Abstract
Hepatocellular carcinoma (HCC) is highly heterogeneous in nature and has been one of the most common cancer types worldwide. To ensure repeatability of identified gene expression patterns and comprehensively annotate the transcriptomes of HCC, we carefully curated 15 public HCC expression datasets that cover around 4000 clinical samples and developed the database HCCDB to serve as a one-stop online resource for exploring HCC gene expression with user-friendly interfaces. The global differential gene expression landscape of HCC was established by analyzing the consistently differentially expressed genes across multiple datasets. Moreover, a 4D metric was proposed to fully characterize the expression pattern of each gene by integrating data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx). To facilitate a comprehensive understanding of gene expression patterns in HCC, HCCDB also provides links to third-party databases on drug, proteomics, and literatures, and graphically displays the results from computational analyses, including differential expression analysis, tissue-specific and tumor-specific expression analysis, survival analysis, and co-expression analysis. HCCDB is freely accessible at http://lifeome.net/database/hccdb.
Keywords: Hepatocellular carcinoma, Database, Transcriptome, Integrative analysis, Meta-analysis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and lethal cancer types. With the development of high-throughput technologies, various types of molecular data have been used to delineate the alterations in HCC at multiple levels [1], [2], [3]. Good curation of these molecular data can lay the foundation for scientific research and drive translation in clinical applications. Until now, some efforts have been made in the data curation about HCC, such as OncoDB.HCC [4] and Liverome [5]. OncoDB.HCC was established as a resource of HCC genomes, to visualize somatic aberrations at levels varying from DNA, RNA to protein, with main focus on HCC tumorigenesis. Liverome was set up to curate liver cancer-related gene signatures and provide interface for gene search and signature comparison.
In the past decades, transcriptomic data have been accumulated vastly and applied widely in molecular classification [6], [7] and biomarker identification [8], [9]. As HCC is a highly heterogeneous disease [10], [11], [12], inference of expression signatures from single gene expression dataset usually faces challenges of low repeatability and high false positive rates. To improve the confidence of high-throughput experiments, gene expression patterns should be carefully assessed in multiple datasets derived from independent studies [6], [7], [13]. However, there is no large-scale collection of HCC transcriptomics data, let alone detailed annotation and intuitive display.
Recent studies have revealed that tissue-specificity matters in tumorigenesis [14]. Different cancer types share a set of common cancer hallmarks [15], although they also have extensive tissue-specific oncogenic processes [14], [16], [17], [18], [19]. HCC has been demonstrated to harbor aberrant expression patterns that are obviously different from those observed in other cancer types [20], [21]. So it is important to consider the tissue-specificity information when annotating gene expression patterns in HCC.
Thus, we first compiled a unique resource by carefully curating 15 public HCC gene expression datasets that cover around 4000 clinical samples to facilitate the hypothesis testing and pattern discovery based on multiple gene expression datasets. Meanwhile, we analyzed the consistently differentially expressed genes across multiple datasets to depict a global differential expression landscape of HCC. A 4D metric was proposed to summarize the expression pattern of individual gene by integrating normal tissues from the Genotype-Tissue Expression (GTEx) [22] and tumors from The Cancer Genome Atlas (TCGA). Aiming at providing a one-stop resource for gene expression atlas in HCC, we developed a web-based database HCCDB. HCCDB provides the visualization for the results from several computational analyses, including differential expression analysis, tissue-specific and tumor-specific expression analysis, survival analysis, and co-expression analysis. And it also provides links to third-party databases on drug, proteomics, and literatures. Users can browse or search these results through a simple web-based interface.
Database content and computation methods
The archived expression datasets
In the current database release, we archived 15 public HCC gene expression datasets containing totally 3917 samples (Table 1). For 13 microarray datasets, probe values (log2 intensity) and probe annotations were extracted from raw files downloaded from the Gene Expression Omnibus (GEO) database. Multiple probes mapped to a single gene (i.e., unique Entrez gene ID) were collapsed as their medians. For the two remaining RNA-seq datasets, Liver Hepatocellular Carcinoma Project of The Cancer Genome Atlas (TCGA-LIHC) and Liver Cancer - RIKEN, JP Project from International Cancer Genome Consortium (ICGC LIRI-JP), we took the normalized read counts for log2 transformation. Among the 15 datasets, 12 datasets contain both tumor and the adjacent normal samples, whereas only tumor samples are available for the three remaining datasets. Clinical information, such as tumor stages and survival time, was also collected if available.
Table 1.
The collected gene expression datasets in HCCDB
| Dataset ID | No. of adjacent tissue samples | No. of HCC samples | No. of other normal tissue samples | Platform | Source |
|---|---|---|---|---|---|
| HCCDB1 | 97 | 100 | Rosetta/Merck Human RSTA Custom Affymetrix 1.0 microarray | GSE22058 | |
| HCCDB3 | 243 | 268 | Healthy 6; Cirrhotic 40 | Rosetta/Merck Human RSTA Affymetrix 1.0 microarray | GSE25097 |
| HCCDB4 | 193 | 240 | Illumina HumanHT-12 V4.0 expression beadchip | GSE36376 | |
| HCCDB6 | 220 | 225 | Affymetrix Human Genome U133A 2.0 Array | GSE14520 | |
| HCCDB7 | 82 | 80 | Human 6k Transcriptionally Informative Gene Panel for DASL | GSE10143 | |
| HCCDB8 | 0 | 91 | Affymetrix Human Genome U133 Plus 2.0 Array | GSE9843 | |
| HCCDB9 | 0 | 164 | Illumina HumanRef-8 WG-DASL v3.0 | GSE19977 | |
| HCCDB11 | 48 | 88 | Illumina Human Whole-Genome DASL HT | GSE46444 | |
| HCCDB12 | 80 | 81 | Agilent-014850 Whole Human Genome Microarray 4x44K G4112F | GSE54236 | |
| HCCDB13 | 168 | 228 | Affymetrix Human Genome U219 Array | GSE63898 | |
| HCCDB14 | 0 | 88 | Illumina HumanHT-12 V4.0 expression beadchip | GSE43619 | |
| HCCDB15 | 49 | 356 | RNA-Seq | TCGA-LIHC | |
| HCCDB16 | 60 | 60 | Healthy 6 | Affymetrix Human Gene 1.0 ST Array | GSE64041 |
| HCCDB17 | 52 | 115 | Illumina HumanHT-12 V4.0 expression beadchip | GSE76427 | |
| HCCDB18 | 177 | 212 | RNA-Seq | ICGC-LIRI-JP | |
| Total | 1469 | 2396 | Healthy 12; Cirrhotic 40 | – | – |
Moreover, we also integrated the expression data of 9755 tumor samples covering 35 tumor types from TCGA and 11,688 normal tissue samples covering 54 tissue types from GTEx (v7) to comprehensively annotate the expression patterns of each gene.
Identification of consistently differentially expressed genes
To identify consistently differentially expressed genes in HCC, the 12 datasets containing both tumor and the adjacent normal samples were used. The function t test in R was employed to detect whether there existed significant difference in gene expression between tumor samples and the adjacent samples in each dataset, followed by the Benjamini–Hochberg correction [23]. Genes with expression measured in ≥8 datasets and significantly differential (adjusted P < 0.001 and > 0.6) in at least half of the datasets containing these genes were identified as consistently differentially expressed genes.
Definition of a 4D metric for summarizing gene expression patterns
We proposed a 4D metric based on log2 fold change (FC) to characterize the expression patterns of each gene. With denoting the gene expression values in samples from a certain resource (indicated in subscript, such as GTEx), four metrics are defined in the following way, with positive value for high specificity and negative value for low specificity, respectively:
1) liver-specific metric, , quantifies the specificity of a gene in liver in comparison with other tissues:
| (1) |
2) deregulation metric, , measures the degree of deregulation of a gene in HCC in comparison with adjacent samples:
| (2) |
3) tumor-specific metric, , quantifies the specificity of a gene in HCC in comparison with other tissues:
| (3) |
4) HCC-specific metric, , denotes the specificity of a gene in HCCs compared with other tumor types:
| (4) |
Prognostic analysis
The prognostic performance of each gene was evaluated using three datasets (HCCDB6, HCCDB15, and HCCDB18) with overall survival time information available. HCC samples in each dataset were classified into high-expression group and low-expression group according to the median expression value of each gene. Then we used log-rank test [24] to compare the survival distribution of samples between the two groups. Genes with adjusted P < 0.001 (Benjamini–Hochberg correction) in ≥1 dataset or adjusted P < 0.01 in ≥2 datasets were considered as prognostic genes. These genes were tagged as “favorable genes” if their Cox coefficients were negative, meaning the higher expression levels and the lower risk extents. Conversely, genes were tagged as “unfavorable genes” if their Cox coefficients were positive.
To reduce the noise of disease-irrelevant deaths, survival time that was greater than five years was truncated to five years and the status of the corresponding patient was set to be “alive”.
Co-expression analysis
For each gene, we computed and displayed its co-expressed genes in HCC, adjacent tissue samples, and normal liver samples, respectively. In each dataset, the following steps were taken. Firstly, the 10% genes with smallest expression variances were considered to be almost invariably expressed and thus excluded in co-expression analysis. We then used the function cor.test in R to compute the Pearson correlations between each of the remaining genes (pivot) and all the other genes and significance was tested followed by the Benjamini–Hochberg correction. For each gene (pivot), its significantly co-expressed genes (candidates) in a certain dataset were detected as those with adjusted P < 0.001.
For co-expression in normal liver (GTEx liver samples), we picked the top 20 strongest co-expressed genes for display on the website. For co-expression in HCC/adjacent samples, we combined multiple datasets (Table 1) to provide a robust result. Suppose one pivot gene had a candidate gene detected in k datasets. If k is smaller than 3, the co-expression relationship would be thought to be unstable and excluded in downstream analysis. For pivot genes with stable co-expression relationships present (k ≥ 3), their candidates’ correlation values were combined with Fisher’s z-transformation, if the candidate was detected in at least one third of k datasets:
| (5) |
| (6) |
| (7) |
where i is the index of dataset and is the sample size. In this way, we could get the consistently co-expressed genes for each pivot gene in multiple HCC or adjacent datasets. Similarly, only the top 20 strongest co-expressed genes are displayed on the website.
Implementation and results
The database overall design
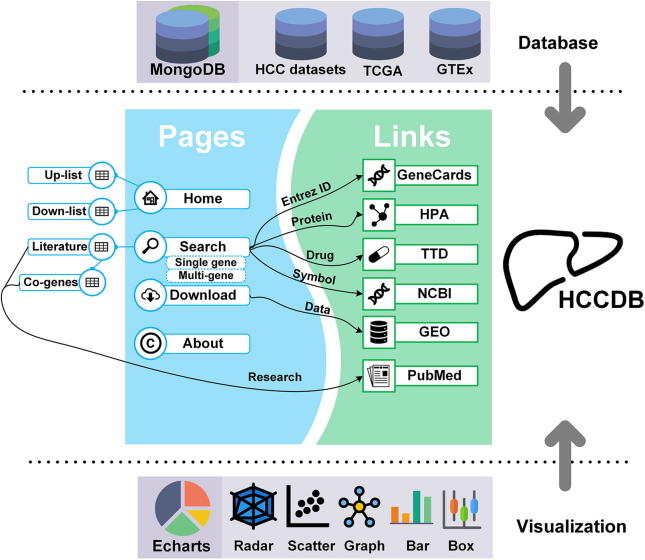
To facilitate a convenient and user-friendly browsing and searching, HCCDB provides both graphical and text-based interfaces. The graphical interface is provided on the database home page by clicking on a certain gene in the differential expression landscape. For the text-based interface, HCCDB offers two search modes: single gene search and multi-gene search. In the single gene search mode, users can query a particular gene with Entrez ID or official gene symbol. All results about the queried gene will be retrieved in seconds, including summary information, expression patterns, survival analysis, and co-expression analysis. As for the multi-gene search mode, summary information of the queried genes will be returned after submitting their symbols or Entrez IDs split by some common separators. The whole design of the database is shown in Figure 1, with the possible jumps among web pages illustrated.
Figure 1.
The design of HCCDB
MongoDB was used to store the data of archived HCC datasets, GTEx, and TCGA in the format of JSON. Echarts was applied for charting and data visualization. HCCDB provides four main pages and serval downstream pages. The possible jumps among HCCDB pages and the third-party links are indicated by arrows. HCC, hepatocellular carcinoma; GTEx, Genotype-Tissue Expression; TCGA, The Cancer Genome Atlas; HPA, Human Protein Atlas; TTD, Therapeutic Target Database; GEO, Gene Expression Omnibus; Up-list, up-regulated gene list; Down-list, down-regulated gene list; Co-genes, co-expressed genes.
HCCDB is freely available to all users without login requirement. The server is driven by the framework of Linux + Apache + MongoDB + PHP. ECharts (http://echarts.baidu.com/), a third-party JavaScript library, is used for charting and data visualization. The design of user interface is optimized by Bootstrap (http://getbootstrap.com/), a third-party HTML5 library.
The home page and overall analysis results
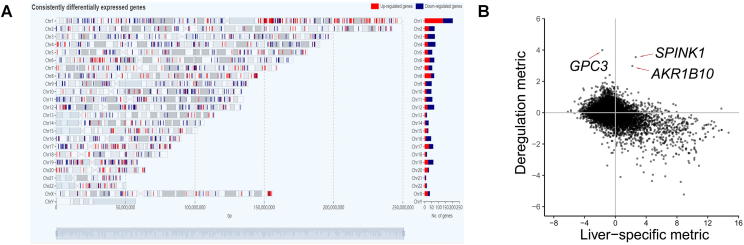
The home page mainly exhibits the differential expression landscape of HCC, provides search interfaces and links to some summarized results. In total, we identified 1259 consistently differentially expressed genes, including 557 up-regulated and 702 down-regulated genes. By mapping these genes onto the respective chromosomes, a differential expression landscape of HCC is depicted at the genome scale (Figure 2A).
Figure 2.
Differential analysis results of HCC
A. Differential expression landscape of HCC. The consistent expression up-regulation and down-regulation of genes on the corresponding genomic locations are shown in red and blue, respectively. The bar plot on the right shows the number of up-regulated (red) and down-regulated (blue) genes on each chromosome. The number and percentage of the up-regulated or down-regulated genes located on a particular chromosome can be shown on the screen when the mouse hovers over the respective bar. B. Relationship between liver-specific metric () and deregulation metric () in HCC.
There are some interesting observations. Among 138 consistently differentially expressed genes on arm 1q, 83% (114 genes) was up-regulated genes. While among 62 consistently differentially expressed genes on arm 4q, 76% (47 genes) was down-regulated genes. This observation coincide with the fact that 1q and 4q exhibit the most frequently detected gain and loss of chromosomal materials, respectively [25], [26]. Therefore, such a global view provides potential evidence that genomic events, such as CNV, could be closely related to expression alterations.
Analyzing the 4D metric values of all genes could also lead to some inspiring results. The relationship between liver specificity () and deregulation degree () is shown in Figure 2B. Liver-specific genes ( > 0) significantly fall into the group of HCC down-regulated () genes (P < , suggesting the dedifferentiation in HCC. This is also consistent with existing observations in pan-cancer analysis [27], [28]. Three genes with expression significantly up-regulated in HCC, including GPC3, SPINK1, and AKR1B10, are highlighted in Figure 2B. GPC3, which encodes clypican-3, an oncofetal proteoglycan anchored to the cell membrane, is not detected in normal liver and benign liver lesions [29], but overexpressed in HCCs at both the gene and protein levels [30]. It has been demonstrated that GPC3 promotes HCC growth and metastasis by activating WNT/-catenin and other signaling pathways [31], [32]. And GPC3 is used as a diagnostic biomarker and immunotherapeutic target [29], [30]. SPINK1, which encodes serine peptidase inhibitor, Kazal type 1, has been found to be table to differentiate between a well-differentiated HCC and a high-grade dysplastic nodule [33], [34]. AKR1B10, which encodes aldo–keto reductase family 1 member B10, may play an important role in liver carcinogenesis and is a promising biomarker to differentiate HCCs from benign liver lesions [35]. The three genes with clinical value can be highlighted as shown in Figure 2B, suggesting the effectiveness and utility of the defined metric to filter HCC biomarkers for follow-up experiments.
The result page for single gene search
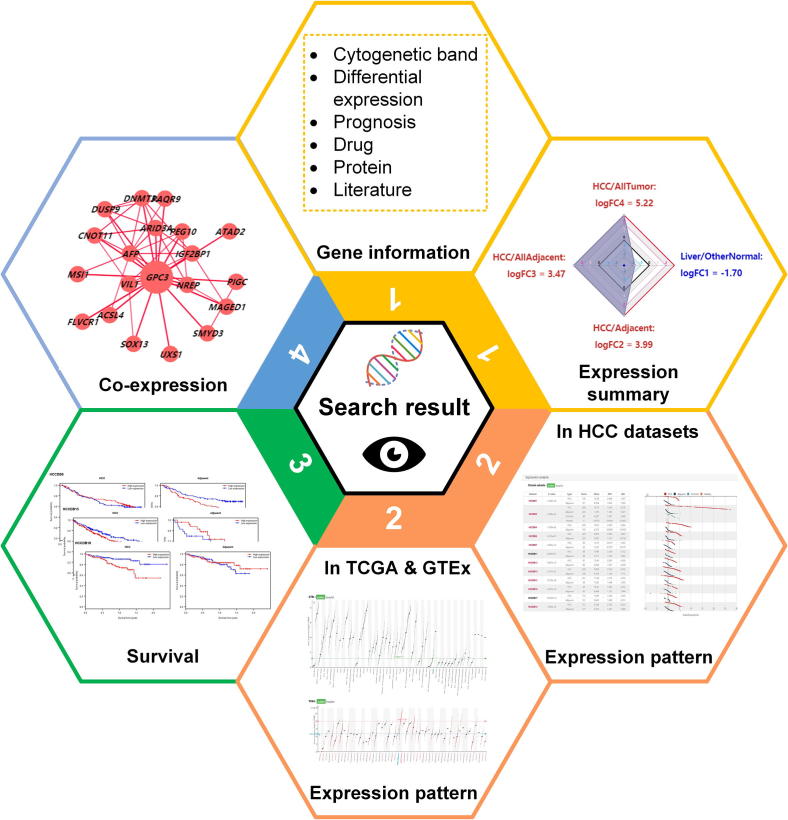
The result page for single gene search has four major views: (1) summary view; (2) expression pattern view; (3) survival view; and (4) co-expression view. Taking GPC3 as an example, we showed the detailed design of gene page in Figure 3.
Figure 3.
The result page for single gene search
This page consists of four views: (1) the summary view showing the gene information and expression pattern summarized by the 4D metric; (2) the expression pattern view displaying the expression patterns in archived HCC datasets, tissue samples in GTEx, and tumor samples in TCGA; (3) the survival view showing the gene’s prognostic performance in HCC for patients with high (above the median) and low (below the median) expression levels; (4) the co-expression view displaying the consistently co-expressed genes of the queried one in HCCs, adjacent samples, and normal liver tissues.
The summary view provides an overview of general information and computation results of the queried gene. The general information covers Entrez ID, official gene symbol, and so on. The computation results include the prognostic performance and the consistency of differential expression in HCCDB. The third-party database links, such as drug, proteomics, and literature mining results, are also provided in this part. The 4D metric summarizes gene expression with a radar chart, with red and blue axis labels representing the positive and negative metric values, respectively. We exemplify the view using GPC3, which is not a liver-specific gene ( = −1.7). While it was highly up-regulated in HCC compared with adjacent samples ( = 3.99), GPC3 was also highly expressed in HCC compared with other normal tissues ( = 3.47) and other kinds of tumors ( = 5.22).
The expression pattern view displays the expression patterns of a specific gene in the archived HCC datasets, tissue samples in GTEx, and tumor samples in TCGA. Users can view the differential gene expression in HCCs compared with adjacent samples by both table and graph visualization. The dataset labels are colored (red or blue) to indicate whether the gene is identified to be significantly up-regulated or down-regulated in the corresponding dataset. In GTEx and TCGA graphs, liver-related data (liver tissue and LIHC) are also highlighted with distinct colors. Users can switch between scatter and boxplot to view the gene expression plot. The expression pattern view corresponds to the 4D metric, which is displayed in numeric form in radar chart of gene summary view.
The survival view shows the prognostic performance of the queried gene (GPC3) in three datasets with the overall survival time information available, that is, HCCDB6, HCCDB15, and HCCDB18. Patients are classified into two groups based on the expression levels of GPC3 in HCC and adjacent tissue samples. The Kaplan–Meier survival curves are shown for the two groups of patients, with P values computed by log-rank test shown above the plots, which could be clicked to zoom in and saved.
The co-expression view is designed to display three kinds of co-expression relationships of the queried gene: meta co-expression in HCCs, meta co-expression in adjacent samples, and co-expression in GTEx liver samples. In Figure 3, meta co-expression network in HCCs of GPC3 is shown as an example. Notably, AFP, a well-known stem marker [9], is co-expressed with GPC3 in HCCs.
Perspectives and concluding remarks
Genome-wide gene expression data are valuable resources for studying the molecular mechanisms and identifying biomarkers of cancers. Given the high heterogeneity nature of HCC, identifying consistent patterns from multiple gene expression datasets would be beneficial for identifying reliable biomarkers of HCC. First of all, we detected consistently differentially expressed genes across multiple HCC expression datasets to provide a global differential landscape. Integrating data from GTEx and TCGA, we defined a 4D metric to comprehensively summarize the gene expression pattern. Taking GPC3 as example, we show the utility of this metric and the idea of integrating multiple-source data. Coupled with the commonly used third-party database links and convenient download links of computational results, HCCDB provides a one-stop online resource for exploring HCC gene expression with user-friendly interfaces.
To better serve the research community of HCC, we will continue to collect related data and update HCCDB regularly in the future. Our next plan is to obtain more public multi-omics data on HCC, such as CNV [36], mutation [37], or DNA methylation [38], mine relationships between expression patterns and genomic events, and provide quick query and graphical presentation about these relationships. We believe HCCDB could serve as a very useful public resource for both bench scientists and computational biologists, and contribute to clinical and translational studies.
Authors’ contributions
QL performed the most analyses and drafted the paper. SW set up the database and participated in analyses. GZ, DW, JT, and GL participated in the data curation. LC and JG contributed to data analysis and website design. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (NSFC) (Grant Nos. 61370035, 81630103, and 61721003) and Tsinghua University Initiative Scientific Research Program. The icons used in Figure 1 were obtained using FLATICON.
Handled by Zhang Zhang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Contributor Information
Lei Chen, Email: chenlei@smmu.edu.cn.
Jin Gu, Email: jgu@tsinghua.edu.cn.
References
- 1.Zucman-Rossi J., Villanueva A., Nault J.C., Llovet J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239.e4. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 2.Allain C., Angenard G., Clement B., Coulouarn C. Integrative genomic analysis identifies the core transcriptional hallmarks of human hepatocellular carcinoma. Cancer Res. 2016;76:6374–6381. doi: 10.1158/0008-5472.CAN-16-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y., Chen L., Gu J., Zhang H., Yuan J., Lian Q. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su W.H., Chao C.C., Yeh S.H., Chen D.S., Chen P.J., Jou Y.S. OncoDB.HCC: an integrated oncogenomic database of hepatocellular carcinoma revealed aberrant cancer target genes and loci. Nucleic Acids Res. 2007;35:D727–D731. doi: 10.1093/nar/gkl845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee L., Wang K., Li G., Xie Z., Wang Y., Xu J. Liverome: a curated database of liver cancer-related gene signatures with self-contained context information. BMC Genomics. 2011;12:S3. doi: 10.1186/1471-2164-12-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Planey C.R., Gevaert O. CoINcIDE: a framework for discovery of patient subtypes across multiple datasets. Genome Med. 2016;8:27. doi: 10.1186/s13073-016-0281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshida Y., Nijman S.M., Kobayashi M., Chan J.A., Brunet J.P., Chiang D.Y. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita T., Forgues M., Wang W., Kim J.W., Ye Q., Jia H. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 9.Baig J.A., Alam J.M., Mahmood S.R., Baig M., Shaheen R., Sultana I. Hepatocellular carcinoma (HCC) and diagnostic significance of A-fetoprotein (AFP) J Ayub Med Coll Abbottabad. 2009;21:72–75. [PubMed] [Google Scholar]
- 10.Ling S., Hu Z., Yang Z., Yang F., Li Y., Lin P. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci U S A. 2015;112:E6496–E6505. doi: 10.1073/pnas.1519556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhai W., Lim T.K., Zhang T., Phang S.T., Tiang Z., Guan P. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat Commun. 2017;8:4565. doi: 10.1038/ncomms14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue R., Li R., Guo H., Guo L., Su Z., Ni X. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology. 2016;150:998–1008. doi: 10.1053/j.gastro.2015.12.033. [DOI] [PubMed] [Google Scholar]
- 13.Guinney J., Dienstmann R., Wang X., de Reynies A., Schlicker A., Soneson C. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider G., Schmidt-Supprian M., Rad R., Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer. 2017;17:239–253. doi: 10.1038/nrc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Schaefer M.H., Serrano L. Cell type-specific properties and environment shape tissue specificity of cancer genes. Sci Rep. 2016;6:20707. doi: 10.1038/srep20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bleeker F.E., Lamba S., Rodolfo M., Scarpa A., Leenstra S., Vandertop W.P. Mutational profiling of cancer candidate genes in glioblastoma, melanoma and pancreatic carcinoma reveals a snapshot of their genomic landscapes. Hum Mutat. 2009;30:E451–E459. doi: 10.1002/humu.20927. [DOI] [PubMed] [Google Scholar]
- 18.Shen Q., Cheng F., Song H., Lu W., Zhao J., An X. Proteome-scale investigation of protein allosteric regulation perturbed by somatic mutations in 7,000 cancer genomes. Am J Hum Genet. 2017;100:5–20. doi: 10.1016/j.ajhg.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J., Cheng F., Zhao Z. Tissue-specific signaling networks rewired by major somatic mutations in human cancer revealed by proteome-wide discovery. Cancer Res. 2017;77:2810–2821. doi: 10.1158/0008-5472.CAN-16-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 21.Kim P., Park A., Han G., Sun H., Jia P., Zhao Z. TissGDB: tissue-specific gene database in cancer. Nucleic Acids Res. 2018;46:D1031–D1038. doi: 10.1093/nar/gkx850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carithers L.J., Moore H.M. The genotype-tissue expression (GTEx) project. Biopreserv Biobank. 2015;13:307–308. doi: 10.1089/bio.2015.29031.hmm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 24.Kaplan E.L., Meier P. Nonparametric-estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 25.Xu H., Zhu X., Xu Z., Hu Y., Bo S., Xing T. Non-invasive analysis of genomic copy number variation in patients with hepatocellular carcinoma by next generation DNA sequencing. J Cancer. 2015;6:247–253. doi: 10.7150/jca.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan X.Y., Fang Y., Sham J.S., Kwong D.L., Zhang Y., Liang Q. Recurrent chromosome alterations in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2000;29:110–116. [PubMed] [Google Scholar]
- 27.Friedmann-Morvinski D., Verma I.M. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15:244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaude E., Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat Commun. 2016;7:13041. doi: 10.1038/ncomms13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y., Liu H., Ding H. GPC-3 in hepatocellular carcinoma: current perspectives. J Hepatocell Carcinoma. 2016;3:63–67. doi: 10.2147/JHC.S116513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou F., Shang W., Yu X., Tian J. Glypican-3: a promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med Res Rev. 2018;38:741–767. doi: 10.1002/med.21455. [DOI] [PubMed] [Google Scholar]
- 31.Capurro M., Martin T., Shi W., Filmus J. Glypican-3 binds to Frizzled and plays a direct role in the stimulation of canonical Wnt signaling. J Cell Sci. 2014;127:1565–1575. doi: 10.1242/jcs.140871. [DOI] [PubMed] [Google Scholar]
- 32.Gao W., Ho M. The role of glypican-3 in regulating Wnt in hepatocellular carcinomas. Cancer Rep. 2011;1:14. [PMC free article] [PubMed] [Google Scholar]
- 33.Rasanen K., Itkonen O., Koistinen H., Stenman U.H. Emerging roles of SPINK1 in cancer. Clin Chem. 2016;62:449–457. doi: 10.1373/clinchem.2015.241513. [DOI] [PubMed] [Google Scholar]
- 34.Marshall A., Lukk M., Kutter C., Davies S., Alexander G., Odom D.T. Global gene expression profiling reveals SPINK1 as a potential hepatocellular carcinoma marker. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matkowskyj K.A., Bai H., Liao J., Zhang W., Li H., Rao S. Aldoketoreductase family 1B10 (AKR1B10) as a biomarker to distinguish hepatocellular carcinoma from benign liver lesions. Hum Pathol. 2014;45:834–843. doi: 10.1016/j.humpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roessler S., Long E.L., Budhu A., Chen Y., Zhao X., Ji J. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957. doi: 10.1053/j.gastro.2011.12.039. –66.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J., Zaidi S., Rao S., Chen J.S., Phan L., Farci P. Analysis of genomes and transcriptomes of hepatocellular carcinomas identifies mutations and gene expression changes in the transforming growth factor-beta pathway. Gastroenterology. 2018;154:195–210. doi: 10.1053/j.gastro.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Y., Huang Q., Ding Z., Liu T., Xue C., Sang X. Genome-wide DNA methylation analysis identifies candidate epigenetic markers and drivers of hepatocellular carcinoma. Brief Bioinform. 2018;19:101–108. doi: 10.1093/bib/bbw094. [DOI] [PubMed] [Google Scholar]