Abstract
Type 2 diabetes (T2D) is generally regarded as a metabolic disorder disease with various phenotypic expressions. Traditional Chinese medicine (TCM) has been widely used for preventing and treating diabetes. In our study, we demonstrated that Cyclocarya paliurus formula extractum (CPE), a compound of TCM, can ameliorate diabetes in diabetic rats. Transcriptome profiles were performed to elucidate the anti-diabetic mechanisms of CPE on pancreas and liver. Pancreatic transcriptome analysis showed CPE treatment significantly inhibited gene expressions related to inflammation and apoptosis pathways, among which the transcription factors (TFs) nuclear factor κB (NF-κB), STAT, and miR-9a/148/200 may serve as core regulators contributing to ameliorate diabetes. Biochemical studies also demonstrated CPE treatment decreased pro-inflammatory cytokines (tumor necrosis factor alpha [TNF-α], interleukin [IL]-1β, and IL-6) and reduced β cell apoptosis. In liver tissue, our transcriptome and biochemical experiments showed that CPE treatment reduced lipid accumulation and liver injury, and it promoted glycogen synthesis, which may be regulated by TFs Srebf1, Mlxipl, and miR-122/128/192. Taken together, our findings revealed CPE could be used as a potential therapeutic agent to prevent and treat diabetes. It is the first time to combine transcriptome and regulatory network analyses to study the mechanism of CPE in preventing diabetes, giving a demonstration of exploring the mechanism of TCM on complex diseases.
Keywords: traditional Chinese medicine, diabetes, transcriptome sequencing, inflammation, apoptosis
Introduction
Type 2 diabetes (T2D) is a complex and heterogeneous disease caused by insulin resistance and pancreatic β cell failure.1 Although insulin resistance exists in all stages from pre-diabetes to overt diabetes, the onset of diabetes and its progression is largely determined by the progressive failure and apoptosis of the pancreatic β cells.2 Growing evidence suggests that islet inflammation plays an important role in β cell failure and apoptosis.3 Currently, although the available therapies for diabetes are some western medications or insulin, management of diabetes with fewer side effects at lower costs is still a big challenge.4, 5 Herbal medications can be a good alternative to replace or at least supplement western medications.6
Traditional Chinese herbal medicines have been used for a long time to treat diabetes.7 Accumulated evidence demonstrates that many of these have beneficial effects on the prevention and treatment of diabetes and its complications.8 In particular, traditional Chinese medicine (TCM) with compound recipes has been widely applied as a clinical medication in diabetes.9, 10 Cyclocarya paliurus formula extractum (CPE), a compound of TCM, consists of 4 crude plant products: Cyclocarya paliurus, Dendrobium, Morus alba L., and Pericarpium Citri Reticulatae. It was reported that the extract of Cyclocarya paliurus significantly decreased fasting blood glucose (FBG) level, protected pancreas islet from injury, and increased the level of serum insulin in diabetic mice.11 The other 3 crude plant products are also effective in improving diabetes. Several studies indicated that Dendrobium decreased blood glucose by stimulating the secretion of insulin in diabetic rats.12 Morus alba L. and Pericarpium Citri Reticulatae exhibit anti-diabetic effects through protecting pancreatic β cells and increasing the content of liver glycogen.13, 14 Therefore, the compound of Cyclocarya paliurus, Dendrobium, Morus alba, and Pericarpium Citri Reticulatae may have more beneficial anti-diabetic effects.
Undoubtedly, the recent development of next-generation sequencing technology, particularly RNA sequencing (RNA-seq) and microRNA sequencing (miRNA-seq), has provided a more comprehensive view of the transcriptional landscape for complex diseases at different layers,15 which offers insights into molecular mechanisms of disease and bridges the gap between genotype and phenotype. Both of the two technologies have been widely used in diabetic research, and they have identified some key factors relevant to diabetes, such as Bcl-216 and miR-375.17 Moreover, transcription factors (TFs) and miRNAs are two key types of gene expression regulators, and their co-regulation has been studied in biological processes and diseases.18 However, their co-regulation of an anti-diabetic mechanism rarely has been studied, especially for TCM.
In this study, we validated that CPE treatment ameliorated diabetes in diabetic rats induced by a combination of high-fat diet (HFD) and dexamethasone (DEX), and transcriptomic profiles based on RNA-seq and miRNA-seq were performed on the pancreas and liver to explore the molecular mechanisms. Our results demonstrated that CPE could be a potential anti-diabetic agent to prevent and treat diabetes and its complications.
Results
CPE Alleviates Hyperglycemia and Improves Glucose Tolerance in Diabetic Rats
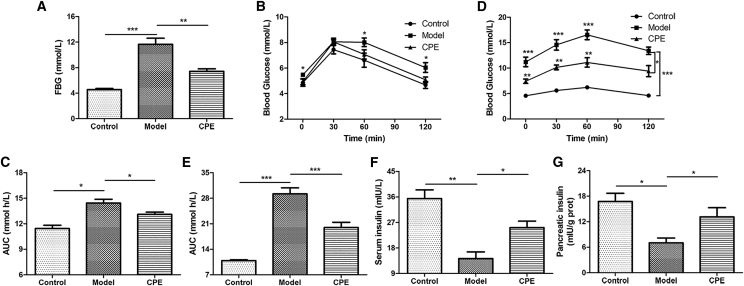
To investigate the anti-diabetic effect of CPE, HFD combined with DEX was used to induce diabetic rats. The dosage of 600 mg/kg body weight was chosen in our subsequent study since the dosage was better in our preliminary experiments (Figure S1A). As indicated, CPE treatment significantly decreased FBG compared to the model group (Figure 1A). At the end of 5 weeks’ treatment, the body weights of the CPE-treated group were much higher than the model group. In contrast, the food intake showed the opposite tendency (Figures S1B and S1C). Results of the oral glucose tolerance test (OGTT) and area under the curve (AUC) showed that glucose intolerance began to occur at the end of 3 weeks in the model group and CPE treatment improved glucose tolerance (Figures 1B and 1C). Especially, CPE treatment decreased blood glucose level at 30, 60, and 120 min after glucose loading compared with that of the model group at the end of 5 weeks’ treatment (Figure 1D). The AUC was also significantly decreased after CPE treatment (Figure 1E).
Figure 1.
CPE Treatment Alleviates Diabetes in Diabetic Rats
(A) The level of FBG at the end of the experiment before sacrifice. (B and D) The levels of blood glucose before oral glucose and after glucose administration at 30, 60, and 120 min in OGTT at the end of 3 weeks (B) and 5 weeks (D). (C and E) The AUC constructed from blood glucose levels of OGTT at the end of 3 weeks (C) and 5 weeks (E). (F and G) Levels of insulin in the serum (F) and pancreas tissue (G). All data are presented as means ± SEM (n = 6). *p < 0.05, **p < 0.01, ***p < 0.001.
To explore how CPE treatment helped to resist hyperglycemia, we detected levels of insulin. There were significant increases in insulin levels both in the serum and pancreas in the CPE-treated group compared with the model group (Figures 1F and 1G). These results suggested that CPE treatment alleviated diabetes in diabetic rats by increasing insulin secretion.
Transcriptome Analysis Demonstrates that CPE Protects the Pancreas by Inhibiting Inflammation and Apoptosis Pathways
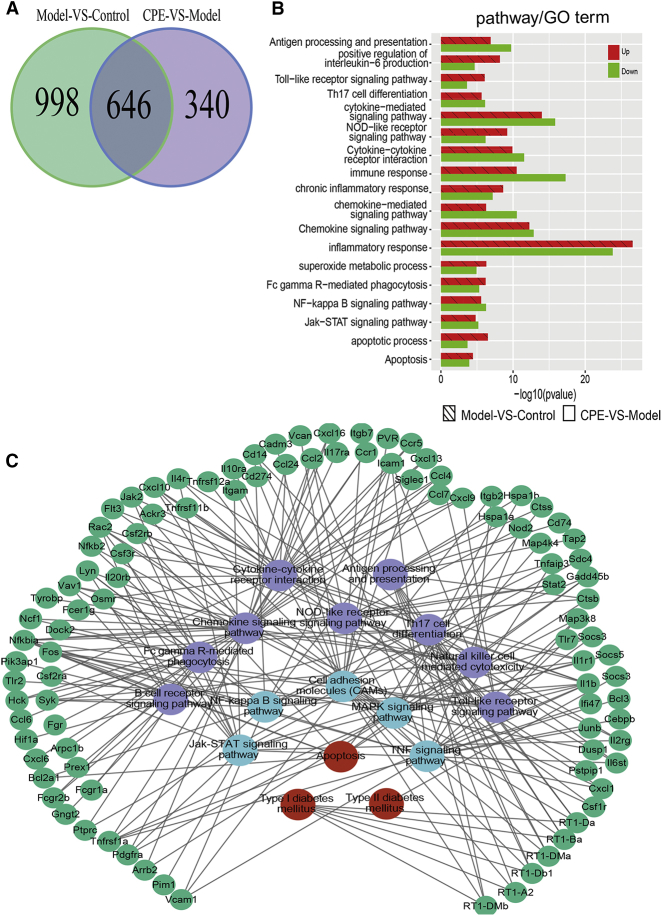
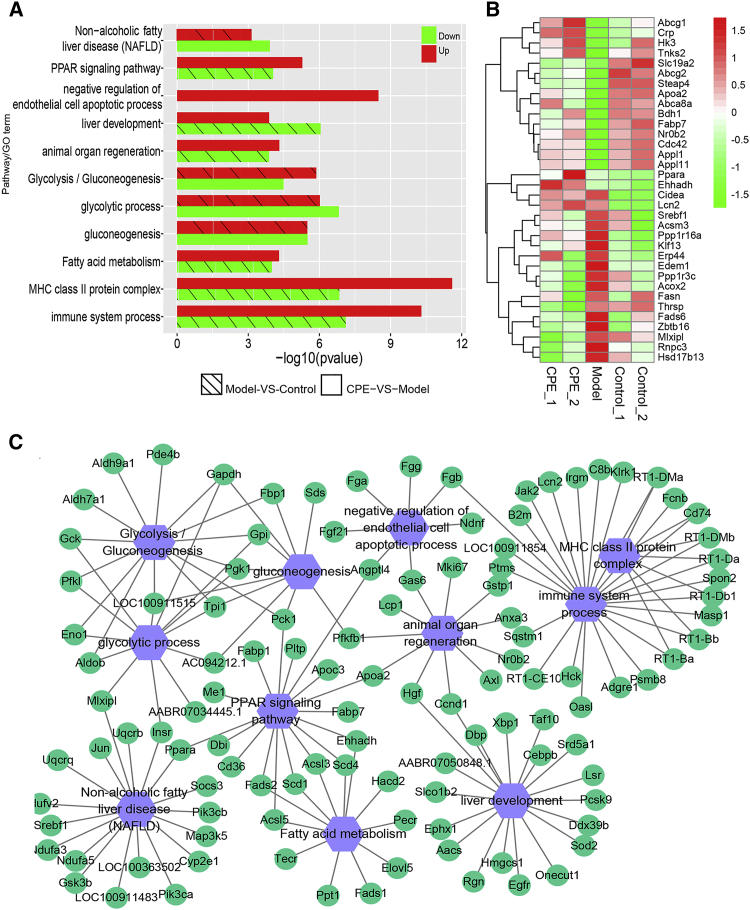
To understand the molecular mechanisms of CPE alleviating diabetes, we investigated the transcriptome profiling of the pancreas and liver in three groups, respectively. In pancreas, 16,109 genes (fragments per kilobase of transcript per million fragments mapped [FPKM] > 1) and 278 miRNAs (TPM [tags per million reads] > 10) were detected (Figures S2A and S2B). We identified 1,644 differentially expressed genes (DEGs) in the comparison of versus model control groups, and 986 DEGs were detected in the CPE-treated versus model groups. Interestingly, 646 common genes emerged in both comparisons (Figure 2A). The majority of the 646 genes were downregulated in the CPE-treated group and upregulated in the model group, and these were mainly related to inflammatory or immune response (cytokine-signaling pathways, NOD-like/Toll-like/nuclear factor κB [NF-κB] pathways) and apoptosis processes (Figure 2B). The results suggested that CPE treatment exhibited anti-diabetic effects probably through inhibiting inflammation and its induced apoptosis. Pathway crosstalk analysis for these genes showed that CPE treatment suppressed the expression of inflammatory factors and their receptors, such as interleukin and interferon family members (interleukin [IL]-1β/IL-1βr/IL-17/IL-17R and interferon [IFN]-γ/IFN-γR). Then it inhibited inflammation-related pathways and the downstream biological processes like Jak-STAT/NF-κB, and it alleviated apoptosis of islet cells to ameliorate diabetes (Figure 2C).
Figure 2.
Transcriptomic Profiling of the Pancreas
(A) DEGs between the comparisons of CPE-VS-Model group and Model-VS-Control group. (B) Functional enrichment of DEGs in the CPE-VS-Model and Model-VS-Control comparisons. (C) The pathway crosstalk of DEGs in CPE-VS-Model and Model-VS-Control comparisons.
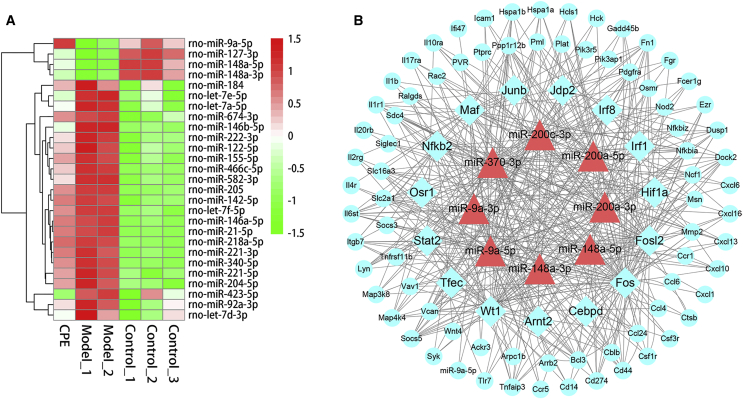
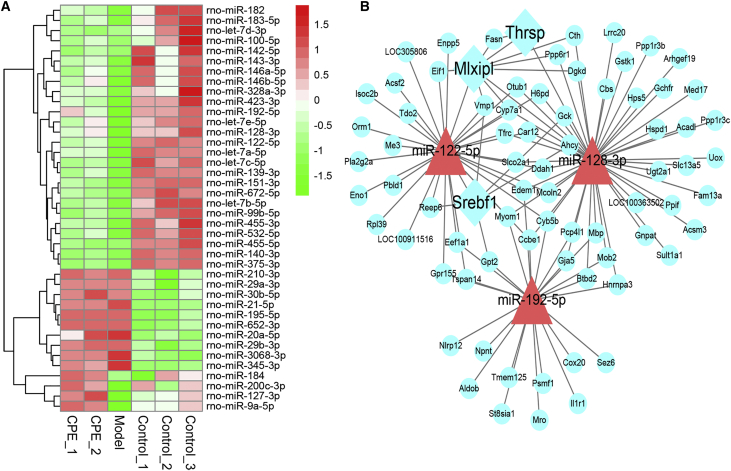
Results of miRNA-seq analysis showed that miR-148/143 and let-7 family members, including let-7a/c/i, accounted for 70% (Figure S2C). Particularly, 27 differentially expressed miRNAs (DEMs, 23 upregulated and 4 downregulated) between the two comparisons were selected for further analysis (Figure 3A). Most of the DEMs have been confirmed to be involved in the development of diabetes. Upregulation of miR-146/21/222 promotes inflammation-mediated β cell damage and apoptosis,19, 20 which were downregulated under CPE treatment (Figure 3A).
Figure 3.
Differentially Expressed miRNAs and miRNA-TF-Gene Analysis of the Inflammation and Apoptosis-Related Genes in the Pancreas
(A) Heatmap of differentially expressed miRNAs with opposite profiling in the comparison of CPE-VS-Model and Model-VS-Control. (B) Core miRNA-TF-gene regulatory network contributed to ameliorate diabetes in the pancreas under CPE treatment. Blue rectangles, TFs; red triangles, miRNAs; blue cycles, genes.
To investigate the roles of miRNAs and TFs participating in the anti-diabetes under CPE treatment, we constructed a miRNA-TF-gene regulatory network using DEMs and DEGs related to inflammation and apoptosis (Figure S3). The network demonstrated that TFs (e.g., STAT and NF-κB) and miRNAs (e.g., miR-200/9a/148a) were the hub nodes (Figure 3B). CPE treatment upregulated miR-148a/9a, which inhibited TFs NF-κB and STAT and downregulated inflammatory and apoptotic genes (Figure 3B). These data were consistent with previous reports that miR-148a-3p and miR-9a inhibited inflammation through inhibiting NF-κB activation and targeting the Jak/STAT-signaling pathway.21, 22 miR-200c can inhibit the NF-κB-signaling pathway and downregulate gene expressions of inflammatory cytokines,23 which were upregulated under CPE treatment. The regulatory network and functional enrichment analysis of DEGs indicated that CPE may improve diabetes by inhibiting β cell apoptosis induced by inflammation.
CPE Treatment Protects the Pancreas against Inflammation and Oxidative Stress Damage
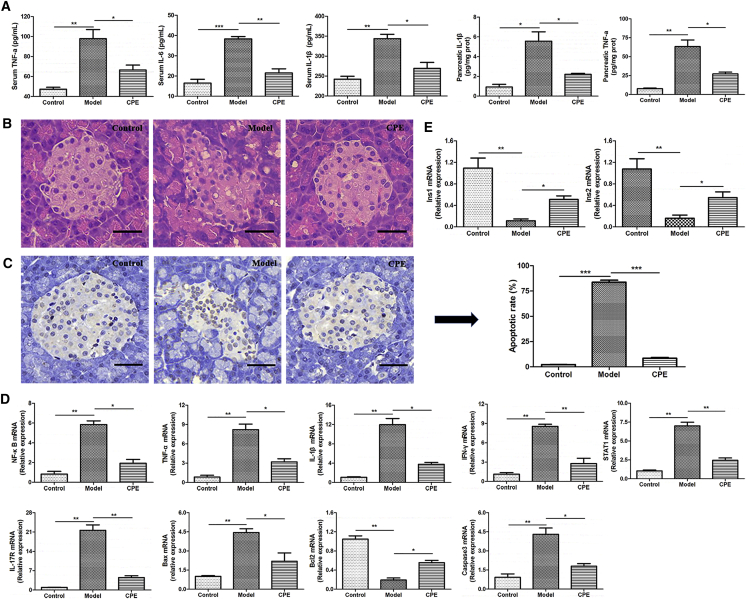
To validate the CPE effects on alleviating inflammation and apoptosis by bioinformatics analysis, inflammatory cytokines IL-1β, IL-6, and tumor necrosis factor alpha (TNF-α) were examined using ELISA kits. Results indicated that CPE treatment reduced the levels of IL-1β and TNF-α both in the serum and pancreas, as well as IL-6 in the serum (Figure 4A). Pancreatic histopathological section showed that pancreas islet of the model group was seriously damaged, characterized with an irregular shape and disorder islet cells, compared to that in the control group (Figure 4B). Improved signs of destruction could be seen in the islets following CPE treatment. An average of 84.2% apoptotic cells was found in the model group, which was decreased to an average of 6.8% after CPE treatment (Figure 4C). As indicated, the expressions of genes related to inflammation and apoptosis, including NF-κB/TNF-α/IL-1β/IFN-γ/STAT1/IL-17R/Caspase-3/Bax/Bcl2, were significantly downregulated after CPE treatment (Figure 4D). At the same time, CPE treatment upregulated gene expressions of Ins1 and Ins2 significantly, which were inhibited in the model group (Figure 4E). These findings were consistent with our RNA-seq and biochemical results that CPE treatment inhibited inflammation and apoptosis and increased insulin levels in the serum and pancreas (Figures 2B, 1F, and 1G). Therefore, our results demonstrated that CPE treatment reduced pancreas islet injury and inhibited β cell apoptosis through reducing inflammation, which guaranteed the normal secretion of insulin to reduce blood glucose.
Figure 4.
The Effects of CPE on the Inflammation and Apoptosis Processes in the Pancreas
(A) Levels of IL-1β, TNF-α, and IL-6 in the serum and pancreas tissue. (B) Histopathological examination of pancreas tissues (H&E staining) (magnification, 200×). Scale bars, 40 μm. (C) TUNEL staining of pancreas tissues (magnification, 200×) and apoptotic rate of pancreas islet cells. Scale bars, 40 μm. (D) Real-time qPCR results for selected genes involved in the inflammation and apoptosis. (E) Real-time qPCR results of Ins1 and Ins2. All data are presented as means ± SEM (n = 6). *p < 0.05, **p < 0.01, ***p < 0.001.
Chronic hyperglycemia is responsible for the oxidative stress observed in the pancreatic β cells.24 β cells are susceptible to oxidative damage because the islet has the lowest intrinsic antioxidant capacity.25 The level of malondialdehyde (MDA), concentration of glutathione, and activity of antioxidant enzymes superoxide dismutase (SOD) were detected (Figure S4). A great increase of serum MDA level and decreases of glutathione concentration and SOD activity in the serum and pancreatic tissue were observed in the model group, whereas CPE treatment decreased the level of MDA and increased glutathione concentration and SOD activity significantly. The results indicated that CPE treatment reduced oxidative stress damage.
Transcriptome Analysis and Biochemical Results Indicate that CPE Treatment Regulates Hepatic Lipid and Glucose Homeostasis
The liver controlling glucose and lipid metabolism homeostasis plays key roles in the development of diabetes.26 Transcriptome-profiling analysis of the liver demonstrated 10,787 genes (FPKM > 1) and 179 miRNAs (TPM > 10) were expressed among the control, model, and CPE-treated groups (Figures S5A and S5B). DEGs between CPE treatment and model groups were mainly assigned to lipid accumulation, including nonalcoholic fatty liver disease (NAFLD) and peroxisome proliferator activated receptor (PPAR) pathways, glycolysis, gluconeogenesis, and organ regeneration-related terms (Figure 5A). CPE treatment decreased the expression levels of genes involved in NAFLD and gluconeogenesis, which were increased in the model group (Figures 5B and 5C). Meanwhile, fatty acid metabolism and organ regeneration processes were strengthened in the CPE-treated group (Figure 5A). In particular, fatty acid β oxidation and lipid transport-associated genes, such as Ehhadh, Fabp7, and Apoa2, were upregulated under CPE treatment; while lipid synthesis-associated genes, including Fasn, Fads6, Acsm3, Cidea, and Acox2, were downregulated (Figure 5B). Therefore, CPE treatment decreased lipid synthesis and increased lipid oxidation and transport, as well as inhibited gluconeogenesis.
Figure 5.
Transcriptomic Profiling of the Liver
(A) Functional enrichment of DEGs in CPE-VS-Model and Model-VS-Control comparisons. (B) Heatmap for the gene expression of lipid synthesis, oxidation, and transport, as well as glucose metabolism. (C) The pathway crosstalk of DEGs in CPE-VS-Model and Model-VS-Control comparisons.
miRNA profiling showed that the expressions of miR-122, miR-143, and miR-21 accounted for nearly 70% of all miRNAs (Figure S5C). Some miRNAs were significantly changed after CPE treatment, such as miR-122/143/192 being significantly upregulated in the CPE-treated group compared with the model group (Figure 6A). To understand the miRNA function in liver, we constructed a miRNA-TF-target regulatory network for DEMs and DEGs enriched in the above functions (Figure 5A; Figure S6). miR-122/128/192 and their target genes were hub nodes in the work (Figure 6B). TFs Srebf1/Mlxipl were the major regulators of de novo lipid synthesis,27, 28 and these were the main hub TFs in this network. miR-122 targeting Srebf1 controlling the lipid accumulation participated in the development of NAFLD.29 miR-122/192 regulated the expression of Srebf1 and its downstream target genes, such as Fasn and Cidea, which were involved in the lipid synthesis.30, 31 CPE treatment upregulated the expression of miR-128, which inhibited the expression of Mlxipl (Figure 6B). Mlxipl targeted the Thrsp gene, which regulated fat synthesis by directly activating some classical lipogenic enzymes, such as Fasn.32, 33
Figure 6.
Differentially Expressed miRNAs and miRNA-TF-Gene Analysis of the Glucose and Lipid Metabolism-Related Genes in the Liver
(A) Heatmap of differentially expressed miRNAs with opposite profiling in the comparison of CPE-VS-Model and Model-VS-Control. (B) Core miRNA-TF-gene regulatory network contributed to ameliorate diabetes in the liver under CPE treatment. Blue rectangles, TFs; red triangles, miRNAs; green cycles, genes.
Thus, CPE treatment may reduce lipid synthesis by the miR-122/192-Srebf1-Fasn/Cidea and miR-128-Mlxipl-/Thrsp-Fasn pathways. NAFLD increases the susceptibility of the liver to acute liver injury and hepatocyte apoptosis.34 However, CPE treatment enhanced liver development and the organ regeneration process (Figure 5A). Our results suggested CPE treatment reduced lipid accumulation and liver injury by enhancing fatty acid oxidation and transport, as well as decreasing triglyceride synthesis.
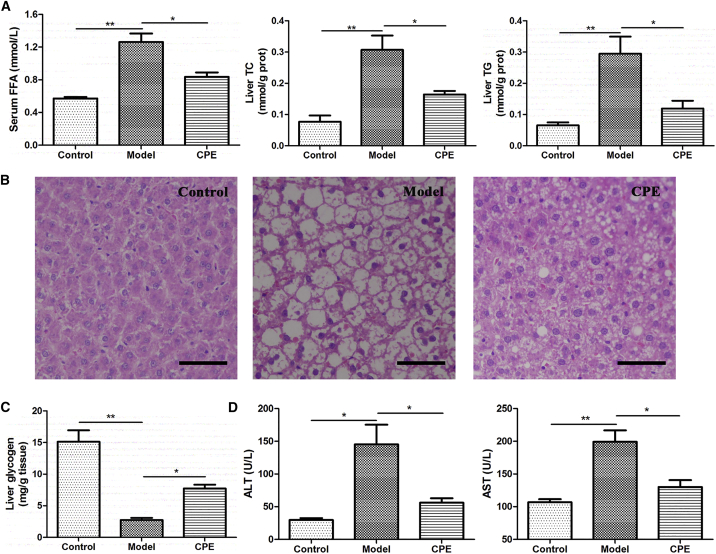
The results of morphological observation and biochemical experiment were consistent with transcriptome profiling. CPE treatment decreased the level of serum free fatty acids (FFAs) and the contents of triglycerides (TGs) and total cholesterol (TC) in the liver, and it increased the content of liver glycogen compared to that in the model group (Figures 7A and 7C). The histopathological section also demonstrated that CPE treatment reduced fat accumulation (Figure 7B). CPE treatment significantly decreased the levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT), markers of hepatic function, which were elevated in the model group (Figure 7D). Therefore, CPE treatment reduced fat accumulation, promoted glycogen biosynthesis, and alleviated liver injury.
Figure 7.
The Effects of CPE on Lipid Accumulation, Liver Injury, and Glycogen Synthesis
(A) Levels of serum FFA and liver TG and TC. (B) Histopathological examination of liver tissues (H&E staining) (magnification, 200×). Scale bars, 40 μm. (C) The content of liver glycogen. (D) Levels of serum ALT and AST. All data are presented as means ± SEM (n = 6). *p < 0.05, **p < 0.01.
Discussion
In T2D, pancreatic β cells fail to produce enough insulin to meet the body’s demand, partly due to an acquired decrease in β cell mass.35 Notably, increased β cell apoptosis is an important factor contributing to β cell loss,36 and protecting pancreatic β cells from apoptosis is beneficial to ameliorate T2D. Meanwhile, the onset and progression of T2D is often associated with dyslipidemia, which indicates a progressive impairment in hepatic control of glucose and lipid fluxes in the liver.37, 38 In the present study, our results demonstrated that CPE treatment attenuated inflammation and pancreas injury and inhibited β cell apoptosis, which ensured the normal secretion of insulin. Supplementation with CPE reduced lipid accumulation, increased FFA oxidation and transport, and eventually decreased liver injury.
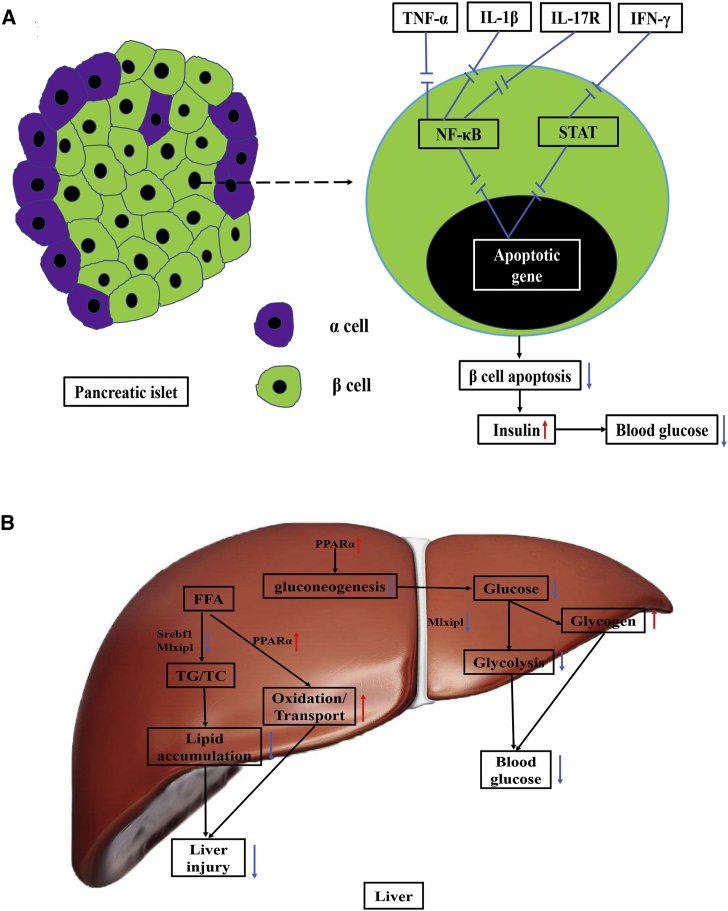
Insulin is the only hormone that decreases blood glucose in the body, which is secreted by pancreatic islet β cells. β cell dysfunction and apoptosis result in a decrease in insulin and lead to hyperglycemia.39 It was reported that islet inflammation played an important role in β cell apoptosis.3 In our study, CPE exhibited anti-inflammatory effects, which reduced β cell apoptosis and then guaranteed the secretion of insulin, contributing to the improving diabetes (Figure 8A). NF-κB and Jak-STAT pathways play vital roles in the β cell apoptosis induced by inflammation cytokines.3 Pro-inflammatory cytokines, such as IL-1β, TNF-α, and IL-17, induce the activation of NF-κB.40 IFN-γ can activate the Jak-STAT pathway.41 CPE treatment inhibited gene expressions of TNF-α/IL-1β/NF-κB/IFN-γ/STAT1/IL-17R and then reduced β cell apoptosis (Figure 4D). On the other hand, CPE treatment enhanced the gene expression of insulin, which increased the levels of insulin (Figures 1F, 1G, and 4E).
Figure 8.
The Potential Schematic Diagram of the Effects of CPE on the β Cell and Liver Tissue
(A) The potential pathway involved in inflammation and apoptosis under CPE treatment in the pancreas of diabetic rats. CPE treatment may significantly downregulate expressions of TNF-α, IL-1β, IFN-γ, and IL-17R, which blocks NF-κB and Jak-STAT pathways and then contributes to reduce β cell apoptosis. (B) The potential mechanisms that CPE treatment improves impaired glucose and lipid metabolism. CPE treatment downregulates Srebf1 and Mlxipl, which decreases the synthesis of TG/TC. Meanwhile, CPE treatment upregulates PPARa, which enhances the lipid transport and oxidation, as well as promotes glycogen synthesis. All the above effects contribute to alleviate liver injury and decrease blood glucose. Red arrow, upregulate; blue arrow, downregulate.
miRNAs have made an important contribution to regulate gene expression observed in dysfunctional β cells.42 CPE treatment downregulated miR-146/21/222, related to inflammation and apoptosis.19 CPE treatment downregulated let-7, which was reported to reduce glucose-induced insulin secretion.43 The feedback loops, miR-200/148a/NF-κB and miR-9a/STAT, as core nodes regulating inflammation-related signaling pathways, contributed to the improving diabetes after CPE treatment (Figure 3B). Given the above results, CPE treatment protected β cells from apoptosis induced by inflammation.
The combination of HFD and DEX induced serious lipid accumulation, which resulted in NAFLD and liver injury.44 NAFLD was caused by the imbalance between lipid synthesis and lipid oxidation and transport, which is associated with an increased risk of developing diabetes.26 In our study, CPE treatment significantly reduced lipid synthesis and enhanced lipid oxidation and transport, as well as promoted glycogen synthesis, and eventually it decreased liver injury and blood glucose (Figure 8B). CPE treatment upregulated the PPARa signal pathway, which activated Cd36, Fabp7, and Ehhadh, contributing to the decrease of cholesterol and the enhancements of β oxidation and transport of fatty acid (Figures 5A and 5B). The upregulation of the PPARa signal pathway also inhibited gluconeogenesis.45 Meanwhile, CPE treatment upregulated miR-143, which inhibited the lipogenesis by suppressing the expression of Fasn.46 Moreover, the regulatory network between miR-122/192/128 and Srebf1/Mlxipl played an important role in improving diabetes, which reduced lipid synthesis and glycolysis in the liver (Figure 6B). On the other hand, CPE treatment enhanced glucose utilization by downregulating gluconeogenesis and promoting glycogen synthesis (Figures 5A, 5C, and 7C). Therefore, CPE treatment ameliorated impaired glucose and lipid homeostasis by reducing lipid accumulation and increasing lipid transport, as well as increasing glucose utilization.
In summary, we validated that CPE treatment could ameliorate diabetes by protecting pancreas islet from injury and inhibiting β cell apoptosis in diabetic rats. It was the protective effects of CPE on β cells that guaranteed the normal secretion of insulin, which decreased the blood glucose. In addition, CPE treatment improved impaired glucose and lipid metabolism by reducing lipid accumulation and promoting glycogen biosynthesis in liver tissue, which contributed to ameliorating diabetes. Therefore, CPE could be an alternative for preventing and treating diabetes. Furthermore, our study indicated that transcriptome profiles based on RNA-seq and miRNA-seq could provide insights for exploring the potential mechanism of TCM on complex diseases.
Materials and Methods
Preparation of the CPE
CPE contained 4 crude plant products, consisting of Cyclocarya paliurus, Dendrobium, Morus alba L., and Pericarpium Citri Reticulatae. Briefly, leaves of Cyclocarya paliurus and stems of Dendrobium, Morus alba, and Pericarpium Citri Reticulatae were mixed in the ratio of 4:2:2:1 (dry weight). The mixture was decocted with 20 vol (v/w) boiling distilled water for 2 hr, after which the filtrate was collected and the residue was decocted once again. The process was repeated three times. Extractions of filtrates were collected and then concentrated in vacuo to obtain CPE. The yield was 28.6% relative to the original crude plant products. It was dissolved in physiological saline before use. The phenol-sulfuric acid colorimetric titration method was used for the determination of polysaccharide content, which was 5.12/100 g CPE. The estimation of total flavonoid content was performed by the aluminum chloride method; it was 2.24/100 g CPE.
Animals
Male Sprague-Dawley rats (200 ± 20 g) were purchased from Hubei Province Center for Disease Control and Prevention. Rats were separated into control, model, and CPE-treated groups, respectively. Rats in the control group were fed with a standard diet for 5 weeks. The model and CPE-treated groups were fed with a standard diet for the first week, and then with a HFD (sucrose 15%, lard oil 10%, egg yolk powder 5%, cholesterol 1%, bile acid sodium 0.2%, and standard diet 68.8%) for the second and third weeks. At the fourth and fifth weeks, the model and CPE-treated groups were injected intraperitoneally with DEX (0.8 mg/kg) daily based on the HFD feeding. CPE-treated groups received an oral daily dose of CPE 600 mg/kg body weight, whereas the control and model groups received only the equivalent amount of physiological saline.
At the end of the experiment, all rats were fasted overnight and anesthetized with pentobarbital sodium. Blood samples were obtained from abdominal aorta, and sera were collected and stored at −20°C in a freezer until use. The pancreas and liver tissues were isolated, washed with physiological saline solution, immediately frozen in liquid nitrogen, and stored at −80°C. The animal experimental protocol was approved by the Animal Experimentation Ethics Committee of Huazhong University of Science and Technology, and it was performed in accordance with guidelines approved by the Science and Technology Department of Hubei Province.
Measurement of Blood Glucose, Insulin, and Oral Glucose Tolerance
FBG was measured in blood collected from the tail. A blood glucose meter (Accu-Check Active 1, Roche, Basel, Switzerland) was used for blood glucose detection. An OGTT was performed at the end of 3 and 5 weeks. After an overnight fast, blood glucose levels were measured immediately before and 30, 60, and 120 min after glucose administration at 2.5 g/kg (Sigma, USA). Insulin levels both in the serum and pancreas were measured with insulin ELISA Kit (Nanjing Jiancheng Bioengineering Institute, China).
Analysis of Inflammatory Cytokines and Oxidative Stress in Serum and Pancreatic Tissue
Levels of TNF-α, IL-1β, and IL-6 were measured with commercial ELISA Kits (Cloud Clone, China). The level of MDA and activities of SOD and glutathione were measured using commercial kits purchased from Nanjing Jiancheng Bioengineering Institute (China). The protein concentration of the pancreas was assayed with a BCA Protein Assay Kit (Beyotime Biotechnology, China).
Measurement of FFA, AST, and ALT in Serum and TG, TC, and Glycogen in the Liver
Liver glycogen was assayed using a liver glycogen assay kit. The level of serum FFA and contents of TG and TC in the liver were detected using corresponding kits. All assay kits used were obtained from Nanjing Jiancheng Bioengineering Institute. The activities of serum AST and ALT were detected with an automatic biochemical analyzer in Wuhan general hospital of Guangzhou military (China). The protein concentration of the liver was assayed with a BCA Protein Assay Kit (Beyotime Biotechnology, China).
Histopathological Examination
Histopathological examinations of pancreas and liver tissues were performed as in the literature.11 Briefly, pancreatic and liver tissues were dehydrated in alcohol, embedded in paraffin, and sectioned at 5-μm thickness using a microtome (RM2235 ccwUS, Leica Microscopy System). The sections were dewaxed in xylene and then in a graded series of ethanol to elute xylene, washed with water, and stained with H&E. Histopathological examinations were carried out under a light microscope (DM3000, Leica Microscopy System, Germany).
TUNEL Staining
Apoptotic pancreatic β cells were visualized by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining according to the manufacturer’s instructions (Bioyear Biological Technology). Briefly, the sections were digested with proteinase K and washed three times. After that, the sections were incubated with terminal deoxynucleotidyl transferase (TdT) buffer at 37°C for 60 min and then incubated with dUTP. At last, streptavidin-labeled horseradish peroxidase (HRP) and diaminobenzidine (DAB) were added for chromogenic reaction.
RNA Isolation, Library Preparation, and Sequencing
The pancreas and liver tissues were collected from each group. Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, NY, USA). The RNA concentrations and purities were measured using a spectrophotometer (Nanodrop 2000c). After establishing cDNA libraries, their quality was evaluated using an Agilent 2100 BioAnalyzer (Agilent, CA, USA). RNA-seq (Ribo-Zero) and miRNA-seq libraries were prepared using TruSeq protocol and sequenced using the Illumina HiSeq2500 (Illumina, CA, USA). Base-calling was performed using the Illumina CASAVA pipeline (version 1.8.2). The raw data have been deposited in the Genome Sequence Archive of the Beijing Institute of Genomics, Chinese Academy of Sciences (http://gsa.big.ac.cn/; Genome Sequence Archive [GSA]: CRA000812 and CRA000814).
miRNA-Seq: Identification, Quantification, and DEM Analysis
Low-quality reads were filtered out according to the following criteria: miRNA-seq read length <15 nt or >45 nt, reads with base N, and reads with an average quality score ≤10. The remaining reads were mapped onto rat canonical pre-miRNA sequences (miRBase version (v.)21.0).47 In addition, other noncoding RNA databases, such as GenBank, Rfam, and Piwi, were used for alignment. The normalized values (TPM) and read count of expressed miRNAs in different samples were merged into matrix, respectively. The DESeq2 and NOISeq packages48 were used to quantify and assess DEMs with a threshold false discovery rate (FDR) < 0.05 and |fold change| > 1.5.
RNA-Seq: Read Processing and DEG Analysis
RNA-seq reads, <35 bp after adaptor trimming or with ploy-N (>7 base) or ratio of low quality (quality value ≤ 5) base > 10%, were removed from raw data by using FLEXBAR.49 The remaining reads were aligned to Rnor_6.0 with HISAT2.50 Transcript reassembly and quantification were processed according to the HISAT-StringTie-ballgown pipeline51 with the GTF file derived from the database of Ensembl (build 83). The resulting transcripts were pooled across samples using the merge function of StringTie,52 discarding all contained or redundant and low-expressed isoforms (FPKM < 0.5). The transcripts gained from the above steps were considered as the high-confident (HC) transcript dataset and used for further studies. Normalized abundance estimates (FPKM) of genes were computed by StringTie with all HC transcripts. Only reliably expressed genes (average across replicates) with FPKM > 1 were included in the DEG analysis, which was performed using ballgown and NOISeq with default parameters under threshold FDR < 0.05 and |fold change| > 2.
Real-Time qPCR
cDNA was synthesized using PrimeScriptTM RT reagent Kit with gDNA Eraser. Then real-time PCR was carried out using SYBR Premix Ex TaqTM II (Tli RNaseH Plus) (Takara Biotechnology, Dalian, China) in a Bio-Rad C1000 detecting system (StepOnePlus_1). β-actin was chosen as a housekeeping gene. mRNA levels of NF-κB, TNF-α, IL-1β, IFN-γ, STAT1, IL-17R, Bax, Bcl2, Ins1, Ins2, and Caspase3 were compared with that of β-actin. Primers used for RT-PCR were according to the literature.53, 54, 55, 56, 57 The fold change for all the samples was calculated by the 2−Ct method.
Statistical and Regulatory Network Analysis
All data were given as mean value and SEM. Statistical analysis was conducted by one-way ANOVA with SPSS software, and p < 0.05 was considered as statistically significant. The hypergeometric tests for pathway and gene ontology enrichment were analyzed by in-house R scripts. The gene list of TFs was downloaded from AnimalTFDB 2.0.58 The method used for regulatory network analysis was described in our previous work.59 Graphs were created by gglot2 and cytoscape.
Author Contributions
Y.Z., X.Y., and A.-Y.G. designed the research and revised the manuscript. J.L. performed the experiments and wrote the manuscript. Q.Z. performed bioinformatics analysis and partially wrote the manuscript. W.Z. and Y.W. assisted with the experiments. M.L. drew some pictures for this paper.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (NSFC) (81573013, 31471247, 31771458, and 31801113), the National Basic Research Program of China (2015CB931802), PCSIRT (IRT13016), the China Postdoctoral Science Foundation (2018M632830), and the National Key Research and Development Program of China (2017YFA0700403).
Footnotes
Supplemental Information includes six figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.024.
Contributor Information
Yanhong Zhu, Email: yhzhu@hust.edu.cn.
An-Yuan Guo, Email: guoay@hust.edu.cn.
Xiangliang Yang, Email: yangxl@hust.edu.cn.
Supplemental Information
References
- 1.Kasuga M. Insulin resistance and pancreatic beta cell failure. J. Clin. Invest. 2006;116:1756–1760. doi: 10.1172/JCI29189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donath M.Y., Böni-Schnetzler M., Ellingsgaard H., Ehses J.A. Islet inflammation impairs the pancreatic beta-cell in type 2 diabetes. Physiology (Bethesda) 2009;24:325–331. doi: 10.1152/physiol.00032.2009. [DOI] [PubMed] [Google Scholar]
- 3.Marzban L. New insights into the mechanisms of islet inflammation in type 2 diabetes. Diabetes. 2015;64:1094–1096. doi: 10.2337/db14-1903. [DOI] [PubMed] [Google Scholar]
- 4.Whiting D.R., Guariguata L., Weil C., Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res. Clin. Pract. 2011;94:311–321. doi: 10.1016/j.diabres.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 5.Liao X., Yang L., Chen M., Yu J., Zhang S., Ju Y. The hypoglycemic effect of a polysaccharide (GLP) from Gracilaria lemaneiformis and its degradation products in diabetic mice. Food Funct. 2015;6:2542–2549. doi: 10.1039/c4fo01185f. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., Wang J., Chan P. Treating type 2 diabetes mellitus with traditional chinese and Indian medicinal herbs. Evid. Based Complement. Alternat. Med. 2013;2013:343594. doi: 10.1155/2013/343594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J.P., Zhang M., Wang W.Y., Grimsgaard S. Chinese herbal medicines for type 2 diabetes mellitus. Cochrane Database Syst. Rev. 2004;(3):CD003642. doi: 10.1002/14651858.CD003642.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seto S.W., Yang G.Y., Kiat H., Bensoussan A., Kwan Y.W., Chang D. Diabetes Mellitus, Cognitive Impairment, and Traditional Chinese Medicine. Int. J. Endocrinol. 2015;2015:810439. doi: 10.1155/2015/810439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu I.M., Tzeng T.F., Liou S.S., Chang C.J. The amelioration of streptozotocin diabetes-induced renal damage by Wu-Ling-San (Hoelen Five Herb Formula), a traditional Chinese prescription. J. Ethnopharmacol. 2009;124:211–218. doi: 10.1016/j.jep.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Li W.L., Zheng H.C., Bukuru J., De Kimpe N. Natural medicines used in the traditional Chinese medical system for therapy of diabetes mellitus. J. Ethnopharmacol. 2004;92:1–21. doi: 10.1016/j.jep.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Xiao H.T., Wen B., Ning Z.W., Zhai L.X., Liao C.H., Lin C.Y., Mu H.X., Bian Z.X. Cyclocarya paliurus tea leaves enhances pancreatic β cell preservation through inhibition of apoptosis. Sci. Rep. 2017;7:9155. doi: 10.1038/s41598-017-09641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H., Zhao T., Sheng Y., Zheng T., Fu L., Zhang Y. Dendrobium officinale Kimura et Migo: A Review on Its Ethnopharmacology, Phytochemistry, Pharmacology, and Industrialization. Evid. Based Complement. Alternat. Med. 2017;2017:7436259. doi: 10.1155/2017/7436259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singab A.N., El-Beshbishy H.A., Yonekawa M., Nomura T., Fukai T. Hypoglycemic effect of Egyptian Morus alba root bark extract: effect on diabetes and lipid peroxidation of streptozotocin-induced diabetic rats. J. Ethnopharmacol. 2005;100:333–338. doi: 10.1016/j.jep.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed O.M., Hassan M.A., Abdel-Twab S.M., Abdel Azeem M.N. Navel orange peel hydroethanolic extract, naringin and naringenin have anti-diabetic potentials in type 2 diabetic rats. Biomed. Pharmacother. 2017;94:197–205. doi: 10.1016/j.biopha.2017.07.094. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Li Q., Chamba Y., Zhang B., Shang P., Zhang H., Wu C. Identification of Genes Related to Growth and Lipid Deposition from Transcriptome Profiles of Pig Muscle Tissue. PLoS ONE. 2015;10:e0141138. doi: 10.1371/journal.pone.0141138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cnop M., Abdulkarim B., Bottu G., Cunha D.A., Igoillo-Esteve M., Masini M., Turatsinze J.V., Griebel T., Villate O., Santin I. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63:1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 17.Lynn F.C., Skewes-Cox P., Kosaka Y., McManus M.T., Harfe B.D., German M.S. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Lu M., Qiu C., Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma X., Becker Buscaglia L.E., Barker J.R., Li Y. MicroRNAs in NF-kappaB signaling. J. Mol. Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihle M.A., Trautmann M., Kuenstlinger H., Huss S., Heydt C., Fassunke J., Wardelmann E., Bauer S., Schildhaus H.U., Buettner R., Merkelbach-Bruse S. miRNA-221 and miRNA-222 induce apoptosis via the KIT/AKT signalling pathway in gastrointestinal stromal tumours. Mol. Oncol. 2015;9:1421–1433. doi: 10.1016/j.molonc.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel V., Carrion K., Hollands A., Hinton A., Gallegos T., Dyo J., Sasik R., Leire E., Hardiman G., Mohamed S.A. The stretch responsive microRNA miR-148a-3p is a novel repressor of IKBKB, NF-κB signaling, and inflammatory gene expression in human aortic valve cells. FASEB J. 2015;29:1859–1868. doi: 10.1096/fj.14-257808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Han Z., Fan Y., Zhang J., Chen K., Gao L., Zeng H., Cao J., Wang C. MicroRNA-9 Inhibits NLRP3 Inflammasome Activation in Human Atherosclerosis Inflammation Cell Models through the JAK1/STAT Signaling Pathway. Cell. Physiol. Biochem. 2017;41:1555–1571. doi: 10.1159/000470822. [DOI] [PubMed] [Google Scholar]
- 23.Chuang T.D., Khorram O. miR-200c regulates IL8 expression by targeting IKBKB: a potential mediator of inflammation in leiomyoma pathogenesis. PLoS ONE. 2014;9:e95370. doi: 10.1371/journal.pone.0095370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihara Y., Toyokuni S., Uchida K., Odaka H., Tanaka T., Ikeda H., Hiai H., Seino Y., Yamada Y. Hyperglycemia causes oxidative stress in pancreatic beta-cells of GK rats, a model of type 2 diabetes. Diabetes. 1999;48:927–932. doi: 10.2337/diabetes.48.4.927. [DOI] [PubMed] [Google Scholar]
- 25.Robertson R.P., Harmon J., Tran P.O., Poitout V. Beta-cell glucose toxicity, lipotoxicity, and chronic oxidative stress in type 2 diabetes. Diabetes. 2004;53(Suppl 1):S119–S124. doi: 10.2337/diabetes.53.2007.s119. [DOI] [PubMed] [Google Scholar]
- 26.Perry R.J., Samuel V.T., Petersen K.F., Shulman G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postic C., Dentin R., Denechaud P.D., Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu. Rev. Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 28.Rudolph M.C., Monks J., Burns V., Phistry M., Marians R., Foote M.R., Bauman D.E., Anderson S.M., Neville M.C. Sterol regulatory element binding protein and dietary lipid regulation of fatty acid synthesis in the mammary epithelium. Am. J. Physiol. Endocrinol. Metab. 2010;299:E918–E927. doi: 10.1152/ajpendo.00376.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung O., Puri P., Eicken C., Contos M.J., Mirshahi F., Maher J.W., Kellum J.M., Min H., Luketic V.A., Sanyal A.J. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H., Wang H., Zhou F., Yang J. [Roles of microRNAs in non-alcoholic fatty liver disease] Chin. J. Hepatol. 2014;22:472–474. [PubMed] [Google Scholar]
- 31.Lin Y., Ding D., Huang Q., Liu Q., Lu H., Lu Y., Chi Y., Sun X., Ye G., Zhu H. Downregulation of miR-192 causes hepatic steatosis and lipid accumulation by inducing SREBF1: Novel mechanism for bisphenol A-triggered non-alcoholic fatty liver disease. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:869–882. doi: 10.1016/j.bbalip.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Sud N., Zhang H., Pan K., Cheng X., Cui J., Su Q. Aberrant expression of microRNA induced by high-fructose diet: implications in the pathogenesis of hyperlipidemia and hepatic insulin resistance. J. Nutr. Biochem. 2017;43:125–131. doi: 10.1016/j.jnutbio.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimada M., Mochizuki K., Goda T. Feeding rats dietary resistant starch reduces both the binding of ChREBP and the acetylation of histones on the Thrsp gene in the jejunum. J. Agric. Food Chem. 2011;59:1464–1469. doi: 10.1021/jf103111u. [DOI] [PubMed] [Google Scholar]
- 34.Ibrahim S.H., Kohli R., Gores G.J. Mechanisms of lipotoxicity in NAFLD and clinical implications. J. Pediatr. Gastroenterol. Nutr. 2011;53:131–140. doi: 10.1097/MPG.0b013e31822578db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donath M.Y., Ehses J.A., Maedler K., Schumann D.M., Ellingsgaard H., Eppler E., Reinecke M. Mechanisms of beta-cell death in type 2 diabetes. Diabetes. 2005;54(Suppl 2):S108–S113. doi: 10.2337/diabetes.54.suppl_2.s108. [DOI] [PubMed] [Google Scholar]
- 36.Rhodes C.J. Type 2 diabetes-a matter of beta-cell life and death? Science. 2005;307:380–384. doi: 10.1126/science.1104345. [DOI] [PubMed] [Google Scholar]
- 37.Staehr P., Hother-Nielsen O., Beck-Nielsen H. The role of the liver in type 2 diabetes. Rev. Endocr. Metab. Disord. 2004;5:105–110. doi: 10.1023/B:REMD.0000021431.90494.0c. [DOI] [PubMed] [Google Scholar]
- 38.Jones J.G. Hepatic glucose and lipid metabolism. Diabetologia. 2016;59:1098–1103. doi: 10.1007/s00125-016-3940-5. [DOI] [PubMed] [Google Scholar]
- 39.Chandra J., Zhivotovsky B., Zaitsev S., Juntti-Berggren L., Berggren P.O., Orrenius S. Role of apoptosis in pancreatic beta-cell death in diabetes. Diabetes. 2001;50(Suppl 1):S44–S47. doi: 10.2337/diabetes.50.2007.s44. [DOI] [PubMed] [Google Scholar]
- 40.Gysemans C., Callewaert H., Overbergh L., Mathieu C. Cytokine signalling in the beta-cell: a dual role for IFNgamma. Biochem. Soc. Trans. 2008;36:328–333. doi: 10.1042/BST0360328. [DOI] [PubMed] [Google Scholar]
- 41.Arif S., Moore F., Marks K., Bouckenooghe T., Dayan C.M., Planas R., Vives-Pi M., Powrie J., Tree T., Marchetti P. Peripheral and islet interleukin-17 pathway activation characterizes human autoimmune diabetes and promotes cytokine-mediated β-cell death. Diabetes. 2011;60:2112–2119. doi: 10.2337/db10-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lovis P., Roggli E., Laybutt D.R., Gattesco S., Yang J.Y., Widmann C., Abderrahmani A., Regazzi R. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frost R.J., Olson E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc. Natl. Acad. Sci. USA. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Videla L.A., Pettinelli P. Misregulation of PPAR Functioning and Its Pathogenic Consequences Associated with Nonalcoholic Fatty Liver Disease in Human Obesity. PPAR Res. 2012;2012:107434. doi: 10.1155/2012/107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bechmann L.P., Hannivoort R.A., Gerken G., Hotamisligil G.S., Trauner M., Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012;56:952–964. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 46.Li W., Li J., Li Y., Ren L., Song W. Inhibition of the lipogenesis of hepatocellular carcinoma HepG2 cells by miR-143 via suppressing FASN. J. Shanghai Jiaotong Univ. 2015;35:337–342. [Google Scholar]
- 47.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarazona S., Furió-Tarí P., Turrà D., Pietro A.D., Nueda M.J., Ferrer A., Conesa A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dodt M., Roehr J.T., Ahmed R., Dieterich C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aloui F., Charradi K., Hichami A., Subramaniam S., Khan N.A., Limam F., Aouani E. Grape seed and skin extract reduces pancreas lipotoxicity, oxidative stress and inflammation in high fat diet fed rats. Biomed. Pharmacother. 2016;84:2020–2028. doi: 10.1016/j.biopha.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 54.Palumbo M.O., Levi D., Chentoufi A.A., Polychronakos C. Isolation and characterization of proinsulin-producing medullary thymic epithelial cell clones. Diabetes. 2006;55:2595–2601. doi: 10.2337/db05-1651. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Z., Chen H., Wang H., Ke B., Zheng B., Li Q., Li P., Su L., Gu Q., Xu X. Improvement of retinal vascular injury in diabetic rats by statins is associated with the inhibition of mitochondrial reactive oxygen species pathway mediated by peroxisome proliferator-activated receptor gamma coactivator 1alpha. Diabetes. 2010;59:2315–2325. doi: 10.2337/db10-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ninkov M., Popov Aleksandrov A., Mirkov I., Demenesku J., Mileusnic D., Jovanovic Stojanov S., Golic N., Tolinacki M., Zolotarevski L., Kataranovski D. Strain differences in toxicity of oral cadmium intake in rats. Food Chem. Toxicol. 2016;96:11–23. doi: 10.1016/j.fct.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 57.Zhu M., Yuan S.T., Yu W.L., Jia L.L., Sun Y. CXCL13 regulates the trafficking of GluN2B-containing NMDA receptor via IL-17 in the development of remifentanil-induced hyperalgesia in rats. Neurosci. Lett. 2017;648:26–33. doi: 10.1016/j.neulet.2017.03.044. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H.M., Liu T., Liu C.J., Song S., Zhang X., Liu W., Jia H., Xue Y., Guo A.Y. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2015;43:D76–D81. doi: 10.1093/nar/gku887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin Y., Zhang Q., Zhang H.M., Liu W., Liu C.J., Li Q., Guo A.Y. Transcription factor and miRNA co-regulatory network reveals shared and specific regulators in the development of B cell and T cell. Sci. Rep. 2015;5:15215. doi: 10.1038/srep15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.