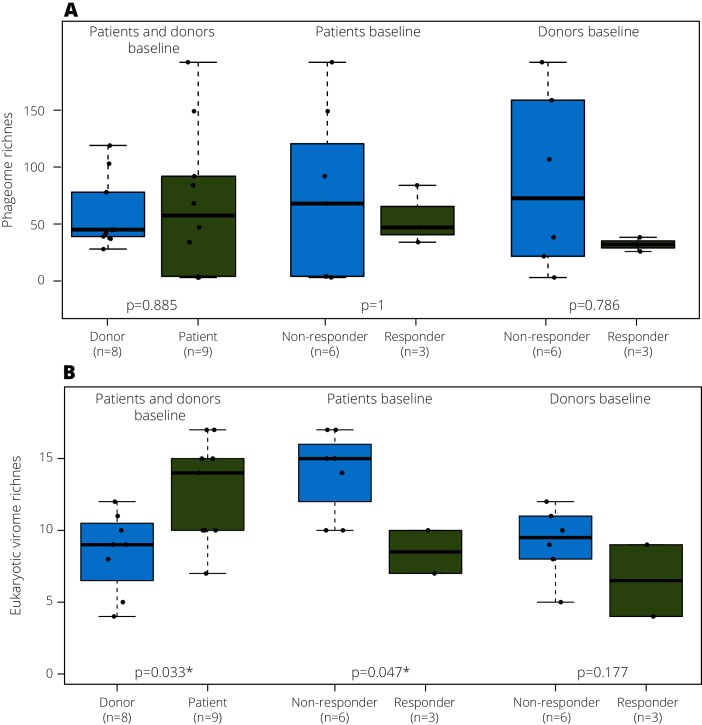
We read with interest the recent Gut study by Zuo and colleagues showing that a good outcome of faecal microbiota transplantation (FMT) in Clostridium difficile infection (CDI) was associated with bacteriophage transfer.1 FMT is an established treatment to restore a balance in disturbed intestinal microbiotas of patients with CDI,2 3 and recent multicentre randomised, double-blind, placebo-controlled trials showed its effectiveness in inducing clinical and endoscopic remission in patients with UC.4 We examined the gut virome dynamics of nine patients with UC who were undergoing FMT (64 patient and 8 healthy donor samples).5 Two patients with UC achieved long-lasting complete remission with mucosal healing (>2 years; figure 1: patient 1 and 3), and one patient reported temporary remission for 6 weeks (figure 1; patient 4).5 The remaining six patients did not respond to treatment. Using the NetoVIR protocol,6 we analysed the faecal virome (both RNA/DNA genomes of phages and eukaryotic viruses) using tailored bioinformatics approaches (see online supplementary data). Irrespective of their outcome, no statistical differences were observed in phageome richness between patients with UC and healthy donors (figure 2A) at any time point, in contrast with the findings of Norman and colleagues, who observed a lower DNA phage richness in healthy controls compared with IBD patients.7 However, for the eukaryotic virome, healthy donors presented a significantly lower richness than patients at baseline (figure 2B, p value=0.033). Furthermore, before FMT, responders already presented a significantly lower eukaryotic viral richness than non-responders (figure 2B, p=0.047). These two observations suggest that eukaryotic virome richness is a potential novel diagnostic marker for UC and UC FMT response. In contrast with the observations for bacteria,5 the richness of donor viromes was not associated with the outcome of therapy (figure 2B, p=0.177). Overall, the most abundant eukaryotic viral families found were Anelloviridae, Circoviridae, Picobirnaviridae and Virgaviridae (see online supplementary data).
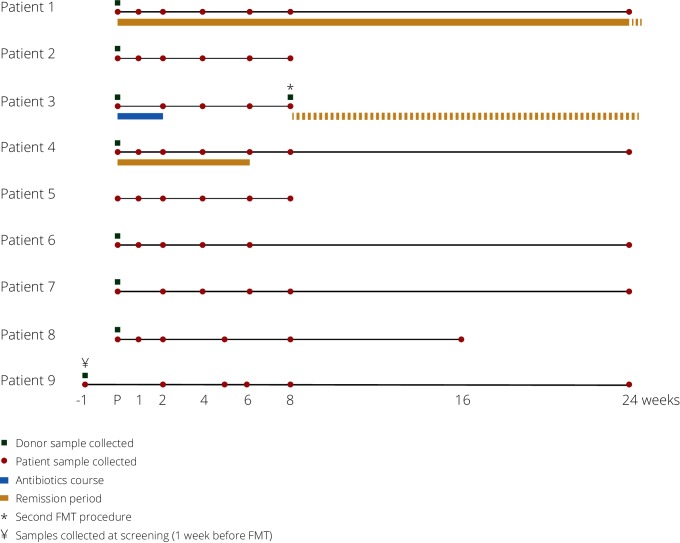
Figure 1.
Overview of the sample collection from patients and donors and their respective remission periods. For the partial responder (patient 4), samples from the remission period were considered as responder in the analyses. Patient 3 vomited after the first FMT, leading to bilateral aspiration and bilateral pneumonia for which the patient was treated for 14 days with broad-spectrum antibiotics. On patient’s request, a second FMT was administered 8 weeks later, and the patient has been in clinical remission ever since. FMT, faecal microbiota transplantation.
Figure 2.
(A) Phageome and (B) eukaryotic virome richness comparisons between: (1) healthy donor and patients; (2) responders and non-responders at baseline and (3) donors of responders and non-responders at baseline.
gutjnl-2017-315281supp001.pdf (427.3KB, pdf)
To investigate donor virome retention, we adapted the bacteriome concept of transferred phylotypes8 and defined a transferred viral operational taxonomic unit (OTU) as being present in the donor sample and introduced or increased in the patient samples post-FMT. While we did not observe any transferred eukaryotic viral families, only the full responder (patient 1) presented nine transferred phage OTUs.
Our results highlight, for the first time, the importance of the eukaryotic virome in UC. It is clear that eukaryotic viruses and phages have different ways of affecting the human host9 and our findings now highlight the importance of analysing the phageome and eukaryotic virome separately. In our study, FMT responders already presented a significant lower eukaryotic virome richness than non-responders at baseline (figure 2A), which is a feature of the healthy donors. Although this study was not designed for diagnostic biomarker discovery and sample size is low, these results might lead towards a potential diagnostic marker for UC to increase treatment success. The published bacteriome analyses on the same cohort showed that responders had significantly higher transferred bacterial phylotypes.5 Moreover, only the full responder presented transferred phage OTUs, suggesting phage transfer concomitant with their respective hosts.
In summary, our study shows that virome prescreening of patients with UC prior to FMT might be a valuable biomarker for response prediction. Further validation in a bigger cohort will enable building a predictive model for response to increase FMT success in UC. These results emphasise that our understanding of human gut viromes and their role in chronic disease is still limited, and further studies are urgently needed to understand their role in UC aetiology. The fact that the phageome was successfully associated with FMT in CDI,1 while eukaryotic viruses seem more relevant in UC, suggests that virome interactions are disease specific and FMT guidelines should be tailored to the pathology at hand.
Footnotes
Contributors: NCN, JR, SV and JM designed the experiments. NCN and KCY conducted the wet lab experiments. NCN and WD performed the bioinformatics analysis with bioinformatics support from PM. NCN, TD and JW ran the ecology analysis. NCN, TD, KM, MJ, JR, SV and JM interpreted the data on viral and disease fields. NCN and JM wrote the manuscript. NCN and TD prepared the figures. JM, MVR, SV and JM coordinated the project. All authors reviewed the manuscript.
Funding: NCN was supported by the government agency flanders innovation & entrepreneurship (VLAIO). KCY was supported by the Interfaculty Council for Development Cooperation (IRO) from the KU Leuven. KM, MJ and JW are supported by postdoctoral fellowships by the Fund for Scientific Research-Flanders, Belgium (FWO-Vlaanderen). SV is a Senior Clinical Researcher from the FWO. This study was supported by a KU Leuven OT-grant (OT-14-113).
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Ethical Commission from the University Hospital of Leuven (UZ Leuven).
Provenance and peer review: Not commissioned; internally peer reviewed.
References
- 1.Zuo T, Wong SH, Lam K, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 2017. [Epub ahead of print: 24 May 2017] 10.1136/gutjnl-2017-313952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013;368:407–15. 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- 3.Austin M, Mellow M, Tierney WM. Fecal microbiota transplantation in the treatment of Clostridium difficile infections. Am J Med 2014;127:479–83. 10.1016/j.amjmed.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 4.Cammarota G, Ianiro G, Tilg H, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017;66:569–80. 10.1136/gutjnl-2016-313017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vermeire S, Joossens M, Verbeke K, et al. Donor species richness determines faecal microbiota transplantation success in inflammatory Bowel disease. J Crohns Colitis 2016;10:387–94. 10.1093/ecco-jcc/jjv203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conceição-Neto N, Zeller M, Lefrère H, et al. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci Rep 2015;5:16532 10.1038/srep16532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norman JM, et al. Disease-specific alterations in the enteric Virome in article disease-specific alterations in the enteric Virome in inflammatory Bowel disease. Cell 2015;160:447–60. 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angelberger S, Reinisch W, Makristathis A, et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol 2013;108:1620–30. 10.1038/ajg.2013.257 [DOI] [PubMed] [Google Scholar]
- 9.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015;479:2–25. 10.1016/j.virol.2015.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2017-315281supp001.pdf (427.3KB, pdf)