Abstract
The RE1-silencing transcription factor (REST) is the major scaffold protein for assembly of neuronal gene silencing complexes that suppress gene transcription through regulating the surrounding chromatin structure. REST represses neuronal gene expression in stem cells and non-neuronal cells, but it is minimally expressed in neuronal cells to ensure proper neuronal development. Dysregulation of REST function has been implicated in several cancers and neurological diseases. Modulating REST gene silencing is challenging because cellular and developmental differences can affect its activity. We therefore considered the possibility of modulating REST activity through its regulatory proteins. The human small C-terminal domain phosphatase 1 (SCP1) regulates the phosphorylation state of REST at sites that function as REST degradation checkpoints. Using kinetic analysis and direct visualization with X-ray crystallography, we show that SCP1 dephosphorylates two degron phosphosites of REST with a clear preference for phosphoserine 861 (pSer-861). Furthermore, we show that SCP1 stabilizes REST protein levels, which sustains REST's gene silencing function in HEK293 cells. In summary, our findings strongly suggest that REST is a bona fide substrate for SCP1 in vivo and that SCP1 phosphatase activity protects REST against degradation. These observations indicate that targeting REST via its regulatory protein SCP1 can modulate its activity and alter signaling in this essential developmental pathway.
Keywords: phosphatase, X-ray crystallography, post-transcriptional regulation, gene silencing, brain-derived neurotrophic factor (BDNF), neurodifferentiation, CTD small phosphatase 1 (CTDSP1), nuclear phosphatase
Introduction
The RE1-silencing transcription factor (REST)4 (1), also known as the neuron-restrictive silencer factor (NRSF) (2), is an important transcriptional regulator that silences neuronal gene expression. REST recognizes a DNA motif called the RE1 element found near target genes and recruits several critical transcriptional co-regulators to silence their expression (3). It has been estimated that expression of up to 10% of neuronal genes are subject to regulation by REST (4). REST helps maintain self-renewal and pluripotency of stem cells via cross-talk with key transcriptional regulators of cell fate determination (5). This transcriptional communication requires high levels of REST during embryonic development to prevent inappropriate terminal differentiation of stem cells into neurons (6–8).
Dysregulation of REST has been implicated in multiple diseases. REST function is critical in embryonic development as elimination of REST leads to neonatal death in mice (9). REST protein levels are significantly up-regulated in several cases of mesenchymal and classical glioma tumors (10). In glioblastomas exhibiting high levels of REST expression, REST appears to be a key factor in maintaining tumor growth as knockdown of REST results in shrinkage of these tumors (10, 11). In addition, up-regulation of REST has been implicated in other brain tumors such as medulloblastomas (12) and neuroblastomas (13). However, in non-neuronal tumors, such as epithelial-derived tumors, REST activity is suppressed possibly because of its potential tumor suppressor function (14, 15). The contradictory role of REST in both oncogenesis and tumor suppression in different cell types suggests its role in gene regulation and cellular phenotype is highly context-dependent.
Maintaining appropriate levels of REST target gene suppression is important for the fitness of cells even in the brain. Basal expression of REST in mature neurons is generally low, but in certain regions of the brain REST levels increase upon aging, whereas a deficiency of REST in these regions is associated with Alzheimer's disease (16). A molecular explanation for this observation is that REST silences genes that promote apoptosis, and as certain brain cells age these genes can become active when REST levels do not rise to maintain silencing and lead to cell death (16). Abnormal expression of REST has also been observed in other neurological diseases including Parkinson's (17), epilepsy (18), and Huntington's disease (19–21), but how changes in REST levels can promote these diseases remains unclear.
The difficulty in studying the molecular mechanisms of REST in stem cells and disease states lies in the lack of effective tools to manipulate REST activity. Genetic tools like shRNA have been frequently used to determine the effects of manipulating REST levels on RE1 gene expression and cellular phenotype (8, 15, 22). Small molecule inhibitors would provide the advantage of instantaneous temporal control, reversibility, and ease of application. Unfortunately, transcription factors like REST, whose activity depends on their ability to participate in protein-DNA and protein-protein interactions, are difficult to target with small molecules. Therefore, the regulatory enzymes of REST represent potential alternative targets to control REST function. In cells, REST has a high turnover rate mediated by phosphorylation of its C-terminal domain that targets it for proteasomal degradation via Skp1-Cul1-F-box β-transducin repeat containing E3 ubiquitin ligase (SCFβ-TrCP)-dependent ubiquitination (23). Cell-focused studies suggest that a human phosphatase, the small C-terminal domain phosphatase 1 (SCP1), promotes REST function through regulation of this pathway (24–26). First, SCP1 was found to be expressed in similar cell types as REST and co-immunoprecipitation experiments indicates they physically associate within cells (24). Furthermore, a positive feedback loop has been identified between SCP1 and REST that discloses a close partnership in their functions (25). Expression of SCP1 in cells results in removal of phosphate marks from a degron region at the C terminus of REST, stabilizing the protein and protecting it from degradation (26). Altogether, there seems to be a strong correlation between the phosphatase activity of SCP1 and the gene silencing function of REST.
In this paper, we demonstrate that SCP1 can enzymatically dephosphorylate the Ser-861 and Ser-864 degron phosphosites of REST and quantify rates of dephosphorylation of each site to determine the preferred substrate for SCP1. We also visualized the physical interactions between these proteins by determining the complex structures of phosphorylated REST peptides bound at the active site of SCP1. Extending our analysis to mammalian cells, we find that reduction of SCP1 with shRNA or overexpression of a dominant-negative SCP1 leads to a decrease in REST protein levels. This reduction in REST protein results in increased expression of genes normally repressed by REST. Overall, our data shows that the phosphatase activity of SCP1 controls REST protein stability. Therefore, control of SCP1 enzymatic activity represents a promising avenue to control REST function.
Results
Kinetic characterization of SCP1 dephosphorylating REST
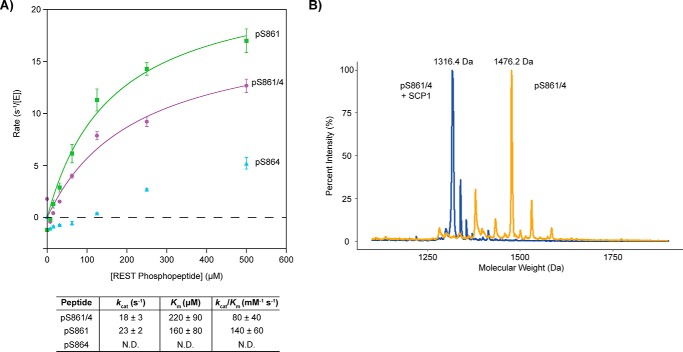
In cells, overexpression of SCP1 increases REST dephosphorylation (26). To test if SCP1 can directly dephosphorylate REST, we obtained 12-mer synthetic peptides with the sequenceEDLSPPSPPLPK from the C-terminal region of REST(858–869), where two degron sites are located (23). The peptides are synthetically defined and phosphorylated at Ser-861 (REST–pS861 peptide), Ser-864 (REST–pS864 peptide), or both (REST-pS861/4 peptide; Table S1). The phosphatase activities of SCP1 against these peptides were quantified by malachite green assays (27) (Fig. 1A). The kcat of SCP1 against the REST–pS861 peptide was 23 ± 2 s−1 and the Km was measured to be 160 ± 40 μm (kcat/Km of 140 ± 60 mm−1 s−1) (Fig. 1A). On the other hand, SCP1 had weak activity when only Ser-864 is phosphorylated (Fig. 1A). These results demonstrate that pSer-861 of REST is the preferred dephosphorylation site for SCP1 compared with pSer-864.
Figure 1.
Dephosphorylation of REST(858–869)-phosphorylated peptides by SCP1. A, SCP1 dephosphorylation rates against the pSer-861 (green), pSer-864 (light blue), and pSer-861/864 (purple). REST peptide reactions were determined in triplicate with error bars indicating S.D. between rate values from the independent reactions. The specificity constants (kcat/Km) from fitting the pSer-861 (green) and pSer-861/864 (purple) curves are indicated in the table below. Kinetic parameters for pSer-864 (light blue) were not determined (N.D.) due to low activity. B, MALDI-TOF spectra of the pSer-861/864 REST peptide untreated (orange) or treated with SCP1 (blue). Mass labels have been indicated for the peaks of maximum intensity in each sample. The predicted molecular weight of the pSer-861/864 peptide is 1477.5 Da, which corresponds to the major peak of the MALDI trace for the untreated peptide (1476.2 Da). There is a loss of 159.8 Da in the major peak of the SCP1-treated reaction compared with the major peak in the untreated reaction (molecular mass of PO3 is ∼80 Da).
When both Ser-861 and Ser-864 are phosphorylated, SCP1 still shows strong phosphatase activity (kcat/Km of is determined to be 80 ± 40 mm−1 s−1). To test whether both phosphorylations are removed by SCP1, we incubated SCP1 with the doubly phosphorylated Ser-861/864 REST peptide and monitored the reaction with matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) MS. MALDI-TOF analysis clearly showed a size difference of ∼160 Da between the major peaks in the untreated and SCP1-treated reactions indicating a loss of two phosphate groups (Fig. 1B). Dephosphorylation of the doubly phosphorylated REST peptide was specific to SCP1 as a single point mutation in SCP1 (D96N) abolishes activity (Fig. S1). Overall, these results clearly indicate that SCP1 can dephosphorylate REST at its C-terminal domain at both Ser-861 and Ser-864, with Ser-861 as the preferred site. Because Ser-861 appears to be the primary site for SCP1 dephosphorylation and mutation of both Ser-861 and Ser-864 sites to alanine abolishes interaction with REST (26), we suspect that phosphorylation of Ser-861 plays a key role in recruiting SCP1 and tethers it to the substrate to dephosphorylate both sites.
X-ray crystal structures of SCP1 bound to phosphorylated REST peptides
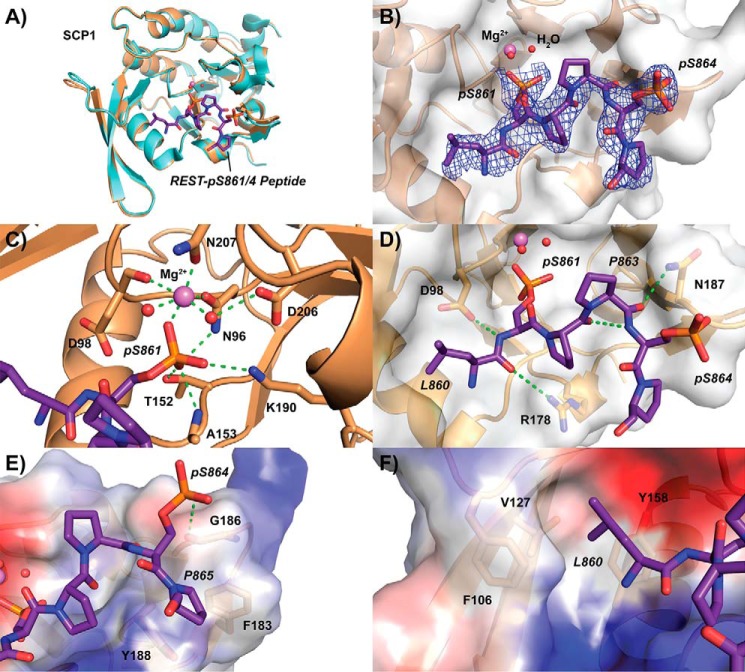
To directly visualize the molecular interaction between REST and SCP1, we crystalized SCP1 in complex with the phosphorylated peptides derived from the C-terminal domain of REST (Fig. 2). Because SCP1 is active against REST peptides as shown in the kinetic assays, we used a catalytically deficient variant of SCP1 in which the nucleophilic aspartate 96 is replaced by asparagine (D96N) to prevent phosphoryl adduct formation (28). This D96N variant allows for effective substrate recognition but prevents nucleophilic attack required for phosphate transfer (27). We obtained SCP1 D96N variant crystals and soaked the phosphorylated REST peptides into the pre-formed crystals. To identify the primary site of recognition using an unbiased approach, we first tried to obtain the complex structure of SCP1 D96N with the doubly phosphorylated Ser-861/864 REST peptide by soaking. The diffraction data were collected in the synchrotron at a resolution of 2.5 Å (Table 1).
Figure 2.
Crystal structure of SCP1 bound to REST pSer-861/864 peptide. A, overall structure of SCP1 (sand) when bound to REST peptide (purple) compared with the apo structure (light blue). Magnesium (magenta) and active site waters (red) are represented as spheres. B, electron density of refined pSer-861/864 bound peptide. The 2mFo − DFc map representing electron density is contoured to 1σ and illustrated as a blue mesh around the REST peptide. The Van der Waals exterior of SCP1 is represented as a transparent gray surface. C, phosphoserine-binding site of SCP1 occupied by pSer-861 of the doubly phosphorylated REST peptide. Hydrogen bonding in the magnesium coordination pocket is represented as dotted green lines. D, tight hairpin configuration of the pSer-861/864 peptide stabilized by internal and external hydrogen bonding. E, binding of the C-terminal residues of the doubly phosphorylated REST peptide to the periphery of the SCP1 pocket. Surface electrostatic potentials are denoted by color (blue, positive; red, negative; gray, neutral). F, hydrophobic pocket binding to the side chain of Leu-860 of the REST peptide.
Table 1.
X-ray crystallography data collection and refinement statistics
| SCP1 + REST-pSer-861/864 peptide | SCP1 + REST-pSer-861 peptide | |
|---|---|---|
| PDB code | 6DU2 | 6DU3 |
| Data collection | ||
| Wavelength (Å) | 0.97648 | 0.97648 |
| Space group | C1 2 1 | C1 2 1 |
| Cell dimensions | ||
| a, b, c (Å) | 125.3, 78.6, 62.9 | 125.4, 78.7, 63.0 |
| α, β, γ (°) | 90.0, 112.1, 90.0 | 90.0, 112.4, 90.0 |
| Resolution (Å) | 50.0–2.50 (2.54–2.50)a | 50.0–2.57 (2.61–2.57)a |
| No. of unique reflections | 19,286 | 17,864 |
| I/σ (I) | 14.3 (1.7) | 14.2 (1.7) |
| Completeness (%) | 98.5 (84.3) | 98.0 (76.6) |
| Redundancy | 3.6 (3.0) | 3.7 (3.0) |
| Rsym | 0.104 (0.486) | 0.082 (0.384) |
| Refinement | ||
| Resolution (Å) | 39.46–2.50 (2.59–2.50) | 39.2–2.58 (2.67–2.58) |
| No. of reflections (test set) | 19268 (1692) | 17864 (1720) |
| Rwork/Rfree (%) | 0.1937/0.2422 | 0.1877/0.2331 |
| No. of atoms | ||
| Protein | 2,908 | 29,06 |
| Mg2+ | 2 | 2 |
| Ligand | 91 | 90 |
| Water | 10 | 18 |
| B-factors (Å2) | ||
| Protein | 47.57 | 37.36 |
| Mg2+ | 47.02 | 37.67 |
| Ligand | 75.38 | 66.88 |
| Water | 44.20 | 32.94 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles (°) | 1.10 | 1.30 |
a Highest resolution shell is shown in parentheses.
Compared with the previously solved apo-SCP1 structure (29), the overall fold of SCP1 shows very few changes upon binding to the doubly phosphorylated REST peptide (Fig. 2A). However, strong positive density for the serine–proline motif was observed at the active site of SCP1, indicating incorporation of the ligand (Fig. S2). This allowed us to confidently build six of the 12 residues of the REST peptide into the electron density (Fig. 2B). Iterative building of the peptide into experimental density made it clear that the preferred phosphoserine at SCP1's active site was pSer-861, which was consistent with our kinetic results showing that SCP1 highly favors pSer-861 over pSer-864. The phosphate group of pSer-861 replaces a water molecule to coordinate a magnesium ion bound at the active site (Fig. 2C). The magnesium ion is coordinated in a dipyrimidal mode with the two conserved aspartate residues from the catalytic DXDXE motif (Asn-96 in the structure) along with Asn-207 from atop and two water molecules to each side (Fig. 2C). In addition to coordinating the magnesium ion, pSer-861 also forms extensive interactions with SCP1. The phosphate group is anchored through hydrophilic interaction with the side chains of Asn-96, Lys-190, and Thr-152 as well as the amide group of Ala-153 (Fig. 2C). The N- and C-terminal residues of the peptide show little density but would seem to extend toward bulk solvent.
The doubly phosphorylated REST peptide forms a tight hairpin around Pro-863, which allows the peptide to conform to the binding pocket of SCP1 (Fig. 2D). An intramolecular hydrogen bond between the carbonyl of Pro-862 and the amide group of pSer-864 appears to stabilize this hairpin structure (Fig. 2D). Furthermore, hydrogen bonding between the carbonyl of Pro-863 and the side chain of Asn-187 (Fig. 2D) as well as phosphate group of pSer-864 and the carbonyl of Gly-186 (Fig. 2E) seem to stabilize the binding mode of the peptide. Pro-865 also sits over a hydrophobic surface at the edge of the SCP1-binding pocket formed by the aromatic side chains of Phe-183 and Tyr-188 (Fig. 2E).
The residues preceding the target phosphoserine of SCP1 substrates have previously been found to be important for recognition by fitting into a unique hydrophobic pocket (27). Indeed, Leu-860 of the REST peptide forms favorable hydrophobic interactions with Phe-106, Leu-127, and Tyr-158 of the SCP1 hydrophobic pocket (Fig. 2F). Tyr-158 of SCP1 is essential for binding of the C-terminal domain of RNA polymerase II (Fig. S3) as replacing this residue with alanine is sufficient to eliminate SCP1's substrate specificity without affecting enzymatic activity (27). Furthermore, this pocket is also critical for SCP1 recognition of the small molecular inhibitor rabeprazole (30) as its sulfoxide group sits over Tyr-158 (Fig. S3). Overall, the structure of SCP1 bound to the doubly phosphorylated REST peptide supports our hypothesis that pSer-861 is the preferred dephosphorylation site and suggests that SCP1 specificity toward REST is dominated by hydrophobic interactions on either side of phosphorylated substrate residue.
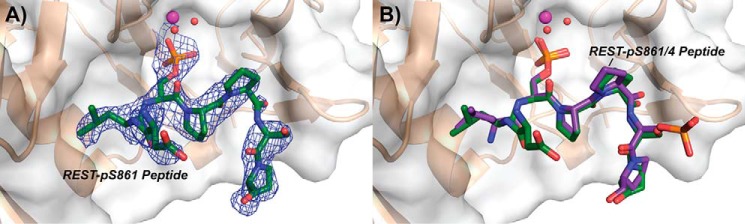
We next attempted to obtain complex structures of SCP1 D96N with REST peptides phosphorylated either at Ser-861 or Ser-864. The conditions used to obtain these protein–ligand complexes were kept highly consistent to make the resultant structures as comparable as possible. The structure of SCP1 bound to the pSer-861 peptide was determined at a resolution of 2.55 Å, which was comparable with the doubly phosphorylated peptide structure (Table 1). The pSer-861 peptide showed strong positive density for the SP motif at the active site (Fig. S2) and could be reliably built into the density (Fig. 3A). An additional residue at the N-terminal end of the pSer-861 peptide, Asp-859, can be built into the density compared with the pSer-861/864 doubly phosphorylated REST peptide (Fig. 3A). The pSer-861 and pSer-861/864 REST peptides shared an almost identical binding mode in SCP1 (Fig. 3B), which is consistent with our kinetics showing that SCP1 prefers recognition of the pSer-861 site compared with the pSer-864 site.
Figure 3.
Crystal structure of SCP1 bound to singly phosphorylated pSer-861 REST peptide. A, structure of SCP1 (sand) bound to pSer-861 peptide (green). Magnesium (magenta) and active site waters (red) are represented as spheres. The 2mFo − DFc map representing electron density is contoured to 1σ and illustrated as a blue mesh around the REST peptide. The Van der Waals exterior of SCP1 is represented as a transparent gray surface. B, binding mode comparison between the pSer-861 and pSer-861/864 (purple) REST peptides.
Despite using identical soaking conditions and obtaining similar resolution data with the pSer-864 peptide, we observed very little positive density for the SP motif let alone the rest of the peptide in any of our data sets even after prolonged soaking. It is surprising that pSer-864 is a much poorer substrate for SCP1 dephosphorylation considering the hydrophobic residues surrounding Ser-864 (Pro-Pro-Ser864-Pro-Pro), which would seem to be favored by the hydrophobic binding pockets close to the SCP1 active site. The shorter side chain of Pro-863 may contribute to this reduced affinity for pSer-864 at the dephosphorylation site, as Leu-860 extends to make extensive hydrophobic contacts in the pSer-861–bound structure (Fig. 2F). Additionally, we suspect that the highly restricted configuration of proline residues on both sides of the pSer-864 site limit flexibility of the pSer-864 peptide, making it a poor substrate for SCP1 association. The high overall flexibility of the C-terminal region of REST may, however, help SCP1 dephosphorylate both target sites, despite pSer-864 being a much poorer substrate compared with pSer-861. Altogether, our structural studies support the kinetic results that pSer-861 of REST is the primary site of SCP1 dephosphorylation and identify structural elements important for the interaction between SCP1 and REST.
SCP1 phosphatase activity maintains the protein levels of REST
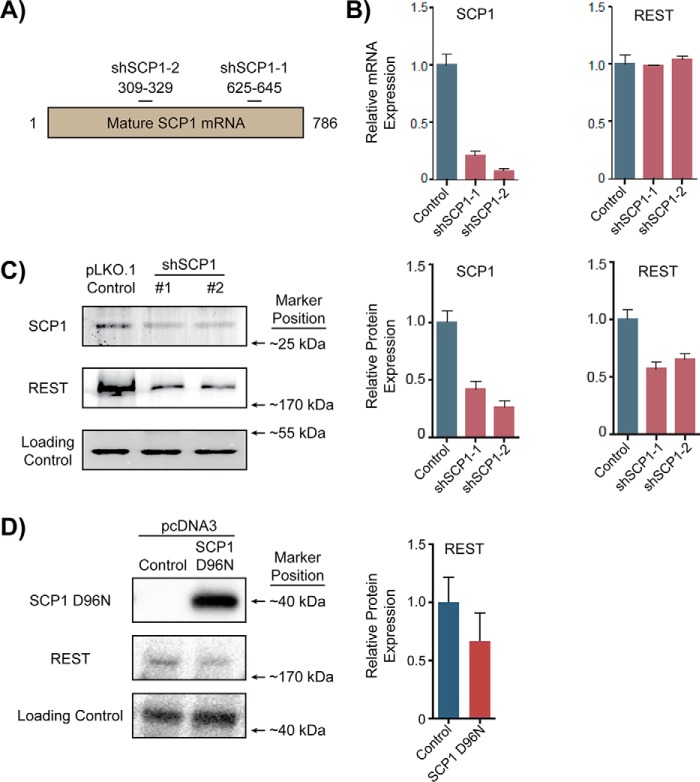
Kinetic and structural analysis of the interaction between SCP1 and the phosphorylated REST peptides elucidated the biophysical characteristics of SCP1-mediated REST dephosphorylation. To understand the biological effect of SCP1 dephosphorylation of REST, we evaluated how SCP1 phosphatase activity affects REST function in cells. First, we knocked down expression of SCP1 in cells and examined the effect on REST mRNA and protein levels. HEK293T cells were transfected with pLKO.1 plasmids that express two commercially availableshRNAs, denoted as shSCP1-1 and shSCP1-2, targeting different regions of the SCP1 mRNA (Fig. 4A and Table S2). We then infected HEK293 cells with the shRNA harboring lentiviruses. Compared with the cells infected with an empty plasmid, knockdown with either shRNA plasmid resulted in greater than 80% reduction in SCP1 mRNA levels (Fig. 4B). Both shSCP1 knockdown plasmids reduced SCP1 protein levels to lessthan 50% compared with the control cells (Fig. 4C). Although both SCP1 mRNA and protein levels dropped in response to shSCP1 transfection, REST mRNA levels remained relatively unchanged (Fig. 4B). On the other hand, we observed ∼40% reductions in REST protein levels with either shSCP1 plasmid (Fig. 4C).
Figure 4.
Effect on REST levels with shSCP1 knockdown or overexpression of SCP1 D96N in HEK293 cells. A, target sites on SCP1 mRNA for shSCP1-1 and shSCP1-2 knockdown. ShSCP1 harboring pLKO.1 plasmids were introduced into cells to reduce SCP1 expression. B, qRT-PCR analysis of SCP1 and REST expression with shSCP1 knockdown vectors. Expression of SCP1 and REST in cells with a pLKO.1 hygromycin vector lacking a shRNA coding region was used as control for comparison. SCP1 and REST mRNA levels (red) were normalized to ACTB mRNA levels (blue) in each cell treatment. qRT-PCR analysis of the knockdown samples were run in biological triplicates with error bars representing S.D. between relative expression values. C, Western blot analysis of SCP1 and REST protein levels with shSCP1 knockdown vectors. ACTB protein levels were used as a loading control for shRNA-treated cell samples. Protein samples from cell extracts were run in triplicate and analyzed by t test using the hygromycin vector levels as a control. The error bars represent S.D. between relative expression values. D, Western blot analysis of SCP1 and REST protein levels with SCP1 D96N overexpression vector. β-Tubulin was used as a loading control for protein samples from SCP1 D96N-transfected cells. Protein samples were run in biological triplicates and compared with the hygromycin control vector.
Knockdown of SCP1 results in reduction of REST protein levels, whereas REST mRNA levels remain relatively constant. Furthermore, REST protein is dephosphorylated by SCP1. Both lines of evidence point toward a regulatory mechanism that SCP1 regulates REST at the post-translational level. We therefore investigated whether SCP1 is important for regulating REST protein levels due to its enzymatic activity. To test whether SCP1 enzymatic activity was required for regulating REST protein levels, we transfected cells with a plasmid expressing a phosphatase activity deficient variant of SCP1 (SCP1 D96N). We predicted that SCP1 D96N would compete with endogenous SCP1 for binding of phosphorylated REST and thus act as a dominant-negative by allowing more REST molecules to remain phosphorylated and be targeted by the cellular degradation machinery. Indeed, overexpression of SCP1 D96N resulted in an ∼40% reduction of REST protein levels (Fig. 4D and Fig. S4), suggesting that SCP1-directed dephosphorylation of REST is important for maintenance of REST levels in cells. This line of evidence supports previous results suggesting that SCP1 dephosphorylation protects REST from degradation (26). These results support a model of SCP1 directly dephosphorylating REST to increase its cellular stability and promote its activity, whereas reduction of SCP1 phosphatase activity diminishes REST protein abundance and function in cells.
Knockdown of SCP1 leads to induction of REST-regulated genes
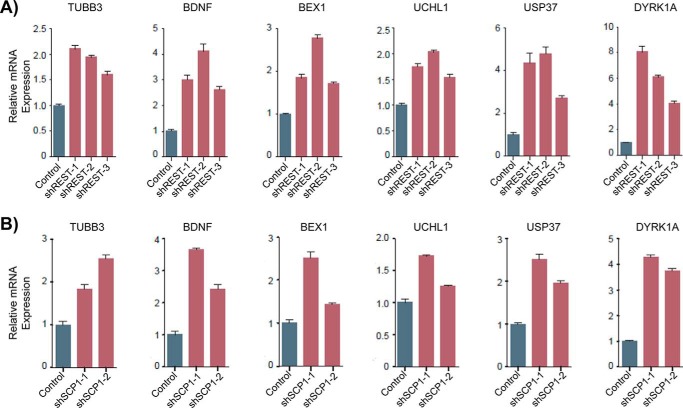
Approximately 2000 genomic loci containing RE1 elements are targeted by REST in non-neuronal cells, many of which are located near genes involved in neuronal development (31, 32). To determine whether modulating SCP1 is sufficient to control REST function in gene silencing, we quantified the expression of genes subject to REST control when SCP1 expression is suppressed. Tubulin β3 (TUBB3, gene ID 10381) has been reported in multiple cell types to be controlled by REST and therefore was used as a positive control in our analysis (33–35). We also chose several genes that are reported to be REST-regulated and have implications in different disease states. Two tumor suppressors called ubiquitin-specific peptidase 37 (USP37, gene ID 57695) and brain-expressed X-linked 1 (BEX1, gene ID 55859) are down-regulated in medulloblastomas (36) and glioblastomas (37) alongside increased REST levels. Expression levels of the brain-derived neurotropic factor (BDNF, gene ID 627) and ubiquitin C-terminal hydrolase L1 (UCHL1, gene ID 7345) are critical for neurodevelopment and dysregulation of either of their genes has been implicated in multiple neurological diseases (38, 39). We also monitored the expression of the dual specificity tyrosine phosphorylation regulated kinase 1A (DYRK1A, gene ID 1859), whose change in expression has been associated with various tumors (40). All six of these genes are associated with a nearby RE1 element and have been reported to be subject to REST silencing.
To test whether reducing REST function is sufficient to activate expression of our target genes, we infected HEK293 cells with lentiviruses that express shRNA against REST and monitored changes in gene expression. REST protein levels dropped below 60% with each of the three shRNAs against REST that we tested (Fig. S5). We used qRT-PCR to monitor the mRNA levels of the six genes selected (Fig. 5 and Table S3). Knockdown of REST increased the expression of TUBB3 by 1.5–2-fold (Fig. 5A), indicating that our REST knockdown system was indeed affecting the expression of REST-regulated genes. We also observed increases in mRNA levels for each of the other five genes tested. DYRK1A exhibited the strongest activation in expression with 4–8-fold higher levels of mRNA after REST knockdown (Fig. 5A). BDNF and USP37 were also strongly induced (2.5–5-fold increase), whereas BEX1 and UCHL1 exhibited moderate increases in expression (1.5–3-fold increase; Fig. 5A). These results show that reduction of REST protein levels indeed leads to induction of REST-silenced genes in HEK293 cells.
Figure 5.
SCP1 activity can modulate the expression level of genes silenced by REST. A, qRT-PCR analysis of REST-regulated gene expression with shREST knockdown vectors. ACTB gene expression was used to normalize mRNA quantification of REST-regulated genes in the differently treated cells. Expression of each gene was quantified in biological triplicates and compared with expression in cells with a pLKO.1 hygromycin vector control, with error bars representing S.D. between relative expression values. B, qRT-PCR analysis of REST-regulated gene expression with shSCP1 knockdown vectors.
After confirming that our test set of genes are under REST control in HEK293 cells, we next asked whether indirectly reducing REST function through elimination of SCP1 could activate the same genes. Therefore, we tested whether knockdown of SCP1 is sufficient to induce expression of this same set of RE1 element-containing genes. We detected an ∼2–2.5-fold increase in mRNA of TUBB3 with our two shSCP1 vectors (Fig. 5B), indicating that targeting SCP1 is a viable method for affecting REST-mediated gene silencing. Furthermore, SCP1 knockdown resulted in up-regulation of ∼3.5–4-fold for DYRK1A, 2.5–3.5-fold for BDNF, 2–2.5-fold for USP37, 1.5–2.5-fold for BEX1, and 1.3–1.7-fold for UCHL1 (Fig. 5B). Knockdown of REST or SCP1 has similar consequences for REST-regulated genes, supporting our hypothesis that SCP1 controls gene expression by modulating REST protein abundance and function. Our in vitro and cell-based experiments revealed both the physical characteristics and biological implications of the REST/SCP1 interaction. These results strongly suggest that controlling SCP1 phosphatase activity is sufficient to alter REST protein levels and modulate expression of genes targeted by REST.
Discussion
Current evidence supports the notion that REST is a bona fide substrate for SCP1, whose phosphatase activity enhances the cellular function of REST. This conclusion is supported by three independent lines of evidence: 1) evidence of SCP1 and REST colocalization in cells, 2) in vivo SCP1 phosphatase activity toward REST, and 3) in vitro SCP1 phosphatase activity against REST. For the first line, SCP1 has been found to colocalize with REST during development using in situ hybridization and has been identified as part of the REST complex through co-immunoprecipitation (24). Second, overexpression of SCP1 in cells increased the dephosphorylation of REST Ser-861/864 (26), whereas knockdown of SCP1 resulted in decreased REST protein levels and induction of REST-controlled genes (Figs. 4C and 5B). We also found that expression of a phosphatase-deficient variant of SCP1, D96N, acts as a dominant-negative in cells, supporting the notion that phosphatase activity of SCP1 in vivo is required for REST regulation. Finally, we have demonstrated direct phosphatase activity of SCP1 against REST and the structural determinants mediating this interaction. SCP1 preferentially dephosphorylates pSer-861 of REST, and phosphorylation of this site seems to promote dephosphorylation of the lower affinity pSer-864 site as well (Fig. 2B). Collectively, these experiments strongly suggest that REST is a bona fide substrate for SCP1 in vivo.
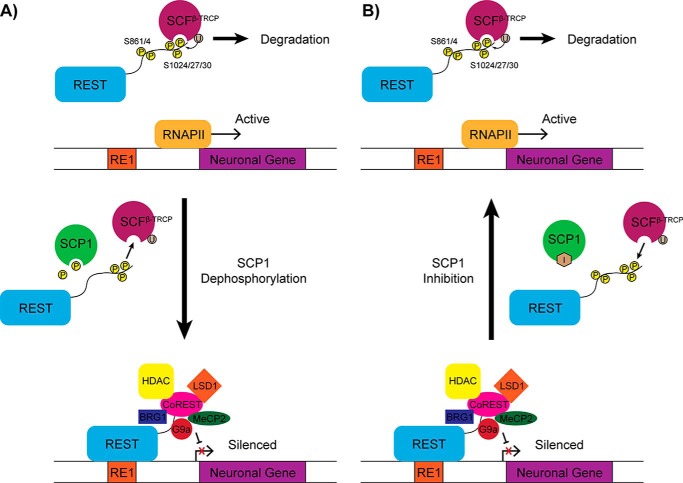
Dephosphorylation of REST by SCP1 appears to be critical for REST function. From our work and the work of others (24, 26, 30), we have created a model detailing how SCP1 can modulate the function of REST through its phosphatase activity (Fig. 6). In this model, SCP1 dephosphorylates REST at its C-terminal degron region and prevents it from being degraded. REST lacking phosphorylation at Ser-861 or Ser-864 can then suppress the expression of genes located near RE1 elements. Inactivation of SCP1 phosphatase activity should thus reduce the amount of REST protein in cells, resulting in loss of REST-mediated silencing and increased expression of many REST-controlled genes. Therefore, control of REST function can likely be achieved by modulating the phosphatase activity of SCP1.
Figure 6.
Model of SCP1 phosphoregulation of REST function. A, when REST is phosphorylated at Ser-861 and/or Ser-864, the SCFβ-TRCP complex targets the Ser-1024/27/30 degron and ubiquitinates REST for degradation. Dephosphorylation at the Ser-861 and Ser-864 sites of REST by SCP1 prevents the SCFβ-TRCP complex from stably associating with the Ser-1024/27/30 degron. Unphosphorylated REST recruits various regulatory factors to RE1 elements and represses neuronal gene expression. B, inactivation of SCP1 by small molecule inhibitors should conversely lead to increased Ser-861 and Ser-864 phosphorylation followed by increased REST degradation and finally induction of RE1-associated neuronal genes.
REST is an important transcriptional regulator in stem cells that controls many neuronal genes (31, 32). However, it has been difficult to discern the precise function of REST using different genetic techniques due to cell-dependent variations in gene regulation. Small molecules targeting SCP1 may help overcome these challenges with instant control, tunable inhibition, and reversible manipulation. Among the 158 phosphatases in humans, SCP1 belongs to a small phosphatase family (containing fewer than 10 members) with active sites and a reaction mechanism that differs from most other phosphatases (41). This allows for specific inhibition of SCP1 with less concern for cross-inhibition against off-target phosphatases. SCP1 has a large substrate-binding pocket that is sufficient to accommodate a variety of drug-like molecules (733 Å2 calculated by the Dogsite program, which is at least 2-fold larger than cysteine based phosphatase-binding pockets; Fig. S6). Specifically, the active site of SCP1 has a unique hydrophobic groove not found in other members of the SCP/FCP phosphatase family (27). Indeed, a compound identified in a pilot screen binds to this pocket (Fig. S6) and shows selective inhibition against SCP1 (Ki of 5 μm) but not any other phosphatases tested (30). Finally, knockdown of SCP1 showed no effect in overall cellular fitness (24, 25). These characteristics make SCP1 a strong drug candidate due to its large active site pocket, unique hydrophobic region, and low risk of toxicity and cross-inhibition in even closely related phosphatases.
Although SCP1 and its homologs (SCP2 and SCPL) represent strong candidates for controlling REST function, it is of concern that reducing SCP activity could affect the other pathways involved. Ser-5 phosphorylation of the RNA polymerase II C-terminal domain can be dephosphorylated by SCP1 in vitro (42). However, knockdown of SCP1 in cells does not alter general transcription (24, 25), suggesting that SCPs do not significantly contribute to global Ser-5 dephosphorylation in vivo. In fact, the only genes whose expression levels have been reported to be altered by knockdown of SCP1 are all controlled by REST, which is consistent with the regulatory action of SCP1 in the REST neuronal silencing program (24). A recent report also showed that SCP1 and its homologues can be targeted to the plasma membrane through palmitoylation in different non-neuronal tumor cell lines (43). However, in non-neuronal tumors there are often significantly decreased amounts of functional REST protein (44, 45), making them poor targets for SCP1 inhibitors. Neuronal tumors such as glioblastomas on the other hand can have abnormally high amounts of REST protein, which correspond with high levels of SCP1 (10, 11), making them ideal targets for SCP inhibitors. SCP1 was also found to suppress tumorigenesis in a lung cancer model (43), but we suspect this function would play a minor role in cancers like glioblastomas due to their aggressive nature. Thus, whereas it is important to explore the possibility of SCP shuttling in neuronal cancers, we still believe that SCP inhibitors could be extremely useful for treating aggressive brain cancers in which REST is highly overexpressed.
Overall, SCP1 is a good small molecule inhibitor target for controlling REST function. Compounds targeting SCPs could be useful for investigating the biological function of REST in various cell types as well as the pathological mechanisms of neurological diseases in which REST is overexpressed. Selective inhibitors have been reported but need further optimizationto obtain higher potency, stronger biological activity, and improved membrane penetration. Nevertheless, enhanced SCP inhibitors will open a new avenue for the modulation of REST function in both academic and clinical settings.
Experimental procedures
Protein expression and purification
The human SCP1(77–261) WT and D96N variants were cloned in a pET28a (Novagen) derivative vector carrying an N-terminal His8 tag with a thrombin linker region. The protein was expressed and purified from Escherichia coli BL21 (DE3) cells as previously reported (46). His-tagged WT SCP1 was used for kinetics, whereas thrombin-cleaved SCP1 D96N was used for crystallography. The vector for expression of dominant-negative SCP1 (D96N) was generated by subcloning cDNA for human SCP1 into pcDNA3FLAG vectors. The D96N point mutation was introduced by site-directed mutagenesis using QuikChange (Stratagene) according to factory directions.
Phosphatase kinetics with peptide substrates
The phosphorylated REST peptides were obtained commercially (AnaSpec) and the peptide sequences are presented in Table S1. Phosphatase reactions were performed with 50 mm Tris acetate, pH 5.5, 10 mm MgCl2, 12 nm WT SCP1, and 0–500 μm REST peptide at 37 °C for 5 min. The reactions were quenched with a 2 times volume of BIOMOL® Green Reagent (Enzo Life Sciences) and allowed to develop for 30 min at room temperature. The A620 of each mixture was measured in a TECAN Infinite M200 plate reader and used to determine the concentration of phosphate released based on a standard curve. Specificity constants were determined by plotting rate against concentration in Kaleidagraph (Synergy Software) and fitting to Michaelis-Menten steady-state kinetics.
MALDI-TOF analysis with peptide substrates
Phosphatase reactions (0.1 mm doubly phosphorylated REST peptide, 10 μm WT SCP1(77–261), 50 mm Tris acetate, pH 5.5, and 10 mm MgCl2) were incubated at 37 °C for 45 min. The samples were then desalted over Ziptip C18 resins (Millipore) using standard protocols. Mass spectrometric analysis of SCP1-treated doubly phosphorylated REST peptide was carried out in an AB Voyager-DE PRO MALDI-TOF with 1:1 DHB matrix. A separate protein sample of known size was used as a standard for calibration.
Protein crystallization and substrate complexation
SCP1 D96N crystals were grown using a vapor diffusion method in sitting-drop at room temperature by mixing 1 μl of ∼10 mg/ml of protein with 1 μl of mother liquor. Optimal SCP1 D96N crystals were obtained in 30% PEG 3350 (w/v) and 0.2 m magnesium acetate. SCP1–REST complex structures were obtained by soaking SCP1 D96N crystals with 1–2 mm REST peptide for 2 h at room temperature in mother liquor. The crystals were then cryo-protected with 25% glycerol, flash frozen, and shipped to the Advanced Photon Source (APS) for data collection. Data were collected on beam line 23-ID-D and processed using HKL2000. SCP1 D96N structures complexed with REST peptides were determined by molecular replacement using a previously solved SCP1 D96N structure (PDB code 2GHT) in Phenix. The molecular replacement solutions for each structure were iteratively built using Coot and Phenix refine. The quality of the final refined structures was evaluated by MolProbity. The final statistics for data collection and structure determination are summarized in Table 1.
Cell culture, virus production, and infection
HEK293T and HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% FBS and 100 units of penicillin/streptomycin. Lentiviral shRNAs used to target human REST (Sigma) (25) and SCP1 were shuttled to pLKO.1-Hygromycin vector (Addgene, number 24150); the target sequences and TRC numbers are presented in Table S2. The empty pLKO.1-Hygromycin vector was used as a control for gene expression in shRNA-transfected cells. For lentiviral packaging, 3 μg of shRNA plasmid and packaging plasmids were co-transfected into 293T cells with Lipofectamine 2000 reagent. Virus was collected twice after 48 and 72 h of transfection. Viral particles were first concentrated by centrifugation at 20,000 rpm for 2 h and then added to HEK293 cells with Polybrene to a final concentration of 8 μg/ml. Dominant-negative experiments were performed in HEK293T cells. pcDNA3FLAG vectors containing SCP1 D96N or empty vector control were transiently transfected into HEK293T cells using calcium phosphate transfection by established protocols. Expression was monitored by Western blotting against the FLAG-tagged SCP1 D96N with maximal expression obtained within 48 to 72 h.
qRT-PCR analysis
Total RNA was extracted using the TRIzol reagent (Life Technologies) and reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Promega, M1701). qPCR analyses were performed using the FastStart Universal SYBR green master mix (Roche, catalogue number 04913914001). All primers used in the study were presented in Table S3.
Western blot analysis
Cells were lysed in a buffer containing 50 mm Tris-HCl, pH 6.8, 2% SDS, and 10% glycerol. Cell extract protein concentration was quantified by Nanodrop. Fifty micrograms of total protein was fractionated on an 8–10% SDS-polyacrylamide gel, and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% fat-free milk for 1 h and washed three times with TBST buffer. The primary antibodies diluted in TBST were further applied and incubated overnight at 4 °C. After thoroughly washing with TBST three times, secondary antibodies were applied for another 1 h at room temperature. Protein bands were detected using the ECL Western blotting substrate kit (Pierce, catalogue number 32106). The following antibodies were used: SCP1 antibody (1:1,000,Proteintech, catalogue number 0952-I-AP), ACTB antibody (1:2,000, Proteintech, catalogue number 60008-I-AP), and REST (1:500, Millipore, catalogue number 07-579).
Author contributions
N. T. B., J. E. M., Y. X., and Y. J. Z. conceptualization; N. T. B., J. E. M., X. Y., S. I., D. K. A., W. M., and Y. J. Z. data curation; N. T. B., J. E. M., X. Y., and S. I. formal analysis; N. T. B. and J. E. M. investigation; N. T. B., X. Y., and Y. J. Z. writing-original draft; N. T. B. and Y. J. Z. project administration; J. E. M. and X. Y. validation; J. E. M. writing-review and editing; F. J. and Y. J. Z. resources; Y. X. methodology; Y. J. Z. supervision; Y. J. Z. funding acquisition.
Supplementary Material
Acknowledgment
Crystallographic data collection was conducted the Advanced Photon Sources (BL23-ID-B), Department of Energy (DOE) National User Facility.
This work was supported by National Institutes of Health Grant R01 GM104896 (to Y. J. Z.), Welch Foundation Grant F-1778 (to Y. J. Z.), Natural Science Foundation of China Grant 31601107 (to X. H. Y.), and the Alzheimer's Drug Discovery Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3 and Figs. S1–S6.
The atomic coordinates and structure factors (codes 6DU2 and 6DU3) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- REST
- RE1-silencing transcription factor
- SCP1
- small C-terminal domain phosphatase 1
- pSer-861
- phosphoserine 861
- NRSF
- neuron-restrictive silencer factor
- SCFβ-TrCP
- Skp1-Cul1-F-box β-transducin repeat containing E3 ubiquitin ligase
- TUBB3
- tubulin β3
- USP37
- ubiquitin-specific peptidase 37
- BEX1
- brain-expressed X-linked 1
- BDNF
- brain-derived neurotropic factor
- UCHL1
- ubiquitin C-terminal hydrolase L1
- DYRK1A
- dual specificity tyrosine phosphorylation regulated kinase 1A
- shRNA
- short hairpin RNA
- qRT
- quantitative reverse transcriptase.
References
- 1. Chong J. A., Tapia-Ramirez J., Kim S., Toledo-Aral J. J., Zheng Y., Boutros M. C., Altshuller Y. M., Frohman M. A., Kraner S. D., and Mandel G. (1995) REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957 10.1016/0092-8674(95)90298-8 [DOI] [PubMed] [Google Scholar]
- 2. Schoenherr C. J., and Anderson D. J. (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363 10.1126/science.7871435 [DOI] [PubMed] [Google Scholar]
- 3. Ballas N., Grunseich C., Lu D. D., Speh J. C., and Mandel G. (2005) REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 10.1016/j.cell.2005.03.013 [DOI] [PubMed] [Google Scholar]
- 4. Otto S. J., McCorkle S. R., Hover J., Conaco C., Han J. J., Impey S., Yochum G. S., Dunn J. J., Goodman R. H., and Mandel G. (2007) A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27, 6729–6739 10.1523/JNEUROSCI.0091-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh S. K., Marisetty A., Sathyan P., Kagalwala M., Zhao Z., and Majumder S. (2015) REST-miR-21-SOX2 axis maintains pluripotency in E14Tg2a.4 embryonic stem cells. Stem Cell Res. 15, 305–311 10.1016/j.scr.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su X., Kameoka S., Lentz S., and Majumder S. (2004) Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol. Cell Biol. 24, 8018–8025 10.1128/MCB.24.18.8018-8025.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gopalakrishnan V. (2009) REST and the RESTless: in stem cells and beyond. Future Neurol. 4, 317–329 10.2217/fnl.09.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mukherjee S., Brulet R., Zhang L., and Hsieh J. (2016) REST regulation of gene networks in adult neural stem cells. Nat. Commun. 7, 13360 10.1038/ncomms13360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aoki H., Hara A., Era T., Kunisada T., and Yamada Y. (2012) Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development 139, 667–677 10.1242/dev.072272 [DOI] [PubMed] [Google Scholar]
- 10. Kamal M. M., Sathyan P., Singh S. K., Zinn P. O., Marisetty A. L., Liang S., Gumin J., El-Mesallamy H. O., Suki D., Colman H., Fuller G. N., Lang F. F., and Majumder S. (2012) REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells 30, 405–414 10.1002/stem.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conti L., Crisafulli L., Caldera V., Tortoreto M., Brilli E., Conforti P., Zunino F., Magrassi L., Schiffer D., and Cattaneo E. (2012) REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS ONE 7, e38486 10.1371/journal.pone.0038486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lawinger P., Venugopal R., Guo Z. S., Immaneni A., Sengupta D., Lu W., Rastelli L., Marin Dias Carneiro A., Levin V., Fuller G. N., Echelard Y., and Majumder S. (2000) The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat. Med. 6, 826–831 10.1038/77565 [DOI] [PubMed] [Google Scholar]
- 13. Liang J., Tong P., Zhao W., Li Y., Zhang L., Xia Y., and Yu Y. (2014) The REST gene signature predicts drug sensitivity in neuroblastoma cell lines and is significantly associated with neuroblastoma tumor stage. Int. J. Mol. Sci. 15, 11220–11233 10.3390/ijms150711220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Westbrook T. F., Martin E. S., Schlabach M. R., Leng Y., Liang A. C., Feng B., Zhao J. J., Roberts T. M., Mandel G., Hannon G. J., Depinho R. A., Chin L., and Elledge S. J. (2005) A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848 10.1016/j.cell.2005.03.033 [DOI] [PubMed] [Google Scholar]
- 15. Wagoner M. P., Gunsalus K. T., Schoenike B., Richardson A. L., Friedl A., and Roopra A. (2010) The transcription factor REST is lost in aggressive breast cancer. PLoS Genet. 6, e1000979 10.1371/journal.pgen.1000979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu T., Aron L., Zullo J., Pan Y., Kim H., Chen Y., Yang T. H., Kim H. M., Drake D., Liu X. S., Bennett D. A., Colaiácovo M. P., and Yankner B. A. (2014) REST and stress resistance in ageing and Alzheimer's disease. Nature 507, 448–454 10.1038/nature13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu M., Cai L., Liang M., Huang Y., Gao H., Lu S., Fei J., and Huang F. (2009) Alteration of NRSF expression exacerbating 1-methyl-4-phenyl-pyridinium ion-induced cell death of SH-SY5Y cells. Neurosci. Res. 65, 236–244 10.1016/j.neures.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 18. Spencer E. M., Chandler K. E., Haddley K., Howard M. R., Hughes D., Belyaev N. D., Coulson J. M., Stewart J. P., Buckley N. J., Kipar A., Walker M. C., and Quinn J. P. (2006) Regulation and role of REST and REST4 variants in modulation of gene expression in in vivo and in vitro in epilepsy models. Neurobiol. Dis. 24, 41–52 10.1016/j.nbd.2006.04.020 [DOI] [PubMed] [Google Scholar]
- 19. Zuccato C., Belyaev N., Conforti P., Ooi L., Tartari M., Papadimou E., MacDonald M., Fossale E., Zeitlin S., Buckley N., and Cattaneo E. (2007) Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J. Neurosci. 27, 6972–6983 10.1523/JNEUROSCI.4278-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rigamonti D., Mutti C., Zuccato C., Cattaneo E., and Contini A. (2009) Turning REST/NRSF dysfunction in Huntington's disease into a pharmaceutical target. Curr. Pharm. Des. 15, 3958–3967 10.2174/138161209789649303 [DOI] [PubMed] [Google Scholar]
- 21. Buckley N. J., Johnson R., Zuccato C., Bithell A., and Cattaneo E. (2010) The role of REST in transcriptional and epigenetic dysregulation in Huntington's disease. Neurobiol. Dis. 39, 28–39 10.1016/j.nbd.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 22. Gao Z., Ure K., Ding P., Nashaat M., Yuan L., Ma J., Hammer R. E., and Hsieh J. (2011) The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772–9786 10.1523/JNEUROSCI.1604-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Westbrook T. F., Hu G., Ang X. L., Mulligan P., Pavlova N. N., Liang A., Leng Y., Maehr R., Shi Y., Harper J. W., and Elledge S. J. (2008) SCFβ-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 10.1038/nature06780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yeo M., Lee S. K., Lee B., Ruiz E. C., Pfaff S. L., and Gill G. N. (2005) Small CTD phosphatases function in silencing neuronal gene expression. Science 307, 596–600 10.1126/science.1100801 [DOI] [PubMed] [Google Scholar]
- 25. Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., Wang G., Wu Q., Wei C., Bi Y., Jiang L., Cai Z., Sun H., Zhang K., Zhang Y., Chen J., and Fu X. D. (2013) Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96 10.1016/j.cell.2012.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nesti E., Corson G. M., McCleskey M., Oyer J. A., and Mandel G. (2014) C-terminal domain small phosphatase 1 and MAP kinase reciprocally control REST stability and neuronal differentiation. Proc. Natl. Acad. Sci. U.S.A. 111, E3929–E3936 10.1073/pnas.1414770111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y., Kim Y., Genoud N., Gao J., Kelly J. W., Pfaff S. L., Gill G. N., Dixon J. E., and Noel J. P. (2006) Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell 24, 759–770 10.1016/j.molcel.2006.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang M., Liu J., Kim Y., Dixon J. E., Pfaff S. L., Gill G. N., Noel J. P., and Zhang Y. (2010) Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein Sci. 19, 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kamenski T., Heilmeier S., Meinhart A., and Cramer P. (2004) Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell 15, 399–407 10.1016/j.molcel.2004.06.035 [DOI] [PubMed] [Google Scholar]
- 30. Zhang M., Cho E. J., Burstein G., Siegel D., and Zhang Y. (2011) Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor. ACS Chem. Biol. 6, 511–519 10.1021/cb100357t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bruce A. W., Donaldson I. J., Wood I. C., Yerbury S. A., Sadowski M. I., Chapman M., Göttgens B., and Buckley N. J. (2004) Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. U.S.A. 101, 10458–10463 10.1073/pnas.0401827101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson D. S., Mortazavi A., Myers R. M., and Wold B. (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 10.1126/science.1141319 [DOI] [PubMed] [Google Scholar]
- 33. Paquette A. J., Perez S. E., and Anderson D. J. (2000) Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc. Natl. Acad. Sci. U.S.A. 97, 12318–12323 10.1073/pnas.97.22.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao S., Zhao X., Lin B., Hu Z., Yan L., and Gao J. (2012) Clinical implications of REST and TUBB3 in ovarian cancer and its relationship to paclitaxel resistance. Tumour Biol. 33, 1759–1765 10.1007/s13277-012-0435-y [DOI] [PubMed] [Google Scholar]
- 35. Lebok P., Öztürk M., Heilenkötter U., Jaenicke F., Müller V., Paluchowski P., Geist S., Wilke C., Burandt E., Lebeau A., Wilczak W., Krech T., Simon R., Sauter G., and Quaas A. (2016) High levels of class III β-tubulin expression are associated with aggressive tumor features in breast cancer. Oncol. Lett. 11, 1987–1994 10.3892/ol.2016.4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dobson T. H. W., Hatcher R. J., Swaminathan J., Das C. M., Shaik S., Tao R. H., Milite C., Castellano S., Taylor P. H., Sbardella G., and Gopalakrishnan V. (2017) Regulation of USP37 expression by REST-associated G9a-dependent histone methylation. Mol. Cancer Res. 15, 1073–1084 10.1158/1541-7786.MCR-16-0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wagoner M. P., and Roopra A. (2012) A REST derived gene signature stratifies glioblastomas into chemotherapy resistant and responsive disease. BMC Genomics 13, 686 10.1186/1471-2164-13-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zuccato C., Ciammola A., Rigamonti D., Leavitt B. R., Goffredo D., Conti L., MacDonald M. E., Friedlander R. M., Silani V., Hayden M. R., Timmusk T., Sipione S., and Cattaneo E. (2001) Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science 293, 493–498 10.1126/science.1059581 [DOI] [PubMed] [Google Scholar]
- 39. Gong B., Cao Z., Zheng P., Vitolo O. V., Liu S., Staniszewski A., Moolman D., Zhang H., Shelanski M., and Arancio O. (2006) Ubiquitin hydrolase Uch-L1 rescues β-amyloid-induced decreases in synaptic function and contextual memory. Cell 126, 775–788 10.1016/j.cell.2006.06.046 [DOI] [PubMed] [Google Scholar]
- 40. Lu M., Zheng L., Han B., Wang L., Wang P., Liu H., and Sun X. (2011) REST regulates DYRK1A transcription in a negative feedback loop. J. Biol. Chem. 286, 10755–10763 10.1074/jbc.M110.174540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang M., Yogesha S. D., Mayfield J. E., Gill G. N., and Zhang Y. (2013) Viewing serine/threonine protein phosphatases through the eyes of drug designers. FEBS J. 280, 4739–4760 10.1111/febs.12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeo M., Lin P. S., Dahmus M. E., and Gill G. N. (2003) A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 278, 26078–26085 10.1074/jbc.M301791200 [DOI] [PubMed] [Google Scholar]
- 43. Liao P., Wang W., Li Y., Wang R., Jin J., Pang W., Chen Y., Shen M., Wang X., Jiang D., Pang J., Liu M., Lin X., Feng X. H., Wang P., and Ge X. (2017) Palmitoylated SCP1 is targeted to the plasma membrane and negatively regulates angiogenesis. Elife 6, e22058 10.7554/eLife.22058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coulson J. M., Edgson J. L., Woll P. J., and Quinn J. P. (2000) A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res. 60, 1840–1844 [PubMed] [Google Scholar]
- 45. Neumann S. B., Seitz R., Gorzella A., Heister A., Doeberitz Mv., and Becker C. M. (2004) Relaxation of glycine receptor and onconeural gene transcription control in NRSF deficient small cell lung cancer cell lines. Brain Res. Mol. Brain Res. 120, 173–181 10.1016/j.molbrainres.2003.10.021 [DOI] [PubMed] [Google Scholar]
- 46. Mayfield J. E., Fan S., Wei S., Zhang M., Li B., Ellington A. D., Etzkorn F. A., and Zhang Y. J. (2015) Chemical tools to decipher regulation of phosphatases by proline isomerization on eukaryotic RAN polymerase II. ACS Chem. Biol. 10, 2405–2414 10.1021/acschembio.5b00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.