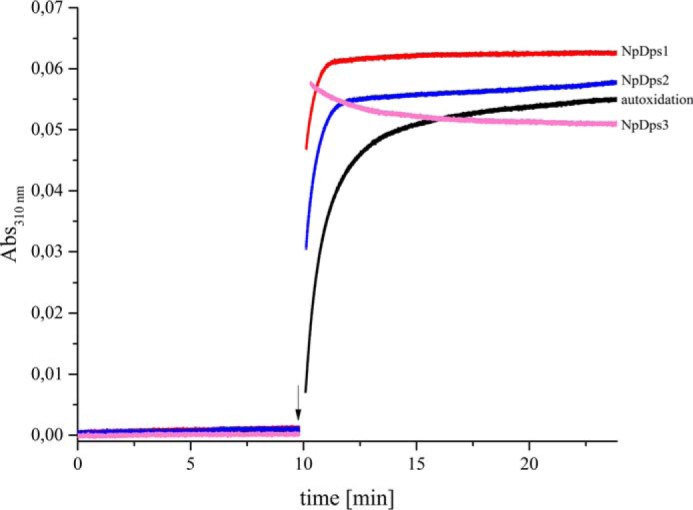
Fe2+ oxidation kinetics in the presence of NpDps1, NpDps2, and NpDps3. Absorbance measured at 310 nm corresponds to the formation of the ferric iron in the presence of NpDps1 (red line), NpDps2 (blue line), NpDps3 (pink line), and for the no protein control (black line). 0.5 μm of each NpDps protein was mixed with 24 μm Fe2+ (48 iron ions per Dps dodecamer), at time 0 min and the absorbance was analyzed for 10 min, before addition of 16 μm H2O2 as indicated by the arrow. Reactions were performed in 5 mm succinate buffer, pH 6.0, 50 mm NaCl, at room temperature and under aerobic conditions.
Under the chosen conditions EcDps could not serve as a control protein due to its quick precipitation upon H2O2 addition, which was indicated by a significant increase of the background scattering and turbidity in the reaction volume. This was in agreement with earlier reported oxidation kinetic experiments on EcDps (3) and the general tendency of EcDps toward self-aggregation (5). Precipitation also occurred for NpDps2, however, at later time point after addition of H2O2 (see Fig. S3). The major results of this study are summarized in Table 2.
Table 2.
Summary of biochemical properties of the three canonical Dps proteins in N. punctiforme
| Protein | DNA-binding capacity | Ferroxidase activity under consumption of H2O2a | Condition for dodecamer stability |
|---|---|---|---|
| NpDps1 | No | Yes | pH 6.0 |
| NpDps2 | Yesb | Yes | pH 6.0 |
| NpDps3 | Yesb | Yes | pH 6.0, pH 8.0 |
a Fe2+ oxidation under the consumption of O2 was not observed.
b Putative DNA-binding motif at the N or C terminus identified (see Fig. S1).
Discussion
Since the first characterization of a Dps protein (27), advances in understanding its molecular and biochemical properties have been achieved through physiological, biochemical studies, and heterologous expression of dps genes in E. coli. These reports have helped to elucidate the role of single Dps proteins in a wide variety of microbes, from E. coli to cyanobacteria. However, the specific functions of the individual Dps and cooperative behavior of multiple Dps proteins within one organism for cellular fitness, including iron homeostasis and oxidative stress tolerance, have not been resolved. As previously mentioned, the working hypothesis of our earlier studies was that multiple Dps proteins within one organism have different and specific function of importance for cell homeostasis (8, 21), and that these differences would be reflected in the biochemical properties of the individual Dps. The results obtained in the present in vitro and in silico studies indeed show clear differences in the individual biochemical properties of the three canonical Dps proteins NpDps1, NpDps2, and NpDps3, as summarized in Table 2.
Differential pH-dependent properties of NpDps1–3 suggest distinct physiological functions
Our results show a clear and protein-specific pH dependence of multimerization and DNA binding of NpDps1–3. In terms of multimerization, the DLS, size-exclusion chromatography and electrophoresis experiments here revealed that whereas all three Dps are able to form multimers, most likely dodecamers as for most Dps proteins, they differ in their pH-sensitivity. NpDps1 and NpDps2 dissociate in alkaline pH, with NpDps2 already dissociating at pH 7.0. The multimeric NpDps3 has a significantly broader pH stability range, and is stable even at pH 8.0. Differential pH-dependence behavior was also observed for DNA binding (discussed below).
These different pH dependences suggest that NpDps1–3 may have specific physiological roles depending on different conditions in the cell environment or compartments. Although there is currently a lack of detailed knowledge about pH conditions in different cellular compartments and in different cells of multicellular cyanobacteria, it is known that considerable pH variations within the cell do exist. For example, inside the thylakoid lumen of cyanobacterium Synechococcus elongatus the pH ranges from pH 4 in light (active photosynthesis) to pH 6 in darkness, whereas the pH of the cytosol ranges from 7 (dark) to 8.5 (light) (28–30). Interestingly it has been proposed that different fractions of one Dps protein could have diverse functions and localization in the cell (31). If these different functions, connected to specific parts of the cell, are regulated by a difference in local pH is yet not known.
There are only few reports on localization of Dps proteins within cyanobacterial cells. One example is the DpsA from Synechococcus 7942 that was found to be associated to the cytosolic side of the thylakoids and suggested to provide iron to the iron-rich photosystem I (31). Whether Dps proteins are required within the thylakoid lumen remains unknown. For more alkaline-sensitive Dps proteins, such as NpDps1 and -2, it might serve a suitable environment to form dodecameric structures. Unfortunately, not much is known about iron homeostasis inside the thylakoid lumen.
Besides the different pH conditions that can be found inside the cell, Dps proteins might also play a role outside the cell. A homologous protein of NpDps3 from Nostoc sp. PCC 7120 was shown to be actively exported out of the cell, as part of the secretome (32). Furthermore, homologs of NpDps1 and NpDps2 were detected in the exoproteome of the same cyanobacterium (33). This is of interest, because Nostoc species have been found being part of cyanobacterial biofilms (34), and the contribution of Dps in biofilm formation has been found for a Dps from Campylobacter jejuni (35).
However, it remains unknown whether multimeric forms of Dps stay stable under the varying environmental conditions that filamentous cyanobacteria are exposed to in this study we found that NpDps3 could indeed be a candidate coping with a wide range of different pH conditions outside the cell and it needs to be tested whether NpDps3 plays a crucial role in biofilm formation.
Different studies have shown that certain multimeric states such as dimers, trimers, and dodecamers can exist under specific conditions and can have distinct functions such as DNA-binding or ferroxidase activity (36–38). The only functional difference between the dodecamer and lower multimeric states might be the capacity to store iron. The observed various multimeric forms of NpDps1–3 at different pH conditions might indicate different forms for specific physiological purposes.
We found a pH-sensitive DNA interaction for NpDps2 and NpDps3, whereas NpDps1 did not bind DNA. For some Dps proteins electrostatic attraction of positively charged amino acids in the C- and N-tails of Dps proteins to DNA is a major determinant for DNA-binding (7), and the prediction of charges on the NpDps tails coincide with the DNA-binding of NpDps2 and NpDps3 (Fig. S1). The DNA-binding properties of NpDps2 and NpDps3 suggest a possible role for these proteins in DNA protection and nucleoid condensation, as previously reported for EcDps (5, 38) and for Staphylococcus aureus Dps (39). NpDps1 did not bind DNA, which was contrary to what was earlier shown for a homolog of NpDps1, namely All0458 in Nostoc sp. PCC 7120. This homolog is located at the nucleoid during stress conditions, suggesting DNA-binding (20). It is interesting to note that DNA-binding itself seems not to be crucial for the protection of DNA from oxidative damage, as demonstrated by several non-DNA-binding Dps (14, 33, 40). We share the hypothesis that has been stated already in 1998 by Grant et al. (23), that the removal of ferrous iron from the solution could be of significance to rescue cells from the oxidative damage and the dangerous product of the Fenton reaction. Therefore, even the non-DNA binding NpDps1 might be important for protection against oxidative damage, by removal of free ferrous iron.
Iron binding sequestration, oxidative protection
The ability for all three NpDps to form multimeric proteins suggests that they have the potential to incorporate iron after heterologous expression in E. coli. Indeed, all the as purified NpDps contained iron, although at different amounts. A variation of iron content after heterologous expression has also been shown for other Dps (41–43).
We suggest that the low amount of bound iron in NpDps1 and NpDps2 is an effect of the purification procedure at pH 8.0 that could have caused a partial dissociation of the multimeric states for the more pH-sensitive NpDps. In contrast, EcDps and NpDps3 appeared to be more resilient to higher pH conditions, which would allow them to keep their multimeric form and thus not lose incorporated iron.
Iron attraction and entry into the NpDps
Our results show that Fe2+ oxidation is enhanced by addition of H2O2 in all three canonical NpDps (Fig. 6). It seems therefore likely that NpDps play an important role in cellular iron homeostasis and for ROS regulation. Involvement in two steps, one physical and one chemical, can be envisaged.
The first step in iron uptake and storage, is attraction and physical transport of Fe2+ via pore structures, to the ferroxidase centers located in the interior of the protein. The predicted electrostatic potential maps of NpDps1–3 suggest the existence of two types of pores that connect the protein exterior with the inside, as earlier shown for EcDps. The ferritin-like pores of the three NpDps showed negative electrostatic potentials, thus indicating it as an entrance for Fe2+. Only a few mechanistic studies have been performed on the Fe2+ translocation into Dps (43), and these propose the ferritin-like pore as the main path for iron entry into ferroxidase centers in heterotrophic bacteria (7), and in the cyanobacterium T. elongatus (13). Furthermore, the variation of electrostatic potential at the ferritin-like pore has been shown to influence iron uptake kinetics (43). For the other pore type, the Dps-type pore, no indication of negative potential for NpDps1 and NpDps2 was found. This is in agreement with what is known about iron binding and translocation in most Dps. The negative electrostatic potential predicted in our model structure for NpDps3 is rarely found, except for Dps2 from Deinococcus radiodurans (44). Although the physiological significance of this difference seen in NpDps3 as compared with NpDps1 and NpDps2 needs further investigations, our results from structural and Fe2+ oxidation kinetics studies indicate biochemical distinctions within the group of the three canonical NpDps.
The second, chemical step, is the Fe2+ oxidation itself. Our data has shown that all three NpDps are potent consumers of H2O2 in vitro when Fe2+ is available, and therefore could play an in vivo role in detoxifying H2O2 during stress conditions. This might seem contradictory to our earlier functional genetics studies in which only one of the three, NpDps2, was shown to have a crucial physiological role in the presence of high exogenously added H2O2 levels (8). However, although the physiological relevance of the iron uptake and iron oxidation by H2O2 is hard to predict, the in vivo roles could to some extent be understood by the determined differential gene and protein expression of NpDps (8, 9, 21, 45, 46). The NpDps-specific expression is distributed among the various cell types, formed during specific cultivation conditions, of N. punctiforme (12), which suggests growth condition-specific functions of the three canonical NpDps proteins.
Conclusion
With this study we gained insights on the protein complex stability and biochemical activity of the three canonical NpDps proteins in the multicellular cyanobacterium N. punctiforme. They show a pH-dependent multimerization that suggests that these three Dps proteins are all involved in iron regulation during different pH conditions indicating differences in spatial and temporal distribution of activity. NpDps1–3 also differ in their DNA-binding pattern, which proposes different protection strategies, by mechanical protection of DNA and/or by iron sequestration to inhibit Fenton chemistry. The division of expression of NpDps proteins within different cell types of N. punctiforme raises questions about cell-specific iron homeostasis and redox regulation in a multicellular prokaryote, which are of importance for the ability to acclimate to environmental fluctuations.
With this study we have taken the first step in the investigation of the protein chemistry to help specify the physiological role of the individual NpDps, and how these proteins work in concert in iron homeostasis and in protection against oxidative stress in multicellular cyanobacteria.
Experimental procedures
Plasmid design for dps expression
One-step isothermal assembly of overlapping DNA fragments (Gibson assembly) (47) was used to construct the plasmids for heterologous expression of the Dps proteins. The Synthetic Biology parts were all obtained from the Registry of Standard Parts. The parts used were Plac promoter (BBa_B0015), ribosome-binding site (BBa_B0034), double terminator (BBa_B0015), as well as the BioBrick plasmid pSB1A3. At the 5′ and 3′ sides of the ribosome-binding site eight and six nucleotides were added, respectively. A Strep-tag II (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) was attached via a short linker (gccgca) to the 3′ end of the dps gene and protein expression was terminated by an added stop codon (taa). The dps genes were amplified from genomic DNA either from E. coli or N. punctiforme and cloned downstream the isopropyl 1-thio-β-d-galactopyranoside-inducible Plac promoter. (for primer sequences see Table S1). All PCR amplification was done using PfuUltra II Fusion HS DNA Polymerase (Stratagene). Constructs were transformed into E. coli DH5α cells and selected against ampicillin (100 μg ml−1). The correct assembly of fragments was confirmed by sequencing.
Bacterial strains, growth conditions, and expression of recombinant protein
All strains were stored in 10% glycerol stocks at −80 °C. E. coli BL21(DE3) (Novagen) was used for heterologous protein expression. Growth of cultures was monitored spectrophotometrically at 600 nm. E. coli BL21(DE3) cells harboring the recombinant plasmids were incubated at 37 °C with shaking (250 rpm) in 600 ml of lysogeny broth (LB) culture medium containing 50 μg ml−1 of ampicillin and 0.1 g liter−1 of FeCl3 and grown to an optical density at 600 nm of 0.6. Isopropyl 1-thio-β-d-galactopyranoside (1 mm) was then added to the cultures, and these were incubated a further 5–6 h for production of the recombinant proteins. Cells were harvested by centrifugation at 4,000 × g for 20 min at 4 °C, and stored overnight at −20 °C. Further treatment was performed on ice except where otherwise stated. The cells were disrupted by sonication in 10 to 15 ml of lysis buffer containing 50 mm Tris-HCl, pH 8.0, 1 m NaCl, 10 mg of lysozyme (Sigma), and 3 mg of avidin (Sigma). Thereafter 10 μl of Protease Arrest (G-Biosciences) and 1 unit of DNase I (Thermo Scientific) were added to the lysate and incubated for 40 min. Lysis buffer was added up to 25 ml, and the lysate was centrifuged at 10,000 × g for 30 min at 4 °C after which the pellet was discarded. All protein purifications were performed at 4 °C on a ÄKTA FPLC (GE Healthcare). The proteins were purified by using a Strep Tactin II column (10 ml) (IBA) equilibrated with degassed equilibration buffer (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 10 mm MgCl2), and the proteins were eluted with degassed equilibration buffer containing an additional 2.5 to 15 mm d-desthiobiotin (Sigma). The eluted proteins were concentrated using 10-kDa centrifugal filter units (Merck) according to the instructions of the manufacturer. The buffer was changed to 50 mm NaCl and 10 mm Tris-HCl, pH 8.0 (4 °C), by using PD-10 columns (GE Healthcare) following the instructions of the manufacturer. The protein concentration was determined based on the Lowry method using the DC protein assay (Bio-Rad).
EMSA
The DNA-binding capacities of all the purified Dps proteins were assessed in electrophoretic mobility shift assays at two different pH values. Before gel electrophoresis 125 ng of pSB1A3 plasmid DNA (2.5 kbp) was incubated with 1 μg of each of the purified proteins at pH 6.0 and 7.0 using MES or BisTris buffer systems, respectively. The incubation volume was 5 μl containing 50 mm NaCl and 100 mm buffer. After an incubation time of 15 min at room temperature, 1 μl of 6× loading buffer containing 60% (v/v) glycerol, 50 mm NaCl, and 0.1% (w/v) bromphenol blue was added. The entire reaction volume was loaded on a 1% (w/v) agarose gel made of running buffer containing 50 mm NaCl and 10 mm MES or BisTris buffer, according to the desired pH. A voltage of 30 V was applied for 45 min at room temperature and the gels were subsequently stained in MES running buffer containing thiazole orange for 1 h under agitation in darkness. The gels were washed twice with dH2O and were documented using the Chemi Doc XRS (Bio-Rad). 1 μg of BSA (Sigma) served as a negative control of interaction with DNA.
SDS-PAGE
Protein expression and purification was confirmed by SDS-PAGE, after Laemmli (48), 0.8 μg of each protein sample was loaded onto any kDa gel (Bio-Rad). The SDS-PAGE Broad Range Molecular mass standards (Bio-Rad) served as the molecular mass marker. Gel electrophoreses were performed at 200 V, and gels were thereafter stained overnight in colloidal Coomassie and de-stained in dH2O (49). The gels were documented using the Chemi Doc XRS (Bio-Rad). The protein bands were cut out from the gel and the identity of the proteins was confirmed by MS as explained under “Protein identification.”
Nondenaturing gel electrophoresis
To investigate the multimeric form of the purified proteins, each protein sample (0.8 μg of Dps and 1.5 μg of BSA (Sigma)) was mixed with loading buffer to set sample conditions to 25 mm NaCl, 30 mm Tris-HCl, pH 6.8 (room temperature), 20% glycerol, and 0.005% (w/v) bromphenol blue. Samples were loaded onto an any kDa gel (Bio-Rad). The running buffer contained 25 mm Tris-HCl and 192 mm glycine set to pH 8.0 at room temperature. The gel exhibited an intrinsic pH of 8.45 according to the manufacturer's guide. The applied voltage was set to 100 V for the first 30 s and then was decreased to 40 V for further separation for 10 h at room temperature. The gel was stained with colloidal Coomassie and documented as described previously for SDS-PAGE analysis.
Protein identification
The proteins were reduced, alkylated, and in-gel digested by trypsin according to a standard operating procedure (50). Thereafter the samples were dried and dissolved in 15 μl of 0.1% formic acid. The peptides were separated in reversed-phase on a C18-column and electrosprayed on-line to Q Exactive Plus mass spectrometer (Thermo Finnigan). Tandem MS was performed applying higher-energy C-trap dissociation. The data were analyzed by using the Sequest algorithm embedded in Proteome Discoverer 1.4 (Thermo Fisher Scientific) toward a FASTA database containing proteins from N. punctiforme ATCC 29133 and E. coli, downloaded from the SwissProt database. The search criteria for protein identification were set to at least two matching peptides of 95% confidence level per protein.
Protein sequences for NpDps1–5 were received from the Cyanobase web tool (http://genome.microbedb.jp/cyanobase/) and SWISS-PROT (https://www.ebi.ac.uk/swissprot/)5 (51) and those of E. coli and Mycobacterium smegmatis from the NCBI. Sequences were aligned with the web-based program Clustal Omega (52, 53).
DLS
The three purified NpDps proteins and EcDps were diluted into 50 mm NaCl and 100 mm buffer: HEPES, pH 3.0; acetate, pH 5.0; succinate, pH 6.0; phosphate, pH 7.0; Tris, pH 8.0; and BisTris propane, pH 9.0, at a concentration between 70 and 100 μg ml−1. All solution components were individually filtered before mixing (0.45 μm Millipore filter HAWP-AO, Merck). Prior to DLS analysis, all samples were centrifuged at 13,000 × g for 5 min at room temperature. DLS analysis was performed at 173 °C (backscattering) on a Zetasizer Nano instrument (Malvern) using 1-cm path length polystyrene cuvettes (VWR). Experiments were performed at 25 °C. At each pH, 5 to 13 data sets (each 100 s in duration) were collected for each protein. The hydrodynamic diameter of each protein species was calculated based on the mean from each data set (number related size distribution). As a negative control, 5 data sets (each 100 s in duration) were collected for solutions at each pH condition without protein material.
Molecular mass determination by size-exclusion chromatography
The molecular mass of the three NpDps and EcDps proteins was determined by size-exclusion chromatography (gel filtration) using a HiLoad 16/600 Superdex 200 prep grade column connected to an Äkta FPLC (GE Healthcare Life Sciences). Analyses were performed at 4 °C at two different pH using either 10 mm Tris-HCl, pH 8.0, containing 150 mm NaCl or 10 mm MES, pH 6.0, containing 150 mm NaCl (pH values at 4 °C) as running buffers. Each Dps protein was equilibrated in the appropriate running buffer by either diluting directly from concentrated stocks (when dilution factor was at least 10 times) or diluting from less concentrated stocks followed by buffer exchange in Vivaspin ultrafiltration devices (Sartorius) with a 30-kDa cutoff. Proteins were allowed to equilibrate in running buffer at 4 °C for at least 30 min prior to analysis. Typically, 500-μl Dps samples (two per protein and pH) at 0.7–1 mg/ml were analyzed by gel filtration. Molecular mass calibration was performed using the High Molecular Weight gel filtration kit (GE Healthcare Life Sciences) according to the manufacturer's instructions.
Dps modeling
The three-dimensional models of the NpDps1, NpDps2, and NpDps3 were created using the intensive modeling mode of Phyre2 (54). The crystal structure of EcDps (PDB 1DPS) served as a template for the dodecameric model builds for NpDps1, NpDps2, and NpDps3 using Swiss-PdbViewer version 4.1.0 (55). The electrostatic potentials of these protein models, as well as for EcDps were calculated using DelPhi (55), as implemented in Discovery Studio 2.1 (Accelrys), to solve the Poisson-Boltzmann equation on the protein system in a cubic lattice (implicit solvent model with solvent dielectric constant 80, solute dielectric constant 2, ionic strength 0.145, Debye-Hückel boundary conditions, 65 grid points per axis). On the basis of the resulting data, visualizations through electrostatic potential maps and isopotential surfaces were generated (Discovery Studio Visualizer 4.5). Prior to the generation of the electrostatic potential maps, the N- and C-terminal random coil tails of the Dps proteins (Fig. S1) were deleted from the sequences. No deletion of the random coiled tails was necessary for the C termini of NpDps2 and EcDps.
Iron quantification assay
The following iron quantification assay is based on the spectrophotometric assay by Artiss et al. (56) with minor modifications to account for the chemical stabilities of Dps proteins and Ferene-S (56, 58). Protein (18 to 70 μg) was mixed in Eppendorf tubes with 20 μl of 37% HCl and boiled for at least 30 min at 100 °C to total evaporation using a heat block in a ventilation hood. The samples were chilled to room temperature. 25 μl of an ascorbic acid (11.5 g liter−1) and TCA (170 g liter−1) solution was added. After the addition of 50 μl of dH2O, the reaction mixture was mixed. 25 μl of a freshly prepared solution of Ferene-S (3.5 g liter−1) in ammonium acetate (333 g liter−1, pH 4.6, at room temperature) and subsequently 8 μl of 10 m NaOH were added to adjust the pH to ∼4.5. The reaction mixture was vortexed and briefly centrifuged on a table centrifuge, at maximum speed. The reaction was incubated for 30 min in a 42 °C water bath and afterward chilled to room temperature and briefly centrifuged again. The spectrum of the supernatant was recorded between 400 and 800 nm on a Cary 50 spectrophotometer using polystyrene cuvettes (Sarstedt). The absorbance at 594 nm, which is the absorption maximum of the formed [Fe(II)(Ferene-S)3]2+ complex, was used for iron quantification. The standard curve was created by using an iron standard (for Atomic Absorption Spectrometry (AAS), Sigma) and an extinction coefficient of 35,000 cm−1 m−1 was obtained. Hemoglobin from bovine blood (Sigma) served as a control protein, and contains approximately four iron ions per protein tetramer according to the manufacturer. BSA (Sigma) was used as negative control and iron could not be detected.
Fe2+ oxidation activity by H2O2
Kinetic experiments of Fe2+ oxidation by O2/H2O2 were performed on a Cary 5000 spectrophotometer (Varian) in 1-cm standard quartz cuvettes at room temperature. The time-dependent absorbance was recorded at 310 nm, which corresponds to Fe3+ formation (57, 59). During the experiment, reaction solutions were maintained in air. 0.5 μm of the purified NpDps protein (assumed dodecamer concentration) in 5 mm succinate and 50 mm NaCl, at pH 7.0, was incubated for 10 min. Freshly prepared and N2-sparged 24 μm FeSO4 (48 μm Fe2+/dodecamer) was added to the solution and the absorbance at 310 nm was followed for 10 min. Thereafter 16 μm H2O2 was rapidly added to the reaction solution and mixed. As a control Fe2+ oxidation by H2O2 was measured under identical conditions, but without proteins. The data were analyzed with OriginPro 2016G software (OriginLab, Northampton, MA). All data presented originated from measurements on two independent protein preparations.
Author contributions
C. H. data curation; C. H. and K. S. formal analysis; C. H., A. N., P. R., and K. S. investigation; C. H., F. H., A. N., and P. R. methodology; C. H., P. R., and K. S. writing-original draft; C. H., F. H., P. R., and K. S. writing-review and editing; F. H. and K. S. supervision; K. S. conceptualization; K. S. funding acquisition; K. S. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Peter Swensson for introduction to the DLS methodology and Prof. Tomas Edvinsson (Uppsala University, Sweden) for allowing us to use the DLS instrument. We also thank the students of our lab for their contributions. Data storage was obtained and supported by BILS (Bioinformatics Infrastructure for Life Sciences).
This work was supported by NordForsk, NCoE Program “NordAqua” project 82845, Swedish Energy Agency project 11674-5, and the Science for Life Laboratory, Mass Spectrometry Based Proteomics Facility in Uppsala. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ROS
- reactive oxygen species
- Bfr
- bacterioferritin
- DLS
- dynamic light scattering
- EMSA
- electrophoretic mobility shift assay
- Dps
- DNA-binding protein from starved cells
- Ftn
- ferritin
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BisTris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- PDB
- Protein Data Bank.
References
- 1. Imlay J. A. (2006) Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 10.1111/j.1365-2958.2006.05028.x [DOI] [PubMed] [Google Scholar]
- 2. Andrews S. C., Robinson A. K., and Rodríguez-Quiñones F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- 3. Zhao G., Ceci P., Ilari A., Giangiacomo L., Laue T. M., Chiancone E., and Chasteen N. D. (2002) Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A “ferritin-like” DNA-binding protein of Escherichia coli. J. Biol. Chem. 277, 27689–27696 10.1074/.M202094200 [DOI] [PubMed] [Google Scholar]
- 4. Calhoun L. N., and Kwon Y. M. (2011) Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J. Appl. Microbiol. 110, 375–386 [DOI] [PubMed] [Google Scholar]
- 5. Ceci P., Cellai S., Falvo E., Rivetti C., Rossi G. L., and Chiancone E. (2004) DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 32, 5935–5944 10.1093/nar/gkh915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chiancone E., and Ceci P. (2010) The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta 1800, 798–805 10.1016/j.bbagen.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 7. Haikarainen T., and Papageorgiou A. C. (2010) Dps-like proteins: structural and functional insights into a versatile protein family. Cell Mol. Life Sci. 67, 341–351 10.1007/s00018-009-0168-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ekman M., Sandh G., Nenninger A., Oliveira P., and Stensjö K. (2013) Cellular and functional specificity among “ferritin-like” proteins in the multicellular cyanobacterium Nostoc punctiforme. Environ. Microbiol. 16, 829–844 [DOI] [PubMed] [Google Scholar]
- 9. Ow S. Y., Noirel J., Cardona T., Taton A., Lindblad P., Stensjö K., and Wright P. C. (2009) Quantitative overview of N2 fixation in Nostoc punctiforme ATCC 29133 through cellular enrichments and iTRAQ shotgun proteomics. J. Proteome Res. 8, 187–198 10.1021/pr800285v [DOI] [PubMed] [Google Scholar]
- 10. Rippka R., Deruelles J., Waterbury J. B., Herdman M., and Stanier R. Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiol. 111, 1–61 10.1099/00221287-111-1-1 [DOI] [Google Scholar]
- 11. Meeks J. C., Campbell E. L., Summers M. L., and Wong F. C. (2002) Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 178, 395–403 10.1007/s00203-002-0476-5 [DOI] [PubMed] [Google Scholar]
- 12. Meeks J. C., and Elhai J. (2002) Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 66, 94–121 10.1128/MMBR.66.1.94-121.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alaleona F., Franceschini S., Ceci P., Ilari A., and Chiancone E. (2010) Thermosynechoccus elongatus DpsA binds Zn(II) at a unique three histidine-containing ferroxidase center and utilizes O2 as iron oxidant with very high efficiency, unlike the typical Dps proteins. FEBS J. 277, 903–917 10.1111/j.1742-4658.2009.07532.x [DOI] [PubMed] [Google Scholar]
- 14. Franceschini S., Ceci P., Alaleona F., Chiancone E., and Ilari A. (2006) Antioxidant Dps protein from the thermophilic cyanobacterium Thermosynechococcus elongatus: an intrinsically stable cage-like structure endowed with enhanced stability. FEBS J. 273, 4913–4928 10.1111/j.1742-4658.2006.05490.x [DOI] [PubMed] [Google Scholar]
- 15. Peña M. M., and Bullerjahn G. S. (1995) The DpsA protein of Synechococcus sp. strain PCC7942 Is a DNA-binding hemoprotein. J. Biol. Chem. 270, 22478–22482 10.1074/jbc.270.38.22478 [DOI] [PubMed] [Google Scholar]
- 16. Peña M. M., Burkhart W., and Bullerjahn G. S. (1995) Purification and characterization of a Synechococcus sp. strain PCC 7942 polypeptide structurally similar to the stress-induced Dps/PexB protein of Escherichia coli. Arch. Microbiol. 163, 337–344 10.1007/BF00404206 [DOI] [PubMed] [Google Scholar]
- 17. Li H., Singh A. K., McIntyre L. M., and Sherman L. A. (2004) Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 186, 3331–3345 10.1128/JB.186.11.3331-3345.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shcolnick S., Summerfield T. C., Reytman L., Sherman L. A., and Keren N. (2009) The mechanism of iron homeostasis in the unicellular cyanobacterium Synechocystis sp. PCC 6803 and its relationship to oxidative stress. Plant Physiol. 150, 2045–2056 10.1104/pp.109.141853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Castruita M., Saito M., Schottel P. C., Elmegreen L. A., Myneni S., Stiefel E. I., and Morel F. M. M. (2006) Overexpression and characterization of an iron storage and DNA-binding Dps protein from Trichodesmium erythraeum. Appl. Environ. Microbiol. 72, 2918–2924 10.1128/AEM.72.4.2918-2924.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato N., Moriyama T., Toyoshima M., Mizusawa M., and Tajima N. (2012) The all0458/lti46.2 gene encodes a low temperature-induced Dps protein homologue in the cyanobacteria Anabaena sp. PCC 7120 and Anabaena variabilis M3. Microbiology 158, 2527–2536 10.1099/mic.0.060657-0 [DOI] [PubMed] [Google Scholar]
- 21. Moparthi V. K., Li X., Vavitsas K., Dzhygyr I., Sandh G., Magnuson A., and Stensjö K. (2016) The two Dps proteins, NpDps2 and NpDps5, are involved in light-induced oxidative stress tolerance in the N2-fixing cyanobacterium Nostoc punctiforme. Biochim. Biophys. Acta 1857, 1766–1776 10.1016/j.bbabio.2016.08.003 [DOI] [PubMed] [Google Scholar]
- 22. Schmidt R., Zahn R., Bukau B., and Mogk A. (2009) ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol. Microbiol. 72, 506–517 10.1111/j.1365-2958.2009.06666.x [DOI] [PubMed] [Google Scholar]
- 23. Grant R. A., Filman D. J., Finkel S. E., Kolter R., and Hogle J. M. (1998) The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 5, 294–303 10.1038/nsb0498-294 [DOI] [PubMed] [Google Scholar]
- 24. Chowdhury R. P., Vijayabaskar M. S., Vishveshwara S., and Chatterji D. (2008) Molecular mechanism of in vitro oligomerization of Dps from Mycobacterium smegmatis: mutations of the residues identified by “interface cluster” analysis. Biochemistry 47, 11110–11117 10.1021/bi801158e [DOI] [PubMed] [Google Scholar]
- 25. Fan R., Boyle A. L., Cheong V. V., Ng S. L., and Orner B. P. (2009) A helix swapping study of two protein cages. Biochemistry 48, 5623–5630 10.1021/bi900387t [DOI] [PubMed] [Google Scholar]
- 26. Bellapadrona G., Stefanini S., Zamparelli C., Theil E. C., and Chiancone E. (2009) Iron translocation into and out of Listeria innocua Dps and size distribution of the protein-enclosed nanomineral are modulated by the electrostatic gradient at the 3-fold “ferritin-like” pores. J. Biol. Chem. 284, 19101–19109 10.1074/jbc.M109.014670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Almirón M., Link A. J., Furlong D., and Kolter R. (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6, 2646–2654 10.1101/gad.6.12b.2646 [DOI] [PubMed] [Google Scholar]
- 28. Belkin S., Mehlhorn R. J., and Packer L. (1987) Proton gradients in intact cyanobacteria. Plant Physiol. 84, 25–30 10.1104/pp.84.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hinterstoisser B., and Peschek G. A. (1987) Fluorimetric pH measurement in whole cells of dark aerobic and anaerobic cyanobacteria. FEBS Lett. 217, 169–173 10.1016/0014-5793(87)80657-9 [DOI] [Google Scholar]
- 30. Falkner G., and Horner F. (1976) pH changes in the cytoplasm of the blue-green alga Anacystis nidulans caused by light-dependent proton flux into the thylakoid space. Plant Physiol. 58, 717–718 10.1104/pp.58.6.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Durham K. A., and Bullerjahn G. S. (2002) Immunocytochemical localization of the stress-induced DpsA protein in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Basic Microbiol. 42, 367–372 10.1002/1521-4028(200212)42:6%3C367::AID-JOBM367%3E3.0.CO%3B2-T [DOI] [PubMed] [Google Scholar]
- 32. Hahn A., Stevanovic M., Brouwer E., Bublak D., Tripp J., Schorge T., Karas M., and Schleiff E. (2015) Secretome analysis of Anabaena sp. PCC 7120 and the involvement of the TolC-homologue HgdD in protein secretion. Environ. Microbiol. 17, 767–780 10.1111/1462-2920.12516 [DOI] [PubMed] [Google Scholar]
- 33. Oliveira P., Martins N. M., Santos M., Couto N. A. S., Wright P. C., and Tamagnini P. (2015) The Anabaena sp. PCC 7120 exoproteome: taking a peek outside the box. Life (Basel) 5, 130–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de los Ríos A., Ascaso C., Wierzchos J., Vincent W. F., and Quesada A. (2015) Microstructure and cyanobacterial composition of microbial mats from the High Arctic. Biodivers. Conserv. 24, 841–863 10.1007/s10531-015-0907-7 [DOI] [Google Scholar]
- 35. Theoret J. R., Cooper K. K., Zekarias B., Roland K. L., Law B. F., Curtiss R. 3rd, and Joens L. A. (2012) The Campylobacter jejuni Dps homologue is important for in vitro biofilm formation and cecal colonization of poultry and may serve as a protective antigen for vaccination. Clin. Vaccine Immunol. 19, 1426–1431 10.1128/CVI.00151-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gupta S., and Chatterji D. (2003) Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J. Biol. Chem. 278, 5235–5241 10.1074/jbc.M208825200 [DOI] [PubMed] [Google Scholar]
- 37. Ceci P., Ilari A., Falvo E., Giangiacomo L., and Chiancone E. (2005) Reassessment of protein stability, DNA binding, and protection of Mycobacterium smegmatis Dps. J. Biol. Chem. 280, 34776–34785 10.1074/jbc.M502343200 [DOI] [PubMed] [Google Scholar]
- 38. Nguyen K. H., and Grove A. (2012) Metal binding at the Deinococcus radiodurans Dps-1 N-terminal metal site controls dodecameric assembly and DNA binding. Biochemistry 51, 6679–6689 10.1021/bi300703x [DOI] [PubMed] [Google Scholar]
- 39. Morikawa K., Ohniwa R. L., Kim J., Maruyama A., Ohta T., and Takeyasu K. (2006) Bacterial nucleoid dynamics: oxidative stress response in Staphylococcus aureus. Genes Cells 11, 409–423 10.1111/j.1365-2443.2006.00949.x [DOI] [PubMed] [Google Scholar]
- 40. Martinez A., and Kolter R. (1997) Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179, 5188–5194 10.1128/jb.179.16.5188-5194.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kauko A., Pulliainen A. T., Haataja S., Meyer-Klaucke W., Finne J., and Papageorgiou A. C. (2006) Iron incorporation in Streptococcus suis Dps-like peroxide resistance protein Dpr requires mobility in the ferroxidase center and leads to the formation of a ferrihydrite-like core. J. Mol. Biol. 364, 97–109 10.1016/j.jmb.2006.08.061 [DOI] [PubMed] [Google Scholar]
- 42. Yamamoto Y., Poole L. B., Hantgan R. R., and Kamio Y. (2002) An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J. Bacteriol. 184, 2931–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ceci P., Di Cecca G., Falconi M., Oteri F., Zamparelli C., and Chiancone E. (2011) Effect of the charge distribution along the “ferritin-like” pores of the proteins from the Dps family on the iron incorporation process. J. Biol. Inorg. Chem. 16, 869–880 10.1007/s00775-011-0784-9 [DOI] [PubMed] [Google Scholar]
- 44. Cuypers M. G., Mitchell E. P., Romão C. V., and McSweeney S. M. (2007) The crystal structure of the Dps2 from Deinococcus radiodurans reveals an unusual pore profile with a non-specific metal binding site. J. Mol. Biol. 371, 787–799 10.1016/j.jmb.2006.11.032 [DOI] [PubMed] [Google Scholar]
- 45. Campbell E. L., Summers M. L., Christman H., Martin M. E., and Meeks J. C. (2007) Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 189, 5247–5256 10.1128/JB.00360-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Christman H. D., Campbell E. L., and Meeks J. C. (2011) Global transcription profiles of the nitrogen stress response resulting in heterocyst or hormogonium development in Nostoc punctiforme. J. Bacteriol. 193, 6874–6886 10.1128/JB.05999-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gibson D. G. (2011) Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349–361 10.1016/B978-0-12-385120-8.00015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 49. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., and Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 10.1002/elps.200305844 [DOI] [PubMed] [Google Scholar]
- 50. Shevchenko A., Wilm M., Vorm O., and Mann M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- 51. Fujisawa T., Narikawa R., Maeda S. I., Watanabe S., Kanesaki Y., Kobayashi K., Nomata J., Hanaoka M., Watanabe M., Ehira S., Suzuki E., Awai K., and Nakamura Y. (2017) CyanoBase: a large-scale update on its 20th anniversary. Nucleic Acids Res. 45, D551–D554 10.1093/nar/gkw1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., and Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modelling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rocchia W., Alexov E., and Honig B. (2001) Extending the applicability of the nonlinear Poisson-Boltzmann equation: multiple dielectric constants and multivalent ions. J. Phys. Chem. B 105, 6507–6514 10.1021/jp010454y [DOI] [Google Scholar]
- 56. Artiss J. D., Vinogradov S., and Zak B. (1981) Spectrophotometric study of several sensitive reagents for serum iron. Clin. Biochem. 14, 311–315 10.1016/S0009-9120(81)91065-1 [DOI] [PubMed] [Google Scholar]
- 57. Ardini M., Fiorillo A., Fittipaldi M., Stefanini S., Gatteschi D., Ilari A., and Chiancone E. (2013) Kineococcus radiotolerans Dps forms a heteronuclear Mn-Fe ferroxidase center that may explain the Mn-dependent protection against oxidative stress. Biochim. Biophys. Acta 1830, 3745–3755 10.1016/j.bbagen.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 58. Hennessy D. J., Reid G. R., Smith F. E., and Thompson S. L. (1984) Ferene: a new spectrophotometric reagent for iron. Can. J. Chem. 62, 721–724 10.1139/v84-121 [DOI] [Google Scholar]
- 59. Smith F. E., Herbert J., Gaudin J., Hennessy D. J., and Reid G. R. (1984) Serum iron determination using ferene triazine. Clin. Biochem. 17, 306–310 10.1016/S0009-9120(84)90613-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.