Abstract
Background and aims
Microbiota alterations are linked with colorectal cancer (CRC) and notably higher abundance of putative oral bacteria on colonic tumours. However, it is not known if colonic mucosa-associated taxa are indeed orally derived, if such cases are a distinct subset of patients or if the oral microbiome is generally suitable for screening for CRC.
Methods
We profiled the microbiota in oral swabs, colonic mucosae and stool from individuals with CRC (99 subjects), colorectal polyps (32) or controls (103).
Results
Several oral taxa were differentially abundant in CRC compared with controls, for example, Streptococcus and Prevotellas pp. A classification model of oral swab microbiota distinguished individuals with CRC or polyps from controls (sensitivity: 53% (CRC)/67% (polyps); specificity: 96%). Combining the data from faecal microbiota and oral swab microbiota increased the sensitivity of this model to 76% (CRC)/88% (polyps). We detected similar bacterial networks in colonic microbiota and oral microbiota datasets comprising putative oral biofilm forming bacteria. While these taxa were more abundant in CRC, core networks between pathogenic, CRC-associated oral bacteria such as Peptostreptococcus, Parvimonas and Fusobacterium were also detected in healthy controls. High abundance of Lachnospiraceae was negatively associated with the colonisation of colonic tissue with oral-like bacterial networks suggesting a protective role for certain microbiota types against CRC, possibly by conferring colonisation resistance to CRC-associated oral taxa and possibly mediated through habitual diet.
Conclusion
The heterogeneity of CRC may relate to microbiota types that either predispose or provide resistance to the disease, and profiling the oral microbiome may offer an alternative screen for detecting CRC.
Keywords: colonic bacteria, colorectal cancer, colorectal cancer screening, diet, tumour markers
Key messages.
What is already known on this subject?
The gut microbiota is associated with colorectal cancer (CRC) development.
Faecal microbiota has potential as a biomarker for CRC.
Putatively oral bacteria are more abundant on CRC biopsies and Fusobacterium nucleatum has been reported to be enriched in IBD.
A ‘Western diet’ contributes to CRC development.
What are the new findings?
We developed an oral and faecal microbiota-based classifier that distinguished individuals with CRC and adenomas from healthy controls. The discriminatory power particularly for adenomas was higher than for currently used tests.
We detected similar networks of oral bacteria at both oral and colonic mucosal surfaces, including in individuals with colonic lesions (on and off the tumour), healthy controls and children with and without Crohn’s disease.
A microbiota rich in Lachnospiraceae was negatively correlated with ‘Western diet’ and colonic colonisation with oral bacteria, including oral pathogens associated with CRC, suggesting a protective role, possibly mediated through habitual diet.
How might it impact on clinical practice in the foreseeable future?
If the suitability of oral microbiota screening for the detection of CRC and polyps can be verified in larger study groups, this could significantly improve current screening programmes.
Introduction
Microbes have been implicated in the pathogenesis of several human cancers, most strikingly in the case of Helicobacter pylori and gastric carcinoma and some gastric lymphomas.1 2 H. pylori is now designated a gastric carcinogen and a preclinical risk factor. Current non-invasive screening approaches for colon cancer such as faecal immune test (FIT) and faecal occult blood test (FOBT) have very low sensitivity for detecting early lesions, and more reliable biomarkers are required. We and others have reported changes in the faecal or colonic mucosal microbiota in patients with colorectal cancer (CRC),3–8 and data from several animal models have implicated the microbiota in the pathogenesis of CRC.9–13 Our finding of a microbiota configuration associated with benign colonic polyps that is intermediate between that of controls and those with cancer suggests that the microbiota might provide a potential biomarker predictive of the risk of later development of cancer. It also suggests that an intervention could theoretically be applicable years before the development of the disease. The additional finding by us and others of microbes that are normally associated with the oral cavity3–8 13 being present in the faecal and mucosal microbiota linked with CRC prompted us to investigate the oral microbiota in colon cancer as a first step in determining if it might serve as a more accessible sampling site for convenient and widespread screening. Previously, several groups reported the applicability of faecal microbiota profiling as a tool for detection of CRCs,4 5 14 particularly in conjunction with the FOBT5 or FIT.4 Moreover, distinct bacterial profiles in the oral cavity have been associated with oral cancers15 16 and with esophageal17 and pancreatic cancers.18 19 A single study identified significant differences in the bacteria present in oral rinse samples from individuals with CRC compared with healthy controls.20
Here, we present the findings of an extended CRC study population and include an assessment of the oral microbiota. We developed a classifier using oral and faecal microbiota profiles with high specificity and sensitivity particularly for the detection of colorectal polyps. Furthermore, we found similar bacterial networks at both oral and colonic mucosal surfaces that were enriched in CRC and also detectable in healthy tissue. However, we could not find a direct link between oral and colonic microbiota such that elevated abundance of bacteria in the oral cavity was predictive of colonic colonisation by the same taxa. Rather, colonic presence and abundance of oral pathogens was negatively associated with the colonic abundance of Lachnospiraceae. We also detected weak negative correlations of Lachnospiraceae with dietary habits reminiscent of a ‘Western diet’. Lastly, in a meta analysis, we found similar networks of oral bacteria to be enriched in colonic tissue of children in a recent Crohn’s disease study. Thus, our data indicate that oral bacterial networks found in the colon already establish at an early age and before disease can be detected. Lachnospiraceae may protect against colonisation with oral bacteria, possibly mediated through dietary habits. Colonic oral bacterial overgrowth is not unique to CRC but associated with Crohn’s disease.
Materials and methods
Sampling, DNA extraction and 16S rRNA gene amplicon sequencing
Faecal samples were self-collected and sent to the laboratory within 1 hour of defecation. Tissue samples were either collected during colorectal surgery or colonoscopy. Oral samples were obtained by rubbing the inside of both cheeks with a swab. Exclusion criteria were a personal history of CRC, IBD and irritable bowel syndrome. A breakdown of the analysed samples is given in table 1. Detailed demographic information for each individual is given in (online supplementary table 1).
Table 1.
Clinical data of the studied individuals
| Sample type | Samples (n) | Age (mean±SD) | BMI (mean±SD) | Males | Tumour size (mean±SD) | Rectal bleeding (%) | Alcohol (1st quantile, third quantile) (units/week) | Currently smoking (%) |
| Tissue controls | 59 | 53.2±13.5 | 27.4±6.1 | 44.1 | NA | 57.7 | 1 (1, 10) | 4 |
| Off CRC | 74 | 66±11.3 | 28.7±5.7 | 66.7 | 3.2±1.8 | 65.6 | 2 (1, 10) | 10.4 |
| Off Polyps | 31 | 61.6±14.8 | 28.8±5 | 71 | NA | 38.7 | 1 (1, 10) | 13.3 |
| On CRC | 65 | 67±11.6 | 28.8±5.8 | 60.9 | 3.6±2 | 64.4 | 2 (0.8, 10) | 5 |
| On Polyps | 2 | 76.5±0.7 | 27.4±0.9 | 100 | NA | 50 | 7.5 (4.2, 10.8) | 0 |
| Stool controls | 62 | 63.9±11.1 | 28.2±5.4 | 50.9 | NA | 55.6 | 1 (0.2, 6.2) | 0 |
| Stool CRC | 69 | 65.3±10.8 | 28.4±6.1 | 66.7 | 3.1±1.7 | 65.5 | 2 (1, 11.2) | 11.3 |
| Stool polyp | 23 | 60.4±13.4 | 29.3±5.4 | 78.3 | NA | 39.1 | 4 (1, 13.5) | 17.4 |
| Swab controls | 25 | 51.5±12.4 | 27.1±5.5 | 37.5 | NA | 57.9 | 1 (1, 6) | 0 |
| Swab CRC | 45 | 65.7±10.9 | 27.1±5 | 56.1 | 3.3±1.9 | 66.7 | 2 (1, 9) | 0 |
| Swab polyp | 21 | 59.2±15.1 | 28.5±5.2 | 71.4 | NA | 38.1 | 1 (1, 10) | 10 |
BMI, body mass index; CRC, colorectal cancer.
gutjnl-2017-314814supp008.pdf (1MB, pdf)
Genomic DNA was extracted using the AllPrep DNA/RNA kit from Qiagen (Hilden, Germany). 16S rRNA gene amplicon sequencing libraries of the V3-V4 region were prepared, and pools of amplicons were sequenced at GATC (Konstanz, Germany) on a MiSeq sequencing instrument (Illumina, San Diego, California, USA) using 2×250 bp chemistry.
16S amplicon sequences from our Irish cohort were processed as previously described.3 We also conducted a meta-analysis with amplicon sequencing data pertaining to Gevers et al21 and processed data associated with this study similarly. In order to compare bacterial operational taxonomic units (OTUs) obtained in the Irish CRC cohort (sequenced region: V3-V4) with OTUs obtained in the Crohn’s disease cohort (V4), we shortened the sequences of the CRC cohort to the sequenced region of the CD cohort using cutadapt,22 and then processed the sequences of the two studies together.
Statistical analysis was carried out in R.23
A more detailed description of the employed protocols is available as (online supplementary information).
gutjnl-2017-314814supp011.docx (22.3KB, docx)
CRC classifier
The Random forest (RF) classifier to determine OTUs suitable as biomarkers of colonic lesions was described elsewhere.4 In brief, we used log-ratio transformed values of OTUs present in at least 5% of individuals as input to the function AUCRF of the AUCRF package.24 Significance of difference between ROC curves was assessed using the function roc.test of the pROC package.25 A schematic is depicted in online supplementary figure 1. We also employed an in-house pipeline for classification that consisted of a two-step procedure: the least absolute shrinkage and selection operator (LASSO) feature selection, followed by RF modelling. The full dataset was preprocessed (ie, filtered to exclude features that were present in less than 5% of individuals). Ten-fold cross-validation (CV) was applied to the data. Within each iteration of the 10-fold CV, feature selection was performed using the LASSO algorithm on 90% of the dataset, which was used as a training set to generate a predictive model within each iteration. LASSO improves accuracy and interpretability of models by efficiently selecting the relevant features, a process which is tuned by the parameter lambda. The model was generated within the 10-fold CV training data by filtering the dataset to include only the features selected by the LASSO algorithm, and RF was used for subsequent modelling of this subset. Both LASSO feature selection and RF modelling were performed within the 10-fold CV, which generates an internally validated list of features and an internal 10-fold prediction in order to generate an estimate of the predictive value of the overall model. We report both the results from the default threshold selected by the model and an Youden optimised result where the threshold has been optimised to improve the sensitivity and specificity. A schematic for this protocol is presented in online supplementary figure 2).
gutjnl-2017-314814supp001.pdf (29.1KB, pdf)
gutjnl-2017-314814supp002.pdf (31.5KB, pdf)
Results
The oral microbiota is significantly different in CRC
We analysed the microbiota from individuals with CRC, colorectal polyps and healthy controls from multiple body sites (table 1) using 16S rRNA gene amplicon sequencing. The 10 most abundant bacterial genera across all oral swab samples were Streptococcus (30.7% of all assigned reads), Haemophilus (14.2%), Neisseria (8.8%), Prevotella (6.6%), Fusobacterium (5.4%), Veillonella (5.4%), Leptotrichia (3.9%), Rothia (3.9%), Actinomyces (2.9%) and Porphyromonas (2.4%) (online supplementary table 2). These proportional abundance values are similar to those reported in previous studies of the microbiota associated with the oral cavity.26 27
gutjnl-2017-314814supp009.pdf (1.3MB, pdf)
Microbiota profiling by sequencing identifies bacterial taxa as sequence-based divisions or OTUs. The overall oral profile of bacterial OTUs (grouped at 97% sequence similarity) was significantly different between individuals with CRC and healthy controls (permutational analysis of variance of the unweighted UniFrac distance, figure 1). Moreover, eight oral microbiota OTUs were differentially abundant between individuals with CRC and healthy controls (ANCOM, FDR<0.05). Differentially abundant OTUs were classified as Haemophilus, Parvimonas, Prevotella, Alloprevotella, Lachnoanaerobaculum, Neisseria and Streptococcus (online supplementary table 2). Almost all differentially abundant OTUs (7/8) were less abundant in individuals with CRC than in healthy individuals. Even though the overall microbiota was similar between individuals with polyps and healthy controls, four individual bacterial OTUs were differentially abundant between the two groups (online supplementary table 2), three of which were also differentially abundant in CRC.
Figure 1.
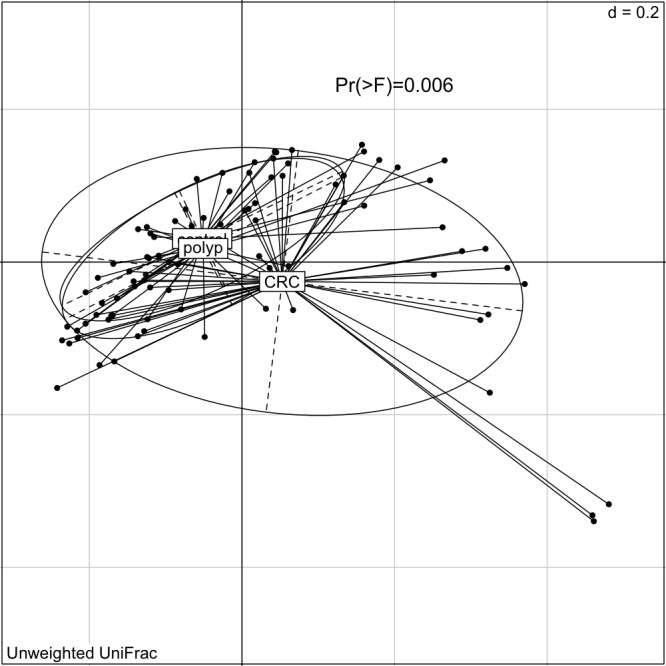
The oral microbiota of individuals with CRC is statistically significantly different from that of healthy individuals. Shown is the PCoA of the unweighted UniFrac distance (significance assessed using PERMANOVA as described in Materials and Methods). CRC, colorectal cancer; PERMANOVA, permutational analysis of variance.
Oral and stool microbiota as biomarkers of CRC
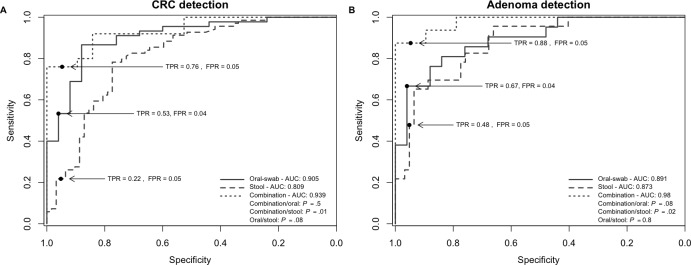
Current non-invasive screening tools for CRC can reliably detect advanced carcinomas based on traces of blood in faeces released by colonic lesions, but these methods suffer from low sensitivity for detecting early lesions.28 Motivated by the findings presented above, we assessed the suitability of oral microbiota as a screening tool for identifying subjects with polyps and CRC by employing a previously established RF classification methodology4 (online supplementary figure 1). The model identified 16 oral microbiota OTUs that distinguish individuals with CRC from healthy controls. The sensitivity of detection was 53% (95% CI (31.11% to 93.33%) with a specificity of 96% (area under the curve (AUC): 0.9; 95% CI (0.83 to 0.9); figure 2 and online supplementary figure 3). The model could also be used to detect individuals with colorectal polyps based on the abundance of 12 oral OTUs (sensitivity 67%; 95% CI (23.81% to 90.48%); AUC: 0.89; 95% CI (0.8 to 0.89); figure 2 and online supplementary figure 4). Our findings are also consistent with previous reports4 5 in that faecal microbiota abundance of selected OTUs is able to distinguish individuals with CRC or polyps from healthy persons (figure 2). However, the sensitivity of our model to use faecal microbiota to detect individuals with CRCs was considerably lower (sensitivity 22%; 95% CI (4.35% to 52.17%); specificity 95%, AUC 0.81; 95% CI (0.73 to 0.81)) than previously reported. A combination of oral and stool microbiota data improved the model sensitivity to 76% (95% CI (59.9% to 92%), AUC: 0.94; 95% CI (0.87 to 0.94) for the detection of CRCs and 88% for polyps (95% CI (68.75% to 100%), AUC: 0.98; 95% CI (0.95 to 0.98) for the detection of polyps (both: specificity 94% or more) (figure 2). Analysis of the abundances of 28 bacterial OTUs were optimal for the differentiation between individuals with polyps and healthy controls (for 12 OTUs, the abundance in the oral cavity was used, while for 16 OTUs, the faecal abundance was used); the model for the detection of CRCs used 63 OTUs (29 oral OTUs and 34 stool OTUs).
Figure 2.
Oral and stool microbiota profiles are potential tools for the detection of CRC. (A and B) Receiver operating characteristic curves (ROC) and area under the curve (AUC) values for the prediction of CRC (A) and polyps (B) using microbiota profiles from oral swabs, stool or a combination of both. AUC values were highest for the combination test. Significance determined after DeLong (Materials and Methods). Sample numbers: swabs: n=25 (healthy controls), n=45 (CRCs), n=21 (polyps); stool: n=62 (healthy controls), n=69 (CRCs), n=23 (polyps); and combination: n=19 (healthy controls), n=25 (CRCs), n=16 (polyps). CRC, colorectal cancer; FPR, false-positive rate; TPR, true-positive rate.
gutjnl-2017-314814supp003.pdf (89.7KB, pdf)
gutjnl-2017-314814supp004.pdf (98.2KB, pdf)
We were able to confirm the predictive value of the oral microbiota for CRC screening by employing an in-house pipeline using a LASSO feature selection step and a RF classifier within a 10-fold CV pipeline (see online supplementary figure 2). This methodology, using the default probability threshold and when applied to the oral swab microbiota dataset, yielded 74% sensitivity and 90% specificity (AUC 0.91) for the prediction of adenomas and 98% sensitivity and 70% specificity (AUC 0.96) for the prediction of CRC, respectively. For a full list of values, please see online supplementary table 3.
gutjnl-2017-314814supp010.pdf (181.9KB, pdf)
Oral bacteria are abundant in the gut microbiota of individuals with CRCs and polyps and form similar coabundance networks on both oral mucosa and colonic tissue
We extended our analysis of oral bacterial networks to the gut because we and others have reported the over-abundance of putatively oral bacteria on CRCs and polyps.3 5 6 13 These bacteria have also been linked to an inflammatory host response and accelerated progression of CRC.3 6 11 29
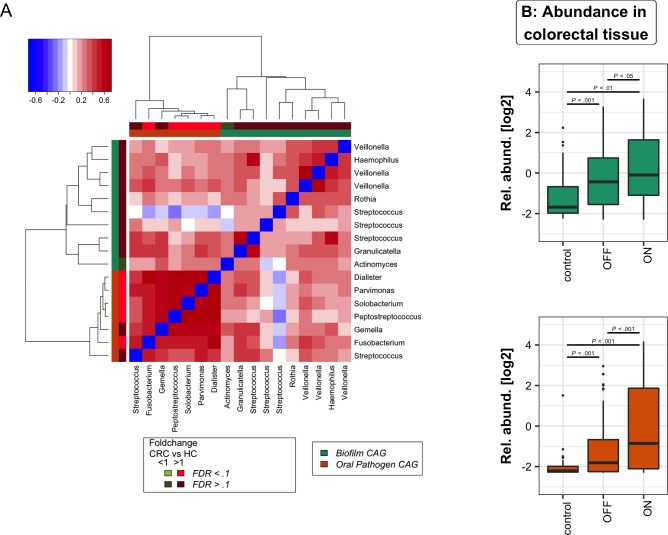
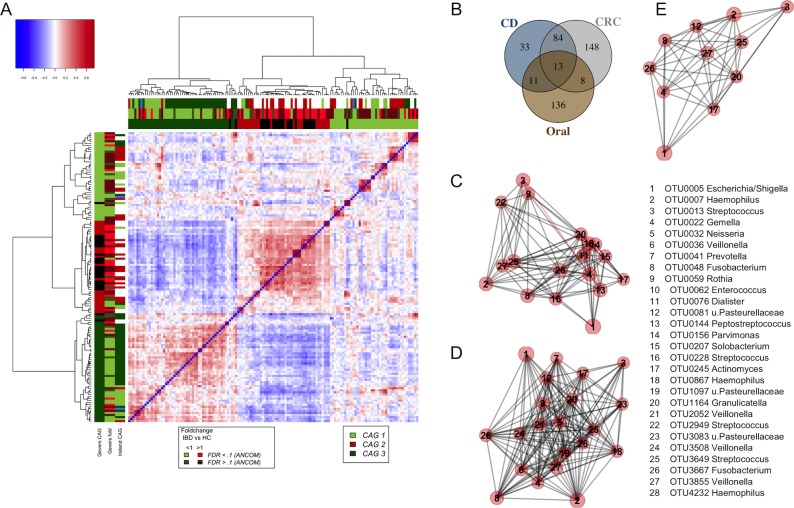
Our analysis focused on the 17 OTUs that were shared between the oral cavity and CRC and polyp samples; that is, OTUs that were detected in 37% of both tissue samples and oral swabs. As with our previous approach to microbiota analysis from cancerous and healthy colon tissue,3 we clustered these bacteria based on their abundance profiles in tumour samples (figure 3A). The two tumour-associated bacterial coabundance groups (CAGs) thus identified comprised (a) oral pathogens previously linked with late colonisation of oral biofilms and with human diseases including CRC (eg, F. nucleatum, Parvimonas micra, Peptostreptococcus stomatis, Dialister pneumosintes and others30–33), designated here as the oral pathogen CAG and comprising seven OTUs in total. The second CAG comprised dominant bacteria in early dental biofilm formation, including Actinomyces, Haemophilus, Rothia, Streptococcus and Veilonella spp.,34 genera also associated with relatively healthy tooth pockets32 (so-called biofilm CAG; 10 OTUs). Collectively, the read counts of OTUs found in the pathogen CAG and the biofilm CAG comprised more than 55% of the average number of sequence reads from oral mucosal surfaces. Bacteria of these two CAGs were significantly more abundant both on and off the tumour compared with healthy controls (figure 3B).
Figure 3.
Oral bacterial networks are detected in colonic mucosa and are enriched in CRC. (A) Clustering of the 17 oral bacterial OTUs associated with tumour tissue into two coabundance groups (CAGs). CAGs were defined on the basis of the clusters in the vertical or horizontal trees and named after their most notable characteristic. Column and row bars indicate bacterial CAGs (as per legend to the bottom right) and fold change between individuals with CRC and healthy controls (as per legend to the bottom left). Legend top left: colour-scale correlation coefficient. (B) The two CAGs comprising typically oral bacteria (oral pathogen CAG and biofilm CAG) were more abundant in colonic microbiota of CRC. Shown are boxplots of relative abundances of the two CAGs in colon tissue. n (controls)=59, n (off)=105, n (tumours)=67. CRC, colorectal cancer; FDR, false discovery rate; HC, healthy controls; OTUs, operational taxonomic units.
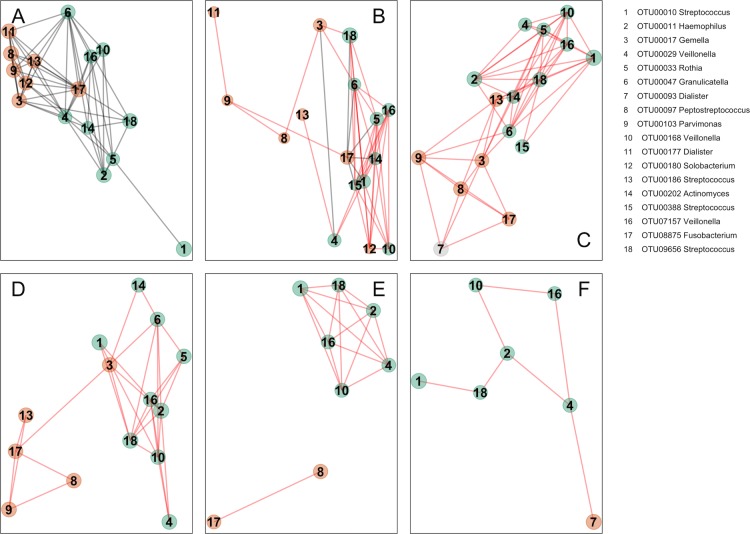
Additionally, we analysed oral bacterial networks detected across different disease stages and different sample types, that is, in (1) oral swabs, (2) in undiseased tissue from individuals with CRC and polyps, (3) in tissue from healthy controls, (4) in stool from individuals with CRC and (5) in stool from healthy controls. Strikingly, the bacterial networks detected on CRC biopsies (figure 3A and figure 4A) were similar in oral swabs (figure 4B), undiseased tissue samples from individuals with CRC and colonic polyps (figure 4C). Moreover, similar networks of bacterial OTUs from the oral cavity were also found in tissue samples from healthy individuals (figure 4D), indicating that these networks exist prior to the development of CRC and could theoretically be involved in the initiation of CRC. These networks were only partially detectable in faecal samples from individuals with CRC or polyps (figure 4E) and faecal microbiota of healthy controls (figure 4F), suggesting tight association with the mucosa and highlighting the limitations of faecal samples for CRC microbiota detection. Details of the overlap of OTUs between the different sample types are presented in the Venn diagram in online supplementary figure 5.
Figure 4.
Bacterial networks detected at colonic mucosal surfaces (panels A,C,D) are similar to those networks detected at oral mucosal surfaces (B) and were not or only partially detected in faecal samples (panels E,F). Shown are network plots of bacterial OTUs found in both the oral cavity and colonic microbiota in different groups of samples: (A) diseased colorectal tissue (ON; 65 individuals with CRC and 2 polyps), (B) oral swab samples (45 individuals with CRC, 21 individuals with polyps and 25 healthy controls), (C) undiseased colorectal tissue (off) from 74 individuals with CRC and 31 individuals with polyps, (D) colorectal tissue from 59 healthy controls, (E) faecal samples from 69 individuals with CRC and 23 individuals with polyps and (F) faecal samples from 62 healthy controls. For each group of samples, the OTUs shared with the oral cavity was determined separately. The size of each node (OTU) correlates to the mean abundance of each OTU across all samples in each respective sample group. The colour of each node corresponds to the CAG determined using diseased colorectal tissue only (figure 3A). One OTU (no. 7, panel C) was only shared between undiseased colorectal tissue and the oral cavity, and it is thus coloured grey. The width of each edge corresponds to the p value of the correlation between each respective node (lower p value, higher line width). The location of each node was determined by a PCoA of the correlation distance as described in Materials and Methods. Only nodes with at least one significant edge are shown. Legend to the right: genus-level classification using RDP reference, version 14 of OTU representative sequences. CRC, colorectal cancer; ON, sample from the cancer or polyp; OTUs, operational taxonomic units; PCoA, Principal Coordinates Analysis; RDP, Ribosomal Database Project.
gutjnl-2017-314814supp005.pdf (518.7KB, pdf)
Low colonic abundance of Lachnospiraceae favours colonisation of gut mucosa by oral pathogens linked to CRC
Given the associations of oral bacteria with the altered microbiota found on CRC biopsies and our current finding that characterising oral microbiota profiles has potential for CRC detection, we hypothesised that the oral microbiota might generally be reflected in gut microbiota composition. However, bacteria typically enriched on colorectal tumours and found in both the oral cavity and the colon, such as Porphyromonas, Parvimonas and Fusobacterium, were less abundant in the oral mucosa of individuals with CRC compared with healthy controls (online supplementary table 2, statistically significant difference for one Parvimonas OTU). Furthermore, the total number of bacteria of the oral pathogen CAG (figure 3) detected in the oral cavity was lower in CRC (p<0.01; Wilcoxon rank-sum test). Surprisingly, we even detected statistically significant negative correlations between oral and colonic abundance of OTUs of the oral pathogen CAG (figure 3) classified as Dialister (Kendall’s =−0.34, p<0.05, n=65 sample pairs), Peptostreptococcus (=−0.28, p<0.05) and Parvimonas (=−0.24, p<0.05).
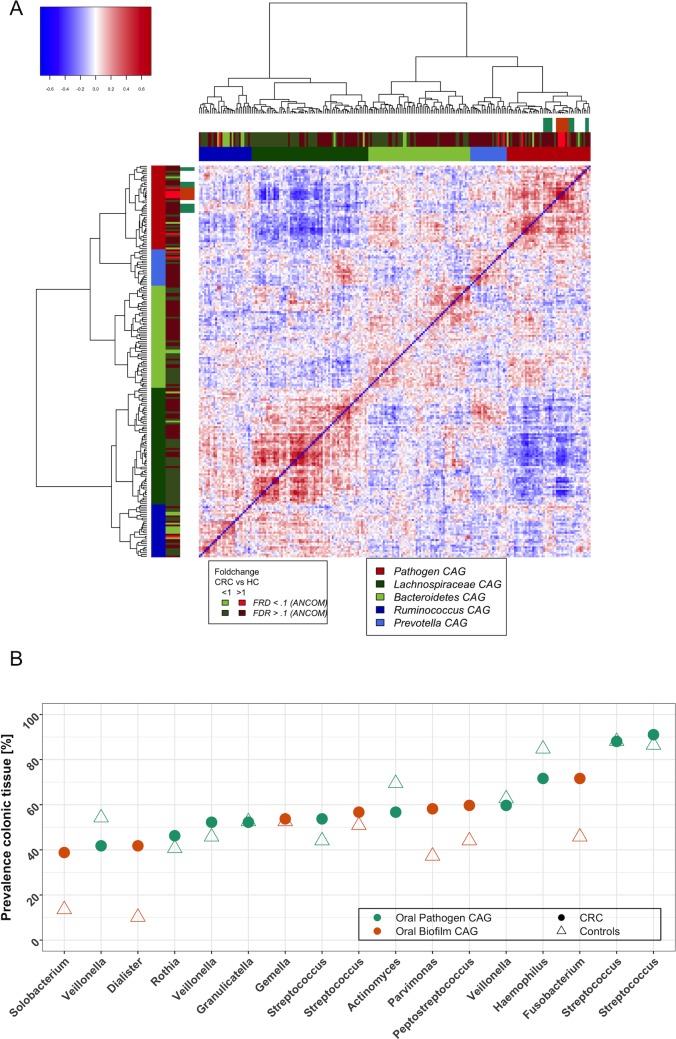
We then asked whether the overall gut microbiota composition of a subject, irrespective of having cancer or being a healthy control, determines whether oral bacteria become part of the gut microbiota. To test this, we first determined bacterial CAGs in this extended dataset for all OTUs (as opposed to the analysis shown in figure 3A, which only considers the 17 OTUs shared between the oral cavity and tumours) at colonic lesions (figure 5A) and in colonic mucosa of healthy individuals (CAG plot not shown). Similar to our previous analyses,3 OTUs with increased relative abundance in CRC were predominantly clustered into Bacteroidetes, Prevotella and oral bacterial CAGs, whereas OTUs in the Lachnospiraceae CAG were mostly less abundant in individuals with CRC (figure 5A). Detailed analysis of the correlations between the abundance of bacteria shared between the oral cavity and colonic tissue (figure 3) and the abundance of other colonic mucosa-associated bacteria revealed that oral pathogen CAG OTUs were strongly negatively correlated with Lachnospiraceae CAG OTUs (figure 5A, dark green-coloured CAG). More specifically, 125 of the 130 (96%) negative correlations (SparCC pseudo-p<0.01) these oral bacterial OTUs had with other bacteria were with OTUs of the Lachnospiraceae CAG. We next analysed the prevalence of oral bacteria on colorectal tumours and found that most bacteria of the oral pathogen and biofilm CAGs, while often statistically significantly more abundant in CRC (figure 4), only colonised ~40%–70% of cancers (figure 5B), a phenomenon also noted by others.29 35 When we compared the colonic abundance of the other bacterial CAGs detected at colorectal lesions (figure 5A) between these two groups, prevalence of bacteria of the oral pathogen CAG was associated with decreased abundance of the Lachnospiraceae CAG in both healthy individuals (p<0.1, Wilcoxon rank-sum test) and individuals with colonic lesions (p<0.01). Lastly, we detected a negative association between the abundance of the Lachnospiraceae CAG and and a ‘Western diet’, even though the signal was weak (see online supplementary information for details). In summary, our data suggest that colonic establishment of a putatively pathogenic oral-like community is more common in individuals with CRC and colonic polyps and in individuals with a low colonic abundance of Lachnospiraceae, a family of butyrate-producing Clostridia already associated with reduced risk of colon cancer. Colonisation of the gut by oral bacteria associated with CRC may be partially mediated or facilitated by consuming a diet high in fat and carbohydrates (typical ‘Western Diet’).
Figure 5.
Oral bacterial colonisation of human CRCs is negatively associated with the colonic mucosal abundance of the Lachnospiraceae CAG. (A) The relative abundance of oral pathogens at colonic lesions (found mostly in bright red CAG) is negatively correlated with the relative abundance of OTUs clustered in a CAG mainly comprising Lachnospiraceae (Lachnospiraceae CAG; dark green CAG). Shown is the heatplot of the correlation values between OTUs detected at colonic mucosal surfaces. CAGs were defined on the basis of the clusters in the vertical or horizontal trees and named after their most notable characteristic. Column and row bars indicate bacterial CAGs (as per legend to the bottom right), fold change between individuals with CRC and healthy controls (as per legend to the bottom left) and bacterial CAGs determined with only the subset of 17 OTUs found both at colonic and oral mucosal surfaces (figure 3A). Legend top left: colour-scale correlation coefficient. (B) Scatterplot of the colonic prevalence of bacterial OTUs associated with oral pathogen and biofilm CAGs (figure 3A). Most OTUs were only detected on a subset of CRCs and polyps (circle) or healthy controls (triangle). CAGs, coabundance groups; CRCs, colorectal cancer; OTUs, operational taxonomic units.
Orally derived bacteria and non-neoplastic colonic disease
To determine if colonic colonisation with orally derived bacteria occurs only in older people and if it occurs in other disorders, we included 16S rRNA sequences from a large microbiota dataset of >300 children with and without CD21 into our analysis. To facilitate comparison of the two datasets, we reanalysed our sequences from the oral cavity and tumour mucosa using only the shared sequence fragment (16S rRNA variable region 4) together with the CD data. We detected similar relationships of bacteria in both CD and CRC, particularly with regards to the Lachnospiraceae, Bacteroidetes and Pathogen CAGs (figure 6 and for comparison figure 5 and online supplementary figures 3 and 4). However, we could not detect a Prevotella CAG in children, which may reflect age and/or workflow-associated specifics. Strikingly, as in CRC, the pathogen CAG-type microbiota in children with CD comprised most of the statistically significantly more abundant bacterial OTUs, including OTUs also found in the oral cavity (figure 6A, onlinesupplementary figures 6 and 7). Detailed comparison of the OTUs found in the oral cavity and at colonic mucosal surfaces of individuals with CRC or CD revealed that the same OTUs classified as Fusobacterium, Haemophilus, Streptococcus, Gemella, Rothia, Actinomyces, Granulicatella and Veilonella were detected in both CD and CRC, whereas OTUs classified as Peptostreptococcus and Parvimonas were only found in CRC (see also figure 6B). Some genera found in both CD and CRC contained OTUs that were unique to either CD or CRC (eg, the genus Dialister was found in both CD and CRC, but the OTUs were different). The same oral OTUs were detected in the two diseases, and the OTUs were organised in similar networks (figure 6, panels C–E). Lastly, similar to CRC, the abundance of oral bacterial OTUs was negatively correlated with the abundance of Lachnospiraceae CAG OTUs in CD (91% of all negative correlations were with OTUs of the Lachnospiraceae CAG).
Figure 6.
Similarity of non-neoplastic and neoplastic colonic disease associated bacterial profiles. (A) Shown is the heatplot of the correlation values between OTUs associated with rectal tissue of children with and without CD.21 CAGs were defined on the basis of the clusters in the vertical or horizontal trees and named after their most notable characteristic. Column and row bars indicate bacterial CAGs (as per legend to the bottom right), fold change between individuals with CD and healthy controls (as per legend to the bottom left). Additionally, two row and column bars indicate the CAG in the Irish CRC cohort (figure 4A) and the fold change between individuals with CRCs/polyps and healthy controls. Legend top left: colour scale correlation coefficient. (B) Venn diagram of bacteria found in colorectal tissue of children with CD, colorectal tumours and oral swabs. (C–E) Network plots of bacterial OTUs found in both the oral cavity and different colonic tissue samples: (C) tumours (ON; 65 individuals with CRC and 2 polyps), (D) mucosa from children with CD (n=201) and (E) mucosa from healthy children (n=122). For each group of samples, the OTUs shared with the oral cavity was determined separately. The size of each node (OTU) correlates to the mean abundance of each OTU across all samples in each respective sample group. The width of each edge corresponds to the p value of the correlation between each respective node (lower p value, higher line width). The location of each node was determined by a PCoA of the correlation distance as described in Materials and Methods. Only nodes with at least one significant edge are shown. Legend to the right: genus-level classification using RDP reference, version 14 of OTU representative sequences. CAGs, coabundance groups; CD, Crohn’s disease; CRCs, colorectal cancer; OTUs, operational taxonomic units; PCoA, Principal Coordinates Analysis; RDP, Ribosomal Database Project.
Discussion
We present for the first time the combined analysis of the microbiota of subjects with CRC using samples from the oral cavity, colonic mucosal tissue and faeces. We show that profiling the bacteria associated with the oral cavity may have value in the detection of CRC. Our data also indicate that many bacterial taxa found in the oral cavity colonise a subset of colorectal tumours and form bacterial coabundance networks similar to those found in the oral cavity. These networks seem to form tight associations with the mucosa, because they are less readily detectable in faecal samples. Many of these bacterial taxa have previously been associated with oral biofilms. Together, these data suggest that oral-like biofilms also form on the mucosa of the colon. These bacteria were typically significantly more abundant on and off colorectal tumours as well as on and off colorectal polyps compared with the mucosa of healthy individuals and have been found to be associated with distinct mucosal gene expression profiles3 6 11 suggesting a role in the development or progression of CRC. The mucosal abundance of Lachnospiraceae CAG microbiota was significantly lower in individuals with CRC and was inversely associated with the presence (and abundance) of CRC-associated, oral-like bacterial OTUs, in both healthy individuals and individuals with CRC and colorectal polyps. Thus, we postulate that Lachnospiraceae CAG-type microbiotas prevent colonic colonisation with CRC-associated, oral-like bacterial biofilms.
The use of microbiome structure as a biomarker of health and disease is gaining momentum particularly with the development of affordable high-throughput DNA sequencing technology. It is now possible to obtain deep knowledge about the microbiota of a sample for less than $10 sequencing cost. Moreover, improved pipelines for in silico analysis of sequencing data enable researchers and clinicians to rapidly turn 16S rRNA amplicon sequencing data into clinically informative data without the need for dedicated large-scale computational facilities. Recent reports have shown the potential suitability of faecal microbiota profiles for screening for colonic lesions using 16S rRNA amplicon sequencing,4 5 14 36 metagenomic sequencing5 and qPCR.14 In addition, diagnostic tests may be improved with a combination of microbiota information and the FIT.4 5 The AUC values we obtained when using a combination of oral and faecal microbiota OTUs for CRC and adenoma detection (0.94 and 0.98, respectively) and the specificity (95% for both) and sensitivity (76% and 88%, respectively) were comparable or higher than those reported in the above-named studies (ranging from 0.64 to 0.93), suggesting that the inclusion of oral microbiota information has the potential to enhance the performance of current diagnostic tests. Particularly promising is the high sensitivity for the detection of adenomas (88%) because of the prognostic and therapeutic importance of early discovery of colonic disease. By comparison, Baxter et al4 reported sensitivities below 20% for the detection of adenomas using either FIT or faecal microbiota composition alone and a sensitivity of below 40% when using a combination (specificity >90%). Our analysis significantly improves on this, and we were able to confirm the value of the oral microbiota to predict colonic lesions with an independent classification strategy employing both LASSO and RF feature selection. We concede, however, that larger prospective studies that account for potential confounders such as age, tumour stage, smoking and alcohol consumption and that combine FIT, faecal microbiota and oral microbiota composition are needed to verify these promising results.
Numerous reports have noted the enrichment of oral-type bacteria on colorectal tumours or in faeces of individuals with CRC or adenomas, particularly Fusobacterium, Peptostreptococcus, Porphyromonas and Parvimonas,3–8 13 14 37 and the association of these bacteria with microscopic inflammation of the colonic mucosa.3 6 11 However, none of these studies included samples from the oral cavity, and direct comparisons were not possible. Dejea et al reported the association of bacterial biofilms with proximal CRCs38 but identified no single genus that was consistently associated with tumour biofilms, not even genus Fusobacterium, nor did they report differences in overall microbiota composition of biofilm-positive tumours compared with biofilm-negative tumours. Interestingly, members of the tumour-associated biofilm CAG that we report here (figure 3A) were indeed more prevalent and trended towards greater abundance in the mucosa of proximal tumours. This sidedness was not detected in healthy individuals indicating that biological specificities of proximal cancers may be responsible.
Previous research indicated a secondary role of F. nucleatum, one of the oral bacterial OTUs also found in this study, in the development of CRC. F. nucleatum had no effect on the proliferation on healthy colon epithelial cells11 and was reported to be enriched on colorectal lesions compared with paired healthy tissue of individuals with CRC,7 8 likely mediated by the overexpression of Gal-GalNAc.39 These findings suggest a secondary involvement of F. nucleatum in the pathology of CRC. We also detected increased abundance of oral bacteria, including F. nucleatum at the site of the tumour, both compared with paired healthy tissue and healthy controls. However, the fact that we detect similar oral bacterial networks both off the tumour and in colonic mucosa of healthy controls makes it at least conceptually possible that such bacteria are indeed involved in the initiation of CRC and are both drivers and passengers of disease40 mediated by thus far undiscovered mechanisms. Recently, Abed et al39 showed that F. nucleatum enrichment is mediated through Fap2 binding to Gal-GalNAc expressed on CRCs and suggested a hematogenous route for translocation of the bacterium from the oral cavity. In addition to F. nucleatum enrichment in individuals with CRCs or polyps, we also detected enrichment of the same bacterial OTU in paediatric CD. It has been shown that a subset of CD and ulcerative colitis subjects (9/20) express Gal-GalNAc.41 It is thus tempting to speculate that a similar mechanism also leads to F. nucleatum enrichment in CD. However, although Gal-GalNAc was shown in one previous study to be expressed on 21 out of 25 of adenocarcinomas,41 we and others11 29 detected F. nucleatum in 50% or less of CRCs, and high abundance is found in only ~10% of CRCs.35 Thus, other mechanisms apparently modulate the abundance of F. nucleatum at the site of CRC. The enrichment of similar oral bacterial networks in both CRC and CD (figure 6) may reflect general microbial community adaptation to a variable host response and it may be secondary in nature.
Our finding that the presence and abundance of oral pathogens both in CRC and in healthy individuals is negatively associated with the abundance of Lachnospiraceae such as Anaerostipes, Blautia and Roseburia suggests that these bacteria also play an important protective role. The concept that the gut microbiota protects against the colonisation of the bowel with environmental bacteria, including pathogens, is well established42 and, according to our data, is also relevant in the context of CRC and CD. Moreover, the association we report here between the abundance of Lachnospiraceae and a healthy diet points to a new thread in the recognised diet–microbiota–disease paradigm, specifically in the concept of CRC, and provides further rationale for promoting a healthy diet to limit lifelong risk of this disease.
gutjnl-2017-314814supp006.pdf (28.3KB, pdf)
gutjnl-2017-314814supp007.pdf (35.7KB, pdf)
Footnotes
Contributors: BF: study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis and study supervision. RDW and MPB: acquisition of data. KC, AD and IBJ: CRC classifier development. EH: study concept and design. MO: study concept and design and acquisition of data. FS and PWO: study concept and design, drafting of the manuscript, critical revision of the manuscript for important intellectual content, obtained funding and study supervision.
Funding: The authors are funded in part by Science Foundation Ireland (APC/SFI/12/RC/2273) in the form of a research centre, the APC Microbiome Institute. IBJ is supported by a Science Foundation Ireland grant (13/SIRG/2128).
Disclaimer: The authors are funded in part by Science Foundation Ireland (APC/SFI/12/RC/2273) in the form of a research centre which is/has recently been in receipt of research grants from the following companies: Cremo, Mead Johnson Nutrition, Kerry, General Mills, GE Healthcare, Friesland Campina, Sigmoid, Alimentary Health, Second Genome, Nutricia, Danone, Janssen, AbbVie, Suntory Morinaga Milk Industry Ltd, Pfizer Consumer Health, Radisens, 4D Pharma, Crucell, Adare Pharma, Artugen Therapeutics, Caelus. FS isa founder shareholder in Atlantia Food Clinical Trials, Tucana Health and Alimentary Health Ltd. PWOT and IBJ are founder shareholders of Tucana Health. These relationships with industry have no bearing on the present work and neither influenced nor constrained it.
Ethics approval: University Ethics Committee.
Provenance and peer review: Not commissioned; internally peer reviewed.
Data sharing statement: All sequencing data will be available upon request.
References
- 1.Polk DB, Peek RM. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer 2010;10:403–14. 10.1038/nrc2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mf G, Smoot DT, pylori H. gastric malt lymphoma, and adenocarcinoma of the stomach. Semin Gastrointest Dis 2000;11:134–41 https://www.ncbi.nlm.nih.gov/pubmed/10950459 [PubMed] [Google Scholar]
- 3.Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017;66:633–43. 10.1136/gutjnl-2015-309595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter NT, Ruffin MT, Rogers MA, et al. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med 2016;8:37 10.1186/s13073-016-0290-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 2014;10:766 doi:10.15252/msb.20145645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren RL, Freeman DJ, Pleasance S, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013;1:16 10.1186/2049-2618-1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292–8. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22:299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 2009;15:1016–22. 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120–3. 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013;14:195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015;6:8727 10.1038/ncomms9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 2017;23:2061–70. 10.1158/1078-0432.CCR-16-1599 [DOI] [PubMed] [Google Scholar]
- 15.Pushalkar S, Ji X, Li Y, et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol 2012;12:144 10.1186/1471-2180-12-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt BL, Kuczynski J, Bhattacharya A, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One 2014;9:e98741 10.1371/journal.pone.0098741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Winckler B, Lu M, et al. Oral Microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of china. PLoS One 2015;10:e0143603 10.1371/journal.pone.0143603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell JJ, Zhang L, Zhou H, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012;61:582–8. 10.1136/gutjnl-2011-300784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres PJ, Fletcher EM, Gibbons SM, et al. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ 2015;3:e1373 10.7717/peerj.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato I, Vasquez AA, Moyerbrailean G, et al. Oral microbiome and history of smoking and colorectal cancer. J Epidemiol Res 2016;2:92–101. 10.5430/jer.v2n2p92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011;17:10–12. doi:10.14806/ej.17.1.200 [Google Scholar]
- 23. Core Team R. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2016. https://www.R-project.org/ [Google Scholar]
- 24.Urrea V, Calle M. AUCRF: Variable selection with random forest and the area under the curve, 2012. https://CRAN.R-project.org/package=AUCRF
- 25.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segata N, Haake SK, Mannon P, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 2012;13:R42 10.1186/gb-2012-13-6-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 2015;6:e00037 10.1128/mBio.00037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hundt S, Haug U, Brenner H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann Intern Med 2009;150:162–9 https://www.ncbi.nlm.nih.gov/pubmed/19189905 10.7326/0003-4819-150-3-200902030-00005 [DOI] [PubMed] [Google Scholar]
- 29.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014;33:1381–90. 10.1007/s10096-014-2081-3 [DOI] [PubMed] [Google Scholar]
- 30.Arora N, Mishra A, Chugh S. Microbial role in periodontitis: have we reached the top? some unsung bacteria other than red complex. J Indian Soc Periodontol 2014;18:9–13. 10.4103/0972-124X.128192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jover-Díaz F, Cuadrado JM, Laveda R, et al. Porphyromonas asaccharolytica liver abscess. Anaerobe 2003;9:87–9. 10.1016/S1075-9964(03)00065-9 [DOI] [PubMed] [Google Scholar]
- 32.Palmer RJ. Composition and development of oral bacterial communities. Periodontol 2000 2014;64:20–39. 10.1111/j.1600-0757.2012.00453.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol 1998;25:134–44 https://www.ncbi.nlm.nih.gov/pubmed/9495612 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- 34.Heller D, Helmerhorst EJ, Gower AC, et al. Microbial Diversity in the Early In Vivo-Formed Dental Biofilm. Appl Environ Microbiol 2016;82:1881–8. 10.1128/AEM.03984-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 2014;74:1311–8. 10.1158/0008-5472.CAN-13-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah MS, DeSantis TZ, Weinmaier T, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 2017. 10.1136/gutjnl-2016-313189 [DOI] [PubMed] [Google Scholar]
- 37.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One 2011;6:e20447 10.1371/journal.pone.0020447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A 2014;111:18321–6. 10.1073/pnas.1406199111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abed J, Emgård JE, Zamir G, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 2016;20:215–25. 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjalsma H, Boleij A, Marchesi JR, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol 2012;10:575–82. 10.1038/nrmicro2819 [DOI] [PubMed] [Google Scholar]
- 41.Said IT, Shamsuddin AM, Sherief MA, et al. Comparison of different techniques for detection of Gal-GalNAc, an early marker of colonic neoplasia. Histol Histopathol 1999;14:351–7 https://www.ncbi.nlm.nih.gov/pubmed/10212796 [DOI] [PubMed] [Google Scholar]
- 42.Zhang C, Derrien M, Levenez F, et al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. Isme J 2016;10:2235–45. 10.1038/ismej.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2017-314814supp008.pdf (1MB, pdf)
gutjnl-2017-314814supp011.docx (22.3KB, docx)
gutjnl-2017-314814supp001.pdf (29.1KB, pdf)
gutjnl-2017-314814supp002.pdf (31.5KB, pdf)
gutjnl-2017-314814supp009.pdf (1.3MB, pdf)
gutjnl-2017-314814supp003.pdf (89.7KB, pdf)
gutjnl-2017-314814supp004.pdf (98.2KB, pdf)
gutjnl-2017-314814supp010.pdf (181.9KB, pdf)
gutjnl-2017-314814supp005.pdf (518.7KB, pdf)
gutjnl-2017-314814supp006.pdf (28.3KB, pdf)
gutjnl-2017-314814supp007.pdf (35.7KB, pdf)